Abstract
Transcriptional activation of the Bacillus subtilis ackA gene, encoding acetate kinase, was previously shown to require catabolite control protein A (CcpA) and sequences upstream of the ackA promoter. CcpA, which is responsible for catabolite repression of a number of secondary carbon source utilization genes in B. subtilis and other gram-positive bacteria, recognizes a cis-acting consensus sequence, designated cre (catabolite response element), generally located within or downstream of the promoter of the repressed gene. Two sites resembling this sequence are centered at positions −116.5 and −56.5 of the ackA promoter and have been termed cre1 and cre2, respectively. Synthesis of acetate kinase, which is involved in the conversion of acetyl coenzyme A to acetate, is induced when cells are grown in the presence of an easily metabolized carbon source such as glucose. In this study, cre2, the site closer to the promoter, and the region upstream of cre2 were shown to be indispensable for CcpA-dependent transcriptional activation of ackA, whereas cre1 was not required. In addition, insertion of 5 bp between cre2 and the promoter disrupted activation, while 10 bp was tolerated, suggesting face-of-the-helix dependence of the position of cre2 and/or upstream sequences. DNase footprinting experiments demonstrated binding of CcpA in vitro to cre2 but not cre1, consistent with the genetic data. Activation of ackA transcription was blocked in a ptsH1/crh double mutant, suggesting involvement of this pathway in CcpA-mediated transcriptional activation.
The mechanism of carbon catabolite regulation in gram-positive bacteria appears to be fundamentally different from that employed by enteric gram-negative bacteria. The absence of detectable amounts of cyclic AMP under normal growth conditions in Bacillus subtilis has long indicated that the regulation of secondary carbon source utilization genes in B. subtilis is different from that of Escherichia coli, which relies on cyclic AMP as an effector of the CAP/CRP protein (see references 17 and 25 for a review). Unlike the positive regulatory mechanism found in E. coli, carbon catabolite regulation in gram-positive bacteria appears to be mediated by transcriptional repression, requiring trans-acting CcpA (catabolite control protein A), a member of the LacI-GalR family of bacterial regulatory proteins (16), and a cis-acting consensus sequence, designated cre (20, 51). The cis-acting site, first identified in the regulation of the gene for α-amylase (amyE) and termed amyO, is a palindromic sequence structurally similar to the E. coli lac and gal operators (38, 51). Elements resembling the consensus sequence have been found in other secondary carbon source utilization genes, including acsA and acu (12), hut (52), gnt (33), xyl (26), bgl (27), lev (29), and mmg (1). Catabolite repression of these genes is relieved upon mutation of the consensus sequences or expression in a ccpA mutant.
CcpA homologs have been identified in a number of other gram-positive organisms, including B. megaterium (21), Staphylococcus xylosus (8), Clostridium acetobutylicum (4), and Lactobacillus casei (35). Western analysis suggests that proteins with related antigenic determinants are widely distributed in gram-positive bacteria (28), although it has not been demonstrated that these proteins are functional homologs of CcpA. The cre element has also been found in a large number of genes in a variety of gram-positive organisms (20), although the significance of most of these sequences has not been tested. A second B. subtilis gene related to ccpA has recently been identified (2); this gene, designated ccpB, appears to play a role in catabolite repression under low-oxygen growth conditions.
CcpA is constitutively expressed in B. subtilis (1a, 17, 34) and is therefore likely to require a signalling pathway to control its activity. The details of this pathway are not completely clear. CcpA has been shown to bind in vitro to the HPr protein of the phosphoenolpyruvate-dependent phosphotransferase system when HPr is phosphorylated at Ser-46, and this interaction affects the DNA binding activity of CcpA (5, 11, 22). The ATP-dependent phosphorylation of Hpr at Ser-46 is catalyzed by the ptsK-encoded kinase, activity of which is stimulated by fructose 1,6-bisphosphate (41). A mutant form of HPr containing an alanine substitution at Ser-46 (ptsH1) results in the complete or partial loss of catabolite repression of several genes that are subject to control by CcpA (3, 6, 29, 42); however, the ptsH1 mutation has no effect on repression of amyE (50), suggesting at least one other mechanism for CcpA activation. Crh, an HPr-like protein that can only be phosphorylated at Ser-46, has recently been identified in B. subtilis (10) and may provide an alternate pathway for control of CcpA activity.
Along with its role as a transcriptional repressor, CcpA is also required for the activation of at least two carbon excretion pathways, acetoin biosynthesis (43) and acetate production (13). Acetyl coenzyme A (acetyl-CoA) and acetate are interconverted by two separate pathways in B. subtilis. Phosphotransacetylase, the product of the pta gene, catalyzes the conversion of acetyl-CoA to the intermediate acetyl phosphate (39), while conversion of acetyl phosphate to acetate is catalyzed by acetate kinase, encoded by ackA (13). Utilization of acetate by its conversion back to acetyl-CoA requires the acsA gene product, acetyl-CoA synthetase (14). The acsA gene contains a cre site downstream of the transcription start site, and transcription is repressed by CcpA in the presence of excess glucose (12, 14). In contrast to its effect on acsA transcription, CcpA activates transcription of ackA during growth in the presence of excess glucose (13).
