Abstract
Medicinal plants have got notable attention in recent years in the field of pharmaceutical and drug research. The high demand of herbal medicine in the rural areas of developing countries and drug industries necessitates correct identification of the medicinal plant species which is challenging in absence of expert taxonomic knowledge. Against this backdrop, we attempted to assess the performance of seven advanced deep learning algorithms in the automated identification of the plants from their leaf images and to suggest the best model from a comparative study of the models. We meticulously trained VGG16, VGG19, DenseNet201, ResNet50V2, Xception, InceptionResNetV2, and InceptionV3 deep neural network models. This training utilized a dataset comprising 5878 images encompassing 30 medicinal species distributed among 20 families. Our approach involved two avenues: the utilization of public data (PI) and a blend of public and field data (PFI), the latter featuring intricate backgrounds. Our study elucidates the robustness of these models in accurately identifying and classifying both interfamily and interspecies variations. Despite variations in accuracy across diverse families and species, the models demonstrated adeptness in these classifications. Comparing the models, we unearthed a crucial insight: the Normalized leverage factor () for DenseNet201 stands at 0.19, elevating it to the pinnacle position for PI with a remarkable 99.64 % accuracy and 98.31 % precision. In the PFI scenario, the same model achieves a of 0.15 with a commendable 97 % accuracy. These findings serve as a guiding beacon for shaping future application tools designed to automate medicinal plant identification at the user level.
Keywords: Artificial intelligence, Species identification, Deep learning, Medicinal plants, CNN
Highlights
-
•
Used seven advanced deep learning algorithms for their comparative performance.
-
•
Employed in total 5878 images of medicinal plants of 30 species that belong to 20 families.
-
•
Models correctly classified inter-species and inter-family variations.
-
•
99.64 % prediction accuracy achieved with 99.5 % recall from DenseNet201.
1. Introduction
Almost 70 % people in developing countries directly depend on traditional medicine for primary health care and treating common ailments [1]. Due to limited government medical facilities and the high cost of allopathic medicine in developing countries, many people rely on herbal medicine [2]. Besides, the emerging drug industries in developed countries depend on medicinal plants to some extent for the pharmaceutical products [3]. In the world, 18 % of the top 150 prescription medications, along with 25 % of modern pharmacopoeia, are plant-based. Asia is one of the major hubs for the world's bioresources [4], and around 50 % of the world's traditional medicine exports are made up of Asian medicinal plants [5]. The recognition of plant medicines among the public improved as a result of the positive and reassuring results of several clinical trials. There are studies that shed light on and spur the therapeutic use of plant-based medicines by examining the many active ingredients of herbs along with their clinical functions [6].
In most of the cases, people who are older than 45 years seem to be better knowledgeable about medicinal plants, likely due to their greater experiences in dealing with these plants grown in their surrounding habitats [7]. In fact, ethnobotanical knowledge is gradually eroding among young people, importantly among the educated youth, that poses a threat to transfer of the knowledge to the future generations. The paucity of standardized preparation techniques and scientific evidences about their effectiveness and possible toxicity would resulted in inefficient use of the plants or misidentification of the species, consequently impacting the potential use of herbal medicine in the future generation [8]. Poisoning from medicinal plants is commonly documented as a result of either incorrect identification of the plant when sold or inadequate preparation and administration by untrained individuals. Without any prior botanical knowledge, finding information about medicinal plants from books or internet can be challenging and time-consuming, especially when dealing with diverse local names for the same species. According to Refs. [9,10], there is a significant disparity of herbal knowledge acquired by people in cities or researchers in compared to the knowledge of tribes or villages. Low-cost, efficient, and accurate identification of medicinal plants can drive a revolution in the field of medical research as well as the conservation of these precious natural resources. Leaves of the plants could perform a significant role in plant identification because of their uniqueness and abundance throughout the seasons.
Adoption of cutting-edge technology reduces the labor involved in expert inspection for detecting signs of disease, nutrient deficiency, and plant identification [11,12]. Significant progress has been seen in recent decades with a variety of well-known architectures, including InceptionV3, GoogleNet, VGG16, AlexNet, and ReseNet for handling a wide range of image classification tasks with the introduction of deep learning techniques. Numerous studies [[11], [12], [13], [14], [15]] have used these pre-trained models for identification of plants and diseases owing to the outstanding performance of these models. Applications of deep learning pre-trained models are more accurate due to the fact that these architectures were designed to identify the 1000 classes in the Imagenet dataset [16].
A number of studies [[17], [18], [19], [20], [21], [22], [23], [24]] have been carried out in recent years to provide tools for the identifications of medicinal plants. Le et al. [25] used modified kernel descriptor and support vector machine for visual identification of Vietnamese medicinal plants. Although, overall performance was satisfactory in their study, the highest accuracy of identifying individual species was around 80 %. Authors in other studies [20,26,27] used ML algorithms like k-nearest neighbour (KNN), naïve Bayes, Support vector machine (SVM), Decision tree, neural networks for the automated medicinal plant identification, the number of species used in their studies varies from 10 to 24 and trained maximum 97 % accuracy. Moreover, a study [18]) obtained 85 % accuracy employing Deep Neural Networks (DNN). Using transfer learning with convolutional neural network for identifying medicinal plants using 10 species obtained 98.7 % accuracy [28]. Pushpa and Rani [29] utilized the Ayur-PlantNet deep convolutional neural network to conduct a comparative analysis involving MobileNetV3Large, Densenet121, Resnet34, VGG16, and Resnet50 for ayurvedic plant identification. Notably, the highest accuracy of 92.27 % was achieved by Ayur-PlantNet. Muneer and Fati [30] leveraged shape and texture features of leaves for the classification of Malaysian leaf types. Their study evaluated the performance of both DNN and ML classifiers on a comprehensive dataset. Additionally, Pushpanathan [31] delved into the identification of twelve locally found perennial herbs in Malaysia, renowned for their medicinal significance. Kan [32] focused on twelve Chinese medicinal plant species, extracting distinctive shape and texture attributes. Their exploration also highlighted the distinctive characteristics of plant photographs and medicinal variations. Amuthalingeswaran [18] employed a DNN to classify medicinal plant species across four distinct plant types, achieving a notable average accuracy. VijayaLakshmi and Mohan [33] introduced a method that harnessed texture, shape, and color attributes for plant leaf type identification, employing a fuzzy relevance vector machine for classification. The authors encountered challenges in distinguishing leaves with similar shapes, handling images with shadows, addressing cluttered backgrounds, and effectively recognizing flawed leaves. Russel and Selvaraj [34] introduced a multiscale parallel DNN for disease detection based on leaf images. Their experimental endeavors were conducted using existing datasets captured in controlled environments with plain backgrounds.
