Abstract
Aims
Denture use may potentially increase the risk of cardiometabolic diseases (CMDs), but the casual relevance and strength of the associations are currently unknown.
Methods and results
A total of 495 938 participants from the UK Biobank were included in the observational analyses. Linkage disequilibrium score (LDSC) regression and Mendelian randomization analyses were employed to estimate genetic correlation and the associations between the genetic liability for denture use with coronary artery disease, myocardial infarction, heart failure (HF), any stroke (AS), ischaemic stroke, haemorrhagic stroke, type 2 diabetes (T2D), and related clinical risk factors. In observational analysis, denture use was associated with 14–25% higher risks of various CMDs. The LDSC analysis found that denture use showed a positive genetic correlation with CMDs (rg 0.21–0.38). Genetic liability for denture use was associated with an elevated risk of HF [odds ratio: 1.49 (1.20–1.83)] and T2D [1.11 (1.01–1.24)]. By integrating genetic summary data of denture use with the sum of decayed, missing, and filled tooth surfaces (DMFS), a clinical measure of dental caries obtained from an independent source, genetically determined denture use/DMFS was also associated with an elevated risk of AS [1.21 (1.04–1.40)]. Furthermore, genetically predicted denture use/DMFS was significantly associated with established cardiometabolic risk factors, including HDL cholesterol, triglycerides, waist circumference, waist-to-hip ratio, and height.
Conclusion
Our study supported potential causal associations between the genetic liability for denture use and risks for HF, AS, T2D, and related clinical risk factors. These findings may inform prevention and intervention strategies targeting dental diseases and CMDs.
Keywords: Oral health, Heart failure, Stroke, Type 2 diabetes, Cardiometabolic disease, Mendelian randomization
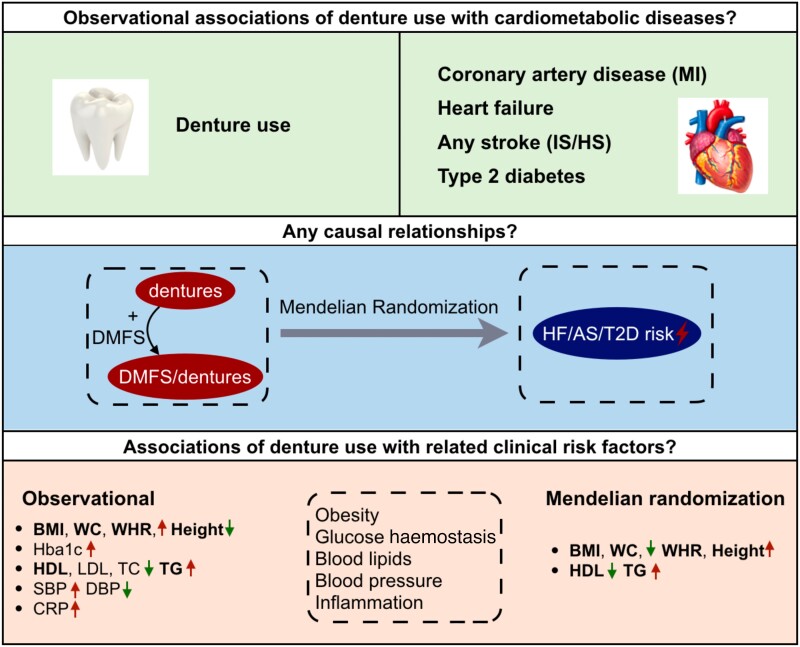
Graphical Abstract
Graphical Abstract.
AS, any stroke; BMI, body mass index; CRP, C-reactive protein; DBP, diastolic blood pressure; DMFS, the sum of decayed, missing, and filled tooth surfaces; Hba1c, haemoglobin A1c; HF, heart failure; HS, haemorrhagic stroke; IS, ischaemic stroke; MI, myocardial infarction; SBP, systolic blood pressure; T2D, type 2 diabetes; TC, total cholesterol; TG, triglycerides; WC, waist circumference; WHR, waist-to-hip ratio.
See the editorial comment for this article ‘The oral-systemic axis: how oral health affects cardiovascular and metabolic health’, by E. Mauri-Obradors and R. Elosua, https://doi.org/10.1093/eurjpc/zwad353.
Introduction
Oral diseases are common and affect almost 3.5 billion individuals globally.1,2 Several observational studies have reported associations of different oral diseases or symptoms, including periodontitis and tooth loss, with an increased risk of cardiovascular diseases (CVDs)3–6 and diabetes.7,8 Likewise, intervention studies have reported the effects of treatments for oral health symptoms or diseases on risk factors of cardiometabolic diseases (CMDs), such as obesity, hypertension, dyslipidaemia, hyperglycaemia, and chronic inflammation, which may mediate the associations between oral health and CVD.9–14 As a strong predictor of oral health–related quality of life, denture problem,15–18 particularly denture stomatitis, has been demonstrated to be associated with an increased risk of CVD and type 2 diabetes (T2D) in observational studies. Due to the high cost of clinical trials and thorough oral health examination, most studies have been limited to small sample sizes, short durations, or specific CVD outcomes; therefore, the causal relationships between oral health and various CMD types remain to be elucidated. Large population-based studies with a validated oral health questionnaire, electronic health data linkage, and genetic data are now able to assess possible causal relationships of dental diseases with CMD outcomes and related clinical risk factors.
Mendelian randomization (MR) approaches use genetic variants as instrumental variables (IVs) to assess the causality of an exposure with an outcome.19 As genetic variants are randomly allocated at conception, their associations with exposures of interest are less likely to be affected by confounding and reverse causation. Recent developments in MR methods together with the increasing availability of large-scale human genetic data, both individual-level genetic data and summary statistics, have enabled investigations of causality.
Using data from UK Biobank (UKB) and public consortia, this study aimed to evaluate and compare the observational and genetic associations of denture use with various types of CMD and related clinical risk factors, including anthropometric measurements, glycaemic traits, blood lipids, blood pressure, and inflammatory markers.
