Abstract
In order to form the catalytic nucleoprotein complex called the invertasome in the Hin-mediated DNA inversion reaction, interactions of the DNA-binding proteins Hin and Fis are required. Assays for these protein-protein interactions have been exploited with protein cross-linkers in vitro. In this study, an in vivo assay system that probes protein-protein interactions was developed. The formation of a DNA loop generated by protein interactions resulted in transcriptional repression of an artificially designed operon, which in turn increased the chance of survival of Escherichia coli host cells in a streptomycin-containing medium. Using this system, we were able to assay the Hin-Hin interaction that results in the pairing of the two recombination sites and protein interactions that result in the formation of the invertasome. This assay system also led us to find that an individual Hin dimer bound on a recombination site can form a stable complex with Fis bound on the recombinational enhancer; this finding has never been observed in in vitro studies. Possible pathways toward the formation of the invertasome are discussed based on the assay results for a previously reported Hin mutant.
Hin invertase from Salmonella typhimurium belongs to the recombinase family, which includes Gin invertase from phage Mu, Cin invertase from phage P1, and resolvases from Tn3 and the transposon γδ (8). Hin promotes the inversion of a chromosomal DNA segment of 996 bp that is flanked by the 26-bp DNA sequences of hixL and hixR (19). Hin-mediated DNA inversion in S. typhimurium leads to the alternative expression of the H1 and H2 flagellin genes known as phase variation. Hin (21 kDa) exists in solution as a homodimer and binds to hix sites as a dimer (7). In addition to Hin and the two hix sites, a cis-acting DNA sequence (recombinational enhancer) and its binding protein (Fis, 11 kDa) are required for efficient inversion in vitro (18).
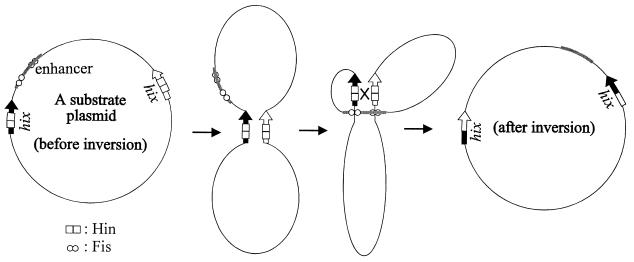
The inversion reaction requires the interaction of DNA-binding proteins at a distance. It has been generally accepted that after DNA binding, a two-step pathway is used to assemble a functional synaptic complex (Fig. 1) (15). The first step is to bring distant hix sites in close proximity through the interaction of hix-bound Hin proteins, forming a “paired-hix” structure (12). Negative supercoiling assists the Hin-Hin interaction to promote the formation of the paired-hix structure (26). The next step is to assemble the paired-hix structure with the Fis-bound enhancer to make a nucleoprotein complex called an invertasome (12). Negative supercoiling is essential for the formation of the invertasome (12, 26). However, no experimental data support the notion that the paired-hix structure is required for the formation of the invertasome.
FIG. 1.
Schematic expression of the Hin-mediated inversion reaction. Negative supercoiling of the substrate plasmid is required for efficient inversion. Negative supercoiling is not included in these drawings to show protein interactions more clearly. Hin binds to the two hix sites and Fis binds to the enhancer in a substrate plasmid. Hin pairs the hix sites, creating two DNA loops. All the necessary proteins and their DNA-binding sites are brought in close proximity to form the invertasome, creating three DNA loops of different sizes. In the invertasome, Hin cleaves the middle of each hix site and exchanges the cleaved DNA ends, followed by ligation. The intervening DNA between the two hix sites is now inverted.
This nucleoprotein complex has been suggested to form at a branch point in negatively supercoiled DNA (21). It is in the invertasome complex that Hin is able to cleave the middle hix sites and exchange the cleaved DNA ends to bring the intervening DNA to an inverted configuration (17). A current hypothesis is that Fis interacting with Hin in the invertasome triggers a conformational change(s) in the dimer interface to initiate DNA cleavage by Hin (11, 25). The region of Fis that is responsible for triggering the change in Hin resides in the N terminus (29) and contains a flexible β-hairpin structure (32). Recently, it was suggested that Hin needs to separate (melt) the two DNA strands at the hix sites after DNA cleavage to perform strand exchange (24). After the strand exchange, the DNA ends can be religated by Hin.
Protein interactions that result in hix pairing and the formation of the invertasome have been assayed in vitro. By use of protein cross-linkers, the paired-hix structure and the invertasome formed on a plasmid DNA were visualized by electron microscopy (12). The same structures were also detected as discrete bands by agarose gel electrophoresis (26). In this study, we devised an in vivo assay system to study protein-protein interactions on DNA at a distance. We were able to show not only that the paired-hix structure and the invertasome are formed and stably maintained in vivo but also that an individual Hin dimer bound on hix can form a stable complex with Fis bound on the enhancer; the latter finding has never been observed in in vitro studies. With the in vivo assay system, the hix-pairing and invertasome-forming activities of Hin mutants, each with a single-amino-acid change in the dimer interface, were analyzed. The consequences of the assay results are discussed in the context of the protein-protein interactions necessary to assemble the invertasome.