Previous studies [26,[35], [36], [37]] have predominantly relied on public datasets or focused on specific backgrounds, leaving a gap in research concerning the identification of medicinal plants from raw data with complex backgrounds that collected from different geographic distribution. In real-world scenarios, variations in leaf size and shape can significantly impact accurate identification. While a few studies have utilized real field datasets [19,38] with complex backgrounds, there is a need for comprehensive and rigorous investigations to validate and establish models for rapid and precise identification of medicinal plants under different geographic field conditions. Furthermore, a thorough comparative study utilizing advanced deep learning algorithms to assess the performance and predictive capabilities of these models across different species and families has not been conducted yet. This research gap needs to be addressed in order to gain a better understanding of the strengths and limitations of different algorithms in identifying medicinal plants.
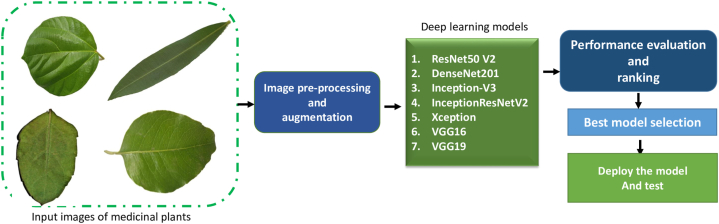
To bridge this research gap, our study aims to analyze the performance of seven advanced deep learning algorithms (VGG16, VGG19, DenseNet201, InceptionV3, ResNet50V2, Xception, InceptionResnetV2) in a family-wise manner using various public data sources and our own real field images of medicinal plants. By utilizing data sources that vary in terms of resolution and contrast, reflecting the challenges encountered in real-world scenarios. These seven DNN algorithms were selected based on their proven performance and success in various image recognition tasks. We also performed their comparative assessment to find out best model for the advancement of drug development whether it be non-pharmacopeial, pharmacopeial, or synthetic drugs or benefiting the local communities.
2. Materials and methods
2.1. Data acquisition and pre-processing
Medicinal plant leaf images from 30 species belonging to 20 families (Table-1) were gathered from the data archive of Kaggle [39] with a plain background (PI) and from local field images (FI) with complex real backgrounds (Fig. 1 (d, f, g, h, k, I, m, p)). The images were saved in JPG format with corresponding scientific names. The Pillow library (Version 8.4.0) was used for resizing the images to 224 × 224 pixels before using a pretrained deep learning model. Resizing a large image reduces computational load on the GPU and potentially speeds up the processing of the model. Fig. 1(a–p) shows some representative images that we used to train the DNN models.
Table 1.
Medicinal plant species list that used for species identifications with their common name in respect of Bangladesh and India.
| Scientific name | Family | Local name | Image number |
|
|---|---|---|---|---|
| Public (PI) | Field image (FI) | |||
| Amaranthus viridis L. | Amaranthaceae | Data shak, Marissag | 122 | 157 |
| Artocarpus heterophyllus Lam. | Anacardiaceae | Jackfruit, Kanthal | 92 | 133 |
| Brassica juncea (L.) Czern | Basellaceae | Shorisha, Indian mustard | 85 | 102 |
| Azadirachta indica (L.) | Apocynaceae | Neem | 95 | 112 |
| Basella alba (L.) | Apocynaceae | Puishaak | 103 | 98 |
| Carissa carandas (L.) | Brassicaceae | Koromcha, Kilakkai | 74 | 107 |
| Citrus limon (L.) Burm. f. (pro. sp.) | Fabaceae | Lebu, Goranebu | 77 | 115 |
| Ficus auriculata Lour. | Fabaceae | Trimmal, Puroi khak | 80 | 90 |
| Ficus religiosa L. | Lamiaceae | Ashvattha, Peepal | 73 | 93 |
| Hibiscus rosa sinensis L. | Lamiaceae | Joba, China rose | 84 | 100 |
| Jasminum officinale L. | Lamiaceae | Jasmine | 71 | 145 |
| Mangifera indica L. | Lythraceae | Aam, Mango | 92 | 135 |
| Mentha piperita L. (pro. sp.) | Malvaceae | Mentha, Pudina | 97 | 97 |
| Moringa oleifera Lam. | Meliaceae | Shojne, Moringa | 77 | 120 |
| Alpinia galanga (L.) Willd | Moraceae | Kulanjan, Blue ginger | 80 | 87 |
| Muntingia calabura L. | Moraceae | Jamaica cherry, Calabura | 86 | 105 |
| Murraya koenigii (L.) Spreng | Moraceae | Curry leaf | 60 | 113 |
| Nyctanthes arbor tristis Linn. | Rutaceae | Har singar, Shiuli | 79 | 100 |
| Santalum album L. | Rutaceae | Sandalwood | 88 | 95 |
| Syzygium jambos (L.) Alston | Rutaceae | Golapjaam, Mountain apple | 78 | 139 |
| Trigonella foenum-graecum L. | Santalaceae | Fenugreek, Methi | 66 | 92 |
| Syzygium cumini (L.) Skeels | Zingiberaceae | Jamun | 78 | 133 |
| Tabernaemontana divaricate (L.) R. Br. ex Roem. & Schult. | Apocynaceae | Thoka tagar, Pinwheel flower | 66 | 89 |
| Plectranthus amboinicus (Lour.) Spreng | Piperaceae | Indian borage, Mexican mint | 85 | 94 |
| Punica granatum L. | Moringaceae | Dalim | 82 | 141 |
| Psidium guajava L. | Muntingiaceae | Guava | 80 | 167 |
| Nerium oleander L. | Myrtaceae | Kaner, Rokto korobi | 70 | 143 |
| Piper betle L. | Myrtaceae | Betel, Paan | 55 | 165 |
| Ocimum tenuiflorum L. | Oleaceae | Kalotulsi, Tulshi | 58 | 119 |
| Pongamia pinnata (L.) Pierre | Oleaceae | Karanja | 61 | 98 |
Fig. 1.
Representative leaf sample images from 30 medicinal plant species used in the study for automated identification. The images denoted by labels d, f, g, h, k, I, m, p represent real field data, while the remaining images represent public data from Kaggle.