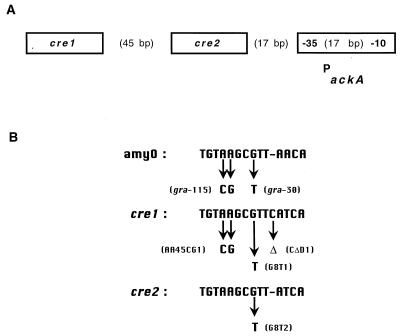
Activation of ackA transcription requires sequences upstream of the ackA promoter (13). Two cre sites, designated cre1 and cre2, are centered at positions −116.5 and −56.5 relative to the transcription start site of ackA (Fig. 1). These sites differ from amyO by a single base substitution of an A to a T; this substitution at amyO has no effect on repression (51). In addition, cre1 contains a one-base insertion. While cre sequences are generally positioned within or downstream of the promoter in genes that are repressed by CcpA, cre1 and cre2 are located upstream of the ackA promoter. In this study, the roles of these sites in the CcpA-dependent activation of ackA transcription were investigated. Mutational analyses demonstrated that cre2 and the region immediately upstream of this site are required for transcriptional activation and that the position of cre2 relative to the −35 region of the ackA promoter is important for function.
FIG. 1.
(A) Structure of the ackA promoter region. The positions of cre1 and cre2 upstream of the ackA promoter are shown. (B) Mutations in cre1 and cre2. The G-to-T substitutions in cre1 and cre2 are labeled G8T1 and G8T2, respectively. Mutation of the upstream region of cre1 is labeled AA45CG1. Δ indicates the base that was deleted in cre1 to generate a sequence identical to cre2. The amyO consensus sequence is included for comparison. gra-115 and gra-30 indicate mutations which eliminate repression at amyO (51).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. Plasmids were propagated in E. coli DH5α grown in LB medium (32). B. subtilis was grown in tryptose blood agar base (Difco), 2XYT, or TSS medium (9) with NH4Cl as the nitrogen source and 1% Casamino Acids as the carbon source, in the presence or absence of 1% glucose. All growth was at 37°C. Antibiotics (Sigma) were used at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 5 μg/ml for selection and 0.1 μg/ml for induction; erythromycin, 1 μg/ml for selection and 0.1 μg/ml for induction; lincomycin, 25 μg/ml; kanamycin, 5 μg/ml; spectinomycin, 200 μg/ml. Amino acids were added to TSS medium at 50 μg/ml as required for auxotrophic strains. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was used at 40 μg/ml as an indicator of β-galactosidase activity.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| B. subtilis | ||
| BR151MA | lys-3 trpC2 | 12 |
| BR151MACcp::spc | lys-3 trpC2 ccpA::Tn917lacΔ(lacZ-erm)::spc | 12 |
| ZB307A | SPβc2del2::Tn917::pSK10Δ6 | 54 |
| ZB449 | trpC2 pheA1 abrB703 (SPβ cured) | 36 |
| 168 | trpC2 | Laboratory stock |
| MIV1 | trpC2 ptsH1 | Transformation of 168 with DNA of SA003 (6) |
| 168crh | trpC2 crh::aphA3 | Transformation of 168 with DNA of QB7096 (I. Martin-Verstraete) |
| MIV1crh | trpC2 ptsH1 crh::aphA3 | Transformation of MIV1 with DNA of QB7096 |
| E. coli DH5α | φ80dlacZΔM15 endA1 recA1 hsdR17 (rk− mk+) thi-1 gyrA96 relA1 Δ(lacZYA-argF)U169 | Bethesda Research Laboratories |
Genetic techniques.
Restriction endonucleases were purchased from New England BioLabs or Promega and used as described by the manufacturer. Preparation of single-stranded M13 template DNA and double-stranded plasmid DNA and dideoxynucleotide sequencing (Sequenase; United States Biochemicals) were performed by using standard techniques. Oligonucleotide primers were purchased from Cruachem and are shown in Table 2. PCR was carried out on a Perkin-Elmer DNA Thermal Cycler 480 using a Gene-Amp kit (Perkin-Elmer Cetus). B. subtilis transformation was carried out as described previously (15).
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence | Use |
|---|---|---|
| AckBam1 | 5′-TTTATGGATCCAGTTGAC-3′ | 5′ deletion |
| AckKpn1 | 5′-GGAGAGGGTACCAAAGCGCCGGGGC-3′ | 5′ deletion |
| AckBam2 | 5′-CGAATTGGATCCAGCTTTG-3′ | 5′ deletion |
| AckBam3 | 5′-GCGTTCATGGATCCAAAACCTATAGTG-3′ | 5′ deletion |
| AckKpn2 | 5′-GTGTCTGAAGGTACCGACTTCTTATTG-3′ | 5′ deletion |
| M13Sal | 5′-CGACGTTGTAAGTCGACGGCCACTG-3′ | 5′ pGEM7Zf(+) polylinker |
| M13Rev | 5′-GATAACAATTTCACACAGGA-3′ | 3′ pGEM7Zf(+) polylinker |
| AckO1 | 5′-TTCTTATTGTAAGCTTTATCAATAC-3′ | cre2 G8T2 |
| AckO2 | 5′-GTATTGATAAAGCTTACAAT-3′ | cre2 G8T2 |
| TH448 | 5′-CAAATTGTAAGCTTTCATCAATACAAAACC-3′ | cre1 G8T1 |
| TH449 | 5′-ATTGATGAAAGCTTACAATTTGCGG-3′ | cre1 G8T1 |
| AA45CG1 | 5′-AATTGTCGGCGTTCATCAATAC-3′ | cre1 AA45CG1 |
| Ack4 | 5′-CGCTCCTTTATACTCTG-3′ | cre1 AA45CG1 |
| AckBRV | 5′-CGCGGATCCTCAACTTGCGTATTG-3′ | Linker insertion |
| AckBam5 | 5′-CGCGGATCCTTGAAAAGCCGACATG-3′ | Linker insertion |
| AckBS | 5′-CGCGGATCCAGGCCTTGAAAAGCCGACATG-3′ | Linker insertion |
| CΔD1 | 5′-AGCGTTATCAATACAAAAC-3′ | CΔD1 mutation |
ackA deletion analysis and operator mutations.