MATERIALS AND METHODS
Construction of plasmids.
Plasmids with an artificial operon were constructed as follows. The rpsL gene isolated on a 0.75-kb HindIII-EcoRI fragment from pHSG664 (9) was cloned into pBluescript II SK+ (Stratagene) digested with HindIII and EcoRI, creating pHL100. An EcoRI site was created in the middle of the Pribnow box of the rpsL gene in pHL100 by changing the adenine residue (base 78 upstream from the ATG start codon of the rpsL gene) (31) to guanine by a site-directed mutagenesis method (23), creating pHL101. The small EcoRI fragment (155 bp, from pHL101) generated as the result of site-directed mutagenesis was replaced with a 341-bp EcoRI fragment containing the promoter ant and the operator hixL-AT (Fig. 2) from pKH37 (K. T. Hughes, University of Washington), creating pHL102. A BamHI fragment from pHL102 containing the entire open reading frame of the rpsL gene, the promoter, and the operator was filled in with the large fragment of E. coli DNA polymerase I (Klenow fragment; Bethesda Research Laboratories) and cloned into the filled-in EcoRI site of low-copy-number plasmid pLG339 (35) in the orientation opposite that of the tetracycline resistance gene in pLG339. The plasmid generated was designated pSingle. pDouble1 was constructed by cloning a synthetic 33-bp DNA containing the hixL-WT (Fig. 2) and EagI sites on both ends into the EagI site of pSingle. This cloning placed the hixL-WT site about 1.1 kb away from hixL-AT and toward ori of pSC101. pDouble2 was generated by replacing a 300-bp EcoRV DNA fragment of pSingle with the synthetic DNA of hixL-WT containing blunt ends. pTriple was generated by replacing a 300-bp EcoRV fragment of pDouble2 with a synthetic 74-bp DNA containing the recombinational enhancer sequence. DNA sequence analysis showed that the 300-bp EcoRV fragment removed from pDouble2 does not have any functional determinants that are essential for the purpose of this study. This cloning placed the enhancer and hixL-WT sites 200 bp away from hixL-AT. pDouble3 was generated by removing the hixL-WT sequence from pTriple by EagI digestion and ligation. To make pDouble3′, a synthetic 32-bp DNA fragment containing the proximal half site of the enhancer (5′-TCGGGTGTCAACAATTGACCAAAATATCGATT-3′) was inserted into the EcoRV site of pSingle. The nucleotide sequences of the recombined promoter, operator, and rpsL structural gene regions of these plasmids were confirmed by a double-stranded DNA sequencing method (28).
FIG. 2.
(A) Five rpsL operon-containing plasmids used to analyze protein interactions in vivo. All the plasmids have the wild-type rpsL gene under the control of the ant promoter from phage P22. The common DNA sequence surrounding the ant promoter is given. RBS, ribosome binding site. The difference between the plasmids is the number or the position of operators. pSingle has a single hix operator, pDouble1, pDouble2, and pDouble3 have two operators, and pTriple has three operators. (B) DNA sequences of the two hix sites used as operators. These two hix sequences differ only in the central 2 bp (bold), preventing inversion. (C) Schematic representation of the Hin-producing plasmid pHinWT. This plasmid was derived from pBluescript II SK+.
A high-copy-number plasmid used to express the Hin protein (pHinWT) was constructed as follows. A DNA fragment harboring both the wild-type hin gene under the control of the tac promoter and the lacIq gene was isolated by partial digestion of pKH66 (14) with HindIII and EcoRI and ligated to HindIII- and EcoRI-digested pBluescript II SK+.
Measuring SF.
Streptomycin-resistant E. coli HB101 (supE44 hsdS20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1) harboring both the rpsL operon-containing plasmid and the Hin-producing plasmid was grown at 37°C for 12 h in 2 ml of Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), and isopropyl-β-d-thiogalactopyranoside (IPTG) (20 μM). Proper serial dilutions of the culture were made and plated on two different agar plates. One contained kanamycin and ampicillin, and the other contained streptomycin (100 μg/ml), kanamycin, and ampicillin. Both plates contained a 20 μM concentration of IPTG. The plates were incubated at 37°C for 24 h. Colony numbers were counted, and the survival frequency (SF) was calculated. SF was defined as the ratio of Strr Ampr Kanr colonies per milliliter to Ampr Kanr colonies per milliliter. Measuring the SF in a Fis-negative Strr strain, CSH50fis::cat (34), was performed as described above. CSH50fis::cat is a fis derivative of CSH50 [ara Δ(pro-lac) thi rpsL].
Recombination on pTriple.