2.2. Deep convolutional neural networks
The study evaluated seven distinct Deep Convolutional Neural Networks (DCNN) (Fig. 2). DCNN models use convolutional layers to extract features from medicinal plant leaf images in a spatial hierarchy way, where the lower layers can learn simple features such as edges and textures and higher layer learn complex features. Leaf input images (224 × 224) comprised of three matrices or color (IC) channels (RGB). The Convolution Layer plays a crucial role in our Deep Convolutional Neural Network models. It is responsible for extracting and learning features from input images, producing a feature map as its output.
Fig. 2.
The overall workflow that employed in this study for medicinal plant species identification using leaf.
After the convolution operation, a non-linear activation function was applied to allow the network to learn more complex representations of the input image. For input image height (IH), width (IW) this CL can be represented according to Eq. (1).
| dim(image) = (IH, IW, IC) = (224,224,3) | (1) |
The kernel or filter (k) must have a same iC of the image. Then, the filter dimension can be calculated using Eq. (2).
| dim(filter) = (k, k, IC) | (2) |
For the input image of medicinal plant leaf (I), padding (p) and stride (s) the tensor dimension calculated based on Eq. (3).
| (3) |
The next step in the process involved a 2D Convolution Layer (CL) with a max pooling function and a Rectified Linear Unit (ReLU) activation. The ReLU activation was used to handle nonlinearities and ensure efficient activation, as it does not activate all neurons simultaneously. The CL utilized convolutional products with filters and an activation function (ψ) as inputs. The lth layer can be expressed as Eq. (4).
| (4) |
Then, in each layer 3 × 3 max pooling was included to prevent overfitting issue and to make efficient and robust performance. For the pooling function φ[l] the pooling layer can be expressed by Eq. (5).
| (5) |
The output is finally processed through a flattened layer, followed by a fully connected layer where each neuron is connected to an activation unit and a 40 % dropout is applied. This fully connected layers received input a[i−1] vector and give back a[i] vector, for the ith layer of jth node, then we can express it by Eq. (6),
| (6) |
The input features were then processed through the ReLU function, which classifies the images into specific labels. The softmax activation function makes the final decision on classification based on the output of the neurons' classification labels.
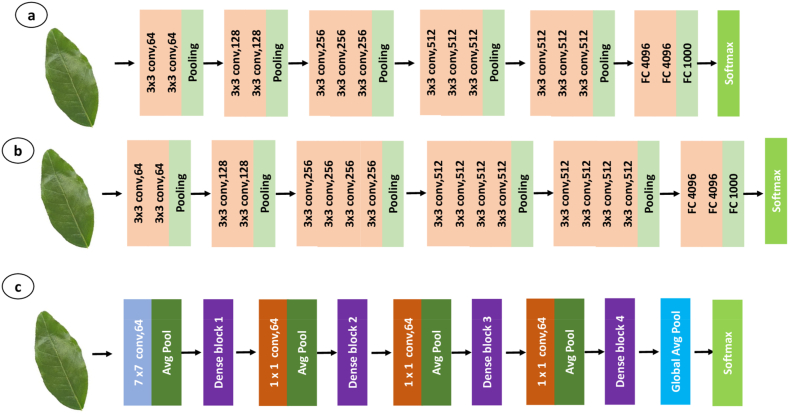
Besides three fully connected layers (FCL) of VGG 16 (Fig. 3a) and VGG 19 (Fig. 3b), they have 16 layers with 13 convolution layers and 19 layers with 16 convolution layers, respectively. Both models use max-pooling in feature maps for reducing the spatial dimensions and enhance their translation invariance. To reduce the volume size the models modified with pre-trained weights by incorporating max-pool function, as well as a Softmax classifier for the output from the previous fully connected layer. These models differ in their deep layer architecture and the use of small convolutional filters, which enable them to learn detailed features from images.
Fig. 3.
Basic deep neural network architecture of (a) VGG16, (b)VGG19, (c) DenseNet201.
DenseNet201 is a CNN architecture that is employed for this leaf image classification tasks. Which is a variation of DenseNet architecture, and dense connections between layers (Fig. 3c). Which contains 201 total layers and a total of 19,628,095 parameters, with both parameters trainable and non-trainable. Additionally, the layers in each block are connected to all preceding layers, facilitating feature learning from all previous layers [12], and enhancing the flow of information and gradients through the network. The model also employs a global pooling and full-connected layer, also implements “transition layers” to decrease the number of channels and spatial resolution between dense blocks.
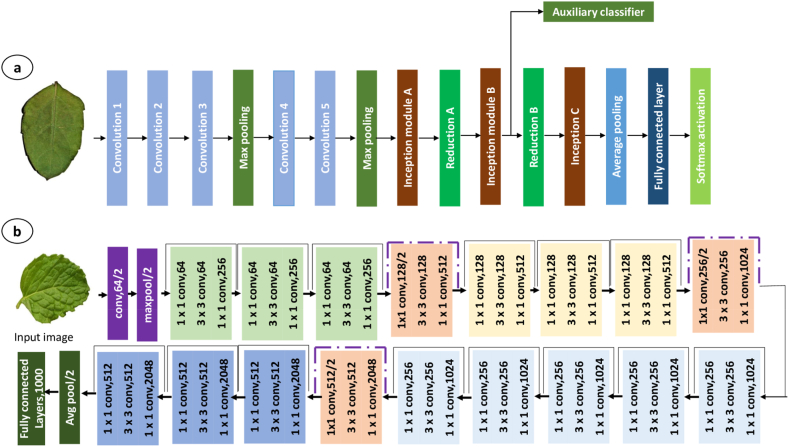
An Inception model comprises of multiple parallel branches, each with a different convolutional filter size (1 × 1, 3 × 3, 5 × 5) with a max-pool layer. The 36 convolutional layers of model act as the foundation for constructing the network architecture (Fig. 4a). In order to enable the model to learn features at various scales and capture various levels of abstraction, these branches are then concatenated along the channel dimension. This also employs a method known as “factorization,” which lowers the computational cost of the 3 × 3 and 5 × 5 convolutional filters by utilizing a 1 × 1 convolutional filter to cut down on the number of input channels before the 3 × 3 or 5 × 5 convolutional filter is applied.
Fig. 4.
Basic deep neural network architecture of (a) Inception, (b)ResNet50.
ResNet50V2 is a reformed variant of ResNet50, incorporating a new residual-unit and featuring 49 convolutional layers, 1 fully connected layer for classification, one average pooling and one max-pool layer (Fig. 4b). ResNet152V2 is similar to ResNet152 and is a modified version of the latter, which comprises 151 convolutional layers with one max-pool, one average pooling, and one full-connected layer for the classification purpose.