Mutations in the region upstream of ackA were generated by PCR-mediated oligonucleotide-directed mutagenesis. All PCRs were performed on a 1.35-kb EcoRI fragment which contained the ackA promoter and 1 kb of DNA upstream of the promoter. The 1.35-kb fragment was inserted into the EcoRI site of plasmid pGEM7Zf(+) (Promega). The EcoRI site in pGEM7Zf(+) is flanked by a KpnI restriction site on the 5′ end and an XbaI restriction site on the 3′ end. Truncation of the region upstream of ackA used preexisting restriction sites or the creation of new sites by PCR. The 3′ end of each deletion fragment made use of the same XbaI site and primer M13Sal. Wild-type activity was based on expression from the 1.35-kb ackA fragment.
The G8T1 mutation in cre1 was generated by PCR using the 1.35-kb EcoRI fragment as the template, with primers M13Rev and TH449 (5′ fragment) and M13Sal and TH448 (3′ fragment) in separate reactions. The products were digested with KpnI plus HindIII and HindIII plus XbaI, respectively, and inserted together into plasmid pFG328 digested with KpnI and XbaI to regenerate the 1.35-kb fragment with a HindIII site at cre1. The G8T2 mutation in cre2 was generated in a similar manner by using primers M13Rev and AckO2 (5′ fragment) and primers M13Sal and AckO1 (3′ fragment). The AA45CG1 mutation in cre1 was generated by mismatched PCR mutagenesis (45). Primers AA45CG1, containing the mutation, and M13Sal were used in one reaction to generate the downstream portion of the ackA fragment, and primers Ack4 and M13Rev were used to generate an overlapping upstream fragment. The PCR fragments were gel purified, mixed in equimolar concentrations, denatured, and annealed to form heteroduplex molecules, and the 3′ ends of the heteroduplex molecules were extended in a second PCR using primers M13Rev and M13Sal. The resulting fragment was then cloned by using KpnI and XbaI. The CΔD1 mutation was generated in a similar manner by using primer CΔD1 in place of AA45CG1.
Insertions of 5 and 10 bp between the ackA promoter and cre2 were obtained by using oligonucleotide primers with BamHI sites. Two separate amplification reactions were set up for each insertion. PCR of the upstream fragment for the two insertions with primers AckBRV and M13Rev and digestion with KpnI and BamHI produced a 1-kb fragment that contained the two cre sites with a BamHI site at the 3′ end. Amplification of the downstream fragments with primers M13Sal and AckBam5 or AckBS for the 5- or 10-bp insertions, respectively, and digestion with BamHI and XbaI resulted in two 350-bp fragments with a BamHI site just upstream of the −35 region of the promoter. Ligation of the AckBRV fragment with the AckBam5 or AckBS fragment resulted in a 5- or 10-bp insertion, respectively, between the ackA promoter and cre2. All mutations were verified by DNA sequencing.
Construction of lacZ fusions.
ackA-lacZ transcriptional fusions were generated by cloning the sequenced ackA fragment into E. coli plasmid pFG328 (13) by using KpnI or BamHI at the 5′ end and XbaI at the 3′ end. The fusions were incorporated into specialized transducing phage SPβ by homologous recombination between the plasmid and the prophage contained in strain ZB307A. The resulting phage were purified by passage through strain ZB449 (SPβ cured) and used to transduce isogenic wild-type and CcpA− strains (12).
β-Galactosidase measurements.
Strains carrying the ackA-lacZ transcriptional fusions were grown in TSS medium in the presence or absence of 1% glucose and maintained in early exponential growth by serial dilutions for adaptation to the growth medium. Samples were then taken at 30-min intervals until 2 h past entry into stationary phase (T0). β-Galactosidase assays were performed as described by Miller (32), by using toluene permeabilization of the cells. All growth experiments were repeated at least twice and showed less than 10% variation.
Acetate production.
Cells were grown in TSS medium with 1% Casamino Acids in the presence or absence of glucose (0.25%). Samples were harvested at 30 min prior to T0, and culture supernatants were assayed for acetate by using a kit purchased from Boehringer Mannheim.
DNase footprinting.
A DNA fragment containing the ackA promoter region between positions −195 and +18 was subcloned into plasmid pUC18 and labeled at the 3′ ends with [α-32P]dATP by using Klenow fragment (Promega). The labelled DNA was gel purified and recovered by electroelution as described previously (23). Purification of the CcpA protein and DNase footprinting were carried out as described by Kim et al. (24).
RESULTS
Deletion analysis.
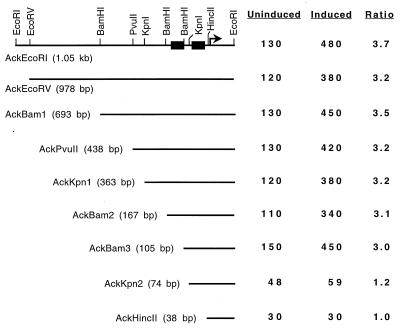
Deletion of the entire region upstream of the ackA promoter eliminates transcriptional activation in the presence of glucose (13). This region contains two sequences resembling amyO, the sequence in the amyE promoter region where the CcpA protein binds to repress transcription during growth in glucose (23, 24, 51). These sequences, designated cre1 and cre2 (Fig. 1), were considered likely targets for CcpA-dependent transcriptional activation of ackA. To precisely identify the sequences required for transcriptional activation, a more detailed deletion analysis was performed. Transcriptional fusions to lacZ using deletion fragments extending from 3 bp to 1 kb upstream of the −35 region of the ackA promoter (Fig. 2) were introduced in single copy into the B. subtilis chromosome by using specialized transducing phage SPβ. Expression of the fusions was monitored in TSS medium containing 1% Casamino Acids in the presence (induced) or absence (uninduced) of 1% glucose.