Even though we used two different hix operators to prevent any recombination, it is possible that a small amount of inversion or deletion between the hix operators in pTriple might have led to the streptomycin-resistant phenotype of HB101/pTriple/pHinWT. Thus, we investigated if any recombination occurred when pTriple (Kanr) coexisted with pHinWT (Ampr) in HB101. First, fresh HB101 was transformed with plasmids isolated from an overnight culture of HB101/pTriple/pHinWT, and transformants were plated on solid media containing kanamycin to select ones that had pTriple. Five hundred Kanr colonies were stabbed on two different solid media, one containing both ampicillin and kanamycin (to look for those transformed with both pTriple and pHinWT) and the other containing kanamycin and streptomycin. All the Kanr but Amps colonies (about 450) were Strs. Thus, this experiment showed that in vivo Hin could not perform either inversion or deletion on pTriple. If either inversion or deletion had occurred on pTriple in HB101/pTriple/pHinWT, fresh HB101 transformed with recombinant pTriple would have been Strr. The same experiment was performed with HB101/pDouble1/pHinWT and HB101/pDouble2/pHinWT, and all the Kanr and Amps colonies were Strs. Again, Hin was not able to carry out any type of recombination on pDouble1 and pDouble2.
Western blot analysis.
The amount of Hin produced in HB101 harboring the Hin-producing plasmid with different concentrations of IPTG was measured by Western blot analysis. Cells grown to an optical density at 600 nm of 0.2 in 20 ml of LB medium with the proper antibiotics were pelleted and washed with 1 ml of 50 mM Tris-HCl (pH 7.5). Cells were resuspended in 50 μl of 1× sodium dodecyl sulfate (SDS) gel loading buffer and boiled for 3 min. Samples were centrifuged at 13,000 × g for 5 min, and 20 μl of supernatant was loaded on an SDS–12.5% polyacrylamide gel. Hin protein bands were visualized with horseradish peroxidase-conjugated secondary antibodies (Amersham).
RESULTS
Rationale for the in vivo assay of protein interactions occurring at a distance.
The initial idea for our in vivo assay system to study protein interactions in the Hin-mediated inversion reaction was based on the following observations. First, for more efficient repression of a promoter, a prokaryotic gene regulation apparatus frequently uses more than one operator. Having more operators increases the local concentration of repressors through protein-protein interactions (reviewed in references 1 and 33). Such interactions often create a DNA loop. In addition, it has been shown that the DNA loop itself represses a promoter more efficiently than does the mere binding of repressors to operators. Second, Escherichia coli strains, such as HB101, having a mutant allele of rpsL (rpsL20, coding for ribosomal protein S12) in their chromosome are resistant to the antibiotic streptomycin. Since the rpsL20 allele is recessive, a merozygotic strain that harbors the wild-type rpsL gene on a plasmid becomes Strs (6, 30).
We constructed a series of plasmids that bear an artificial rpsL operon on a low-copy-number plasmid, pLG339 (Kanr) (35), derived from pSC101 (5). The artificial operon was built such that hix sites and the recombinational enhancer site served as operators (Fig. 2A). To prevent recombination (deletion or inversion) between the hix sites in the plasmids, we used two hix sites that are different only in the central two bases (hixL-WT for the second operator [O2] and hixL-AT for the first operator [O1]) (Fig. 2B). Inversion between these two hix sites is blocked because the central two bases become noncomplementary after strand exchange (13). We confirmed that, in vivo, Hin cannot perform either inversion or deletion on the substrate plasmids (see Materials and Methods). The original promoter for the rpsL structural gene was removed, and the ant promoter from phage P22 (2, 14) was inserted instead. The Hin protein that served as a repressor in this system was supplied from a high-copy-number plasmid, pHinWT (Ampr) (Fig. 2C), that was derived from pBluescript II SK+. These two plasmids were compatible in E. coli. Another trans-acting protein, Fis, was provided from the fis gene in the chromosome of the host HB101 (16, 22).
We hypothesized that the growth of HB101 harboring both the Hin-producing plasmid pHinWT and one of the rpsL operon-containing plasmids in the presence of streptomycin would be dependent on how tightly transcription from the wild-type rpsL gene is repressed. The state of repression would depend on what kind of operators are present in the rpsL operon and how they are organized. Therefore, we expected that we would be able to analyze macromolecular interactions, such as those of Hin-hix, Hin-Hin, and Hin-Fis, by measuring the numbers of colonies of HB101 cells harboring both pHinWT and an rpsL operon-containing plasmid on agar plates containing streptomycin.
Assay for streptomycin sensitivity.
First of all, the streptomycin sensitivity of HB101 harboring the artificial rpsL operon in a plasmid was tested when the hin gene was not present. A single colony of HB101 having both pBluescript II SK+ (Ampr) and one of the rpsL operon-containing plasmids (Kanr) was grown in 2 ml of LB medium containing ampicillin and kanamycin for 12 h. Cells were plated on agar plates containing kanamycin and ampicillin and agar plates containing those antibiotics plus streptomycin, and the SF in the presence of streptomycin (ratio of Strr Ampr Kanr colonies per milliliter to Ampr Kanr colonies per milliliter) was calculated. The SFs of HB101 cells having both rpsL operon-containing plasmids and pBluescript were within the range of 10−7 (Table 1), showing that HB101, originally Strr, became Strs when it harbored the wild-type rpsL gene in these plasmids. HB101 carrying the promoterless rpsL gene in a plasmid was Strr (data not shown). These results indicated that the inserted ant promoter successfully drives the transcription of the rpsL structural gene when Hin, the repressor, is not present.