Methods
Study population
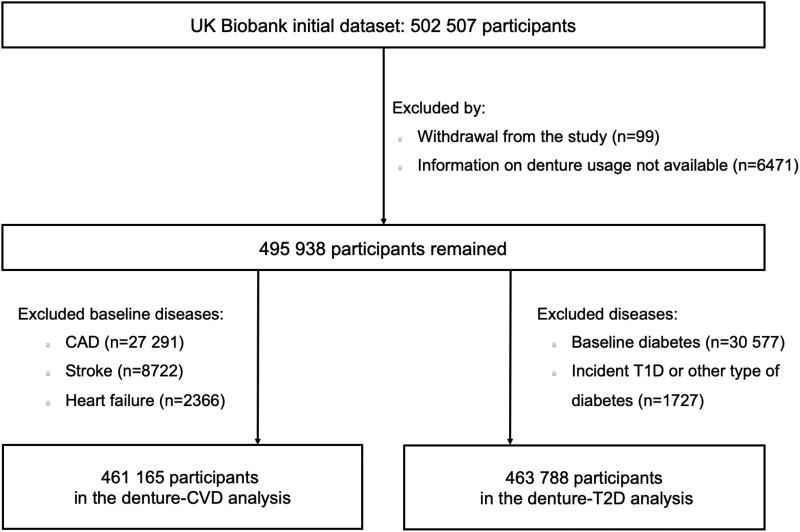
The study design and methods of UKB have been reported previously.20,21 In brief, UKB is a large-scale prospective study with over 500 000 participants aged 37–73 years who were recruited in 2006–2010. We excluded participants who withdrew from the study (n = 99), had missing data on self-reported denture use (n = 6471), and those with CVD (n = 34 773) or diabetes (32 150) at baseline, leaving 461 165 or 463 788 participants for the denture–CVD or denture–T2D analysis, respectively. Further details of the study population are provided in Figure 1.
Figure 1.
Flow diagram for study cohort of observational analysis. CAD, coronary artery disease; CVD, cardiovascular diseases; T1D, type 1 diabetes; T2D, type 2 diabetes.
UK Biobank received ethical approval from the North West Multi-centre Research Ethics Committee (REC reference: 11/NW/03820). All participants gave written informed consent before enrolment in the study, which was conducted in accordance with the principles of the Declaration of Helsinki.
Exposure and outcome definition
Self-reported denture usage (yes or no) was assessed by a multichoice question: ‘Do you have any of the following? (You can select more than one answer)’, and possible answers included ‘dentures’, ‘bleeding gums’, ‘painful gums’, ‘loose teeth’, ‘toothache’, and ‘mouth ulcers’. We defined incident coronary artery disease (CAD), myocardial infarction (MI), heart failure (HF), any stroke (AS), ischaemic stroke (IS), haemorrhagic stroke (HS), and T2D using hospital inpatient records, death register, and self-reported health conditions from the UKB assessment clinics with the end time of 28 February 2021. The definition and International Classification of Diseases (ICD) code used for each CMD are provided in Supplementary material online, Table S1.
Genetic variants as instrumental variables
We performed genome-wide association studies (GWASs) of denture use among participants of European ancestry in the UKB, using a linear mixed model in BOLT-LMM (version 2.3.4). In light of the strong genetic correlation (Rg = 0.82, se = 0.087, P = 4.1 × 10−21) of self-reported denture use in UKB with the sum of decayed, missing, and filled tooth surfaces (DMFS) previously reported in the Gene-Lifestyle Interactions in Dental Endpoints (GLIDE) study, we also combined them as denture use/DMFS using the fixed effects Z-score method in METAL22 to strengthen data analysis and interpretation. Instrumental variables were obtained from the previously reported variants for denture use/DMFS23 and periodontitis9,24,25 (see Supplementary material online, Tables S2 and S3) or identified with the following criteria for denture use in UKB: (i) genome-wide significant (P < 5 × 10−8); (ii) linkage disequilibrium r2 < 0.05; and (iii) P for pleiotropy < 0.01, which was assessed by heterogeneity in dependent instrument (HEIDI)-outlier method (detect and eliminate genetic instruments that have apparent pleiotropic effects on both risk factor and disease) of generalized summary-data-based Mendelian randomization (GSMR)26 in GCTA (version 1.93.2).
We used publicly available genome-wide databases to obtain summary-level genetic data for CMDs and related clinical risk factors (see Supplementary material online, Table S4), including anthropometric measurements [i.e. body mass index (BMI), waist circumference (WC), waist-to-hip ratio (WHR), and height], glycaemic traits [i.e. haemoglobin A1c (HbA1c)], blood lipids (i.e. LDL cholesterol, HDL cholesterol, triglycerides, and total cholesterol], and blood pressure traits [i.e. systolic blood pressure (SBP) and diastolic blood pressure (DBP)]. For each of the single nucleotide polymorphisms (SNPs) associated with exposure of interest, we retrieved summary-level data for either the same SNP or for a proxy SNP in high linkage disequilibrium (r2 > 0.8) from the outcome data set.
Statistical analysis
Observational analysis
Baseline characteristics of 495 938 participants were described as means (standard deviations) and medians (interquartile ranges) for continuous variables or numbers (percentages) for categorical variables. We conducted logistic and linear regressions to estimate the associations of denture use with incident CMDs and related clinical risk factors (obesity, glycaemic traits, blood lipids, hypertension, and inflammatory markers). The multivariate model incorporated sociodemographic factors (age, sex, ethnicity, and education), socioeconomic status (Townsend deprivation index), family history of CVD/diabetes, smoking status, alcohol intake, physical activity, BMI, medication use (antihypertensive or cholesterol-lowering drugs), SBP, and triglycerides where appropriate.