FIG. 2.
Deletion analysis of the region upstream of the ackA promoter. Each number in parentheses indicates the distance of the deletion end point from the transcription start point (arrow). The two black boxes indicate the positions of the cre sites. Cells were grown in TSS medium containing 1% Casamino Acids (uninduced) or TSS medium containing 1% Casamino Acids and 1% glucose (induced). β-Galactosidase activity is expressed in Miller units (32). Values are shown for samples taken 30 min prior to entry into stationary phase. Results for AckHincII were previously reported (13). The ratio indicates the level of induction during growth in the presence of glucose.
Deletion of the region upstream of cre1 had no effect on induction of ackA-lacZ expression (Fig. 2). The shortest deletion fragment, AckHincII (13), made use of a HincII restriction site just upstream of the −35 region of the ackA promoter. This mutant was previously shown to exhibit reduced basal activity and no induction by growth in the presence of glucose. Based on these results, two additional deletion mutants were generated by using sites in the region separating the two cre sites. AckKpn2, which included sequences 11 bp upstream of cre2, resulted in loss of transcriptional activation of ackA-lacZ in the presence of glucose. AckBam3, which contained cre2 along with the entire region separating the two sites (42 bp upstream of cre2), retained full transcriptional activation. This region encompasses 70 bp upstream of the −35 region of the ackA promoter. These results indicated that sequences between cre1 and the AckKpn2 deletion end point are necessary for transcriptional activation of ackA in the presence of glucose.
Mutation of Ccp boxes.
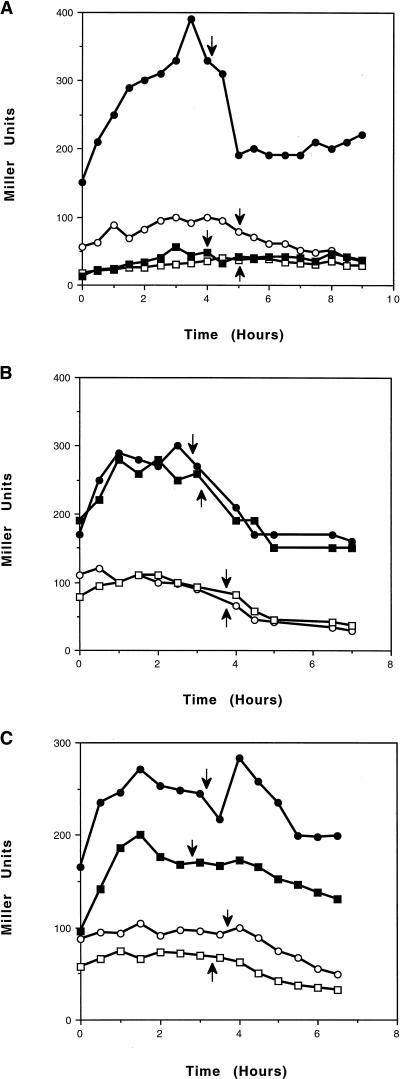
The results of the deletion analysis indicated that cre2 is not sufficient for transcriptional activation and that cre1 is not required. To specifically test the role of these elements, substitutions were made which corresponded to mutations known to eliminate repression at amyO (51; Fig. 1B). As shown in Fig. 3A, the wild-type ackA-lacZ fusion reaches maximal induction toward the end of exponential growth. A single base substitution in cre2, termed G8T2, eliminated transcriptional activation of ackA in the presence of glucose. The level of activity was less than the basal activity of the wild-type fusion and was comparable to the activity obtained with the deletion of both elements (Fig. 2). This indicates that cre2 is necessary for transcriptional activation and also plays a role in basal expression in the absence of glucose.
FIG. 3.
Effect of mutations in cre sites on expression of ackA-lacZ transcriptional fusions. Cells were grown in TSS medium containing 1% Casamino Acids (open symbols) or TSS medium containing 1% Casamino Acids and 1% glucose (filled symbols). β-Galactosidase activity is expressed in Miller units (32). The vertical arrows indicate time of entry of the culture into stationary phase. (A) Effect of the G8T2 cre2 mutant (squares) compared to the wild-type fusion (circles). (B) Effect of the G8T1 cre1 mutant (squares) compared to the wild-type fusion (circles). (C) Effect of the CΔD1 mutation in cre1 (squares) compared to the wild-type fusion (circles).
Two different mutations in cre1 were tested. The G8T1 mutation, which was identical to the mutation made in cre2, had no effect on ackA-lacZ expression (Fig. 3B), in contrast to the drastic effect of this substitution in cre2. These substitutions targeted the downstream region of the elements. To test the possibility that only a portion of cre1 is necessary, a second mutation in the upstream portion of cre1, termed AA45CG1, was generated. This mutation also had no effect on transcriptional activation of ackA (data not shown). These results, in conjunction with the deletion analysis, demonstrate that cre2, but not cre1, is required for transcriptional activation of ackA.
cre1 differs from cre2 by the presence of an extra cytosine 5 bp from the 3′ end (Fig. 1B). A deletion of this extra base, termed CΔD1, was tested to determine if the presence of a second cre2 sequence would increase transcriptional activation of ackA. This mutation resulted in reduction of both the basal and induced levels of expression but had no effect on the induction ratio (Fig. 3C). The presence of a “perfect” element at the position of cre1 apparently interferes with the normal function of cre2, possibly by sequestering the CcpA protein in a nonfavorable interaction.
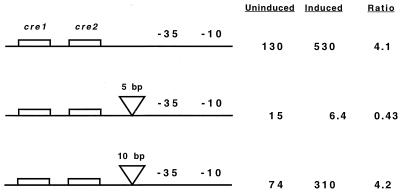
Linker insertions.
