Abstract
Neuroblastoma (NB) is a challenging pediatric extracranial solid tumor characterized by a poor prognosis and resistance to chemotherapy. Identifying targets to enhance chemotherapy sensitivity in NB is of utmost importance. Increasing evidence implicates long noncoding RNAs (lncRNAs) play important roles in cancer, but their functional roles remain largely unexplored. Here, we analyzed our RNA sequencing data and identified the upregulated lncRNA ZNF674-AS1 in chemotherapy non-responsive NB patients. Elevated ZNF674-AS1 expression is associated with poor prognosis and high-risk NB. Importantly, targeting ZNF674-AS1 expression in NB cells suppressed tumor growth in vivo. Further functional studies have revealed that ZNF674-AS1 constrains cisplatin sensitivity by suppressing pyroptosis and promoting cell proliferation. Moreover, ZNF674-AS1 primarily relies on CA9 to fulfill its functions on cisplatin resistance. High CA9 levels were associated with high-risk NB and predicted poor patient outcomes. Mechanistically, ZNF674-AS1 directly interacted with the RNA binding protein IGF2BP3 to enhance the stability of CA9 mRNA by binding with CA9 transcript, leading to elevated CA9 expression. As a novel regulator of CA9, IGF2BP3 positively upregulated CA9 expression. Together, these results expand our understanding of the cancer-associated function of lncRNAs, highlighting the ZNF674-AS1/IGF2BP3/CA9 axis as a constituting regulatory mode in NB tumor growth and cisplatin resistance. These insights reveal the pivotal role of ZNF674-AS1 inhibition in recovering cisplatin sensitivity, thus providing potential therapeutic targets for NB treatment.
Subject terms: Long non-coding RNAs, Paediatric cancer, Cell death
Introduction
Neuroblastoma (NB) is the most common extracranial solid tumor in children, accounting for approximately 12–15% of pediatric tumor-related mortality [1, 2]. NB commonly begins in one of the adrenal glands but can also develop in the chest, neck, abdomen, spine, or brain. The majority of NB cases (90%) are diagnosed in children under the age of 10, with an average age of approximately 18 months [3]. The exact cause of NB is still unclear, but it is believed to be associated with improper differentiation of neural crest cells into mature neurons of the sympathetic nervous system [4]. NB is highly heterogeneous and therapeutically challenging, where patients are assigned to low-risk, intermediate-risk, and high-risk subgroups on the basis of disease stage, age at diagnosis, and non-random chromosomal aberrations [5]. The majority of low-risk patients experience spontaneous regression, while those with high-risk subgroups have a survival rate of only 50% despite intensive therapy [6]. Therefore, there is a critical need to utilize biological markers to minimize inappropriate treatments and develop novel therapeutic strategies for NB patients in order to improve survival.
In cancer therapy, drug resistance remains a major challenge, leading to relapse and even mortality. Chemotherapeutic drugs primarily exert their antitumor effects by inhibiting cell proliferation and inducing regulated cell death (RCD), which limits tumor growth and causes tumor cell death [7, 8]. Recently, apart from apoptosis, other forms of RCD induced by chemotherapeutic drugs, such as pyroptosis, have been discovered [9, 10]. Pyroptosis is a newly identified form of RCD caused by pore-forming effector proteins called gasdermins, leading to cell swelling, the formation of large membrane bubbles, and perforation of the plasma membrane [11]. Recent studies have shown that chemotherapy drugs induce pyroptosis in tumor cells mediated by gasdermin E (GSDME), a key protein within the gasdermin family that is cleaved by activated caspase-3 [9, 12, 13]. In addition, the expression level of GSDME determines whether pyroptosis or apoptosis occurs in response to chemotherapy [9, 14]. Cells with high levels of GSDME undergo pyroptosis, whereas cells with low levels undergo apoptosis upon chemotherapy treatment. The expression of GSDME varies in different tissues and cell types. Interestingly, recent evidence demonstrates that the neuroblastoma SH-SY5Y cell line expresses high levels of GSDME. Furthermore, an analysis of the Cancer Genome Atlas (TCGA) cohort unveiled that neuroblastoma exhibits significantly higher levels of GSDME when compared with 31 other types of tumor tissues, second only to brain tumors [9, 15]. Considering the importance of GSDME activation in chemotherapy-induced tumor cell death, these findings raise the possibility that antitumor drugs like cisplatin (a first-line drug of NB chemotherapy) could induce pyroptosis in NB cells. However, the role of pyroptosis in the antitumor effect of cisplatin in NB, as well as the underlying regulatory mechanisms, remain poorly investigated.
In recent decades, mounting evidence has documented the vital roles played by the non-coding portion of the genome in many cancers. Long noncoding RNA (lncRNA) is a class of RNA transcripts with longer than 200 nucleotides without protein-coding potential [16]. It has been demonstrated that lncRNAs play central roles in a variety of fundamental biological processes and human diseases [17]. By interacting with proteins, lncRNAs regulate gene expression at various levels, including transcriptional, post-translational, and translational regulation, as well as protein activation or degradation [18]. In the context of NB, aberrant expression of certain lncRNAs has been shown to interfere with cell proliferation, cell death, migration, invasion, and even tumor initiation and progression [19–22]. Increasing evidence demonstrates that lncRNAs may also play important roles in determining NB chemotherapy sensitivities. For example, the depletion of lncRNA NBAT1 provides resistance to genotoxic drugs by limiting p53 accumulation in the nucleus and mitochondria by altering the function of CRM1 [23]. SNHG16 contributes to cisplatin resistance in NB by modulating the miR-338-3p/PLK4 pathway [24]. Furthermore, a growing body of studies reveals that lncRNAs are emerging regulators of cell pyroptosis [25–27]. Nonetheless, whether and how lncRNAs modulate chemotherapy drug-induced pyroptosis of NB and whether they resist the antitumor effects of chemotherapy drugs in NB have not been reported yet and require further investigation.
In this study, we identified an up-regulated lncRNA ZNF674-AS1 in non-response patients to chemotherapy, and its high expression was correlated with poor prognosis. Previous studies have demonstrated that ZNF674-AS1 is downregulated in non-small cell lung cancer and liver cancer [28, 29]. These studies have investigated the role of ZNF674-AS1 in tumor inhibition, such as impeding cell proliferation and tumor metastasis. However, the specific role of ZNF674-AS1 in regulating NB progression and its response to drug treatment has not been elucidated yet.
Here, we demonstrated that lower expression levels of ZNF674-AS1 are associated with improved sensitivity to cisplatin-induced pyroptosis, a clinically relevant genotoxic drug. Additionally, ZNF674-AS1 inhibition limited tumor growth both in vitro and in vivo. Moreover, mechanism studies revealed that ZNF674-AS1 interacted with IGF2BP3 to regulate the transcriptional level of Carbonic Anhydrase IX (CA9), a key enzyme involved in tumor growth and correlated with the clinical prognosis of NB patients [30–32]. Importantly, the presence of ZNF674-AS1 enhanced the association between IGF2BP3 and its target, CA9 transcripts. This study, therefore, identifies ZNF674-AS1 as a crucial target for optimizing genotoxic drug therapy in NB patients and targeting ZNF674-AS1 may represent a potential treatment strategy for enhancing chemo-sensitization of anticancer drugs.
Results
LncRNA ZNF674-AS1 is associated with a poor prognosis and chemotherapeutic resistance in neuroblastoma
To identify key long noncoding RNAs associated with chemotherapeutic sensitivity in NB, we performed high-throughput RNA sequencing profiling of 35 NB patient tissues. Among these samples, 22 were nonresponsive to chemical therapy, while 13 showed a positive response. Differential expression analysis showed 100 differentially expressed lncRNAs, with 18 up-regulated and 82 downregulated lncRNAs (Fig. 1A). The top 10 upregulated candidates, including ZNF674-AS1, were identified (Fig. S1A). ZNF674-AS1 stood out with the most significant P-value (Fig. 1A). Kaplan–Meier survival analysis of publicly available data from human NB tissues was conducted using the SEQC-RPM-seqcnb1 dataset from the R2 platform (http://r2.amc.nl) and revealed that high expression levels of ZNF674-AS1 were associated with a poor prognosis in NB patients (Figs. 1B and S1B). In addition, ZNF674-AS1 expression was found to be higher in high-risk neuroblastoma tumors (Fig. 1C). The results above indicate that ZNF674-AS1 may play an important role in NB development and therapy.
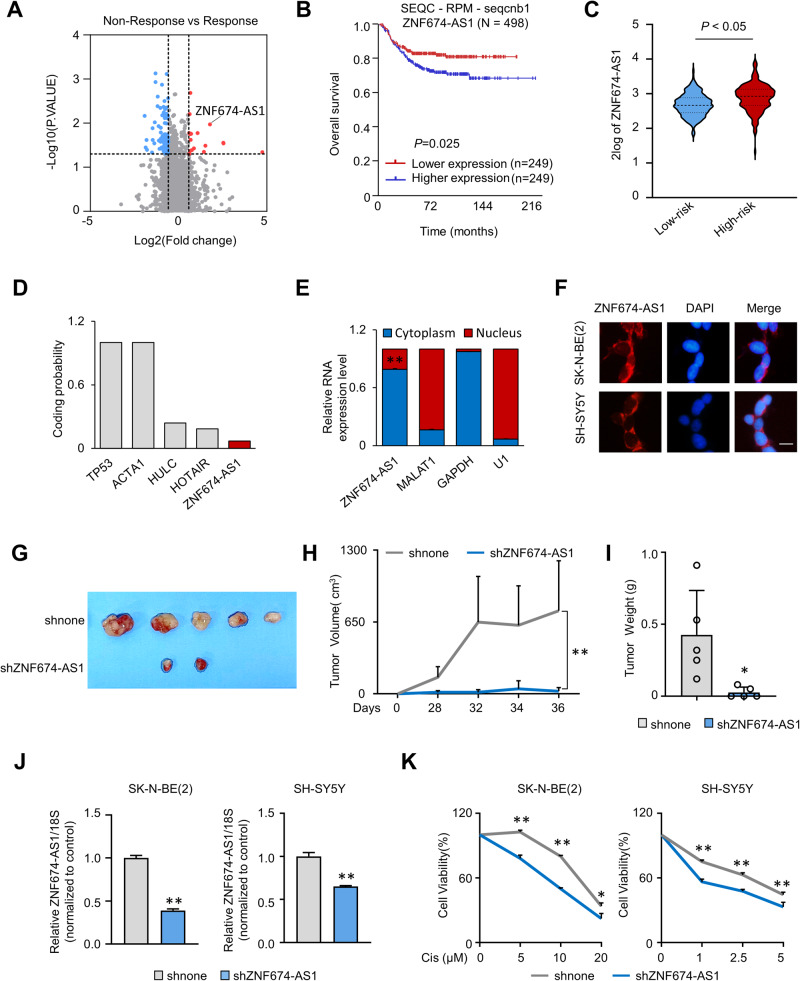
Fig. 1. LncRNA ZNF674-AS1 has a high expression associated with chemotherapeutic resistance and promotes neuroblastoma tumor growth.