TABLE 1.
SFs with different concentrations of IPTGa
| rpsL-containing plasmid | SF of the following strain with the indicated IPTG concn (μM)
|
|||
|---|---|---|---|---|
| HB101/pBluescript II SK+ (0) | HB101/pHinWT
|
|||
| 0b | 10 | 20 | ||
| pSingle | 10−7 | 4 × 10−4 | 2 × 10−3 | 2 × 10−2 |
| pDouble1 | 10−7 | 2 × 10−4 | 5 × 10−3 | 2 × 10−2 |
| pDouble2 | 10−7 | 7 × 10−5 | 5 × 10−4 | 0.22 |
| pDouble3 | 10−7 | 5 × 10−4 | 3 × 10−2 | 0.27 |
| pTriple | 10−7 | 5 × 10−4 | 6 × 10−2 | 0.57 |
The SFs are averages from three independent experiments.
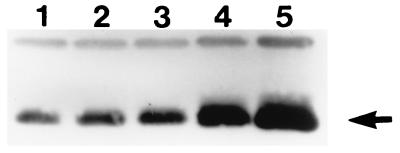
A Western blot (see Fig. 3) showed that a small amount of Hin could be made without the gratuitous inducer.
Assay for Hin binding to hix sites.
Our in vivo assay for Hin binding to hix was performed with an rpsL operon-containing plasmid, pSingle (Fig. 2A), that has a single hix operator (O1) between the ant promoter and the wild-type rpsL structural gene. We anticipated that the binding of Hin to hix would result in the repression of transcription from the ant promoter, which in turn would increase the SF of the host cells. Because the wild-type hin gene in the plasmid pHinWT is under the control of the tac promoter, the SF of HB101 harboring both pHinWT and pSingle (HB101/pHinWT/pSingle) was investigated with increasing amounts of the gratuitous inducer IPTG. As the concentration of IPTG increased, the SF of HB101/pSingle improved up to 10−2 (Table 1). Western blot analysis of HB101/pHinWT showed that as the IPTG concentration increased from 0 to 40 μM, Hin production also increased accordingly (Fig. 3). These results showed that the more Hin is induced, the more the ant promoter is repressed; they also showed that the binding of Hin to the hix operator can increase the SF of the host cells. These results indicated that the experimental design was valid.
FIG. 3.
Western blot analysis showing the amount of Hin expressed in HB101/pTriple/pHinWT with no IPTG (lane 1), 5 μM IPTG (lane 2), 10 μM IPTG (lane 3), 20 μM IPTG (lane 4), and 40 μM IPTG (lane 5). The arrow indicates the Hin protein. A preliminary experiment indicated that overexpression of Hin even with 80 μM IPTG was detrimental to host cells (data not shown). The unknown protein bands above Hin show that the same amounts of total protein were loaded in all lanes.
Assay for hix-pairing activity of Hin.
The next step in Hin-mediated inversion is to bring the two hix sites close together (hix pairing). This action was shown in vitro to be accomplished by the interaction between the Hin dimers bound to each hix site without the participation of Fis (12, 26). To assay the hix-pairing activity of Hin, an additional hix site was inserted in pSingle as a second operator (O2) 1.1 kb away from O1, generating pDouble1 (Fig. 2A). In the chromosome of S. typhimurium, the two hix sites are separated by a 993-bp DNA segment. We hypothesized that the ant promoter in pDouble1 would be more efficiently repressed than that in pSingle because of the DNA loop generated by the interaction between Hin dimers bound to hix operators. Thus, a higher SF was expected for HB101 harboring pDouble1. However, with the IPTG concentrations tested, the SFs of HB101/pHinWT/pDouble1 were the same as those of HB101/pHinWT/pSingle (Table 1), suggesting that the expected cooperative effect between the two operators did not occur. Two possibilities were considered. One is that the paired-hix structure in vivo is so transient (so is the DNA loop) that it cannot repress the promoter more than the mere binding of Hin to O1. The other possibility is that the paired-hix structure is stable in vivo but that somehow the DNA loop is not effective in transcriptional repression.
It has been demonstrated that the DNA loop per se in the galactose operon is important for more effective repression (3). Choy et al. (4) showed that as the size of the DNA loop generated by the interactions of repressor proteins bound to distant operators increased from 114 to 614 bp, the gal promoters were relieved from repression. Based on these observations, we contemplated that the size of the DNA loop formed in pDouble1 (1.1 kb) might have been too large. Thus, we reduced the distance between the hix operators from 1.1 kb (in pDouble1) to 200 bases and generated the plasmid pDouble2 (Fig. 2A). Indeed, the SF of HB101/pHinWT/pDouble2 with 20 μM IPTG was 10 times higher than that of HB101/pHinWT/pDouble1 or that of HB101/pHinWT/pSingle (Table 1). This result demonstrated that the paired-hix structure is formed and stably maintained in vivo. More importantly, this result suggested that hix-pairing activity can be measured in vivo and was consistent with the model that the inflexibility of the gal promoters enclosed by the DNA loop impedes transcription initiation by preventing RNA polymerase from unwinding DNA for open complex formation (4).