The center of cre2 is located 21.5 bp upstream of the −35 region of the ackA promoter. To determine whether the position of the cre site is important in transcriptional activation of ackA, insertions of 5 and 10 bp were made between cre2 and the −35 region of the ackA promoter. The 5-bp insertion, which positioned cre2 on the opposite face of the DNA helix from its normal position, eliminated glucose induction and decreased expression approximately 10-fold (Fig. 4). Insertion of 10 bp, which is predicted to position cre2 on the same face of the DNA helix as in the wild type but 10 bp further upstream, resulted in normal expression and glucose activation. These results demonstrate a face-of-the-helix dependence between the location of cre2, and/or upstream sequences, and the ackA promoter.
FIG. 4.
Effect of linker insertions between cre2 and the ackA promoter. The −35 and −10 regions of the ackA promoter are indicated. Cells were grown in TSS medium with 1% Casamino Acids as the carbon source (uninduced) or in TSS medium with 1% Casamino Acids and 1% glucose (induced). Values shown are for samples taken 30 min prior to entry into stationary phase and indicate β-galactosidase activity expressed in Miller units (32). The ratio reflects the level of induction during growth in the presence of glucose.
Expression of ackA in a CcpA mutant.
The reduction in basal expression caused by mutations of the ackA upstream region could be due to loss of normal CcpA-dependent activation during growth in the absence of glucose or to other factors. As previously shown (13), mutation of ccpA resulted in reduced expression of ackA and loss of activation during growth in glucose (Table 3). Mutation of cre2 or deletion of the region just upstream of cre2 reduced expression twofold more than the CcpA mutation alone, in both the wild-type and CcpA mutant strains; this suggests that while most of the reduction in basal expression of these mutants is likely to be due to loss of interaction with CcpA, there may be additional factors. The insertion of 5 bp between cre2 and the −35 region of the promoter dramatically reduced basal expression. The reduction is apparently not due to improper positioning of CcpA on the wrong face of the DNA helix, since it was also observed in the CcpA mutant. Since sequences upstream of cre2 are also required for activation, this reduction in basal activation could be caused by effects of this upstream region on regulatory events other than CcpA binding.
TABLE 3.
Expression of ackA-lacZ fusions in a ccpA mutant
| ackA-lacZ fusion | Mutation | β-Galactosidase activitya
|
|||
|---|---|---|---|---|---|
| BR151MA
|
BR151MACcp::spc
|
||||
| Uninduced | Induced | Uninduced | Induced | ||
| AckEcoRI | None (wild type) | 75 | 280 | 32 | 39 |
| G8T2 | cre2 | 26 | 39 | 19 | 25 |
| AckKpn2 | Δ-74 | 23 | 31 | 18 | 17 |
| LS | 5-bp insertion | 6.4 | 3.4 | 4.8 | 3.7 |
Cells were grown in TSS medium containing 1% Casamino Acids (uninduced) or 1% Casamino Acids and 1% glucose (induced). β-Galactosidase activities are expressed in Miller units (32) and indicate activity at 30 min prior to entry of the culture into stationary phase (T0).
DNase footprinting.
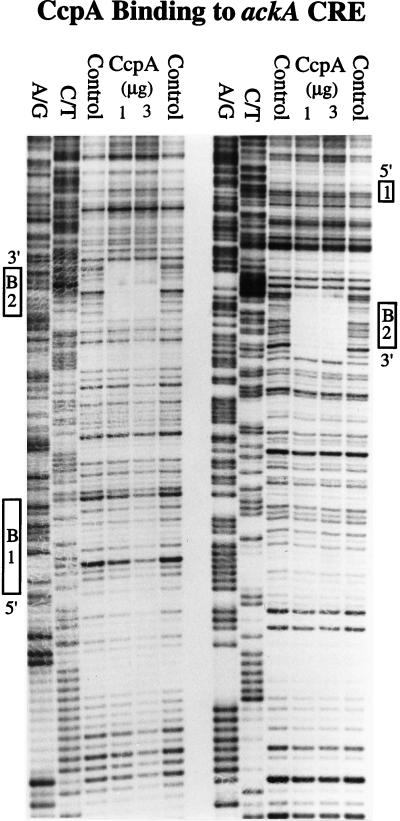
Binding of the CcpA protein to the ackA promoter region was directly tested by in vitro DNase footprinting (Fig. 5). Protection of cre2 was readily observed, with an affinity comparable to that of amyO (24); in contrast, little if any protection of cre1 was detected. These results are consistent with the genetic data indicating that cre2 is the critical element for CcpA-dependent activation. In addition, binding of CcpA to the ackA cre2 element in vitro was not dependent on any other protein.
FIG. 5.
DNase footprinting of the ackA promoter region. End-labeled ackA DNA (0.5 nM) was incubated with purified CcpA protein in TGED buffer (10 mM Tris-HCl [pH 7.4], 1 mM dithiothreitol, 50 mM KCl, 5% glycerol, 2 μg of bovine serum albumin) with 2 μg of poly(dI-dC)-poly(dI-dC) as competitor DNA for 15 min at room temperature. DNase I footprinting was carried out as previously described (24), and digested DNA was resolved on a 6% DNA sequencing gel. A/G and C/T are DNA sequencing ladders; the control contained no CcpA protein. Strand polarities and cre sites (B1, cre1; B2, cre2) are labeled.
Role of HPr/Crh in ackA transcriptional activation.