A Volcano plot revealed the differentially expressed lncRNAs between chemotherapy response and non-response neuroblastoma tissues. B Kaplan-Meier curve showed the overall survival of neuroblastoma patients according to the expression level of ZNF674-AS1 in the SEQC-RPM-seqcnb1 dataset from the R2 platform (http://r2.amc.nl). C The expression levels of ZNF674-AS1 in low- and high-risk neuroblastoma tissues. D The coding potential score of TP53 and ACTA1 (coding genes), HULC and HOTAIR (well-established long non-coding RNAs and ZNF674-AS1 were identified by the online bioinformatic tool (Coding Potential Calculation, http://cpc.cbi.pku.edu.cn). E The expression of ZNF674-AS1, MALAT1, GAPDH, and U1 in cytoplasm and nucleus were calculated by qRT-PCR after cell fraction separation. MALAT1 and U1 were used as nuclear markers, and GAPDH was a cytoplasmic marker. F The distribution of ZNF674-AS1 in SK-N-BE(2) and SH-SY5Y cells was determined by RNA FISH assay. Scale bar, 50 μm. G–I Tumor volumes at the indicated dates (G), as well as images (H) and the tumor weights (I) for shnone and shZNF674-AS1 xenografts. The average values are presented as bar graphs (means ± SD) (n = 5 for each group). J The KD efficiency of ZNF674-AS1 in SK-N-BE(2) and SH-SY5Y cell lines were calculated by qRT-PCR. K The relative cell survival rates of SK-N-BE(2) (left) and SH-SY5Y (right) stable ZNF674-AS1 knockdown (KD) cells were measured after the indicated concentration cisplatin treatment. Data are derived from three independent experiments and presented as mean ± SD in the bar graphs. Values of controls were normalized to 1 (J, K). * P < 0.05; ** P < 0.01.
Based on coding potential calculation, ZNF674-AS1 showed a coding potential value of 0.069, slightly lower than well-characterized lncRNAs and significantly lower than protein-coding genes (Fig. 1D). Moreover, ZNF674-AS1 was predominantly detected in the cytoplasm rather than the nucleus, by using cytoplasmic mRNA GAPDH, nuclear rRNA U1 and nuclear lncRNA MALAT1 as controls (Fig. 1E). This observation was further confirmed by fluorescence in situ hybridization (FISH) (Fig. 1F).
To understand the function of ZNF674-AS1 in NB, we conducted xenograft experiments using stable knockdown (KD) ZNF674-AS1 cells implanted into NSG mice. As shown in Fig. 1G–I, the reduction of ZNF674-AS1 dramatically suppressed tumor growth. Furthermore, as expected, ZNF674-AS1 KD significantly enhanced the susceptibility of NB cells to cisplatin-induced cell death (Fig. 1J, K). These findings strongly suggest that ZNF674-AS1 promotes NB formation and progression and impedes the sensitivity of NB to chemical therapies.
Cisplatin induces pyroptosis via caspase-3/GSDME in neuroblastoma cells
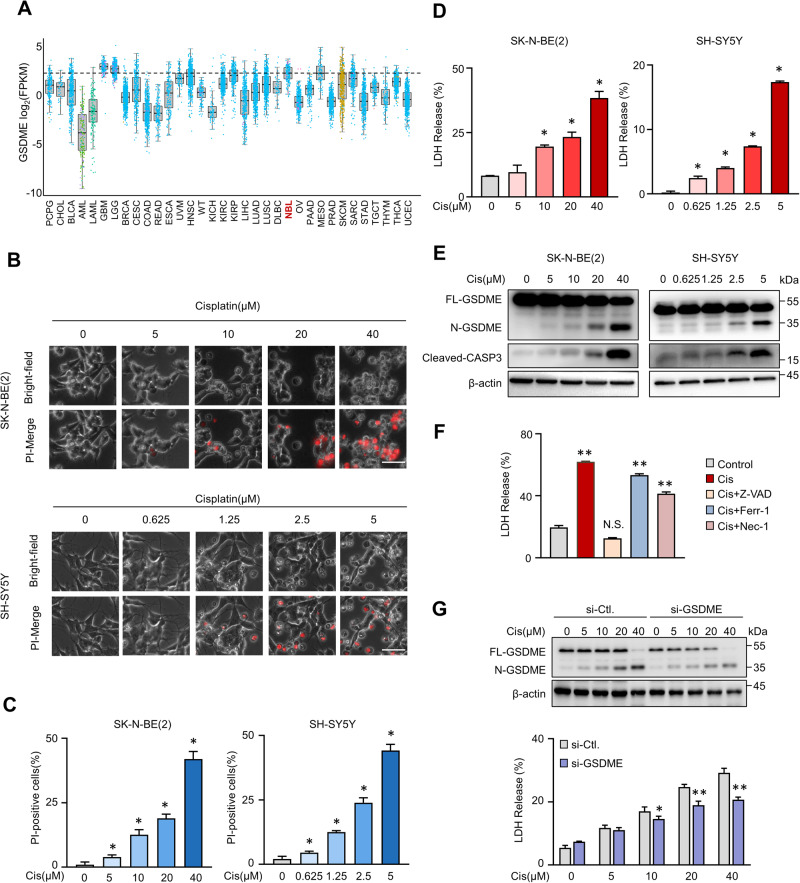
After treating cells with cisplatin, we observed the morphology of dead cells induced by cisplatin differed from apoptosis. In contrast to apoptosis, the majority of dead cells exhibited swelling with plasma membrane blowing large bubbles, which is the typical characterization of cell pyroptosis (Fig. S2A). Previous studies have reported that chemical drugs can induce neuroblastoma cell SH-SY5Y to develop cell pyroptosis, which is associated with high-level endogenous GSDME expression [9]. The GSDME level determines whether cell death occurs in the form of apoptosis or pyroptosis [14]. Based on these clues, we analyzed the expression level of GSDME in various human tumors from TCGAportal (www.tcgaportal.org). Intriguingly, the GSDME mRNA was highly expressed in brain tumor, NB, and pleura among 34 tumor types (Fig. 2A). However, the expression of GSDMD, another pyroptosis inducer, showed a completely different profile (Fig. S2B). We then examined whether NB cells could undergo pyroptosis with cisplatin treatment. To this end, three NB cell lines were incubated with gradient concentrations of cisplatin. Pyroptosis levels were assessed by PI staining, LDH release, and N-GSDME cleavage detection. As expected, cisplatin-induced dose-dependent pyroptosis in SK-N-BE(2), SH-SY5Y, and IMR-32 cell lines (Fig. 2B–E and Fig. S2C–F). As GSDME can be specifically cleaved by active caspase-3 into N-GSDME fragment, the levels of N-GSDME were found to be correlated with cisplatin-triggered caspase-3 activation (Figs. 2E and S2F). More importantly, only the caspase inhibitor Z-VAD completely recovered cisplatin-induced lytic cell death (Fig. 2F). In addition, GSDME KD significantly attenuated cisplatin-induced cell death compared with control groups (Fig. 2G). Taken together, these data collectively suggest that cisplatin induces pyroptotic cell death in NB cells through the cleavage of N-GSDME by active caspase-3.
Fig. 2. Cisplatin induces pyroptosis in neuroblastoma cells.
A GSDME mRNA expression in various organ human tumor tissues in the TCGA cohort was applied from the TCGAportal database (www.tcgaportal.org). The center line in the box was the median, and NBL represented neuroblastoma. B–D Representative phase contrast images (B), PI-positive cells quantification (C), and LDH releasement (D) of SK-N-BE(2) and SH-SY5Y cells treated with indicated concentration cisplatin were presented. Scale bar, 50 μm. E Full length, N-terminal GSDME, and cleavage caspase-3 protein levels were detected with indicated concentration cisplatin treatment. F LDH releasement rates were analyzed after being treated with cisplatin with or without Z-VAD, Ferrostatin-1, and Necrostatin-1. G GSDME KD efficiencies and LDH release were determined in control (si-NC) and GSDME KD (si-GSDME) cells after the indicated concentration cisplatin treatment. Data are derived from three independent experiments and presented as mean ± SD in the bar graphs. * P < 0.05; ** P < 0.01.