Assay for invertasome formation.
Formation of the invertasome requires two proteins, Hin and Fis, and three DNA-binding sites in the same plasmid, two hix sites and one enhancer site. The 65-bp enhancer is composed of two Fis-binding sites separated by 48 bp (12). Presumably, a Fis dimer binds to each binding site. After establishing that the DNA loop formed between cis-acting elements separated by at least 200 bp (as in pDouble2) provides more effective repression of the ant promoter, plasmid pTriple (Fig. 2A) was constructed to measure invertasome formation in vivo. pTriple has the enhancer sequence inserted in pDouble1 200 bp away from the hix operator O1. The relative locations of the three cis-acting DNA sites in pTriple were almost identical to those found in the H region of the Salmonella chromosome. Thus, if the invertasome is formed as shown by electron microscopy (12), we hypothesize that the smaller DNA loop, established between the enhancer and O1, would be the active component for transcriptional repression. The other DNA loop, generated by the enhancer and O2, would not participate in the repression, because the ant promoter is located in the smaller loop.
HB101/pHinWT/pTriple showed the highest SF (Table 1). With 20 μM IPTG, 57% of cells were able to survive in the presence of streptomycin, whereas for HB101/pHinWT/pDouble2, 22% of cells survived. The SF of HB101/pHinWT/pTriple with 20 μM IPTG was identical to that of HB101/pHinWT harboring only the vector pLG339 (Kanr and no rpsL operon), suggesting that the ant promoter in pTriple was so efficiently repressed that hardly any transcripts of the rpsL gene were made.
Interaction between the Fis-enhancer complex and individual Hin-hix complexes.
The results with pTriple raised the question of whether or not Fis bound on the enhancer can interact with Hin bound on the O1 complex without the presence of the Hin-O2 complex. This question was tested by removing O2 from pTriple. The resulting plasmid, pDouble3, has only two operators, the enhancer and O1 (Fig. 2A). If there is a Fis-Hin interaction, then SF of HB101/pHinWT/pDouble3 will be higher than that of HB101/pHinWT/pSingle. Indeed, with 20 μM IPTG, the SF of the pDouble3-containing strain was 10 times higher than that of the pSingle-containing strain, suggesting that the Fis-enhancer complex can interact with individual Hin-hix complexes without the formation of the paired-hix structure and that the resulting nucleoprotein complex can be as stable as the paired-hix structure in vivo.
Because there are two Fis-binding sites in the enhancer, we tested whether both sites are necessary to make the stable complex between Hin and Fis, as shown for pDouble3. One of the Fis-binding sites in the enhancer was removed from pDouble3, and the resulting plasmid (pDouble3′) was assayed. The SF of HB101 harboring pDouble3′ and pHinWT with 20 μM IPTG was almost the same as that of HB101/pHinWT/pDouble3, suggesting that a Fis dimer bound on the half enhancer could make a stable complex with a Hin dimer bound on the hix site.
As a control experiment, pDouble3, pDouble3′, and pTriple were assayed in a Fis-negative Strr strain (CSH50fis::cat). The SFs of CSH50fis::cat/pHinWT harboring each of these plasmids were the same as that for Hin binding on O1, as in HB101/pHinWT/pSingle. These results suggested not only that Fis specifically acts upon the enhancer sequence but also that it is Fis that interacts with Hin.
Analysis of dimer interface mutants with the in vivo assay system.
With the established in vivo assay system, Hin mutants that have been characterized in vitro were analyzed. Their abilities for hix binding, hix pairing, and invertasome formation were analyzed with pSingle, pDouble2, and pTriple, respectively. The interactions between individual Hin-hix and Fis-enhancer complexes were measured with pDouble3. Previously, we substituted each amino acid residue from M101 to H107 with cysteine (25). These residues were shown to exist in a long α helix that forms the dimer interface of Hin. Among the mutants, M101C and H107C had 100 and 40% of the inversion activity of the wild type, respectively. However, the DNA-binding activity of H107C could not be measured (25). R103C and F104C were binding competent but inversion incompetent (25). All the assays were performed with 20 μM IPTG. The amounts of Hin proteins induced from these mutants by 20 μM IPTG were more or less the same (data not shown). The results are summarized in Table 2.
TABLE 2.
SFs of dimer interface mutants of Hina
| Strain | SF in the presence of the following rpsL-containing plasmid:
|
|||
|---|---|---|---|---|
| pSingle (hix binding) | pDouble2 (hix pairing) | pDouble3 (Hin-Fis) | pTriple (invertasome) | |
| Wild type | 0.02 | 0.22 | 0.27 | 0.57 |
| M101C | 0.02 | 0.15 | 0.33 | 0.57 |
| R103C | 1 × 10−4 | 0.06 | 0.22 | 0.07 |
| F104C | 2 × 10−3 | 0.06 | 0.4 | 0.56 |
| H107C | 4 × 10−7 | 0.01 | 3 × 10−8 | 0.09 |
The SFs are averages from three independent experiments.