Phosphorylation of the HPr and Crh proteins has been suggested to play a key role in CcpA-mediated repression of carbon source utilization genes (6, 10). The effects of the ptsH1 mutation, which specifically blocks ATP-dependent phosphorylation of HPrSer-46, and a crh null mutation on ackA transcription were therefore tested (Table 4). While neither single mutation had any effect, the double mutant exhibited a phenotype similar to that of a CcpA mutant. As a control, the effect of the ptsH1 mutation on glucose repression of gluconate kinase activity was tested; the results were identical to those previously reported (6), confirming that the mutation is intact (data not shown). These results suggest that CcpA-mediated activation of ackA requires either HPrSer-46-P or Crh-P.
TABLE 4.
Expression of ackA-lacZ fusion in ptsH1/crh mutant strains
| Strain | β-Galactosidase activitya
|
Ratio | |
|---|---|---|---|
| Uninduced | Induced | ||
| Wild type | 92 | 330 | 3.6 |
| ptsH1 | 120 | 360 | 3.0 |
| crh::aphA3 | 53 | 250 | 4.7 |
| ptsH1/crh::aphA3 | 38 | 40 | 1.1 |
Cells were grown in TSS medium containing 1% Casamino Acids (uninduced) or 1% Casamino Acids and 1% glucose (induced). β-Galactosidase activities are expressed in Miller units (32) and indicate activity at 30 min prior to entry of the culture into stationary phase (T0).
Acetate production.
The effect of ackA transcriptional activation on production of acetate was tested by measurements of acetate accumulation in the culture supernatant. B. subtilis excretes large amounts of acetate during growth in media containing glucose (48). Acetate production is dependent on acetate kinase, and growth of an ackA null mutant is inhibited by addition of glucose to the medium (13). Both wild-type and CcpA mutant strains produced very low levels of acetate (0.04 g/liter) during growth in TSS medium in the absence of glucose; addition of 0.25% glucose resulted in acetate accumulation to 0.84 g/liter in the wild-type strain and to 0.35 g/liter in the CcpA mutant 30 min prior to T0. Normal acetate production is therefore apparently dependent on CcpA-directed activation of ackA transcription.
DISCUSSION
The B. subtilis ackA gene encoding acetate kinase is involved in the production of acetate from acetyl phosphate during growth in media containing large amounts of glucose. Transcription of ackA is induced during growth in glucose. This induction was previously shown to require CcpA and sequences upstream of the promoter, which include two cre-like sequences (13). In this study, the role of these two sites in the CcpA-dependent transcriptional activation of ackA was investigated.
Deletion analysis of the region upstream of the ackA promoter localized sequences necessary for transcriptional activation to 107 bp upstream of the transcription start site. Mutation of cre2, which is centered 56.5 bp upstream of the transcription start site, eliminated transcriptional activation and identified cre2 as a key regulatory sequence. cre1, which is centered at −116.5, is outside of the region identified by deletion analysis as being sufficient for transcriptional activation. The presence of a sequence identical to cre2, except for a single additional base, six helical turns upstream and with no apparent function is somewhat surprising. cre1 may be necessary under growth conditions other than those employed in this study and may assume a role similar to that suggested for two CcpA auxiliary sequences in the xyl operon (11). cre1 could also be required for regulation by a different regulatory factor, such as CcpB, which is apparently active under conditions of low aeration (2); the effect of a ccpB mutation on ackA expression has not been tested. The genetic data on the relative effects of cre1 and cre2 are consistent with DNase I footprinting analyses of CcpA binding to the ackA promoter region in vitro; CcpA bound to cre2 with much higher affinity than to cre1.
The position of the CcpA binding site at −56.5 is intermediate between the standard position of class I activators (−61.5 or farther upstream, in 10-bp increments) and class II activators (−41.5); the best-studied example is the E. coli CAP (CRP) protein (9, 25). The E. coli Cra protein, a CcpA homolog, also acts as both a repressor and an activator of target genes (40, 46); its binding site for activation of the ppsA gene is centered at −45.5 (37). A variety of molecular mechanisms for transcriptional activation have been reported (18, 19). The most common mechanisms involve interactions with the C-terminal domain of the α subunit (α-CTD) of RNA polymerase (RNAP), for class I activators, and interactions with the ς subunit, for class II activators, or both. The only transcriptional activator of this type characterized in detail in B. subtilis is the phage φ29 p4 protein, which binds at −82 and interacts with α-CTD (30, 31).
Transcriptional activation of ackA was lost with a 5-bp insertion between cre2 and the promoter and was reestablished with a 10-bp insertion, demonstrating a position effect suggestive of face-of-the-helix dependence. A similar effect has been demonstrated for class I activators in E. coli where the position of the binding site relative to the transcription start site can vary as long as the sites for activator and RNAP binding remain on the same face of the DNA helix (7, 49). CcpA could activate transcription via a direct interaction with RNAP or could be required for binding or correct positioning of a second factor which, in turn, interacts with RNAP. Alternatively, the insertion of 5 bp could disrupt interactions between RNAP and sequences upstream of cre2 or factors bound to these sequences. The similarity of the phenotype of the ptsH1/crh mutant to that of the CcpA mutant suggests that these proteins play a role in the control of CcpA activity, as has been proposed for genes repressed by CcpA. The reduced levels of β-galactosidase activity observed with the 5-bp insertion compared to that of the wild-type fusion expressed in a CcpA (or ptsH1/crh) mutant, or the G8T2 fusion expressed in the wild type versus a CcpA mutant strain, suggest that elements in addition to CcpA and HPr/Crh are involved. Transcriptional activation dependent on multiple proteins has been reported for a number of systems, including the E. coli ansB, malK, and nir promoters, and can employ a variety of mechanisms (19, 44, 47, 53). It will be of great interest to characterize the role of the region upstream of cre2 and the molecular mechanism of transcriptional activation.
ACKNOWLEDGMENTS
This work was supported by grant MCB-9723091 from the National Science Foundation (T.H.) and by National Institutes of Health grant GM34324 (G.C.).