ZNF674-AS1 modulation regulates cisplatin-induced pyroptosis
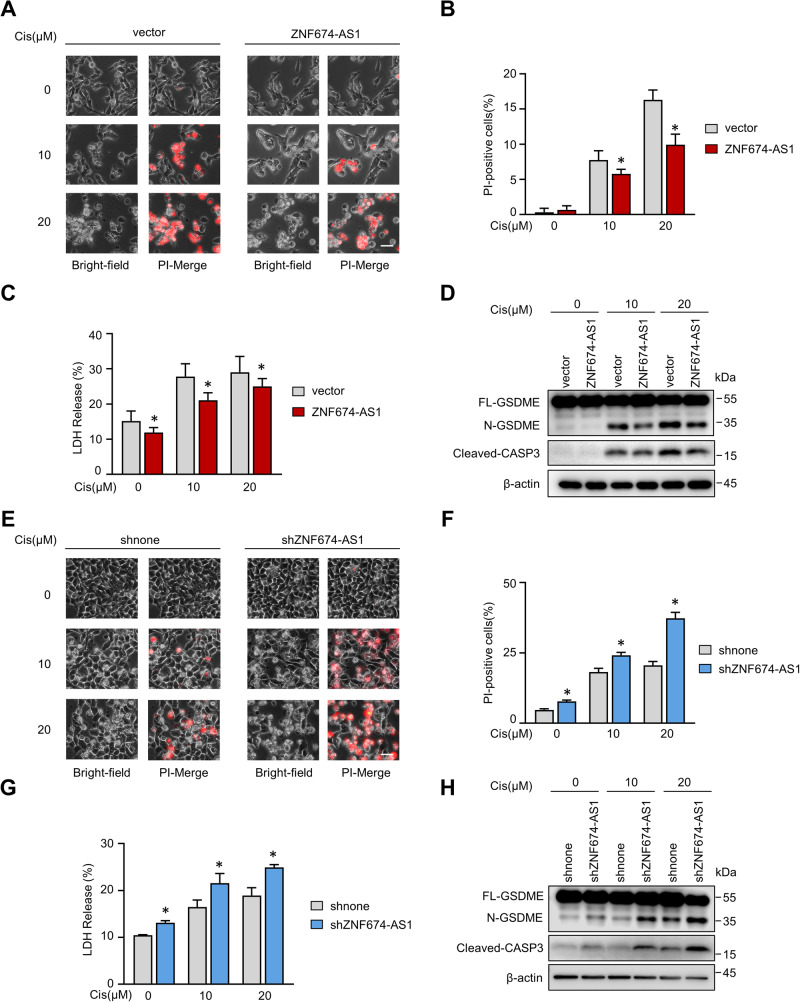
After observing the correlation between ZNF674-AS1 and cisplatin sensitivity (Fig. 1K), we investigated whether ZNF674-AS1 influences cisplatin-induced cell pyroptosis. Control and stable ZNF674-AS1 overexpressed cells were treated with cisplatin in both SK-N-BE(2) (MYCN amplified) and SH-SY5Y (non-MYCN amplified) cells (Fig. S3A). Intriguingly, both bright-field and PI-merge images showed that ZNF674-AS1 protected cells from cisplatin-induced cell death (Fig. 3A, B). In addition, the release of LDH, as well as N-GSDME levels, were markedly decreased upon ZNF674-AS1 overexpression (Figs. 3C, D, and S3B). In contrast, endogenous ZNF674-AS1 inhibition enhanced cisplatin-triggered pyroptosis (Figs. 3E–H and S3C, D). Collectively, these findings demonstrate that ZNF674-AS1 inhibits cell pyroptosis induced by cisplatin, which is likely a common event independent of MYCN.
Fig. 3. ZNF674-AS1 represses cisplatin-induced cell pyroptosis.
A–C Representative phase contrast images (A) PI positive statistic (B) and LDH releasement (C) of ZNF674-AS1 overexpressed and control groups were showed after treated with 10 μM or 20 μM cisplatin. D Expression levels of full-length, N-terminal GSDME and cleavage caspase-3 protein of vector and ZNF674-AS1 overexpressed cells were analyzed by western blot following indicated concentration cisplatin treatment. E–G Representative phase contrast images (E), PI positive statistic (F), and LDH releasement (G) of ZNF674-AS1 KD and control groups were showed after treated with 10 μM or 20 μM cisplatin. H Expression levels of full-length, N-terminal GSDME and cleavage caspase-3 protein of control and ZNF674-AS1 KD cells were analyzed by western blot following indicated concentration cisplatin treatment. Data are derived from three independent experiments and presented as mean ± SD in the bar graphs. Scale bar, 50 μm. * P < 0.05.
ZNF674-AS1 promotes neuroblastoma cell proliferation
The tumor growth rate is a critical factor that determines responses to therapy and resistance. As a widely used and effective chemotherapeutic drug for NB, cisplatin is known to inhibit the proliferation and viability of NB cells [33]. Therefore, we next investigated the role of ZNF674-AS1 in cell growth. As shown in Fig. 4A, ectopic overexpression of ZNF674-AS1 in SK-N-BE(2) or SH-SY5Y cells led to a substantial increase in cell proliferation and colony formation (Fig. 4C–E). Conversely, ZNF674-AS1 KD restrained clonogenic capacity and cell proliferation rate both in SK-N-BE(2) and SH-SY5Y cells (Fig. 4B, F–H, I, J). Consistent with the in vivo model (Fig. 1G–I), these data demonstrate the significant role of ZNF674-AS1 in neuroblastoma cell proliferation and tumorigenesis.
Fig. 4. ZNF674-AS1 promotes neuroblastoma cell proliferation.
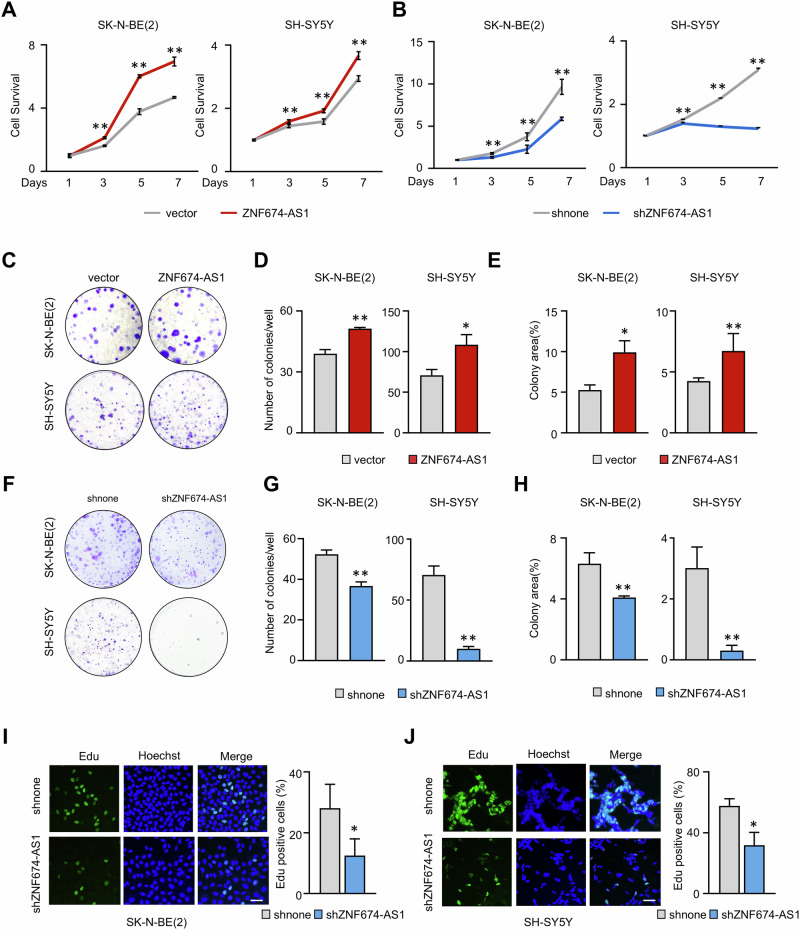
A, B The growth curves of ZNF674-AS1-overexpressing (A) and ZNF674-AS1 KD (B) stable SK-N-BE(2) (left) and SH-SY5Y (right) cells were assessed by MTT assays. C–E Representative micrographs (C), colony numbers (D), and colony area (E) quantifications for ZNF674-AS1-overexpressing stable SK-N-BE(2) (upper) and SH-SY5Y (below) cells. F–H Representative micrographs (F), colony numbers (G), and colony area (H) quantifications for ZNF674-AS1-overexpressing stable SK-N-BE(2) (upper) and SH-SY5Y (below) cells. I, J Representative immunofluorescence images (left) and average Edu positive rates (right) of ZNF674-AS1 KD SK-N-BE(2) (I) and SH-SY5Y (J) cells were presented by Edu incorporation assays. Scale bar, 50 μm. Data are derived from three independent experiments and presented as mean ± SD in the bar graphs. Values of controls were normalized to 1 (A, B). * P < 0.05; ** P < 0.01.