Mendelian randomization analysis
We estimated the genetic correlation for the presence of denture use with CMDs or risk factors using the linkage disequilibrium score (LDSC)27 on the GWAS summary statistics. Generalized summary Mendelian randomization was applied to analyse the genetic associations of denture use with various CMDs, using the ‘HEIDI-outlier’ method to detect and eliminate genetic instruments that show apparent pleiotropic effects.26 In addition to the inverse variance weighted (IVW) method, we also applied weighted median, weighted mode, and MR–Egger28,29 methods to examine potential pleiotropic effects that may bias the causal estimates. Moreover, we tested the potential impacts of sample overlap, i.e. by excluding the studies contributing towards both the SNP-to-exposure and SNP-to-outcome through ‘deMeta’. Bidirectional MR was conducted using SNPs associated with HF/T2D/AS to test whether genetic predisposition for HF/T2D/AS might be a causal factor for denture use. Finally, we performed GSMR to investigate whether genetically predicted denture use/DMFS was also associated with related clinical risk factors, including obesity, blood lipids, blood pressure, and glycaemic traits.
All statistical analyses were performed in R (version 3.6.3), and a two-sided P < 0.05 was considered statistically significant, with a 5% false discovery rate to correct for multiple testing.
Results
Observational associations of denture use with cardiometabolic diseases
The demographic and clinical characteristics of study participants are shown in Table 1. Compared with the total population or participants with the absence of denture use, people with denture use tended to be more deprived, have higher BMI, triglycerides, and C-reactive protein, and are more likely to use antihypertensive or cholesterol-lowering drugs.
Table 1.
Baseline characteristics of participants by the presence or absence of denture use in UK Biobank
| Absence of denture | Presence of denture | All | |
|---|---|---|---|
| Number of participants | 412 742 | 83 196 | 495 938 |
| Demographic factors | |||
| Male, % | 186 277 (45.1) | 39 777 (47.8) | 226 054 (45.6) |
| Age, years | 55.6 (8.1) | 61.4 (6.3) | 56.5 (8.1) |
| Ethnicity, white, % | 388 595 (94.5) | 79 273 (95.6) | 467 868 (94.3) |
| Townsend deprivation index | −2.24 (−3.70, 0.32) | −1.60 (−3.34, 1.51) | −2.15 (−3.65, 0.53) |
| Education, university/college, % | 146 660 (35.9) | 14 153 (17.3) | 160 813 (32.4) |
| Lifestyle factors | |||
| Current smoking, % | 38 644 (9.4) | 13 494 (16.2) | 52 138 (10.5) |
| Current drinking, % | 381 763 (92.5) | 73 796 (88.7) | 455 559 (91.9) |
| Physical activity, MET-h/week | 29.2 (13.3, 58.2) | 31.1 (13.2, 65.9) | 29.5 (13.3, 59.2) |
| Family history of diseases, n (%) | |||
| Family history of CVD | 303 872 (73.6) | 61 039 (73.4) | 364 911 (73.6) |
| Family history of diabetes | 88 698 (21.5) | 18 667 (22.4) | 107 365 (21.6) |
| Medical history, n (%) | |||
| Use of antihypertensive drugs | 77 338 (18.9) | 26 271 (31.8) | 103 609 (20.9) |
| Use of cholesterol-lowering drugs | 62 792 (15.3) | 23 798 (28.8) | 86 590 (17.5) |
| Coronary artery disease | 18 222 (4.4) | 9069 (10.9) | 27 291 (5.5) |
| Myocardial infarction | 7821 (1.9) | 4066 (4.9) | 11 887 (2.4) |
| Heart failure | 1527 (0.4) | 839 (1.0) | 2366 (0.5) |
| Any stroke | 6009 (1.5) | 2713 (3.3) | 8722 (1.8) |
| Ischaemic stroke | 889 (0.2) | 372 (0.4) | 1261 (0.3) |
| Haemorrhagic stroke | 963 (0.2) | 333 (0.4) | 1296 (0.3) |
| Type 2 diabetes | 16 563 (4.0) | 6589 (7.9) | 23 152 (4.7) |
| Clinical measurements | |||
| BMI, kg/m2 | 27.3 (4.8) | 28.2 (4.9) | 27.4 (4.8) |
| WC, cm | 89.7 (13.4) | 93.1 (13.4) | 90.3 (13.5) |
| WHR | 0.87 (0.80, 0.93) | 0.89 (0.83, 0.95) | 0.87 (0.80, 0.94) |
| Height, cm | 168.7 (9.3) | 167.3 (9.3) | 168.5 (9.3) |
| SBP, mmHg | 137.1 (18.5) | 142.0 (18.9) | 137.9 (18.7) |
| DBP, mmHg | 82.2 (10.2) | 82.4 (10.1) | 82.3 (10.1) |
| Random glucose, mmol/L | 4.92 (4.59, 5.29) | 4.99 (4.65, 5.42) | 4.93 (4.60, 5.31) |
| Hba1c, % | 5.35 (5.13, 5.59) | 5.48 (5.25, 5.75) | 5.37 (5.15, 5.62) |
| HDL, mmol/L | 1.46 (0.38) | 1.40 (0.37) | 1.45 (0.38) |
| LDL, mmol/L | 3.57 (0.86) | 3.52 (0.92) | 3.56 (0.87) |
| Triglycerides, mmol/L | 1.45 (1.02, 2.11) | 1.64 (1.16, 2.32) | 1.48 (1.05, 2.15) |
| TC, mmol/L | 5.71 (1.13) | 5.62 (1.22) | 5.69 (1.14) |
| C-reactive protein, mg/L | 1.26 (0.62, 2.61) | 1.75 (0.87, 3.57) | 1.33 (0.66, 2.76) |
Values are shown as numbers (percentages) for categorical variables and means (SDs) or medians (IQRs) for continuous variables.
BMI, body mass index; CVD, cardiovascular disease; DBP, diastolic blood pressure; Hba1c, haemoglobin A1c; IQR, interquartile range; MET, metabolic equivalent of task; SBP, systolic blood pressure; SD, standard deviation; TC, total cholesterol; WC, waist circumference; WHR, waist-to-hip ratio.