Xception is an effective architecture for classifying images. It is distinguished by its use of residual connections and depth wise separable convolutions, both of which increase computing efficiency. Which enhances the network's gradients and information flow. This also uses “residual connections”, where the input of a layer is added to its output before passing it through the next layer. This architecture is especially appropriate for embedded and mobile devices because they have constrained processing resources.
2.3. Hyperparameter setting
We employed two kinds of data to train our DCNN models. The first kind was using public data (PI) exclusively, while the second kind (PFI), combined public data with field data (PI + FI) that had complex backgrounds. For the training process, we utilized 85 % of the dataset in both types. Additionally, 5 % of the data was allocated for validation during each epoch to monitor the model performance and prevent overfitting. Furthermore, we ensured an unbiased evaluation of the model's performance by using a completely separate dataset for testing, 10 % from both PI and FI dataset. This test dataset contained data that the model had never encountered before, replicating its performance on new and real-world data scenarios. Through this approach, we could meter how well the model generalized to previously unseen samples, which is the ultimate objective of a DNN model.
During the training phase, we used batch size 32 which refers to the number of input images the model processes at one time before updating the model's parameters. While a bigger batch size can lead to more precise gradient estimations, it also necessitates more memory [40]. Moreover, average pooling is used in DCNN to reduce the spatial dimensions of feature maps that work by taking the average values in the feature map region, resulting in a smaller output feature map. “softmax” function is also employed in the output layer for mapping the output of the final layer for improving the interpretability of the model's predictions. To ensure the reproducible results of the models, the random generator's seed is set to random state 42. The parameters of the model are modified during the training phase based on input data and the optimization algorithm. This makes it possible to train and evaluate the model consistently, which is crucial for comparing its performance across several runs or implementations.
For turning both rows and columns horizontally, the “horizontal flip” option is set to “True” in the ImageDataGenerator constructor. This technique is used to increase the amount of data available for training from the existing ones. This allows the model to be more resistant to translations in the test data by enabling the model to learn features that are invariant to horizontal translations. For multiclass classification, the “categorical” class mode was employed for labelling the image data with one-hot encoded categorical labels. When there are more than two classes, this mode is helpful as it enables the models to understand the relationships between classes and forecast numerous classes. Additionally, each image was randomly rotated between [32, +32] degrees with 20 % shifting along the X-axis and Y-axis. We used shearing and zoom range 0.2–20 % zoom in and 20 % zoom out of the image, respectively. Additionally, pixel intensity was arbitrarily scaled between [0.5, 1.5]. We employed an “Adam” optimizer function, which is an extension of the stochastic gradient descent optimization algorithm. This function adapts the learning rate for each parameter individually [41], that improves the stability and performance of the model during training, also corrects the estimator bias of the first and second moments of the gradient. In this study, we evaluate the performance of DCNN models using the sensitivity and accuracy based on [11]. Due to the interdependency of various evaluation parameters, ranking AI models based solely on accuracy becomes challenging. To address this issue, we implemented the “Dey & Ahmed method” [12,42] (supplementary file S3).
3. Results
3.1. Training and validation of DCNN models
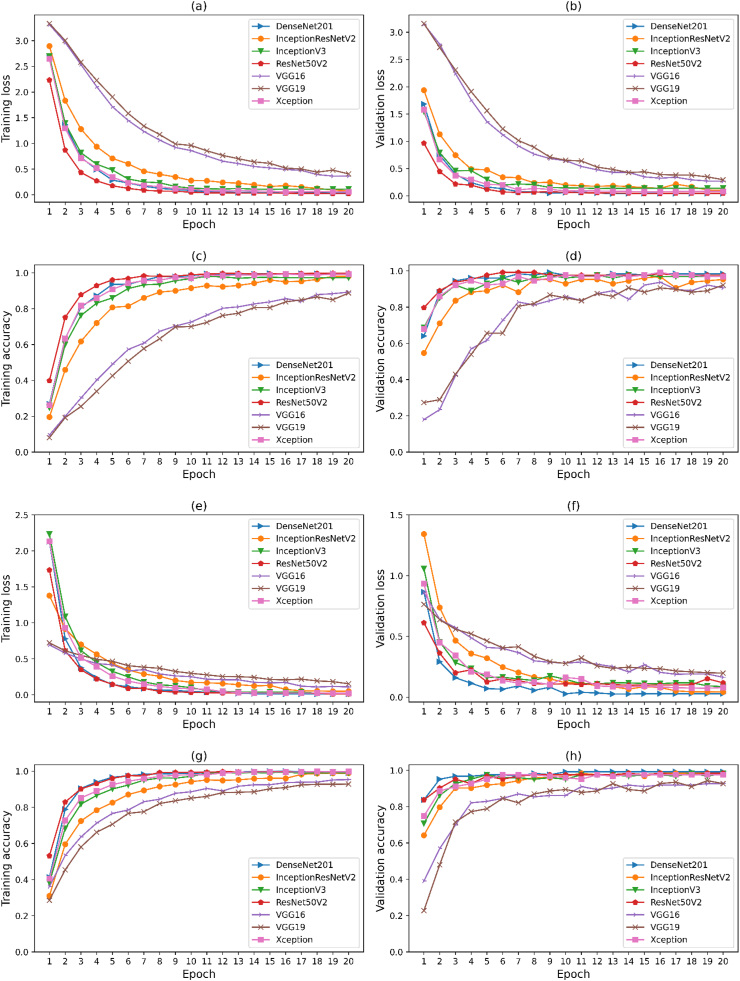
A comparison experiment was conducted using seven different models, namely VGG16, VGG19, ResNet50V2, Xception, InceptionResnetV2, DenseNet201, and InceptionV3, based on PI and PFI. Fig. 5 presents the accuracy and loss curves for all seven models. Initially, during the early epochs, the losses of the PFI dataset were higher (Fig. 5a) than those of the PI dataset (Fig. 5e), such differences can be attributed to the complexity of the backgrounds in the field data. Among the models, VGG19 showed the highest training losses for the PFI dataset and InceptionV3 for the PI. As the number of epochs increased, the gap between the training and validation accuracy decreased, indicating that the models were becoming more robust (Fig. 5 c-d,g-h). Initially, during the first four epochs (Fig. 5 a-b,e-f), the training loss was higher for most models. However, it gradually reduced after the 10th epoch, indicating improved model learning and eventually, all models started to converge. Fig. 5 (c,g) demonstrates that the models reached their peak performance on the training data, and further training is unlikely to significantly improve performance. However, for the validation dataset, the increasing validation accuracy over time suggests that the models were generalizing well to new, unseen data and were not overfitting to the training data (Fig. 5 d,h). Additionally, the high training accuracy and similarly high validation accuracy indicate that the models were not overfitted to the training data. This balance between training and validation accuracy shows that the models were able to generalize effectively to new data.