ZNF674-AS1 acts through CA9 by interacting with IGF2BP3
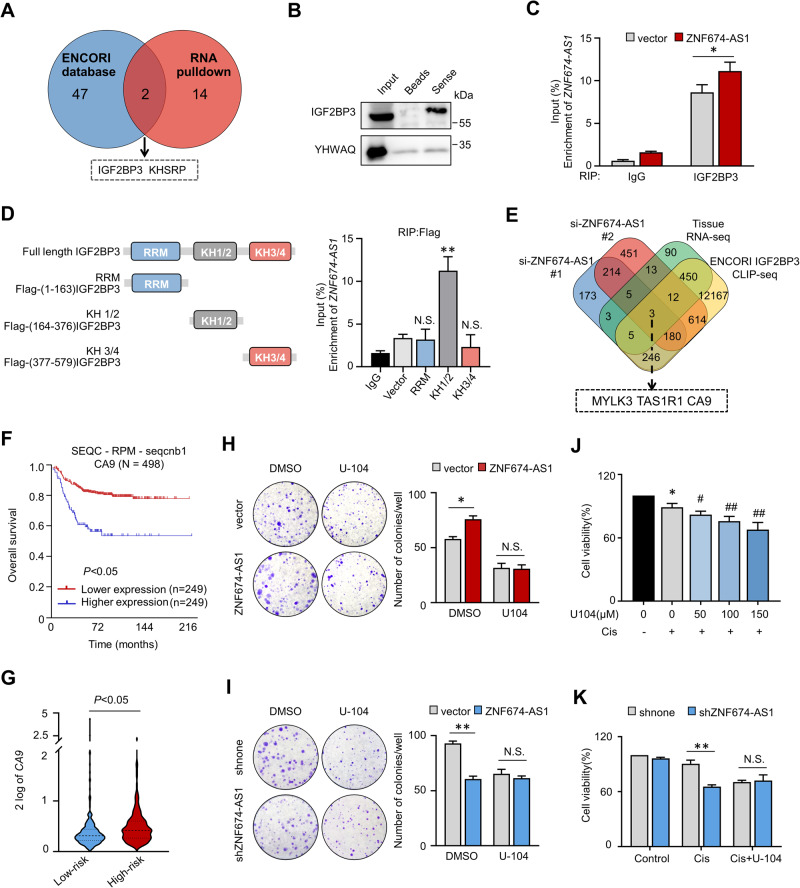
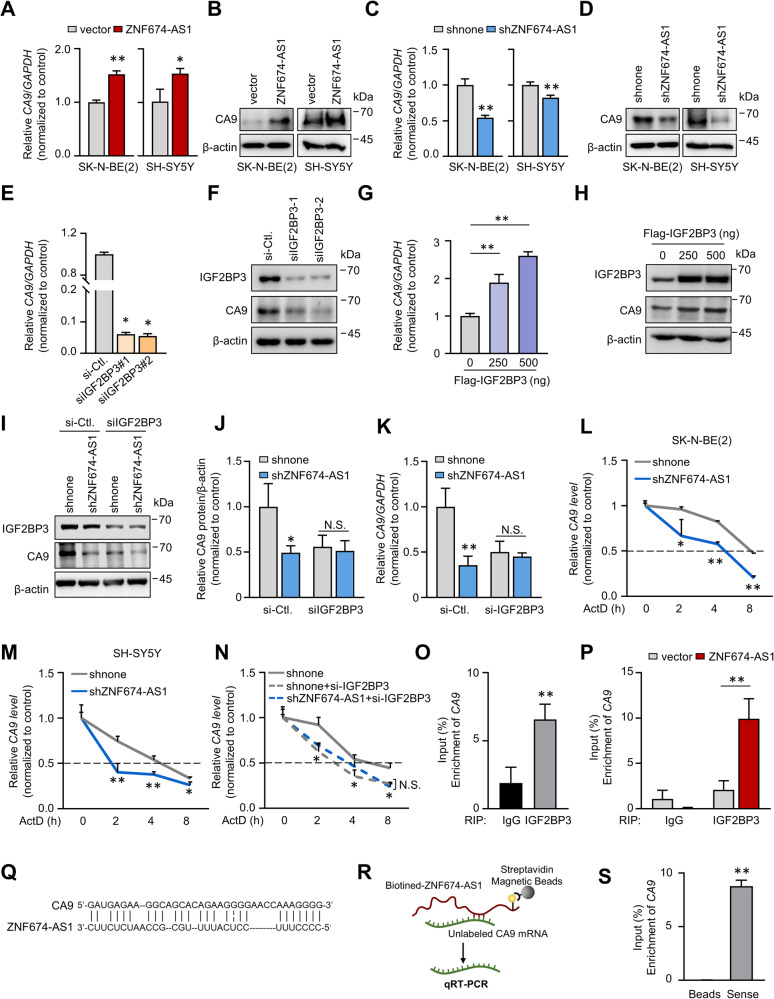
The cellular localization of lncRNAs is closely associated with their molecular mechanisms. Cell fraction and FISH results demonstrated that ZNF674-AS1 is mainly distributed in the cytoplasm (Fig. 1E, F), suggesting that ZNF674-AS1 may exert its function by interacting with cytoplasmic protein. Therefore, a biotin RNA-protein pull-down assay followed by mass spectrometry analysis was applied to identify proteins that potentially interact with ZNF674-AS1. By comparing with the paired antisense control group, we selected 16 candidates which were reviewed in the UniProtKB database based on the number of unique peptides (more than one) obtained from the MS identification, as listed in Supplementary Fig. S4A. In addition, we profiled ZNF674-AS1 interacting RBPs via CLIP-sequence data from the ENCORI database. Among these 49 RBPs, we focused on IGF2BP3 due to its predominantly cytoplasmic distribution rather than the nuclear-localized protein KHSRP (Figs. 5A and S4B). More importantly, the high expression of IGF2BP3 predicted poor prognosis of NB patients (Fig. S4C, D). The interaction between IGF2BP3 and ZNF674-AS1 was validated by western blot. IGF2BP3 co-precipitated with biotin-labeled ZNF674-AS1 in fresh cell lysate but not in the control beads group (Fig. 5B). In contrast, YWHAQ, which got the highest coverage score in the MS analysis result (Fig. S4A), did not bind to ZNF674-AS1 (Fig. 5B). In addition, RNA immunoprecipitation (RIP) assay was applied to validate the physical interaction between ZNF674-AS1 and IGF2BP3 in living cells. Compared with IgG, endogenous ZNF674-AS1 exhibited significant interaction with IGF2BP3, and ZNF674-AS1 overexpression led to increased binding with IGF2BP3 (Fig. 5C). We examined which domain in IGF2BP3 contributes to the interaction with ZNF674-AS1. Different IGF2BP3 truncations were generated, as indicated in Fig. 5D. Our data revealed that KH1/2 bound with ZNF674-AS1 efficiently (Fig. 5D). Supportively, we found that IGF2BP3 inhibition improved caspase-3 activation and N-GSDME cleavage, and also completely abolished the impact of ZNF674-AS1 (Fig. S5A). These data suggest that ZNF674-AS1 fulfills its functions by interacting with IGF2BP3.
Fig. 5. ZNF674-AS1 plays oncogenic roles via CA9.
A Venn diagram showing potential binding target genes by overlapping ZNF674-AS1 RNA pull-down data and IGF2BP3 CLIP-sequence data. B IGF2BP3 protein levels in biotin-labeled ZNF674-AS1 precipitates were analyzed by WB. YWHAQ was used as a negative control. C RIP assay was used to analyze the interaction between IGF2BP3 and ZNF674-AS1 following IGF2BP3 immunoprecipitation in the vector and stable ZNF674-AS1 overexpressing cells. IgG was used as a negative control. D ZNF674-AS1 levels were detected by qRT-PCR following Flag RIP assay in SK-N-BE(2) cells after transfection of Flag-tagged truncations IGF2BP3, as indicated in the diagram shown left. E Venn diagram showing the potential target genes after integrating RNA-seq data of ZNF674-AS1 KD by two independent siRNAs, RNA-seq data of neuroblastoma tissues, and IGF2BP3 CLIP-seq results. F The Kaplan-Meier curve showed the overall survival rate of neuroblastoma patients correlating with CA9 mRNA expression. G The mRNA expression levels of CA9 in low- and high-risk neuroblastoma tissues through the R2 platform. H, I The colonies image for control and ZNF674-AS1 OE (H) /ZNF674-AS1 KD (I) cells with or without U-104 treatment were presented (left), and the quantification of colonies numbers was shown in the bar graph (right). J The cell survival rates were detected after 10 μM cisplatin treatment cultured with indicating concentration U-104. K The cell survival rates of control and ZNF674-AS1 KD cells were analyzed after cisplatin treatment with or without 100 μM U-104 for 24 h. Data are derived from three independent experiments and presented as mean ± SD in the bar graphs. Values of controls were normalized to 1 (J, K). * P < 0.05; ** P < 0.01; N.S., not significant.
Next, to investigate the target genes that could be regulated by both ZNF674-AS1 and IGF2BP3, we performed RNA sequencing by using two independent siRNA targeting ZNF674-AS1. Among the transcripts, a total of 224 genes were downregulated in both two ZNF674-AS1 KD groups compared with the control group (Fig. 5E). Furthermore, by combining the ZNF674-AS1 correlation genes in NB tissues and IGF2BP3 target genes obtained from CLIP-seq in the ECORI database, MYLK3, TAS1R1 and CA9 were identified as three potential candidate genes (Fig. 5E). CA9, but not MYLK3 and TAS1R1, was finally selected as its high expression level was strongly associated with unfavorable prognosis and high-risk in NB patients (Figs. 5F, G and S5B).
In light of this discovery, we investigated whether the protumor effect or cisplatin insensitivity induced by ZNF674-AS1 depended on CA9. To this end, CA9 was inhibited by a specific targeting inhibitor, U-104 (SLC-0111), which has been used in clinical trials [31]. In line with a previous report, U-104 significantly inhibited NB cell growth and accelerated caspase3 activation, which triggered N-GSDME cleavage (Figs. 5H, I and S5C-F). Importantly, the effect of ZNF674-AS1 on cell proliferation was completely abrogated upon CA9 inhibition (Fig. 5H, I). Furthermore, considering that U-104 has been reported to enhance the effectiveness of chemotherapeutic drugs in killing cancer cells [32, 34], we wondered whether CA9 inhibition could improve the cisplatin sensitivity of NB cells. As shown in Fig. 5J, cell viability was slightly reduced by low levels of cisplatin, whereas U-104 treatment improved the cell viability inhibitory effects and pyroptosis levels (Fig. S5E) in a cisplatin dose-dependent manner. Consistent with the previous report, these data indicate that CA9 may be a potential target for enhancing the efficiency of chemical therapies. Furthermore, the effects of ZNF674-AS1 KD on cisplatin-induced cell death were completely abolished by U-104 (Fig. 5K). Collectively, these data suggest that ZNF674-AS1 alters cell proliferation and cisplatin-induced pyroptosis, mainly depending on CA9.