As shown in Table 2, denture use was associated with a higher risk of CAD [1.15 (95% confidence interval: 1.12–1.18)], MI [1.18 (1.13–1.24)], HF [1.25 (1.19–1.31)], AS [1.19 (1.13–1.25)], IS [1.19 (1.12–1.27)], HS [1.14 (1.04–1.25)], and T2D [1.16 (1.11–1.21)].
Table 2.
Observational and genetic associations of denture use with risk of cardiometabolic diseases
| Outcome | Observational | Genetics | |||||
|---|---|---|---|---|---|---|---|
| Dentures | Dentures | Dentures/DMFSa | |||||
| Risk coefficient (95% CI) | SNPs | Risk coefficient (95% CI) | P adjusted | SNPs | Risk coefficient (95% CI) | P adjusted | |
| Coronary artery disease | 1.15 (1.12, 1.18) | 41 | 1.13 (0.85, 1.46) | 0.449 | 55 | 1.14 (0.98, 1.33) | 0.138 |
| Myocardial infarction | 1.18 (1.13, 1.24) | 41 | 1.10 (0.80, 1.46) | 0.554 | 56 | 1.10 (0.93, 1.30) | 0.300 |
| Heart failure | 1.25 (1.19, 1.31) | 39 | 1.49 (1.20, 1.83) | 3.7 × 10−3 | 54 | 1.23 (1.08, 1.39) | 5.4 × 10−3 |
| Any stroke | 1.19 (1.13, 1.25) | 39 | 1.27 (0.96, 1.63) | 0.156 | 56 | 1.21 (1.04, 1.40) | 0.028 |
| Ischaemic stroke | 1.19 (1.12, 1.27) | 41 | 1.63 (0.93, 2.70) | 0.156 | 56 | 1.21 (0.88, 1.65) | 0.300 |
| Haemorrhagic stroke | 1.14 (1.04, 1.25) | 36 | 2.68 (0.57, 14.3) | 0.245 | 44 | 1.50 (0.64, 3.54) | 0.355 |
| Type 2 diabetes | 1.16 (1.11, 1.21) | 42 | 1.11 (1.01, 1.24) | 0.109 | 54 | 1.41 (1.17, 1.72) | 3.1 × 10−3 |
Multivariable logistic regression was used to estimate the risk of incident CMDs among individuals with the presence of denture use or not after adjusting sociodemographic factors (age, sex, ethnicity, and education), socioeconomic status (Townsend deprivation index), family history of CVD or diabetes, smoking status, alcohol intake, physical activity, BMI, baseline diabetes or CVD, medication use (antihypertensive or cholesterol-lowering drugs), SBP, and triglycerides where appropriate. We implement GSMR in GCTA (version 1.93.2) to calculate the causal estimates of denture use or denture use/DMFS with CMDs. Estimates expressed as odds ratio (95% CI) represent the effect of the presence of denture use or a 1 SD increase in levels of denture use/DMFS on outcomes of interest, respectively. A 5% false discovery rate was used to correct for multiple testing in MR analysis.
BMI, body mass index; CI, confidence interval; CMD, cardiometabolic disease; CVD, cardiovascular disease; DMFS, the sum of decayed, missing, and filled tooth surfaces; GLIDE, Gene-Lifestyle Interactions in Dental Endpoints; GSMR, generalized summary-data-based Mendelian randomization; SBP, systolic blood pressure; SD, standard deviation.
aDenture use/DMFS: a combination of summary statistics of denture use in UK Biobank and DMFS in GLIDE using a Z-score genome-wide meta-analysis.
Genetic associations of denture use with cardiometabolic diseases
Genome-wide association analysis identified 41 independent genetic variants associated with denture use (P < 5 × 10−8, Supplementary material online, Figure S1). There were modest positive genetic correlations between denture use and CMDs ranging between 0.21 and 0.38 (P < 0.01, Supplementary material online, Table S5).
To explore whether the positive correlations in both observational and genetic correlations analyses are due to the pathway effects where dental diseases are risk factors for CMDs, we further conducted MR analysis. We selected 36–42 SNPs as IVs for denture use in UKB and a total of 44–56 SNPs as IVs for denture use/DMFS, and 4 IVs were obtained for periodontitis. Using the GSMR approach, we found that genetically predicted denture use was associated with increased risks of HF [1.49 (95% confidence interval: 1.20–1.83)] and T2D [1.11 (1.01–1.24)] (Table 2). Genetic liability for denture use was not significantly associated with risk of CAD [1.13 (0.85–1.46)], MI [1.10 (0.80–1.46)], and HS [2.68 (0.57–14.3)] but turned to be associated with increased AS risk [1.27 (0.96–1.63), Padjusted = 0.156] and IS risk [1.63 (0.93–2.70), Padjusted = 0.156]. No significant association was identified for periodontitis with CMDs (see Supplementary material online, Table S6). Analysis integrating genetic summary statistics of denture use and DMFS showed genetically determined denture use/DMFS was also associated with a higher risk of HF [1.23 (1.08–1.39)], AS [1.21 (1.04–1.40)], and T2D [1.41 (1.17–1.72)], the result of which also showed adequate statistical power (see Supplementary material online, Table S7). Sensitivity analyses using IVW, weighted median, weighted mode, and MR–Egger methods showed similar findings, with no evidence of unbalanced pleiotropy (see Supplementary material online, Table S8). The results were also not materially changed by excluding sample overlap (see Supplementary material online, Figure S2 and Tables S9 and S10).
Bidirectional single-variable analyses showed no evidence of potential reverse causal relationships of genetic liability for HF/AS/T2D as risk factors and denture use/DMFS use as outcome (see Supplementary material online, Table S11).