Fig. 5.
Plots of loss and accuracy of training and validation according to epoch of seven neural network models. Where, a,b,c,d for mix dataset (PFI) and e,f,g,h for public dataset (PI).
3.2. Evaluation of DCNN models
The comprehensive evaluation of the seven DCNN models is detailed in Table 2. The accuracy ratings for each model varied between 0.90 and 0.996 for both PI and PFI. However, it was evident that the PFI dataset exhibited comparatively lower performance than the PI dataset in terms of accuracy, precision, recall, and F1-score (Table 2). In the PFI, the InceptionResNetV2 model achieved an impressive recall score of 0.97, signifying its ability to correctly identify 97 % of all positive instances. This was the highest recall score among all the models, and it also achieved 97 % accuracy and 94 % precision. On the other hand, in the PI, the ResNet50V2 model achieved a precision of 0.9732, indicating a low false positive rate and accurate identification of 97.32 % of medicinal plant species. Among all the DCNN models, DenseNet201 stood out with the highest precision and accuracy for both the PI and PFI datasets (Table 2). DenseNet201 surpassed all other models tried in the study.
Table 2.
The performance scores of seven deep convolutional neural network models were evaluated for identifying medicinal plants in our dataset. In this evaluation, was used as the normalized leverage factor.
| Public data (PI) |
Public field mix data (PFI) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1 | Rank | Accuracy | Precision | Recall | F1 | Rank | |||
| Resnet50V2 | 0.986 | 0.973 | 0.984 | 0.977 | 0.176 | 2 | 0.951 | 0.940 | 0.947 | 0.953 | 0.118 | 2 |
| DenseNet201 | 0.996 | 0.983 | 0.995 | 0.973 | 0.191 | 1 | 0.974 | 0.968 | 0.970 | 0.962 | 0.152 | 1 |
| VGG16 | 0.946 | 0.949 | 0.931 | 0.921 | 0.104 | 6 | 0.919 | 0.917 | 0.891 | 0.911 | 0.074 | 6 |
| VGG19 | 0.911 | 0.918 | 0.894 | 0.901 | 0.071 | 7 | 0.902 | 0.886 | 0.902 | 0.894 | 0.063 | 7 |
| InceptionV3 | 0.978 | 0.982 | 0.964 | 0.969 | 0.166 | 3 | 0.940 | 0.933 | 0.927 | 0.931 | 0.098 | 3 |
| Xception | 0.975 | 0.963 | 0.967 | 0.972 | 0.155 | 4 | 0.959 | 0.944 | 0.959 | 0.960 | 0.130 | 4 |
| Inception ResNetV2 |
0.963 | 0.968 | 0.945 | 0.958 | 0.138 | 5 | 0.968 | 0.943 | 0.976 | 0.962 | 0.140 | 5 |
It is important to consider multiple performance metrics, to evaluate the effectiveness of the adopted DCNN models thoroughly. These metrics provide different perspectives on the model's performance and help ensure accurate predictions and an overall measure of its effectiveness. According to normalized leverage factor (NLF), DenseNet201 attained a weight of 0.19, ranking it as the best-performing model, while the weight of NLF for ResNet50V2 was 0.1752, placing it as the second-best model in the ranking (supplementary file S3). For both PI and PFI, though each model shows different performance matrices, the overall rankings were the same.
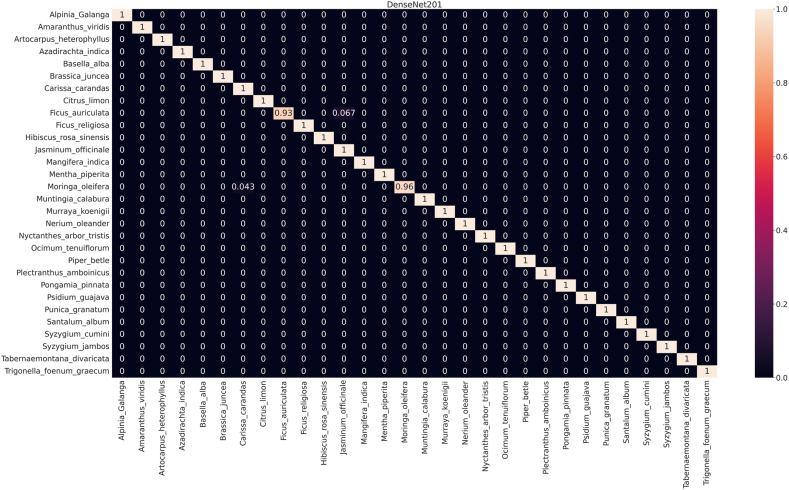
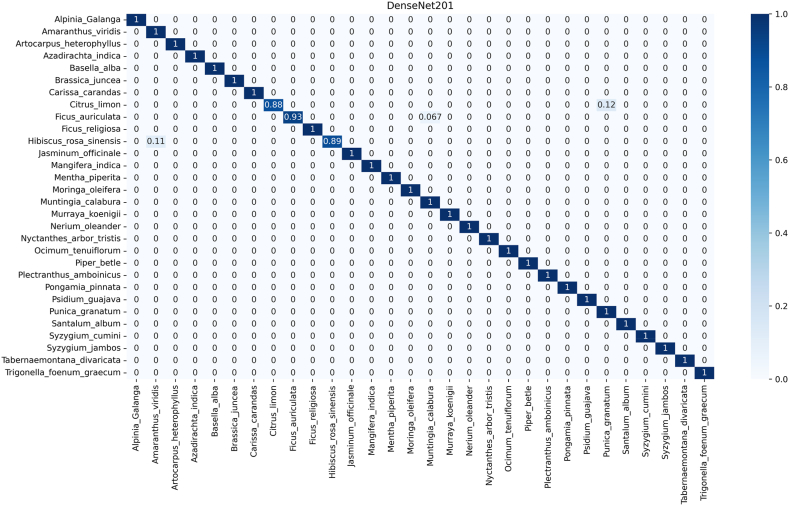
The confusion matrix (CM) describes the functional abilities of the seven implemented DCNN models used to predict each of the 30 medicinal species (Fig. 6, Fig. 7). The horizontal and vertical axes of the matrix represent true and predicted labels, where the normalized diagonal values represent the correct prediction numbers for each species, ranging from 0 to 1. This also represents the true positive of the specific species. As same as performance result, the rate of misclassification is comperatively high in PFI. In DenseNet201 of PI, 28 species were identified with 100 % accuracy while identification accuracy of the other two species namely Ficus auriculata and Moringa oleifera were 93 % and 96 % respectively (Fig. 6). On the contrary, in PFI identification performance of DenseNet201was not as strong as PI, where classification accuracy of 3 species ranged from 88 to 93 % and the rest was100 % (Fig. 7), though both were best performing model based on their respective dataset.