ZNF674-AS1 cooperates IGF2BP3 to promote CA9 expression
To explore how CA9 is involved in ZNF674-AS1 function, we examined the impact of ZNF674-AS1 on CA9 mRNA and protein levels. Intriguingly, ZNF674-AS1 overexpression increased both CA9 mRNA and protein expressions, while ZNF674-AS1 KD inhibited their levels (Fig. 6A–D). Recent evidence has shown that IGF2BP3 could modulate RNA stability and improve protein expression of target genes [35]. Since CA9 was identified as a potential binding target of IGF2BP3, we examined the effect of IGF2BP3 ectopic overexpression or KD on the mRNA and protein levels of CA9. As expected, IGF2BP3 overexpression resulted in an induction of CA9 mRNA and protein, while IGF2BP3 KD decreased their levels (Fig. 6E–H). More importantly, IGF2BP3 KD abrogated the ability of ZNF674-AS1 to affect CA9 expression (Fig. 6I–K). Given that IGF2BP3 is known to bind to mRNA transcripts and control their stability, we then investigated whether ZNF674-AS1/IGF2BP3 was involved in the stabilization of CA9. Transcription inhibitor actinomycin D (ActD) was used to assess the CA9 stability. As data shown in Fig. 6L, M, ZNF674-AS1 KD accelerated the decay of CA9 mRNA. Similarly, IGF2BP3 KD also led to faster degradation of CA9 mRNA and abrogated the effect of ZNF674-AS1 on CA9 mRNA half-life (Fig. 6N). Furthermore, we determined whether ZNF674-AS1 affected the interaction between IGF2BP3 and CA9. The binding of IGF2BP3 and CA9 mRNA was confirmed by RIP assay. Compared with IgG, CA9 mRNA was significantly enriched in IGF2BP3 antibody immunoprecipitate (Fig. 6O). Moreover, ectopic ZNF674-AS1 overexpression dramatically enhanced the association between IGF2BP3 and CA9 (Fig. 6P), demonstrating that the interaction between IGF2BP3 and CA9 was regulated by ZNF674-AS1. We further explored how ZNF674-AS1 fulfills such an ability and identified whether ZNF674-AS1 could interact with CA9 mRNA through unbiased prediction of RNA-RNA interactions by using IntraRNA [36]. As Fig. 6Q shows, the CA9 transcript was recognized as a target of ZNF674-AS1. Furthermore, the result of an RNA-RNA binding assay confirmed this in silico prediction (Fig. 6R, S) [37]. These data suggest that ZNF674-AS1 binds with CA9 mRNA bridges and strengthens the interaction of IGF2BP3 with CA9 mRNA, which contributes to increased CA9 mRNA stability.
Fig. 6. ZNF674-AS1 elevates CA9 mRNA stabilization through IGF2BP3.
A–D The mRNA (A, C) and protein (B, D) levels of CA9 were detected by qRT-PCR and WB in SK-N-BE(2) (left) and SH-SY5Y (right) stable ZNF674-AS1 OE (A, B) or KD (C, D) cells. E, F The mRNA (E) and protein (F) levels of CA9 were measured by qRT-PCR and WB after IGF2BP3 KD with two independent siRNA oligos. G, H CA9 mRNA (G) and protein (H) expression levels were analyzed after indicating IGF2BP3 overexpressing plasmids transfected. I, J The mRNA (I) and protein (J) expression levels of CA9 were measured by WB in ZNF674-AS1 KD stable cells, with or without IGF2BP3 KD. K, L The half-lives of CA9 mRNA in SK-N-BE(2) (K) and SH-SY5Y (L) ZNF674-AS1 KD cells were measured by qRT-PCR in the presence of ActD. M, N qRT-PCR analyzed the CA9 mRNA half-lives in SK-N-BE(2) (M) and SH-SY5Y (N) ZNF674-AS1 KD cell lines with or without IGF2BP3 KD. O CA9 levels enriched by IGF2BP3 were detected by qRT-PCR following RIP assay. P qRT-PCR analyzed the enrichment of CA9 by IGF2BP3 immunoprecipitating in vector and ZNF674-AS1 OE cells. Q Schematic of predicted interaction between ZNF674-AS1 and CA9 according to IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp). R Schematic diagram of RNA-RNA binding assay. S CA9 mRNA coprecipitate by biotin-ZNF674-AS1 determined by qRT-PCR after an RNA-RNA interaction assay. Data are derived from three independent experiments and presented as mean ± SD in the bar graphs. Values of controls were normalized to 1 (A, C, E, G, J, M–P). * P < 0.05; ** P < 0.01.
ZNF674-AS1 is clinically correlated with CA9 in human neuroblastoma tissues
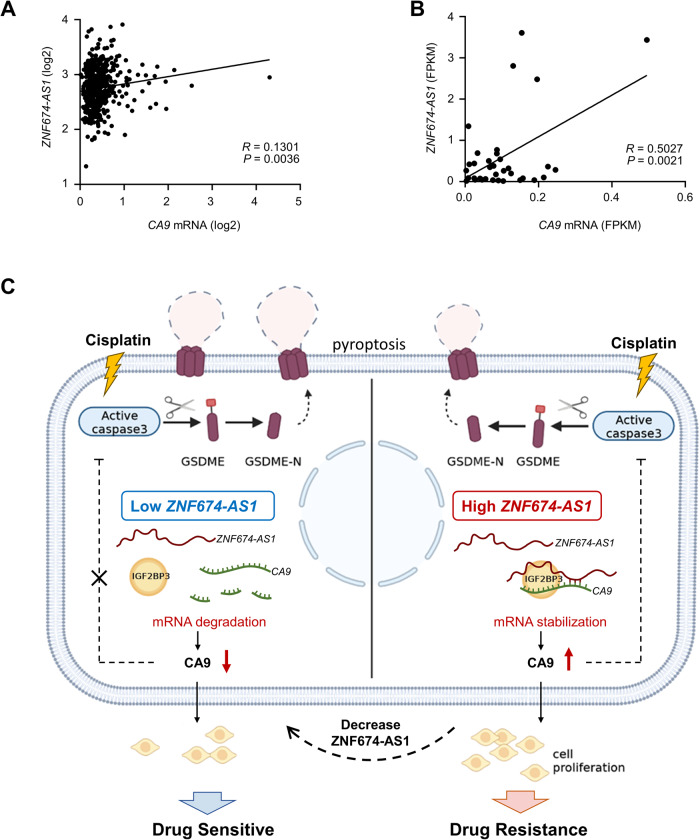
In finding a novel ZNF674-AS1-CA9 axis, we assessed the clinical relevance between ZNF674-AS1 and CA9 in human neuroblastoma tissues. Intriguingly, ZNF674-AS1 levels were positively correlated with increased CA9 mRNA levels both in publicly available neuroblastoma tissues and in our own sequencing data (Fig. 7A, B).
Fig. 7. The association of ZNF674-AS1 with CA9 in human neuroblastoma tissues.
A Correlation analysis of ZNF674-AS1 with CA9 mRNA in 498 human neuroblastoma tissues from the R2 platform. B Correlation analysis of ZNF674-AS1 with CA9 mRNA in 35 human neuroblastoma tissues. C Schematic diagram of the proposed model for ZNF674-AS1/IGF2BP3/CA9 axis modulating NB cisplatin sensitivity. Cytoplasmic ZNF674-AS1 binds with CA9 mRNA, recruits and elevates the capacity of IGF2BP3, and cooperates with IGF2BP3 to regulate their target transcript, CA9, leading to promote NB tumor development and limiting cisplatin-induced tumor cell pyroptosis. Such activity of ZNF674-AS1 is restrained in NB cells due to si-ZNF674-AS1 mediated ZNF674-AS1 inhibition or ZNF674-AS1 down-regulation, which recovers the sensitivity to cisplatin of NB cells. The diagram was created with BioRender.com.
Overall, our results demonstrate that lncRNA ZNF674-AS1, which significantly up-regulated in chemotherapy non-response NB patients, plays a crucial role in promoting NB tumor development and inhibiting cisplatin-induced tumor cell pyroptosis through up-regulating CA9 by binding IGF2BP3 (Fig. 7C). These findings suggest that targeting the ZNF674-AS1/CA9 axis may hold promise for developing more effective chemical therapeutic strategies.
Discussion
The long-term survival rates of neuroblastoma patients, particularly those classified as high-risk, remain a significant concern. It is crucial to improve chemosensitivity and prevent or bypass chemoresistance to enhance the prognosis of NB patients [38]. Therefore, developing novel potential targets related to chemotherapeutic treatment response rates, as well as revealing the underlying mechanisms, is essential for optimizing clinical chemotherapeutical schemes and the treatment of NB. Emerging evidence has highlighted the involvement of lncRNAs, including NB, in tumor development [39]. By analyzing our RNA sequencing data, we identified lncRNA ZNF674-AS1 as the most considerably up-regulated lncRNA in the chemotherapeutic non-response group compared to the chemotherapeutic response group. Remarkably, previous studies have suggested that ZNF674-AS1 acts as a tumor suppressor and has been implicated in tumorigenesis in non-small-cell lung cancer [28]. However, the role of ZNF674-AS1 in pediatric tumors, especially in neuroblastoma, remains unexplored. Analysis of publicly available databases revealed a strong correlation between high levels of ZNF674-AS1 and poor patient outcomes, as well as the high-risk subgroup. Combining our sequencing data, these findings suggest that ZNF674-AS1 may play a crucial role in the tumorigenesis of neuroblastoma and may contribute to the reduced efficacy of chemical therapies. It seems contradictory with the tumor suppressor function of ZNF674-AS1 as described in published works. In fact, it is worth noting that functional molecules, including lncRNA, can exhibit discrepant functions in different cancer types [39]. For example, NEAT1 drives prostate cancer progression through transcriptional regulation of prostate cancer-specific genes [40]. Intriguingly, NEAT1 suppresses tumorigenesis in acute promyelocytic leukemia (APL) by accelerating APL cell differentiation [41]. These examples highlight the complexity of lncRNA functions and emphasize the need for a more comprehensive investigation into their underlying mechanisms, especially across different cancer types. These intriguing findings have encouraged us to explore the potential roles of ZNF674-AS1 in modulating NB progression further.