Observational and genetic associations of denture use/the sum of decayed, missing, and filled tooth surfaces with related clinical risk factors
In observational analyses, participants with denture use were associated with 0.003 (0.003–0.004) higher WHR, 0.042 mmol/L (0.034–0.050 mmol/L) higher triglycerides, and 0.034 mmol/L (0.031–0.037 mmol/L) lower HDL. In the GSMR analysis, significant associations remained for WHR, triglycerides, and HDL (all P < 0.05, details in Table 3). In addition, genetically predicted denture use/DMFS was also associated with 0.051 kg/m2 (0.018–0.084 kg/m2) lower BMI, 0.044 cm (0.006–0.081 cm) lower WC, and 0.040 cm (0.013–0.067 cm) higher height, which were in opposite directions as these in the observational associations.
Table 3.
Observational and genetic associations of denture use/the sum of decayed, missing, and filled tooth surfaces with related clinical risk factors
| Outcome | Observational | Genetics | |
|---|---|---|---|
| Risk coefficient (95% CI)a | Risk coefficient (95% CI)b | P adjusted | |
| Body mass index (kg/m2) | 0.34 (0.31, 0.37) | −0.051 (−0.084, −0.018) | 0.010 |
| Waist circumference (cm) | 0.13 (0.08, 0.18) | −0.044 (−0.081, −0.006) | 0.041 |
| Waist-to-hip ratio | 0.003 (0.003, 0.004) | 0.048 (0.013, 0.083) | 0.018 |
| Height (cm) | −0.45 (−0.50, −0.40) | 0.040 (0.013, 0.067) | 0.012 |
| Systolic blood pressure (mmHg) | 0.37 (0.22, 0.52) | 0.13 (−0.14, 0.40) | 0.379 |
| Diastolic blood pressure (mmHg) | −0.13 (−0.22, −0.05) | 0.15 (−0.01, 0.30) | 0.103 |
| Hba1c (%) | 0.007 (0.005, 0.010) | −0.013 (−0.029, 0.003) | 0.151 |
| HDL (mmol/L) | −0.034 (−0.037, −0.031) | −0.049 (−0.087, −0.010) | 0.030 |
| LDL (mmol/L) | −0.016 (−0.023, −0.009) | 0.006 (−0.035, 0.048) | 0.765 |
| Triglycerides (mmol/L) | 0.042 (0.034, 0.050) | 0.060 (0.023, 0.096) | 0.009 |
| Total cholesterol (mmol/L) | −0.045 (−0.054, −0.035) | 0.033 (−0.009, 0.074) | 0.164 |
| C-reactive protein (mg/L) | 0.25 (0.22, 0.27) | — | — |
Linear regression models were conducted for the presence of denture use with cardiometabolic risk factors in observational analyses, with sociodemographic factors (age, sex, ethnicity, and education), socioeconomic status (Townsend deprivation index), family history of diabetes or CVD, smoking status, alcohol intake, physical activity, BMI, baseline diabetes, medication use (antihypertensive or cholesterol-lowering drugs), SBP, and triglycerides adjusted where appropriate (see Supplementary material online, Methods for full description of each analysis). All causal estimates were calculated by using GSMR in GCTA (version 1.93.2) based on denture use/DMFS and other summary-level genetic data retrieved from publicly available genome-wide databases. A 5% false discovery rate was used to correct for multiple testing in MR analysis.
CI, confidence interval; DMFS, the sum of decayed, missing, and filled tooth surfaces; GSMR, generalized summary-data-based Mendelian randomization; Hba1c, haemoglobin A1c; MR, Mendelian randomization.
aRisk estimates expressed as the differences of values for continuous variables among individuals with the presence of denture use or not after adjusting the corresponding covariates.
bThe regression coefficients of denture use/DMFS with cardiometabolic risk factors were rescaled to the same scale used of denture use, with the interpretation as per log odds of denture use (details in Supplementary material online, Methods).
Discussion
In this large study of UK participants with self-reported oral symptoms, we observed a positive association for denture use with CMD risks and potentially causal associations between genetic liability for denture use and the risk for HF, AS, and T2D in MR analysis. In addition, genetically predicted denture usage was also associated with higher levels of WHR, height, HDL, and triglycerides, respectively.
This large MR study demonstrated a possible causal relationship between denture use and CMDs. Many observational studies have reported positive associations between self-reported or clinically measured oral health issues, such as dental caries,4,8 periodontitis,5,7,30 missing teeth or tooth loss,4,6 and dentures,15–18 with risks of CVDs and diabetes. Moreover, improved oral hygiene care such as frequent tooth brushing and regular dental visits for professional cleaning procedures has been inversely associated with the risk of major cardiovascular events and diabetes.4,31–33 Among the few studies exploring potential causality,23,34–39 Shungin et al.23 observed that genetically predicted denture use/DMFS was related to a higher risk of stroke, which became non-significant after correcting for multiple testing. In our study, genetically predicted denture use showed strong associations with increased risk of HF, AS, and T2D, which remained by further incorporating data on denture use/DMFS. The strength of the observed association between genetic liability to denture use and CVDs was comparable to well-established CVD risk factors such as smoking and obesity according to existing studies. For example, a 49% higher risk for HF was similar to smoking initiation (ever smoked regularly, 53%)40 or 4 kg/m2 increase in BMI (48%).41 In general, dentures are considered the end stage of oral health problems and can be an indicator of tooth loss. In the UK, self-reported dentures were likely to be removable dentures. According to the Adult Dental Survey conducted in 2009/2010,42 20% of the UK population wore some form of removable dentures. These may be colonized by diverse pathogenic microbial biofilms,18 which probably leads to the generation of visible denture plaques, the simulation of immune-inflammatory responses, and the different salivary and gut microbiome compositions that collectively affect metabolic health.15,43 Besides, the potential hazards of denture usage for diabetes were consistent with the findings of a 12-month randomized trial that periodontal treatment reduced HbA1c and plasma glucose concentrations.10 However, further research with thorough measurement for oral status or well-designed clinical trials are required to explore other possible mechanisms for the observed associations and the heterogeneities among CVD subtypes.