Fig. 6.
Confusion matrix for automatic identifying medicinal plants using DenseNet201 for public dataset (PI).
Fig. 7.
Confusion matrix of DenseNet201 that employed in this study for the medicinal plant identifications for mix dataset (PFI).
3.3. Model deploy
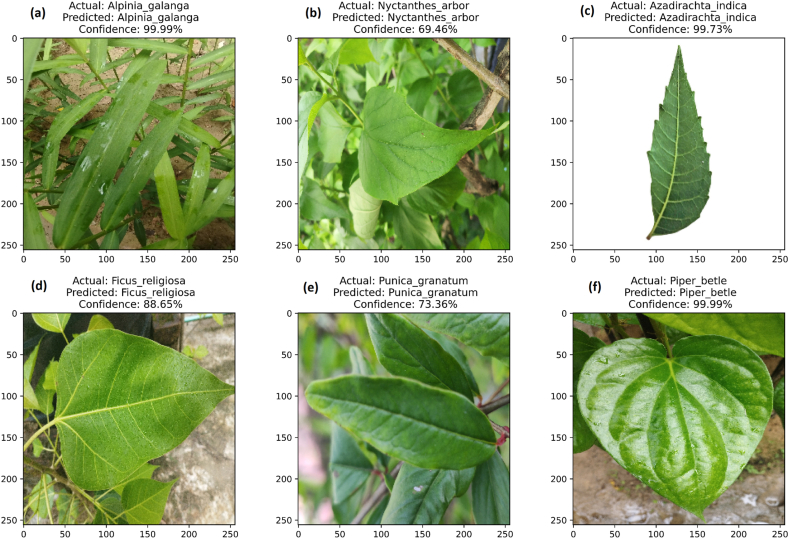
As DenseNet201 outperformed other DCNN models, it was saved in the.h5 file for testing new images in a Python environment. The model was used to make predictions on the new images, which were not used during the training or validation process. Fig. 8(a–f) illustrated that performance of trained model, where each image was correctly identified. Therefore, DenseNet201 deployed for identifying medicinal plant species and demonstrated the robustness of the trained model.
Fig. 8.
Test performance of the leaf based automated medicinal plant detection using trained DenseNet201model.
4. Discussion
While automatic identification has exhibited favorable outcomes, specific constraints persist, demanding attention and resolution in the forthcoming years. Our investigation into the plant species parts and images that influence accuracy can serve as a compass for prospective refinements. Remarkably, our findings unveiled that the proficiency of identification remained consistent for both solitary images extracted from our source and observations amassed from the field, often encompassing multiple images. It is noteworthy that the precision in identifying both uncommon and prevalent species across distinct regions remained unwavering, and the primary species habitat did not exert influence on outcomes. In contrast to our anticipations, the analogous performance observed between database images and field observations could plausibly be ascribed to the meticulous curation of the observation database, ensuring images were discernible to experts. This equitable performance accentuates the application's potential for versatile utility, encompassing tasks such as identifying endangered plants or conducting comprehensive biodiversity evaluations, particularly for frequently encountered species. The accomplished performance of this application implies its potential for extended employment. Furthermore, it harbors the prospect of aiding in the identification of the burgeoning trove of digitized plant image collections, contingent upon the database's image quality adhering to reasonable standards.
4.1. Intra-family, inter-family, and species-level performance
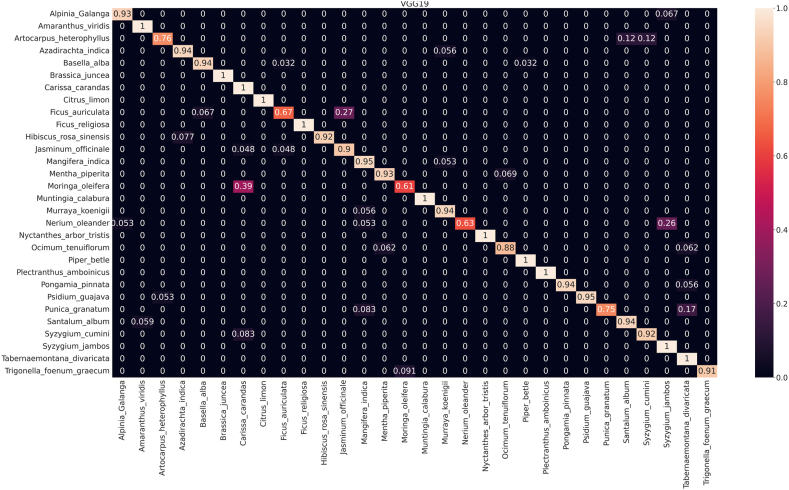
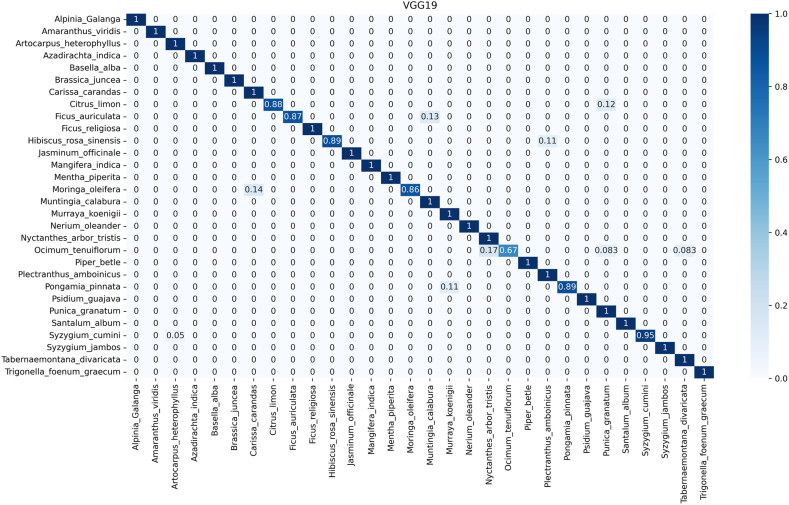
To reveal the model's ability to distinguish broader taxonomic categories, it is crucial to scrutinize family-level identification. This analysis complements species-level evaluation by highlighting potential challenges in classifying higher-level groups. Addressing errors at the family level can enhance the model's understanding of underlying patterns and improve its accuracy across different taxonomic levels, ensuring a more comprehensive and reliable classification system. The confusion matrix (CM) of least performing model (VGG19) shown in Fig. 9 demonstrates the analyses of inter-family and intra-family performance, while rest of the CM are reported in supplementary files S1 and S2. Among the models evaluated, DenseNet201 is the best performer. However, it recognized Ficus auriculata (Fabaceae family) with an accuracy of 93 % (Fig. 6) which was misidentified as Jasminum officinale (Lamiaceae family) in around 7 % cases. Just above 4 % Moringa oleifera of Meliaceae family was misclassified as Carissa carandas of Brassicaceae by this model.