In this study, we found that ZNF674-AS1 inhibition effectively limited NB tumorigenesis and sensitized NB cells to cisplatin-induced pyroptosis. The intracellular distribution of lncRNAs plays a decisive role in its molecular mechanism, lncRNAs can act as molecular decoys for RNA-binding proteins (RBPs), which cooperate with RBPs to regulate their targets [19]. Our data found that ZNF674-AS1 predominantly localized within the cytoplasm of NB cells. Consistent with our findings, ZNF674-AS1 has been reported to localize in the cytoplasm and regulate glycolysis and proliferation of granulosa cells by interacting with aldolase A (ALDOA) [42]. Our RNA pull-down assay and RIP assay provided evidence of the potential interaction between ZNF674-AS1 and the RBP IGF2BP3. IGF2BP3 is an oncofetal protein related to pro-tumorigenesis in various cancer types by regulating target mRNA at the post-transcriptional level. Recent evidence suggests that IGF2BP3 may play crucial roles in regulating the proliferation and migration of NB cells [43, 44]. Additionally, high IGF2BP3 levels were related to NB patients’ poor prognosis (Fig. S4C, D). Consistently, our data suggest that ZNF674-AS1 may play its regulatory role by recruiting IGF2BP3. Previous studies have reported the cooperative interactions between lncRNAs and IGF2BP3 in evaluating the mRNA stability of IGF2BP3 target genes [45]. In accordance with this idea, we identified Carbonic anhydrase IX (CA9) as the candidate target gene co-modulated by ZNF674-AS1 and IGF2BP3. Importantly, we observed a positive correlation between the expression levels of both ZNF674-AS1 and IGF2BP3 and the expressions of CA9. Furthermore, our findings provide evidence that ZNF674-AS1 not only interacts with IGF2BP3 but also enhances its binding capacity to CA9 by binding with the CA9 transcript. However, the intricate molecular mechanism governing the interplay among these three molecular complexes needs to be further investigated. In addition, the upstream regulation mechanism of ZNF674-AS1 is also important; whether ZNF674-AS1 expression could be altered under different conditions, such as chemotherapeutic drug treatment, warrants further investigation. These will provide more information about chemotherapy insensitivity and create new opportunities for therapeutic intervention by repressing ZNF674-AS1 in NB patients.
CA9 is a critical factor in tumorigenesis and has been identified as a potential therapeutic target for various cancers, including NB, due to its up-regulated expression in tumor tissues [46]. Additionally, increased CA9 expression in NB patients is inversely associated with overall survival and event-free survival, indicating its prognostic significance [30]. Several studies have demonstrated that CA9 up-regulation was associated with drug resistance, while the application of carbonic anhydrase inhibitors has shown promising effects in enhancing chemosensitivity [32, 46]. However, the association between CA9 and chemotherapy drug sensitivity in neuroblastoma has not been elucidated. Recently, Bayat et al. proposed that pre-inhibited carbonic anhydrases significantly potentiate the reduction of NB tumorigenesis by the HDAC inhibitor MS-275, indicating that CA9 inhibitor could be a promising therapeutic approach for NB patients [47]. Our study discovered that CA9 inhibition restrained cell proliferation and increased cisplatin-mediated pyroptosis in neuroblastoma cells by facilitating caspase3 activation. The connection between CA9 and caspase3 activation in NB cells remains to be investigated; whether CA9 may bind with other proteins to cooperate in modulating caspase3 activation warrants further investigation.
We identified lncRNA ZNF674-AS1 as a novel epigenetic modulator of CA9 by prolonging its mRNA stability. The regulation of CA9 expression has been mainly studied at the transcriptional level, particularly involving the transcriptional factor hypoxia-induced factor (HIF-1), which recognizes the hypoxia response element (HRE) of CA9 promoter and activates CA9 expression under hypoxia conditions [48]. Furthermore, various epigenetic regulators have been implicated in controlling CA9 expression. It has been reported that DNA methylation predominantly regulated CA9 expression in gastric cancer, while histone modifications [49], mediated by MORC2 and HDAC4, have been documented to control CA9 transcriptional activation through histone H3 deacetylation [50]. However, the involvement of other epigenetic regulators, such as noncoding RNAs, in CA9 regulation remains poorly understood. Sabrina et al. demonstrated that miR-34a accelerated CA9 mRNA degradation via base pairing with 3′UTR [51]. Notably, our findings provide additional regulators of CA9 that improve the stability of its mRNA. This discovery shed light on novel regulatory mechanisms that provide potential avenues for controlling CA9 expression.
Taken together, our findings highlight the crucial role of ZNF674-AS1 in the regulation of cisplatin’s antitumor effects by inhibiting pyroptosis and promoting cell growth. Inhibition of ZNF674-AS1 demonstrates a suppressive effect on NB initiation, indicating that targeting ZNF674-AS1 could be a promising therapeutic strategy.
Material and methods
Cell culture and chemicals
Human neuroblastoma cell lines SK-N-BE2, SH-SY5Y, and IMR-32 were purchased from ATCC and cultured in Dulbecco’s modified Eagle’s medium (DMEM), MEM-F12, or MEM (contained 10 mM NEAA) supplemented with 10% (v/v) fetal bovine serum (Biological industries). All cells were maintained at 37°C in a humidified incubator (Thermo Scientific) with 5% CO2. Critical chemicals used in this study were shown as follows: Cisplatin (MedChemExpress, HY-17394), U-104 (MedChemExpress, HY-13513), and Actinomycin D (MedChemExpress, HY-17559). The cisplatin treatment concentrations of SK-N-BE(2), SH-SY5Y, and IMR-32 cells were based on their IC50s of cisplatin. The dosing time of cisplatin treatment was 24 h.
Tissue samples
In our study, 35 cases of human neuroblastoma tissues were collected from the Beijing Children’s Hospital of Capital Medical University. All patients had undergone chemotherapy before surgery. The assessments of chemotherapeutic response were based on the 2015 neuroblastoma diagnosis and treatment expert consensus classification criteria. According to these classifications, 12 patients were classified as chemo-sensitive, while 23 patients were classified as chemo-resistant. The present study was approved by the Ethics Committees of Beijing Children’s Hospital. A total of 40 surgical specimens were immediately snap‑frozen in liquid nitrogen for subsequent total RNA extraction. Detailed clinical data are summarized in Table 1.
Table 1.
Clinical characteristics of patients with neuroblastoma enrolled in this study (n = 35).
| Parameter | Number of patients (%) |
|---|---|
| Age at diagnosis (years) | |
| <1.5 | 12 (34.29%) |
| 1.5–3 | 10 (28.57%) |
| 3–7 | 11 (31.43%) |
| >7 | 2 (5.71%) |
| Sex | |
| Male | 19 (54.29%) |
| Female | 16 (45.71%) |
| MYCN status | |
| Amplified | 5 (14.29%) |
| Non-Amplified | 25 (71.42%) |
| Not clear | 5 (14.29%) |
| Tumor stage (INSS stage) | |
| I | 0 |
| II | 1 (2.85%) |
| III | 11 (31.43%) |
| IV | 22 (62.86%) |
| IVS | 1 (2.86%) |
| MYCN, MYCN proto-oncogene | |
Lentiviral production and stable cell establishment
The shRNA targeting ZNF674-AS1 and the control sequences were 5′-CCGGGACTGGAATCCACCACTTACTCGAGTAAGTGGTGGATTCCAGTCTTTTTG-3′ and 5′-CCGGGCTGTGGCTCTAGACACTAAACTCGAGTTTAGTGTCTAGAGCCACAGCTTTTTG-3′) were synthesized by Tsingke Biotechnology (Beijing, China). The oligos were then subcloned into the PLKO.1 lentiviral vector. A full-length ZNF674-AS1 transcript was synthesized and subcloned into a lentiviral expression vector, as previously reported [52]. Then, the lentivirus was packaged using the established protocol [53]. Briefly, both control and recombinant plasmids were co-transfected with two helper plasmids into 293 T cells. The lentivirus was collected 48 h after transfection and used to infect the target cells. Stable cells were selected with puromycin.
Plasmids construction and siRNAs transfection
The Flag-IGF2BP3 plasmid was kindly provided by Prof. Sven Diederichs [54]. The indicated IGF2BP3 truncations were subcloned into a pcDNA3.1-3×Flag vector. The siRNA oligos used to target IGF2BP3 and GSDME were listed as follows: si-IGF2BP3-1(GGAATTGACGCTGTATAAT) and si-IGF2BP3-2(GAATCTTCAAGCACATTTA) for IGF2BP3; si-GSDME-1 (GCGGTCCTATTTGATGATGAA) and si-GSDME-2(GATGATGGAGTATCTGATCTT) for GSDME. All siRNAs were purchased from Tsingke Biotechnology (Beijing, China). These oligos were transfected into neuroblastoma cells by using Lipofectamine 2000 following the manufacturer’s instructions. After 48 h or 72 h of incubation, cells were collected for subsequent analysis.
RNA extraction and real-time PCR
TriZol reagent (Takara) was used to extract total RNA by following the manufacturer’s protocol. mRNAs were reverse transcribed by The PrimeScript reverse transcription (TaKaRa, #RR047A) reagent kit with gDNA Eraser. Real-time PCR analysis was carried out by using QuantiNova™ SYBR Green PCR reagent (Qiagen, Duesseldorf, Germany) in the LightCycler® 480 System (Roche, Basel, Switzerland). Gene expression levels relative to 18 S snRNA or GAPDH were calculated using the 2−ΔΔCT method. The primer sequences used in qRT-PCR are listed in Supplementary Table 1.