A causal relationship between genetic liability for denture usage and AS and HF risk was strengthened by the associations between dentures and WHR, height, HDL, and triglycerides, which are among the major risk factors of CMD. These data are consistent with common clinical observations that poor oral health status is closely associated with poor metabolic status.8,9,23,44 Among existing clinical trials, periodontal treatment has been associated with improved serum inflammatory biomarkers,12,13 blood lipids,12,13 glycaemic profiles,10 blood pressure,9 and endothelial function,11 as well as other surrogate measures of CMD. Our findings are consistent with previous studies that denture use/DMFS was causally linked with WHR and triglycerides (P < 0.05),23 and provide further evidence on the possible role of denture use on metabolic health. Surprisingly, the associations BMI, WC, and height with denture use/DMFS were in opposite directions between observational and MR analyses, which may reflect the influence of confounding factors in the observational analyses or poor nutritional status for people with oral health issues.45,46
The strength of UKB included detailed data from questionnaires, biomedical tests, genetic background, and medical records in a large sample, which enabled us to systematically evaluate causal relationships for denture use with various types of CMDs and related clinical risk factors. Our study also has several limitations. First, denture usage was evaluated by self-reported symptoms, which may lead to misclassification of exposures and unclear interpretation of the result. However, it may be implausible to conduct a thorough dental examination in such a large cohort study, while existing validation studies demonstrated that some of the self-reported oral health status may reflect clinically confirmed oral diseases in several independent populations.47–49 Second, participants of the UKB were included in genetic summary data sets of the exposure (denture use) as well as the outcome (HF) in the primary MR analysis, which tended to bias the causal estimates. However, bias due to sample overlap could be reasonably small owing to the large sample sizes of the denture use/DMFS and HF GWASs and the high power of analysis (power ≈ 0.99, F-statistics = 2221.96). In addition, we conducted a series of sensitivity analyses to explore the potential impact of sample overlap, and the results remained unchanged. Third, our estimates could have been biased if there was possible pleiotropy, in which the genetic instruments are associated with the outcome through a phenotype independent of the exposure under consideration. We used various statistical methods (such as MR–Egger) to account for this, which produced similar results to the main analysis. Fourth, MR studies assess lifelong associations; as such, the magnitude of effect might not be comparable to effects that are observed in short-term intervention studies. Fifth, descriptions of the types of dentures (i.e. removable dentures, dental implants, and fixed bridges) were not available in UKB, which may limit data interpretation, though previous studies showed that dentures were more likely to be removable dentures in the UK.42 In addition, information on the duration of denture wearing was not available, although potential impacts could be alleviated through MR analysis. Future studies with detailed evaluations for denture use or other oral status are needed for more robust results. Finally, the majority of the participants in UKB were of European ancestry. Hence, the generalizability of the results to other ethnic groups or countries warrants further investigation.
This large study supported potential causal associations between the genetic liability for denture use and the risk for HF, AS, T2D, and related clinical risk factors. Further research including appropriately designed randomized controlled trials is required to demonstrate the clinical relevance of modifying dental health on long-term cardiometabolic health.
Supplementary Material
Acknowledgements
This research was conducted using the UK Biobank resource under application number 55005. We are thankful to all the participants who have agreed to contribute to this project and to the CARDIoGRAMplusC4D, DIAGRAM, GERFHS I/II, GIANT, GLCC, GLIDE, GOCHA, HERMES, ICBP, ISGC, MAGIC, MEGASTROKE, and METASTROKE consortia for making their data available.
Contributor Information
Yunan Liu, CAS Key Laboratory of Nutrition, Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China.
Haiqiang Qin, Department of Neurology, Beijing Tiantan Hospital, Capital Medical University, Beijing 100050, China.
Tongtong Li, CAS Key Laboratory of Nutrition, Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China.
Chengwu Feng, CAS Key Laboratory of Nutrition, Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China.
Han Han, CAS Key Laboratory of Nutrition, Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China.
Yaying Cao, CAS Key Laboratory of Nutrition, Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China.
Yang Su, CAS Key Laboratory of Nutrition, Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China.
Haihao He, CAS Key Laboratory of Nutrition, Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China.
Changzheng Yuan, Department of Big Data in Health Science, Zhejiang University School of Public Health, Hangzhou, Zhejiang 310058, China; Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA 02115, USA.
Meng Sun, NHS Blood and Transplant (NHSBT) Blood Donor Centre, John Radcliffe Hospital, Oxford OX39BQ, UK.
Robert Clarke, Clinical Trial Service Unit and Epidemiological Studies Unit, Nuffield Department of Population Health, University of Oxford, Old Road Campus, Headington, Oxford OX37LF, UK; Medical Research Council, Population Health Research Unit, University of Oxford, Old Road Campus, Headington, Oxford OX37LF, UK.
Wei Gan, Clinical Trial Service Unit and Epidemiological Studies Unit, Nuffield Department of Population Health, University of Oxford, Old Road Campus, Headington, Oxford OX37LF, UK; Medical Research Council, Population Health Research Unit, University of Oxford, Old Road Campus, Headington, Oxford OX37LF, UK; Genetics Department, Novo Nordisk Research Centre Oxford, Innovation Building, Old Road Campus, Headington, Oxford OX37LQ, UK.
Maurizio Tonetti, Shanghai PerioImplant Innovation Center, Department of Oral and Maxillo-facial Implantology, Shanghai Key Laboratory of Stomatology, National Clinical Research Centre for Oral Diseases, Shanghai Ninth People Hospital, Shanghai Jiao Tong University School of Medicine, 639 Zhizaoju Road, Shanghai 200011, China; European Research Group on Periodontology, WTC Tower Genoa, Via De Marini, 1-16149 Genoa, Italy.
Geng Zong, CAS Key Laboratory of Nutrition, Metabolism and Food Safety, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China; Department of Endocrinology and Metabolism, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, 600 Yishan Road, Shanghai 200233, China.