We thank I. Martin-Verstraete for providing the crh mutant.
REFERENCES
- 1.Bryan E M, Beall B W, Moran C P., Jr A ςE-dependent operon subject to catabolite repression during sporulation in Bacillus subtilis. J Bacteriol. 1996;178:4778–4786. doi: 10.1128/jb.178.16.4778-4786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Chambliss, G. H. Unpublished data.
- 2.Chauvaux S, Paulsen I T, Saier M H., Jr CcpB, a novel transcription factor implicated in catabolite repression in Bacillus subtilis. J Bacteriol. 1998;180:491–497. doi: 10.1128/jb.180.3.491-497.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahl M K, Hillen W. Contribution of XylR, CcpA and HPr to catabolite repression of the xyl operon in Bacillus subtilis. FEMS Microbiol Lett. 1995;132:79–83. [Google Scholar]
- 4.Davison S P, Santangelo J D, Reid S J, Woods D R. A Clostridium acetobutylicum regulator gene regA affecting amylase production in Bacillus subtilis. Microbiology. 1995;141:989–996. doi: 10.1099/13500872-141-4-989. [DOI] [PubMed] [Google Scholar]
- 5.Deutscher J, Kuster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 6.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH1 gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebright R H. Transcription activation at class I CAP-dependent promoters. Mol Microbiol. 1993;8:797–802. doi: 10.1111/j.1365-2958.1993.tb01626.x. [DOI] [PubMed] [Google Scholar]
- 8.Egeter O, Bruckner R. Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol Microbiol. 1996;21:739–749. doi: 10.1046/j.1365-2958.1996.301398.x. [DOI] [PubMed] [Google Scholar]
- 9.Fisher S H, Rosenkrantz M S, Sonenshein A L. Glutamine synthase gene of Bacillus subtilis. Gene. 1984;32:427–438. doi: 10.1016/0378-1119(84)90018-0. [DOI] [PubMed] [Google Scholar]
- 10.Galinier A, Haiech J, Kilhoffer M, Jaquinod M, Stulke J, Deutscher J, Martin-Verstraete I. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosseringer R, Kuster E, Galinier A, Deutscher J, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium results from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 12.Grundy F J, Turinsky A J, Henkin T M. Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J Bacteriol. 1994;176:4527–4533. doi: 10.1128/jb.176.15.4527-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundy F J, Waters D A, Allen S H G, Henkin T M. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J Bacteriol. 1993;175:7348–7355. doi: 10.1128/jb.175.22.7348-7355.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundy F J, Waters D A, Takova T Y, Henkin T M. Identification of genes involved in the utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 15.Henkin T M, Chambliss G H. Genetic mapping of a mutation causing an alteration in Bacillus subtilis ribosomal protein S4. Mol Gen Genet. 1984;193:364–369. doi: 10.1007/BF00330694. [DOI] [PubMed] [Google Scholar]
- 16.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 17.Henkin T M. The role of the CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 18.Hochschild A, Dove S L. Protein-protein contacts that activate and repress prokaryotic transcription. Cell. 1998;92:597–600. doi: 10.1016/s0092-8674(00)81126-5. [DOI] [PubMed] [Google Scholar]
- 19.Hochschild A, Joung J. Synergistic activation of transcription in Escherichia coli. Nucleic Acids Mol Biol. 1997;11:101–114. [Google Scholar]
- 20.Hueck C J, Hillen W, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 21.Hueck C J, Kraus A, Schmiedel D, Hillen W. Cloning, expression and functional analyses of the catabolite control protein CcpA from Bacillus megaterium. Mol Microbiol. 1995;16:855–864. doi: 10.1111/j.1365-2958.1995.tb02313.x. [DOI] [PubMed] [Google Scholar]
- 22.Jones B E, Dossonnet V, Kuster E, Hillen W, Deutscher J, Klevit R E. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J Biol Chem. 1997;272:26530–26535. doi: 10.1074/jbc.272.42.26530. [DOI] [PubMed] [Google Scholar]
- 23.Kim J H, Chambliss G H. Contacts between Bacillus subtilis catabolite regulatory protein CcpA and the amyO target site. Nucleic Acids Res. 1997;25:3490–3496. doi: 10.1093/nar/25.17.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J H, Guvener Z T, Cho J Y, Chung K, Chambliss G H. Specificity of DNA binding activity of the Bacillus subtilis catabolite control protein CcpA. J Bacteriol. 1995;177:5129–5134. doi: 10.1128/jb.177.17.5129-5134.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 26.Kraus A, Hueck C, Gartner D, Hillen W. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J Bacteriol. 1994;176:1738–1745. doi: 10.1128/jb.176.6.1738-1745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruger S, Hecker M. Regulation of the putative bglPH operon for aryl-β-glucoside utilization in Bacillus subtilis. J Bacteriol. 1995;177:5590–5597. doi: 10.1128/jb.177.19.5590-5597.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuster S, Luesink E J, de Vos W M, Hillen W. Immunological crossreactivity to catabolite control protein CcpA from Bacillus megaterium is found in many gram-positive bacteria. FEMS Microbiol Lett. 1996;139:109–115. doi: 10.1111/j.1574-6968.1996.tb08188.x. [DOI] [PubMed] [Google Scholar]