In a comparison with the SFs of the wild type, M101C showed no impairment of hix binding, hix pairing, and invertasome formation. R103C showed an SF for binding of 10−4, which is 2 orders of magnitude lower than that of the wild type, suggesting that the DNA-binding activity of R103C was imperfect. So was the hix-pairing activity of R103C. The SF for invertasome formation by R103C was 0.07, significantly lower than that of the wild type. Thus, the inversion-incompetent phenotype of R103C was partly due to a defect in protein interactions. However, R103C could make a stable contact with the Fis-enhancer complex. Although the SF for the DNA-binding activity of F104C was 10% of that of the wild type, invertasome formation by this mutant was shown to be as efficient as that of the wild type. These results suggested that F104C was defective in one of the steps after invertasome formation.
The SF of H107C with pSingle (4 × 10−7) suggested that it was completely defective in DNA binding, but the SF for hix pairing (0.01) indicated that H107C was able to make the paired-hix structure, albeit less efficiently. Thus, the defect in DNA binding seemed to be corrected by hix-pairing activity. H107C was shown to exist as a homodimer in solution (25). Thus, it is likely that interactions between H107C dimers during their brief stay on hix stabilized DNA binding. The SF of H107C with pDouble3 was 3 × 10−8, suggesting that the unstable DNA binding of H107C dimers, however, could not be stabilized by the Fis-enhancer complex. The SF of H107 with pTriple was 0.09. Taken together, these results suggested that H107C was able to make the invertasome with less stability (see Discussion); therefore, the lower inversion activity of H107C (25) was a result of the unstable invertasome structure. The difference between R103C and H107C, both of which showed the same SFs with pTriple, seemed to be that the invertasome formed by R103C was defective in inversion but that the invertasome formed by H107C was able to make the inverted product.
DISCUSSION
SF measures the relative stabilities of nucleoprotein structures.
The increase in the SF from 10−7 to 10−2 for HB101/pSingle when Hin was induced with 20 μM IPTG showed that the binding of Hin to O1 was certainly effective in transcriptional repression of the ant promoter. The assay results for the four dimer interface mutants with pSingle were consistent with our previous results (25) and showed subtle differences in the binding stability of the Hin mutants. Thus, hix-binding activity can be quantitatively measured with pSingle. The same idea of using hix as an operator has been used to measure the hix-binding activity of Hin in an in vivo assay system called challenge phage (14). A further 10-fold increase in the SF for HB101/pDouble2 indicated synergy between the two hix operators as a result of a Hin-Hin interaction. Based on the differences in SF between pDouble1 and pDouble2, which are different only in the distance between the hix operators, we concluded that the size of the DNA loop is important in transcriptional repression. Furthermore, the structural difference between pDouble1 and pDouble2 eliminates the possibility that the synergistic effect shown for pDouble2 was caused by Hin binding to quasi-binding sites somewhere in pDouble2.
The addition of the third operator (O3, the enhancer) to pDouble2 increased the SF of host cells from 0.22 (pDouble2) to 0.57 (pTriple). We believe that the difference in SF is the consequence of how stable the nucleoprotein structures formed on pDouble2 (paired-hix structure) and pTriple (invertasome) are, because the positions and sizes of the DNA loops engaged in transcriptional repression in pDouble2 and pTriple are the same. Therefore, the twofold difference in SF between pTriple (0.57) and pDouble3 (0.27) is also the result of a difference in the stability of the nucleoprotein structures of the invertasome and the Fis-Hin complex. Differences in stability between the nucleoprotein structures of the invertasome and the Fis-enhancer and Hin-hix complexes became evident when the Hin mutants were analyzed. For example, the SF of pHinR103C/pTriple was 0.07, and that of pHinR103C/pDouble3 was 0.22. Thus, it can be suggested that, unlike the wild type, R103C forms an invertasome that is less stable than the Fis-Hin complex.
Transcriptional repression in pTriple.
In reactions with substrate plasmids containing hixL-WT, hixL-AT, and the enhancer (as in pTriple), Hin made extensive knots instead of an inverted product (13). It was proposed that in the invertasome formed in these plasmids, multiple rounds of strand exchange, each of which occurred through a 180° clockwise rotation of one set of Hin subunits, resulted in the generation of extensive knots. One might argue that the most efficient repression in pTriple might have been accomplished by the DNA knots generated in the invertasome rather than the invertasome structure itself formed on pTriple. However, the assay results for F104C with pTriple argue against this possibility. Purified F104C protein showed no DNA cleavage activity (data not shown), suggesting that there could not be any strand exchange or formation of DNA knots in the invertasome formed by F104C. The SF of pHinF104C/pTriple was the same as that of pHinWT/pTriple, suggesting that the invertasome formed by F104C was as stable as that formed by the wild type. Therefore, these arguments support the notion that the stable DNA loop formed between O3 and O1 as a result of invertasome formation is solely responsible for the repression in pTriple. It is likely that the formation of an O3-O1 DNA loop of about 200 bp is aided by the HU protein, as shown in vitro (10). These data also suggest, contrary to the proposal by Kanaar et al. (20), that Fis should stay in the invertasome after DNA cleavage and even during the multiple strand exchange in pTriple to maintain repression.