Western blot assay
Total cell lysates were isolated using the urea buffer (8 M Urea, 1 M Thiourea, 0.5% CHAPS, 50 mM DTT, and 24 mM Spermine). Then, equal amounts of proteins (20 μg) were separated by SDS-PAGE and transferred onto a PVDF membrane. After incubating with indicated primary and secondary antibodies, the signals were detected using an enhanced chemiluminescence ECL kit (Boster, CA, USA). Antibodies used in the study were listed in Supplementary Table 2.
RNA immunoprecipitation (RIP) assay
Cells were crosslinked with 1% formaldehyde for 10 min at room temperature; then, cells were harvested using lysis buffer supplemented with Protease Inhibitor Cocktail (MCE) and RNase inhibitor (Invitrogen). After sonication and centrifugation, the supernatants were collected and incubated with IgG or indicated antibodies overnight at 4°C. The complexes were then pulled down by Protein A/G after 2 h of incubation at room temperature. After five washes with wash buffer containing Protease Inhibitor Cocktail and RNase inhibitor, the immunoprecipitated RNAs were purified using Trizol and subjected to reverse transcription. The enrichment rate of the target RNA was analyzed by qRT-PCR.
RNA pull-down assay and RNA–RNA binding assay
Biotin-labeled RNA was synthesized in vitro using the T7 promoter by biotin RNA labeling Mix (Roche, 11685597910). After removing DNA templates with RNase-free DNase I, the biotin-labeled RNA was subjected to secondary structure recovery for RNA pull-down assay. Then, it was incubated with fresh cell lysates for 1 h at 4 °C after being captured with streptavidin magnetic beads (MCE, HY-K0208). Beads were then collected and washed with a washing buffer 5 times. Finally, the bound proteins were denatured using an SDS loading buffer and then analyzed by mass spectrum or WB. For RNA-RNA binding assay, biotin-labeled ZNF674-AS1 was captured with streptavidin magnetic beads for 2 h at 4 °C, then incubated with CA9 mRNA for overnight at 4 °C. After washing for 5 times, the amount of CA9 was detected by qRT-PCR.
RNA fluorescence in situ hybridization (FISH) assay
The FISH assay for ZNF674-AS1 was performed by using a lncRNA FISH kit obtained from Genepharma (Shanghai, China) following the manufacturer’s protocol. Briefly, cells adhered to the slides were fixed with 4% paraformaldehyde solution and then permeabilized using triton X-100. After blocking for 30 min at 37 °C, cells were incubated overnight at 37 °C with 1 μM Cy3-labeled probes in a hybridization buffer. After hybridization, cells were washed once with hybridization buffer and six times with 2× SSC buffer; the nuclei were then stained with DAPI for 5 min. Finally, images were acquired with a confocal microscope (Leica).
Immunofluorescence staining
Immunofluorescence staining assay was carried out as our previous protocol described [55]. Briefly, after 4% paraformaldehyde fixing, 0.1% Triton X-100 permeabilizing, and 3% BSA blocking, cells were incubated with the antibody overnight at 4 °C. After that, cells were incubated with fluorescently labeled secondary antibody for 1 h and nucleus was stained by DAPI for 3 min. Images were captured by a confocal microscope (Leica).
Cell proliferation and survival assays
Cell proliferation was measured using colorimetric MTT (MCE, HY-15924), colony formation, and Edu incorporation assays. For the MTT assay, control cells and stable ZNF674-AS1 OE or KD SK-N-BE(2) and SH-SY5Y cells were seeded in 96-well plates. The cell proliferation rates at indicated time points were assessed by measuring the absorbances of formazans at 490 nm. Colony formation assay was assessed by seeding 1 × 103 cells stable ZNF674-AS1 overexpression or KD SK-N-BE(2) and SH-SY5Y cells in 6-well plates. After culturing for 14 days, cells were stained using 0.5% crystal violet. The numbers of colonies (consisting of at least 50 cells) were counted by microscopy. Edu incorporation assays were performed following the manufacturer’s instructions. Briefly, stable ZNF674-AS1 OE or KD cells were incubated with Edu (50 μM) for 2 h. After fixation with 4% formaldehyde, cells were stained with Apollo® 488 and Hoechst 33342 to label Edu and the nucleus.
After drug treatment, cell survival was monitored using the CCK8 kit (MCE, HY-K0301) to indicate time. Cells were incubated with a 10% CCK8 solution for 2 h at 37 °C, and the absorbances were measured at 450 nm.
Cellular fraction assay
The cytoplasmic and nuclear fractions of treated cells were isolated using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (ThermoFisher Scientific) following the manufacturer’s protocol. Briefly, cells were lysed with cold CER I for 10 min, followed by the addition of CER II for a 1-min incubation on ice. After centrifugation, the supernatant was collected as the cytoplasmic component, while the pellet corresponded to the nuclear fraction.
LDH release assay
The levels of LDH in the cell culture supernatants were analyzed using the LDH assay kit (Beyotime Institute of Biotechnology) according to the manufacturer’s protocol. Briefly, the supernatants were collected and incubated with LDH detection reagents in the dark for 30 min. After incubation, the absorbance was determined by a spectrophotometric microplate reader at 450 nm.
PI staining assay
After treatment with the indicated concentration of cisplatin, cells were incubated with PI (10 μg/ml) in a cell incubator for 15 min. Then, the cells were photographed using an inverted fluorescence microscope (Olympus, Tokyo, Japan).
RNA sequencing analysis
Total RNA was extracted from cells transfected with two independent siRNA targeting ZNF674-AS1 and control cells and was subjected to high-throughput sequencing by Oebiotech (Shanghai, China). The strand-specific RNA libraries were constructed using TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Finally, these libraries were sequenced on the HiSeq2500 Illumina sequencing platform, and 125 bp paired-end reads.
In vivo tumor xenografts
Tumor xenografts assay was conducted by following our previous protocol [56]. All animal procedures were performed according to the Rules for Animal Experiments published by the Chinese Government (Beijing, China) and approved by the Research Ethics Committee of Qingdao University, China. Briefly, 5 × 106 control (shnone) or ZNF674-AS1 KD (shZNF674-AS1) stable cells were subcutaneously inoculated into female M-NSG mice (4 and 5 weeks old). The tumor volumes were monitored every 3 days and calculated using the formula: length × width2 × 0.5. At the end of the experiment, mice were anesthetized and culled. M-NSG mice (Cat. NO. NM-NSG-001) were purchased from Shanghai Model Organisms Center, Inc.
Statistical analysis
Statistical analysis was done using GraphPad software, version 8. Data are presented as means ± standard deviation (SD). A two-sided student t-test or One-way analysis of variance (ANOVA) was applied to assess the statistical significance. Correlations were calculated using Spearman or Pearson correlation coefficients. A P value less than 0.05 was considered statistically significant.
Supplementary information
Author contributions
KZ and DY proposed the study and wrote the paper. XN, YG, and DY organized and revised the paper. KZ, XW, and YJ performed all of the experiments and analyzed the data. XZ generated plasmids and performed the experiments. JL reviewed and revised the paper. TZ, YY, XJ, and YC analyzed the data. YJ, YG, and XN collected clinical samples, performed the bioinformatic analysis, and analyzed the data.
Funding
This work was funded by the National Natural Science Foundation of China (82204087 and 81973075) and the Natural Science Foundation of Shandong Province (ZR2021QH278).
Data availability
All data are present in the manuscript and supplementary files. Additional data related to this paper may be requested from the corresponding author.
Competing interests
The authors declare no competing interests.
Ethics
The study for human neuroblastoma samples was approved by the Ethics Committees of Beijing Children’s Hospital, China. All animal procedures were performed according to the Chinese Government published Rules for Animal Experiments (Beijing, China) and approved by the Research Ethics Committee of Qingdao University, China.
Footnotes
Edited by Barak Rotblat
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These author contributed equally: Kunming Zhao, Xinyi Wang, Yaqiong Jin.
Change history
2/3/2025
The original online version of this article was revised: In this article fig. 4 has been updated.
Change history
2/26/2025
A Correction to this paper has been published: 10.1038/s41419-025-07371-z
Contributor Information
Xin Ni, Email: nixin@bch.com.cn.
Yongli Guo, Email: guoyongli@bch.com.cn.
Dianke Yu, Email: dianke.yu@qdu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-023-06394-8.