- 29.Martin-Verstraete I, Stulke J, Klier A, Rapoport G. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol. 1995;177:6919–6927. doi: 10.1128/jb.177.23.6919-6927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mencia M, Monsalve M, Rojo F, Salas M. Transcription activation by phage Φ29 protein p4 is mediated by interaction with the α subunit of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci USA. 1996;93:6616–6620. doi: 10.1073/pnas.93.13.6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mencia M, Monsalve M, Rojo F, Salas M. Substitution of the C-terminal domain of the Escherichia coli RNA polymerase α subunit by that from Bacillus subtilis makes the enzyme responsive to a Bacillus subtilis transcriptional activator. J Mol Biol. 1998;275:177–185. doi: 10.1006/jmbi.1997.1463. [DOI] [PubMed] [Google Scholar]
- 32.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 33.Miwa Y, Nagura K, Eguchi S, Fukuda H, Deutscher J, Fujita Y. Catabolite repression of the Bacillus subtilis gnt operon exerted by two catabolite-responsive elements. Mol Microbiol. 1997;23:1203–1213. doi: 10.1046/j.1365-2958.1997.2921662.x. [DOI] [PubMed] [Google Scholar]
- 34.Miwa Y, Saikawa M, Fujita Y. Possible function and some properties of the CcpA protein of Bacillus subtilis. Microbiology. 1994;140:2567–2575. doi: 10.1099/00221287-140-10-2567. [DOI] [PubMed] [Google Scholar]
- 35.Monedero V, Gosalbes M J, Perez-Martinez G. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J Bacteriol. 1997;179:6657–6664. doi: 10.1128/jb.179.21.6657-6664.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano M M, Zuber P. Cloning and characterization of srfB, a regulatory gene involved in surfactin production and competence in Bacillus subtilis. J Bacteriol. 1989;171:5347–5353. doi: 10.1128/jb.171.10.5347-5353.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Negre D, Oudot C, Prost J-F, Murakami K, Ishihama A, Cozzone A J, Cortay J-C. FruR-mediated transcriptional activation at the ppsA promoter of Escherichia coli. J Mol Biol. 1998;276:355–365. doi: 10.1006/jmbi.1997.1548. [DOI] [PubMed] [Google Scholar]
- 38.Nicholson W L, Park Y, Henkin T M, Won M, Weickert M J, Gaskell J A, Chambliss G H. Catabolite repression-resistant mutations of the Bacillus subtilis alpha-amylase promoter affect transcription levels and are in an operator-like sequence. J Mol Biol. 1987;198:609–618. doi: 10.1016/0022-2836(87)90204-x. [DOI] [PubMed] [Google Scholar]
- 39.Rado T A, Hoch J A. Phosphotransacetylase from Bacillus subtilis: purification and physiological studies. Biochim Biophys Acta. 1973;321:114–125. doi: 10.1016/0005-2744(73)90065-x. [DOI] [PubMed] [Google Scholar]
- 40.Ramseier T M, Negre D, Cortay J, Scarabel M, Cozzone A J, Saier M H., Jr In vitro binding of the pleiotropic transcriptional regulatory protein, FruR, to the fru, pps, ace, pts, and icd operons of Escherichia coli and Salmonella typhimurium. J Mol Biol. 1993;234:28–44. doi: 10.1006/jmbi.1993.1561. [DOI] [PubMed] [Google Scholar]
- 41.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stulke J, Karamata D, Saier M H, Jr, Hillen W. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 42.Reizer J, Romano A H, Deutscher J. The role of phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, in regulation of carbon metabolism in gram-positive bacteria. J Cell Biochem. 1993;51:19–24. doi: 10.1002/jcb.240510105. [DOI] [PubMed] [Google Scholar]
- 43.Renna M C, Najimudin N, Winik L R, Zahler S A. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J Bacteriol. 1993;175:3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richet E, Sogaard-Andersen L. CRP induces the repositioning of MalT at the Escherichia coli malKp promoter primarily through DNA bending. EMBO J. 1994;13:4558–4567. doi: 10.1002/j.1460-2075.1994.tb06777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rollins S M, Grundy F J, Henkin T M. Analysis of the cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol Microbiol. 1997;25:411–421. doi: 10.1046/j.1365-2958.1997.4851839.x. [DOI] [PubMed] [Google Scholar]
- 46.Saier M H, Jr, Ramseier T M. The catabolite repressor/activator (Cra) protein of enteric bacteria. J Bacteriol. 1996;178:3411–3417. doi: 10.1128/jb.178.12.3411-3417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott S, Busby S, Beacham I. Transcriptional co-activation at the ansB promoters: involvement of the activating regions of CRP and FNR when bound in tandem. Mol Microbiol. 1995;18:521–531. doi: 10.1111/j.1365-2958.1995.mmi_18030521.x. [DOI] [PubMed] [Google Scholar]
- 48.Speck E L, Freese E. Control of metabolite secretion in Bacillus subtilis. J Gen Microbiol. 1973;78:261–275. doi: 10.1099/00221287-78-2-261. [DOI] [PubMed] [Google Scholar]
- 49.Ushida C, Aiba H. Helical phase dependent action of CRP: effect of the distance between CRP site and the −35 region on promoter activity. Nucleic Acids Res. 1990;18:6325–6330. doi: 10.1093/nar/18.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voskuil M I, Chambliss G H. Significance of HPr in catabolite repression of α-amylase. J Bacteriol. 1996;178:7014–7015. doi: 10.1128/jb.178.23.7014-7015.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wray L W, Jr, Pettengill F K, Fisher S H. Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting site located downstream of the transcriptional initiation site. J Bacteriol. 1994;176:1894–1902. doi: 10.1128/jb.176.7.1894-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu H-C, Tyson K L, Cole J A, Busby S J W. Regulation of transcription initiation at the Escherichia coli nir operon promoter: a new mechanism to account for co-dependence on two transcription factors. Mol Microbiol. 1998;27:493–505. doi: 10.1046/j.1365-2958.1998.00699.x. [DOI] [PubMed] [Google Scholar]
- 54.Zuber P, Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987;169:2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]