Protein interactions at a distance for assembly of the invertasome.
It was surprising that individual Hin-hix complexes could make a stable complex with the Fis-enhancer complex, because a protein-protein interaction between Hin and Fis has never been observed. This result raised the possibility that the invertasome can be formed by assembly of each Hin-hix complex onto the Fis-enhancer complex, bypassing the formation of the paired-hix structure. However, the assay results for H107C argue against this possibility. The SF of H107C with pDouble3 is 3 × 10−8, suggesting that a DNA loop between O3 and O1 cannot be formed. (This result does not necessarily imply that H107C has lost its ability to enter into a stable interaction with Fis but rather that the interaction is transient due to the unstable hix-binding activity of H107C.) However, considering that the transcriptional repression on pTriple is achieved by the small DNA loop formed between O3 and O1, the O3-O1 DNA loop must have been assembled in the invertasome formed by H107C in order to acquire the SF of 0.09 for pTriple. Otherwise, the SF of pHinH107C/pTriple would have been that for binding, which is 10−7 or less. Thus, the O3-O1 DNA loop, which cannot be made on pDouble3 in H107C, can be made passively during the process of invertasome formation. These results strongly suggest that the invertasome can be assembled without a preformed O3-O1 DNA loop.
There remain two other routes toward the assembly of the invertasome. One is that, as generally accepted so far without any experimental data, the formation of the paired-hix structure precedes the formation of the invertasome. The other is the simultaneous joining of all three cis-acting elements (the two hix sites and the enhancer). If a binding-positive but pairing-negative mutant were isolated, the invertasome formation assay of the mutant with pTriple would provide evidence for the first route. In fact, such a mutant has been isolated. The mutant was not a protein but a Hin-binding site called hixC that has 13-bp perfect symmetry. Wild-type Hin bound to hixC as well as to wild-type hix. However, in reactions with hixC sites, Hin showed a 16-fold-lower rate of inversion, and in vitro studies showed that Hin bound on hixC sites had difficulty forming the paired-hix structure (27).
Interactions between DNA-binding proteins at a distance have been a core mechanism for explaining site-specific recombination and transcription. So far, these interactions have been analyzed in vitro with purified proteins. The in vivo assay system reported in this study could provide a simple way to probe these interactions under many different conditions and could have the potential to be applicable to a wide variety of biological systems.
ACKNOWLEDGMENTS
We thank H. E. Choy and C. Park for thorough review of the manuscript.
This research was supported by KOSEF grant 961-0502-015-2 and by grant 97-4431 from the Basic Science Research Program, Ministry of Education, Korea, awarded to H. M. Lim.
S.Y.L. and H.J.L. contributed equally to this research.
REFERENCES
- 1.Adhya S. Multipartite genetic control elements: communication by DNA loop. Annu Rev Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- 2.Benson N, Sugiono P, Bass S, Mendelman L V, Youderian P. General selection for specific DNA-binding activities. Genetics. 1986;114:1–14. doi: 10.1093/genetics/114.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choy H E, Adhya S. Control of gal transcription through DNA looping: inhibition of the initial transcribing complex. Proc Natl Acad Sci USA. 1992;89:11264–11268. doi: 10.1073/pnas.89.23.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choy H E, Park S-W, Parrack P, Adhya S. Transcription regulation by inflexibility of promoter DNA in a looped complex. Proc Natl Acad Sci USA. 1995;92:7327–7331. doi: 10.1073/pnas.92.16.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen S N, Chang A C Y, Boyer H W, Helling R B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci USA. 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean D. A plasmid cloning vector for the direct selection of strains carrying recombinant plasmids. Gene. 1981;15:99–102. doi: 10.1016/0378-1119(81)90108-6. [DOI] [PubMed] [Google Scholar]
- 7.Glasgow A C, Bruist M F, Simon M I. DNA-binding properties of the Hin recombinase. J Biol Chem. 1989;264:10072–10082. [PubMed] [Google Scholar]
- 8.Glasgow A C, Hughes K T, Simon M I. Bacterial inversion systems. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 637–659. [Google Scholar]
- 9.Hashimoto-Gotoh T, Kume A, Masahashi W, Takeshita S, Fukuda A. Improved vector, pHSG664, for direct streptomycin-resistance selection: cDNA cloning with G:C-tailing procedure and subcloning of double-digest DNA fragments. Gene. 1986;41:125–128. doi: 10.1016/0378-1119(86)90275-1. [DOI] [PubMed] [Google Scholar]