Author contributions
G.Z., W.G., and M.T. participated in the project conception and development of research methods. H.Q., G.Z., and W.G. obtained funding. Y.L. and H.Q. analysed data, performed analysis, and drafted the article with input from all authors. T.L., Hai.H., C.F., Han.H., and Y.C. helped with statistical analysis. C.Y., M.S., R.C., W.G., and M.T. helped with data interpretation. C.Y., Y.S., R.C., W.G., M.T., and G.Z. made critical revision of the manuscript for key intellectual content. All gave final approval and agreed to be accountable for all aspects of work, ensuring integrity and accuracy.
Supplementary material
Supplementary material is available at European Journal of Preventive Cardiology online.
Funding
This study was supported by the National Science Fund for Excellent Young Scholars (81922060), National Natural Science Foundation of China (82271299), Beijing Research and transformation application of clinical characteristic diagnosis and treatment technology (Z221100007422051), and the Talent Introduction Programme of Chinese Academy of Sciences. The British Heart Foundation (CH/1996001/9454), UK Medical Research Council (MC_UU_00017/1, MC_UU_12026/2, MC_U137686851), and Cancer Research UK (C16077/A29186, C500/A16896) provide core funding to the Clinical Trial Service Unit and Epidemiological Studies Unit at Oxford University for the project.
Data availability
Individual-level data from the UK Biobank are not publicly available due to their policy, but the data will be made available after the application of the UK Biobank (https://www.ukbiobank.ac.uk/). Genetic summary data were obtained from publicly available genome-wide databases.
References
- 1. GBD 2017 Oral Disorders Collaborators; Bernabe E, Marcenes W, Hernandez CR, Bailey J, Abreu LG, et al. Global, regional, and national levels and trends in burden of oral conditions from 1990 to 2017: a systematic analysis for the global burden of disease 2017 study. J Dent Res 2020;99:362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peres MA, Macpherson LMD, Weyant RJ, Daly B, Venturelli R, Mathur MR, et al. Oral diseases: a global public health challenge. Lancet 2019;394:249–260. [DOI] [PubMed] [Google Scholar]
- 3. Lockhart PB, Bolger AF, Papapanou PN, Osinbowale O, Trevisan M, Levison ME, et al. Periodontal disease and atherosclerotic vascular disease: does the evidence support an independent association?: a scientific statement from the American Heart Association. Circulation 2012;125:2520–2544. [DOI] [PubMed] [Google Scholar]
- 4. Park SY, Kim SH, Kang SH, Yoon CH, Lee HJ, Yun PY, et al. Improved oral hygiene care attenuates the cardiovascular risk of oral health disease: a population-based study from Korea. Eur Heart J 2019;40:1138–1145. [DOI] [PubMed] [Google Scholar]
- 5. Ryden L, Buhlin K, Ekstrand E, de Faire U, Gustafsson A, Holmer J, et al. Periodontitis increases the risk of a first myocardial infarction: a report from the PAROKRANK study. Circulation 2016;133:576–583. [DOI] [PubMed] [Google Scholar]
- 6. Vedin O, Hagstrom E, Budaj A, Denchev S, Harrington RA, Koenig W, et al. Tooth loss is independently associated with poor outcomes in stable coronary heart disease. Eur J Prev Cardiol 2016;23:839–846. [DOI] [PubMed] [Google Scholar]
- 7. Laniado N, Khambaty T, Hua S, Kaplan R, Llabre MM, Schneiderman N, et al. Periodontal disease and incident prediabetes and diabetes: the Hispanic community health study/study of Latinos. J Clin Periodontol 2022;49:313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ojima M, Amano A, Kurata S. Relationship between decayed teeth and metabolic syndrome: data from 4716 middle-aged male Japanese employees. J Epidemiol 2015;25:204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Czesnikiewicz-Guzik M, Osmenda G, Siedlinski M, Nosalski R, Pelka P, Nowakowski D, et al. Causal association between periodontitis and hypertension: evidence from Mendelian randomization and a randomized controlled trial of non-surgical periodontal therapy. Eur Heart J 2019;40:3459–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. D'Aiuto F, Gkranias N, Bhowruth D, Khan T, Orlandi M, Suvan J, et al. Systemic effects of periodontitis treatment in patients with type 2 diabetes: a 12 month, single-centre, investigator-masked, randomised trial. Lancet Diabetes Endocrinol 2018;6:954–965. [DOI] [PubMed] [Google Scholar]
- 11. Tonetti MS, D'Aiuto F, Nibali L, Donald A, Storry C, Parkar M, et al. Treatment of periodontitis and endothelial function. N Engl J Med 2007;356:911–920. [DOI] [PubMed] [Google Scholar]
- 12. Orlandi M, Graziani F, D'Aiuto F. Periodontal therapy and cardiovascular risk. Periodontol 2000 2020;83:107–124. [DOI] [PubMed] [Google Scholar]
- 13. Herrera D, Molina A, Buhlin K, Klinge B. Periodontal diseases and association with atherosclerotic disease. Periodontol 2000 2020;83:66–89. [DOI] [PubMed] [Google Scholar]
- 14. Sanz M, Del Castillo A M, Jepsen S, Gonzalez-Juanatey JR, D'Aiuto F, Bouchard P, et al. Periodontitis and cardiovascular diseases: consensus report. J Clin Periodontol 2020;47:268–288.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liang X, Chou OHI, Cheung BMY. The association between denture use and cardiovascular diseases. The United States National Health and Nutrition Examination Survey 2009–2018. Front Cardiovasc Med 2023;9:1000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan JQ, Lv YB, Kraus VB, Gao X, Yin ZX, Chen HS, et al. Number of natural teeth, denture use and mortality in Chinese elderly: a population-based prospective cohort study. BMC Oral Health 2020;20:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jang JH, Kim JL, Kim JH. Association between denture use, chewing ability, and all-cause mortality in middle-aged and older adults who exercised regularly in Korea. Sci Rep 2021;11:6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grischke J, Szafrański SP, Muthukumarasamy U, Haeussler S, Stiesch M. Removable denture is a risk indicator for peri-implantitis and facilitates expansion of specific periodontopathogens: a cross-sectional study. BMC Oral Health 2021;21:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018;362:k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shungin D, Haworth S, Divaris K, Agler CS, Kamatani Y, Keun Lee M, et al. Genome-wide analysis of dental caries and periodontitis combining clinical and self-reported data. Nat Commun 2019;10:2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Munz M, Richter GM, Loos BG, Jepsen S, Divaris K, Offenbacher S, et al. Meta-analysis of genome-wide association studies of aggressive and chronic periodontitis identifies two novel risk loci. Eur J Hum Genet 2019;27:102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Munz M, Willenborg C, Richter GM, Jockel-Schneider Y, Graetz C, Staufenbiel I, et al. A genome-wide association study identifies nucleotide variants at SIGLEC5 and DEFA1A3 as risk loci for periodontitis. Hum Mol Genet 2017;26:2577–2588. [DOI] [PubMed] [Google Scholar]