Fig. 9.
Confusion matrix for automatic identifying medicinal plants using VGG19 for public dataset (PI).
On contrary, in the VGG19 model for PI (Fig. 9), predicts incorrectly around 5 % Jasminum officinale as Ficus religiosa, both of which belong to Lamiaceae family. On the other hand, 6 % Ocimum tenuiflorum (Oleaceae), 5.6 % Pongamia pinnata (Oleaceae), 1.7 % Punica granatum (Moringaceae) misclassified as Tabernaemontana divaricata (Apocynaceae). Additionally, ResNet50V2 did not show intrafamily misclassification issue like DenseNet50V2. Such as, 8.7 % Moringa oleifera of Meliaceae family identified as Carissa carandas that belongs to the Brassicaceae family in the ResNet50V2 (Figure S1.5). In the case of PFI, we observed inter-family misclassification rather than intra-family misclassification. Specifically, the Fabaceae (Citrus limon, Ficus auriculata) family demonstrated a tendency to be misclassified with the Moraceae (Muntingia calabura) and Moringaceae (Punica granatum) families (Fig. 10). Conversely, the Moraceae and Moringaceae families were frequently confused with the Oleaceae family. This indicates that all models were not efficient for prediction or classification of species or families based on leaf features. Additionally, Table-2, and Fig. 6, Fig. 7 clearly suggest that implementation of DenseNet201 can solve the problem of medicinal plant identification that is comparatively less biased and free from intrafamily and interspecies misidentification.
Fig. 10.
Confusion matrix of VGG19 that employed in this study for the medicinal plant identifications for mix dataset (PFI).
4.2. Reason of misidentification of species
Misclassification of medicinal plants may arise due to diverse factors inherent to the classification process. A comprehensive comprehension of these underlying causes is of paramount significance for the enhancement and refinement of the accuracy inherent in the identification model. Medicinal plants frequently exhibit analogous morphological attributes encompassing leaf configuration, pigmentation, and dimensions [43]. Such shared resemblances can engender perplexity during image-oriented classification endeavors, particularly when discerning between closely akin species. The intrinsic variability in growth conditions, soil composition, and climatic nuances introduces dissimilarity in plant morphology and foliar manifestation [11]. These environmental intricacies may engender incongruities in feature extraction, subsequently impeding precise identification efforts. Certain medicinal plants boast elaborate and intricate leaf structures, presenting intricate challenges for automated systems that necessitate intricate feature extraction methodologies to effectively encapsulate pertinent information [44]. Within a single species, considerable diversity in leaf characteristics may prevail. Aspects such as age, well-being, and genetic heterogeneity contribute to this multifaceted variability, thereby complicating the generalized depiction of plant attributes. The absence of contextual insights, encompassing factors such as habitat, geographical coordinates, and correlated flora, may serve as impediments to the precise discernment of species.
4.3. Significance of the study
Local guides or books on medicinal plants are common sources for identification, however, they can be difficult to use and are sometimes unreliable. Since key books are beneficial when the user is aware with the observable plant family, which the purpose accomplished most of the cases, it has the ability to direct the observer in the right direction and help bring people closer to plants. Combining the AI program and the key book could be a useful tool for fieldwork for observers with limited experience. Recent approaches for the similar identification strategy achieved more reliable results (Table-3), which is also in line with our results. This also indicates the robustness of our study as we obtained almost 98 % accuracy. Our fine-tuned DenseNet50v2 an updated variant of DenseNet50, has the ability to efficiently propagate gradients and reduce the number of parameters compared to traditional convolutional neural networks.
Table 3.
Scenarios of performance for medicinal plant identification based on findings from some recent studies.
| Dataset | Model | Accuracy | Reference |
|---|---|---|---|
| VNPLANT-200 DATASET | VGG16 | 76 % | [21] |
| InceptionV3 | 82.50 % | ||
| MobileNetV2 | 87.92 % | ||
| Xception | 88.26 % | ||
| Densenet121 | 88 % | ||
| Morocco AMPs | ResNet50 | 90 % | [45] |
| Dynamic CNN | 97 % | ||
| Iran dataset | CNN | 97.6 % | [37] |
| AyurLeaf | CNN | 95 % | [36] |
| SVM | 96.7 % | ||
| Bangladeshi medicinal plant dataset | CNN | 71 % | [46] |
| PlantCLEF 2015 | EfficientNet-B1 | 87 % | [24] |
| Private dataset | Ayur-PlantNet | 92.7 % | [29] |
Our findings of automated medicinal species identifications through deep convolutional neural network approaches using images offer excellent opportunity in the fields of research, academia, pharmaceuticals industry, pharmacognosy, phytochemistry, and horticulture. However, it never eliminates the need for taxonomists; rather, it only speeds up the process of accurate species identification by background staff who are not taxonomists. For analyzing ecosystem, extinctions, population changes and ecological monitoring the importance of appropriate species identifications has been highlighted in several studies [47] as misinterpretation resulted from the misidentifications in many field studies [47,48].