- 10.Haykinson M J, Johnson R C. DNA looping and the helical repeat in vitro and in vivo: effect of HU protein and enhancer location on Hin invertasome assembly. EMBO J. 1993;12:2503–2512. doi: 10.1002/j.1460-2075.1993.tb05905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haykinson M J, Johnson L M, Soong J, Johnson R C. The Hin dimer interface is critical for Fis-mediated activation of the catalytic steps of site-specific DNA inversion. Curr Biol. 1996;6:163–177. doi: 10.1016/s0960-9822(02)00449-9. [DOI] [PubMed] [Google Scholar]
- 12.Heichman K A, Johnson R C. The Hin invertasome: Protein-mediated joining of distance recombination sites at the enhancer. Science. 1990;249:511–517. doi: 10.1126/science.2166334. [DOI] [PubMed] [Google Scholar]
- 13.Heichman K A, Moskowitz I P G, Johnson R C. Configuration of DNA strands and mechanism of strand exchange in the Hin invertasome as revealed by analysis of recombinant knots. Genes Dev. 1991;5:1622–1634. doi: 10.1101/gad.5.9.1622. [DOI] [PubMed] [Google Scholar]
- 14.Hughes K T, Youderian P, Simon M I. Phase variation in Salmonella: analysis of Hin recombinase and hix recombination site interaction in vivo. Genes Dev. 1988;2:937–948. doi: 10.1101/gad.2.8.937. [DOI] [PubMed] [Google Scholar]
- 15.Johnson R. Mechanism of site-specific DNA inversion in bacteria. Curr Opin Genet Dev. 1991;1:1–15. doi: 10.1016/s0959-437x(05)80307-7. [DOI] [PubMed] [Google Scholar]
- 16.Johnson R C, Ball C A, Pfeffer D, Simon M I. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc Natl Acad Sci USA. 1988;85:3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson R C, Bruist M F. Intermediates in Hin-mediated inversion: a role for Fis and the recombinational enhancer in the strand exchange reaction. EMBO J. 1989;8:1581–1590. doi: 10.1002/j.1460-2075.1989.tb03542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson R C, Bruist M F, Simon M I. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986;46:531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- 19.Johnson R C, Simon M I. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985;41:781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- 20.Kanaar R, Klippel A, Shekhtman E, Dungan J M, Kahmann R, Cozzarelli N R. Processive recombination by the phage Mu Gin system: implications for the mechanism of DNA strand exchange, DNA alignment, and enhancer action. Cell. 1990;62:353–366. doi: 10.1016/0092-8674(90)90372-l. [DOI] [PubMed] [Google Scholar]
- 21.Kanaar R, van de Putte P, Cozzarelli N R. Gin-mediated DNA inversion: product structure and the mechanism of strand exchange. Proc Natl Acad Sci USA. 1988;85:752–756. doi: 10.1073/pnas.85.3.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch C, Vandekerckhove J, Kahmann R. Escherichia coli host factor for site-specific DNA inversion: cloning and characterization of the fis gene. Proc Natl Acad Sci USA. 1988;85:4237–4241. doi: 10.1073/pnas.85.12.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;84:4767–4771. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim H M, Lee H J, Jaxel C, Nadal M. Hin-mediated inversion on positively supercoiled DNA. J Biol Chem. 1997;272:18434–18439. doi: 10.1074/jbc.272.29.18434. [DOI] [PubMed] [Google Scholar]
- 25.Lim H M. Analysis of subunit interaction by introducing disulfide bonds at the dimerization domain of Hin recombinase. J Biol Chem. 1994;269:31134–31142. [PubMed] [Google Scholar]
- 26.Lim H M, Simon M I. The role of negative supercoiling in Hin-mediated site-specific recombination. J Biol Chem. 1992;267:11176–11182. [PubMed] [Google Scholar]
- 27.Lim H M, Hughes K T, Simon M I. The effects of symmetrical recombination site hixC on Hin recombinase function. J Biol Chem. 1992;267:11183–11190. [PubMed] [Google Scholar]
- 28.Lim H M, Pene J J. Optimal conditions for supercoiled DNA sequencing with the E. coli DNA polymerase I large fragment. Gene Anal Tech. 1989;5:32–39. doi: 10.1016/0735-0651(88)90024-6. [DOI] [PubMed] [Google Scholar]
- 29.Osuna R, Finkel S, Johnson R C. Identification of two functional regions in Fis: the N-terminus is required to promote Hin-mediated DNA inversion but not λ excision. EMBO J. 1991;6:1593–1603. doi: 10.1002/j.1460-2075.1991.tb07680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozaki M, Mizushima S, Nomura M. Identification and functional characterization of the protein controlled by the streptomycin-resistant locus in E. coli. Nature. 1969;222:333–339. doi: 10.1038/222333a0. [DOI] [PubMed] [Google Scholar]
- 31.Post L E, Nomura M. DNA sequences from the str operon of Escherichia coli. J Biol Chem. 1980;255:4660–4666. [PubMed] [Google Scholar]
- 32.Safo M K, Yang W-Z, Corselli L, Cramton S E, Yuan H S, Johnson R C. The transactivation region of the Fis protein that controls site-specific DNA inversion contains extended mobile β-hairpin arms. EMBO J. 1997;16:6860–6873. doi: 10.1093/emboj/16.22.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 34.Schneider R, Travers A, Muskhelishvili G. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol Microbiol. 1997;26:519–530. doi: 10.1046/j.1365-2958.1997.5951971.x. [DOI] [PubMed] [Google Scholar]
- 35.Stoker N G, Fairweather N F, Spratt B G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982;18:335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]