- 26. Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 2018;9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J; Schizophrenia Working Group of the Psychiatric Genomics Consortium , et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Norhammar A, Kjellstrom B, Habib N, Gustafsson A, Klinge B, Nygren A, et al. Undetected dysglycemia is an important risk factor for two common diseases, myocardial infarction and periodontitis: a report from the PAROKRANK study. Diabetes Care 2019;42:1504–1511. [DOI] [PubMed] [Google Scholar]
- 31. de Oliveira C, Watt R, Hamer M. Toothbrushing, inflammation, and risk of cardiovascular disease: results from Scottish Health Survey. BMJ 2010;340:c2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choi SE, Sima C, Pandya A. Impact of treating oral disease on preventing vascular diseases: a model-based cost-effectiveness analysis of periodontal treatment among patients with type 2 diabetes. Diabetes Care 2020;43:563–571. [DOI] [PubMed] [Google Scholar]
- 33. Chang Y, Woo HG, Park J, Lee JS, Song TJ. Improved oral hygiene care is associated with decreased risk of occurrence for atrial fibrillation and heart failure: a nationwide population-based cohort study. Eur J Prev Cardiol 2020;27:1835–1845. [DOI] [PubMed] [Google Scholar]
- 34. Haworth S, Kho PF, Holgerson PL, Hwang LD, Timpson NJ, Renteria ME, et al. Assessment and visualization of phenome-wide causal relationships using genetic data: an application to dental caries and periodontitis. Eur J Hum Genet 2021;29:300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou M, Dong J, Zha L, Liao Y. Causal association between periodontal diseases and cardiovascular diseases. Genes (Basel) 2021;13:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang YB, Yan SY, Li XH, Huang Q, Luo LS, Wang YY, et al. Causal association between periodontitis and type 2 diabetes: a bidirectional two-sample Mendelian randomization analysis. Front Genet 2022;12:792396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shah PD, Schooling CM, Borrell LN. Impact of liability to periodontitis on glycemic control and type II diabetes risk: a Mendelian randomization study. Front Genet 2021;12:767577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma C, Wu M, Gao J, Liu C, Xie Y, Lv Q, et al. Periodontitis and stroke: a Mendelian randomization study. Brain Behav 2023;13:e2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bell S, Gibson JT, Harshfield EL, Markus HS. Is periodontitis a risk factor for ischaemic stroke, coronary artery disease and subclinical atherosclerosis? A Mendelian randomization study. Atherosclerosis 2020;313:111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Larsson SC, Mason AM, Back M, Klarin D, Damrauer SM, Million Veteran P, et al. Genetic predisposition to smoking in relation to 14 cardiovascular diseases. Eur Heart J 2020;41:3304–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larsson SC, Bäck M, Rees JMB, Mason AM, Burgess S. Body mass index and body composition in relation to 14 cardiovascular conditions in UK Biobank: a Mendelian randomization study. Eur Heart J 2020;41:221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. White DA, Tsakos G, Pitts NB, Fuller E, Douglas GV, Murray JJ, et al. Adult Dental Health Survey 2009: common oral health conditions and their impact on the population. Br Dent J 2012;213:567–572. [DOI] [PubMed] [Google Scholar]
- 43. Hannah VE, O'Donnell L, Robertson D, Ramage G. Denture stomatitis: causes, cures and prevention. Prim Dent J 2017;6:46–51. [DOI] [PubMed] [Google Scholar]
- 44. Shungin D, Cornelis MC, Divaris K, Holtfreter B, Shaffer JR, Yu YH, et al. Using genetics to test the causal relationship of total adiposity and periodontitis: Mendelian randomization analyses in the Gene-Lifestyle Interactions and Dental Endpoints (GLIDE) Consortium. Int J Epidemiol 2015;44:638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Azzolino D, Passarelli PC, De Angelis P, Piccirillo GB, D'Addona A, Cesari M. Poor oral health as a determinant of malnutrition and sarcopenia. Nutrients 2019;11:2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martinon P, Fraticelli L, Giboreau A, Dussart C, Bourgeois D, Carrouel F. Nutrition as a key modifiable factor for periodontitis and main chronic diseases. J Clin Med 2021;10:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deng K, Pelekos G, Jin L, Tonetti MS. Diagnostic accuracy of self-reported measures of periodontal disease: a clinical validation study using the 2017 case definitions. J Clin Periodontol 2021;48:1037–1050. [DOI] [PubMed] [Google Scholar]
- 48. Tonetti MS, Deng K, Christiansen A, Bogetti K, Nicora C, Thurnay S, et al. Self-reported bleeding on brushing as a predictor of bleeding on probing: early observations from the deployment of an internet of things network of intelligent power-driven toothbrushes in a supportive periodontal care population. J Clin Periodontol 2020;47:1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abbood HM, Hinz J, Cherukara G, Macfarlane TV. Validity of self-reported periodontal disease: a systematic review and meta-analysis. J Periodontol 2016;87:1474–1483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual-level data from the UK Biobank are not publicly available due to their policy, but the data will be made available after the application of the UK Biobank (https://www.ukbiobank.ac.uk/). Genetic summary data were obtained from publicly available genome-wide databases.