References
- 1.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev cancer. 2003;3:203–16. [DOI] [PubMed] [Google Scholar]
- 2.Johnsen JI, Dyberg C, Wickström M. Neuroblastoma—a neural crest derived embryonal malignancy. Front Mol Neurosci. 2019;12:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma (Primer). Nat Rev: Dis Prim. 2016;2:16078. [DOI] [PubMed] [Google Scholar]
- 4.Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, et al. Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol. 2015;33:3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, et al. Advances in Risk Classification and Treatment Strategies for Neuroblastoma. J Clin Oncol. 2015;33:3008–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park JR, Bagatell R, London WB, Maris JM, Cohn SL, Mattay KM, et al. Children’s Oncology Group’s 2013 blueprint for research: neuroblastoma. Pediatr Blood Cancer. 2013;60:985–93. [DOI] [PubMed] [Google Scholar]
- 7.Peng F, Liao M, Qin R, Zhu S, Peng C, Fu L, et al. Regulated cell death (RCD) in cancer: key pathways and targeted therapies. Signal Transduct Target Ther. 2022;7:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loftus LV, Amend SR, Pienta KJ. Interplay between cell death and cell proliferation reveals new strategies for cancer therapy. Int J Mol Sci. 2022;23:4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Gao W, Shi X, Ding J, Liu W, He H, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C-C, Li C-G, Wang Y-F, Xu L-H, He X-H, Zeng Q-Z, et al. Chemotherapeutic paclitaxel and cisplatin differentially induce pyroptosis in A549 lung cancer cells via caspase-3/GSDME activation. Apoptosis. 2019;24:312–25. [DOI] [PubMed] [Google Scholar]
- 11.Yu P, Zhang X, Liu N, Tang L, Peng C, Chen X. Pyroptosis: mechanisms and diseases. Signal Transduct Target Ther. 2021;6:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovacs SB, Miao EA. Gasdermins: effectors of pyroptosis. Trends Cell Biol. 2017;27:673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8:14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Zhang Y, Xia S, Kong Q, Li S, Liu X, et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579:415–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan C-Y, Ye F-H, Peng M, Dong J-J, Chai W-W, Deng W-J, et al. Endogenous HMGB1 regulates GSDME-mediated pyroptosis via ROS/ERK1/2/caspase-3/GSDME signaling in neuroblastoma. Am J Cancer Res. 2023;13:436. [PMC free article] [PubMed] [Google Scholar]
- 16.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Long Non Coding RNA Biol. 2017;1008:47–74. [DOI] [PubMed]
- 19.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer cell. 2016;29:452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao X, Li D, Huang D, Song H, Mei H, Fang E, et al. Risk-associated long noncoding RNA FOXD3-AS1 inhibits neuroblastoma progression by repressing PARP1-mediated activation of CTCF. Mol Ther. 2018;26:755–73. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Liu PY, Tee AE, Milazzo G, Hannan KM, Maag J, Mondal S, et al. The long noncoding RNA lncNB1 promotes tumorigenesis by interacting with ribosomal protein RPL35. Nat Commun. 2019;10:5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitra S, Muralidharan SV, Di Marco M, Juvvuna PK, Kosalai ST, Reischl S, et al. Subcellular distribution of p53 by the p53-responsive lncRNA NBAT1 determines chemotherapeutic response in neuroblastoma. Cancer Res. 2021;81:1457–71. [DOI] [PubMed] [Google Scholar]
- 24.Xu Z, Sun Y, Wang D, Sun H, Liu X. SNHG16 promotes tumorigenesis and cisplatin resistance by regulating miR-338-3p/PLK4 pathway in neuroblastoma cells. Cancer Cell Int. 2020;20:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan P, Su W, Zhang Y, Li Z, Deng C, Li J, et al. LncRNA H19 initiates microglial pyroptosis and neuronal death in retinal ischemia/reperfusion injury. Cell Death Differ. 2020;27:176–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai J, Qu T, Yin D, Cui Y, Zhang C, Zhang E, et al. LncRNA LINC00969 promotes acquired gefitinib resistance by epigenetically suppressing of NLRP3 at transcriptional and posttranscriptional levels to inhibit pyroptosis in lung cancer. Cell Death Dis. 2023;14:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan H, Luo B, Wu X, Guan F, Yu X, Zhao L, et al. Cisplatin induces pyroptosis via activation of MEG3/NLRP3/caspase-1/GSDMD pathway in triple-negative breast cancer. Int J Biol Sci. 2021;17:2606–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Huang R, Xie D, Lin X, Zheng L. ZNF674-AS1 antagonizes miR-423-3p to induce G0/G1 cell cycle arrest in non-small cell lung cancer cells. Cell Mol Biol Lett. 2021;26:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D, Xie Y, Sun J, Zhang L, Jiang W. LncRNA ZNF674-AS1 hinders proliferation and invasion of hepatic carcinoma cells through the glycolysis pathway. J Oncol. 2022;2022:8063382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ameis HM, Drenckhan A, Freytag M, Izbicki JR, Supuran CT, Reinshagen K, et al. Carbonic anhydrase IX correlates with survival and is a potential therapeutic target for neuroblastoma. J Enzym Inhib Med Chem. 2016;31:404–9. [DOI] [PubMed] [Google Scholar]
- 31.Bozdag M, Ferraroni M, Ward C, Carta F, Bua S, Angeli A, et al. Carbonic anhydrase inhibitors based on sorafenib scaffold: design, synthesis, crystallographic investigation and effects on primary breast cancer cells. Eur J Med Chem. 2019;182:111600. [DOI] [PubMed] [Google Scholar]
- 32.Xu X, Wang Z, Liu N, Cheng Y, Jin W, Zhang P, et al. Association between SOX9 and CA9 in glioma, and its effects on chemosensitivity to TMZ. Int J Oncol. 2018;53:189–202. [DOI] [PubMed] [Google Scholar]
- 33.Meczes E, Pearson A, Austin C, Tilby M. Schedule-dependent response of neuroblastoma cell lines to combinations of etoposide and cisplatin. Br J Cancer. 2002;86:485–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarnella A, Ferrara Y, Auletta L, Albanese S, Cerchia L, Alterio V, et al. Inhibition of carbonic anhydrases IX/XII by SLC-0111 boosts cisplatin effects in hampering head and neck squamous carcinoma cell growth and invasion. J Exp Clin Cancer Res. 2022;41:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanniford D, Ulloa-Morales A, Karz A, Berzoti-Coelho MG, Moubarak RS, Sánchez-Sendra B, et al. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell. 2020;37:55–70.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mann M, Wright PR, Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45:W435–w439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Q, Liu T, Wang X, Hou G, Xiang Z, Zhang W, et al. Long noncoding RNA HITT coordinates with RGS2 to inhibit PD-L1 translation in T cell immunity. J Clin Investig. 2023;133:e162951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature. 2019;575:299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Wang X, Song W, Xu H, Huang R, Wang Y, et al. Oncogenic properties of NEAT1 in prostate cancer cells depend on the CDC5L–AGRN transcriptional regulation circuit. Cancer Res. 2018;78:4138–49. [DOI] [PubMed] [Google Scholar]
- 41.Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L, et al. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC cancer. 2014;14:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li D, Wang X, Li G, Dang Y, Zhao S, Qin Y. LncRNA ZNF674-AS1 regulates granulosa cell glycolysis and proliferation by interacting with ALDOA. Cell Death Discov. 2021;7:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu K, Gao T, Wang Z, Zhang L, Tan K, Lv Z. RNA N6-methyladenosine reader IGF2BP3 interacts with MYCN and facilitates neuroblastoma cell proliferation. Cell Death Discov. 2023;9:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia C, Tang H, Yang Y, Yuan S, Han T, Fang M, et al. Ubiquitination of IGF2BP3 by E3 ligase MKRN2 regulates the proliferation and migration of human neuroblastoma SHSY5Y cells. Biochem Biophys Res Commun. 2020;529:43–50. [DOI] [PubMed] [Google Scholar]
- 45.Gu Y, Niu S, Wang Y, Duan L, Pan Y, Tong Z, et al. DMDRMR-mediated regulation of m6A-modified CDK4 by m6A reader IGF2BP3 drives ccRCC progression. Cancer Res. 2021;81:923–34. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Jiang L, Chew SH, Hirayama T, Sekido Y, Toyokuni S. Carbonic anhydrase 9 confers resistance to ferroptosis/apoptosis in malignant mesothelioma under hypoxia. Redox Biol. 2019;26:101297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayat Mokhtari, Baluch R, Ka Hon Tsui N, Kumar S M, S. Homayouni T, Aitken K, et al. Acetazolamide potentiates the anti-tumor potential of HDACi, MS-275, in neuroblastoma. BMC Cancer. 2017;17:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonald PC, Chafe SC, Brown WS, Saberi S, Swayampakula M, Venkateswaran G, et al. Regulation of pH by carbonic anhydrase 9 mediates survival of pancreatic cancer cells with activated KRAS in response to hypoxia. Gastroenterology. 2019;157:823–37. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura J, Kitajima Y, Kai K, Hashiguchi K, Hiraki M, Noshiro H, et al. Expression of hypoxic marker CA IX is regulated by site-specific DNA methylation and is associated with the histology of gastric cancer. Am J Pathol. 2011;178:515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao Y, Li Y, Zhang J, Liu D, Liu F, Zhao Y, et al. Involvement of histone deacetylation in MORC2-mediated down-regulation of carbonic anhydrase IX. Nucleic Acids Res. 2010;38:2813–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Carolis S, Bertoni S, Nati M, D’Anello L, Papi A, Tesei A, et al. Carbonic anhydrase 9 mRNA/microRNA34a interplay in hypoxic human mammospheres. J Cell Physiol. 2016;231:1534–41. [DOI] [PubMed] [Google Scholar]
- 52.Huang X, Pan L, Zuo Z, Li M, Zeng L, Li R, et al. LINC00842 inactivates transcription co-regulator PGC-1α to promote pancreatic cancer malignancy through metabolic remodelling. Nat Commun. 2021;12:3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao K, Wang X, Zhao D, Lin Q, Zhang Y, Hu Y. lncRNA HITT inhibits lactate production by repressing PKM2 oligomerization to reduce tumor growth and macrophage polarization. Research. 2022;2022:9854904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutschner T, Hämmerle M, Pazaitis N, Bley N, Fiskin E, Uckelmann H, et al. Insulin‐like growth factor 2 mRNA‐binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology. 2014;59:1900–11. [DOI] [PubMed] [Google Scholar]
- 55.Zhao K, Wang X, Xue X, Li L, Hu Y. A long noncoding RNA sensitizes genotoxic treatment by attenuating ATM activation and homologous recombination repair in cancers. PLoS Biol. 2020;18:e3000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Li L, Zhao K, Lin Q, Li H, Xue X, et al. A novel LncRNA HITT forms a regulatory loop with HIF-1α to modulate angiogenesis and tumor growth. Cell Death Diff. 2020;27:1431–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are present in the manuscript and supplementary files. Additional data related to this paper may be requested from the corresponding author.