Due to the enormous number of potential medicinal species in the world, especially in Asia, their identification faces the biggest hurdle. Over 38,660 different types of medicinal plants are found throughout Asia [49] and species local name depends on indigenous communities and geographic locations. Including geographic data on a species' range in the identification process is one potential solution to enrich local database [50]. Initial studies suggest that regionally constrained identification is probably more effective in automatic identification [51]. Our study successfully used (Fig. 8) the leaves of different species with extent variety of family that proved to be feasible to use in medicinal species identification, which would be more helpful in the drug industries. Medicinal plants in the natural environment are 3D, and 2D images possibly have drawbacks to capture variations. Quality of images in the real exposed environment are typically captured under a variety of environmental factors. Users should be aware that image quality is required for a trustworthy automatic identification. Therefore, it is necessary to provide clear instruction to users about the image taking process.
5. Conclusion
Our deep learning-based models exhibit enhanced accuracy and precision in identifying medicinal plants, reducing human errors that are usual in manual approaches. Notably, DenseNet201 achieved the highest classification accuracy, fine-tuning this model with localized data could result in optimal performance. Advancing the user interface and refining identification algorithms are pivotal for future enhancements. Leveraging deep learning algorithms holds the potential for effective real-world identification solutions.
It is important to acknowledge and elaborate on certain limitations that we encountered during our research. Sometimes it's hard to automatically identify plant species, especially in places like Bangladesh and India with many different species. One way to make it better is by using geographical information from national or international sources, like where these plants are usually found. But this introduces its own problems because species can move to new places, and the same species can look different depending on their age and where they grow. Having more pictures of the same species is important for accurate identification, and the leaves change throughout the year, which makes it even more complicated. Addressing these limitations through further study holds the potential to refine and enhance the robustness of our approach.
However, our study has demonstrated the feasibility of using leaves from a wide variety of plant species, spanning different botanical families, for medicinal species identification. This finding holds significant promise for the pharmaceutical industry, as it can facilitate the identification and utilization of plant species with medicinal properties.
Code availability
Codes are available in this repository https://github.com/biplobforestry/Automated-medicinal-plant-identification-.git.
Data availability statement
Data will be made available on request.
Funding
This research did not receive any funding from public, private, or not-for-profit organizations.
CRediT authorship contribution statement
Biplob Dey: Conceptualization, Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Jannatul Ferdous: Formal analysis, Visualization, Writing – original draft. Romel Ahmed: Conceptualization, Investigation, Project administration, Resources, Supervision, Writing – review & editing. Juel Hossain: Data curation, Resources, Validation, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e23655.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Jeelani S.M., Rather G.A., Sharma A., Lattoo S.K. In perspective: potential medicinal plant resources of Kashmir Himalayas, their domestication and cultivation for commercial exploitation. Journal of Applied Research on Medicinal and Aromatic Plants. 2018;8:10–25. doi: 10.1016/J.JARMAP.2017.11.001. [DOI] [Google Scholar]
- 2.Nankaya J., Nampushi J., Petenya S., Balslev H. Ethnomedicinal plants of the loita Maasai of Kenya. Environ. Dev. Sustain. 2020;22:2569–2589. doi: 10.1007/s10668-019-00311-w. [DOI] [Google Scholar]
- 3.Chapman K., Chomchalow N. Production of medicinal plants in Asia. Acta Hortic. 2005;679:45–59. doi: 10.17660/ACTAHORTIC.2005.679.6. [DOI] [Google Scholar]
- 4.Kala C.P., Dhyani P.P., Sajwan B.S. Developing the medicinal plants sector in northern India: challenges and opportunities. J. Ethnobiol. Ethnomed. 2006;2:32. doi: 10.1186/1746-4269-2-32. [DOI] [Google Scholar]
- 5.Vasisht K., Sharma N., Karan M. Current perspective in the international trade of medicinal plants material: an update. Curr. Pharmaceut. Des. 2016;22:4288–4336. doi: 10.2174/1381612822666160607070736. [DOI] [PubMed] [Google Scholar]
- 6.Martínez-Aledo N., Navas-Carrillo D., Orenes-Piñero E. Medicinal plants: active compounds, properties and antiproliferative effects in colorectal cancer. Phytochemistry Rev. 2020;19:123–137. doi: 10.1007/s11101-020-09660-1. [DOI] [Google Scholar]
- 7.Mbuni Y.M., Wang S., Mwangi B.N., Mbari N.J., Musili P.M., Walter N.O., Hu G., Zhou Y., Wang Q. Medicinal plants and their traditional uses in local communities around cherangani hills, western Kenya. Plants. 2020;9:331. doi: 10.3390/PLANTS9030331. 9 (2020) 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng W.Y., Hung L.Y., Lam Y.H., Chan S.S., Pang K.S., Chong Y.K., Ching C.K., Mak T.W.L. Poisoning by toxic plants in Hong Kong: a 15-year review. Hong Kong Med. J. 2019;25:102–112. doi: 10.12809/HKMJ187745. [DOI] [PubMed] [Google Scholar]
- 9.Roopashree S., Anitha J., Mahesh T.R., Vinoth Kumar V., Viriyasitavat W., Kaur A. An IoT based authentication system for therapeutic herbs measured by local descriptors using machine learning approach. Measurement. 2022;200 doi: 10.1016/J.MEASUREMENT.2022.111484. [DOI] [Google Scholar]
- 10.Herdiyeni Y., Wahyuni N.K.S. 2012 International Conference on Advanced Computer Science and Information Systems. ICACSIS); 2012. Mobile application for Indonesian medicinal plants identification using fuzzy local binary pattern and fuzzy color histogram; pp. 301–306. [Google Scholar]
- 11.Dey B., Masum Ul Haque M., Khatun R., Ahmed R. Comparative performance of four CNN-based deep learning variants in detecting Hispa pest, two fungal diseases, and NPK deficiency symptoms of rice (Oryza sativa) Comput. Electron. Agric. 2022;202 doi: 10.1016/j.compag.2022.107340. [DOI] [Google Scholar]
- 12.Dey B., Ahmed R., Ferdous J., Haque M.M.U., Khatun R., Hasan F.E., Uddin S.N. Automated plant species identification from the stomata images using deep neural network: a study of selected mangrove and freshwater swamp forest tree species of Bangladesh. Ecol. Inf. 2023;75 doi: 10.1016/J.ECOINF.2023.102128. [DOI] [Google Scholar]
- 13.Ferentinos K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018;145:311–318. doi: 10.1016/J.COMPAG.2018.01.009. [DOI] [Google Scholar]
- 14.Mohanty S.P., Hughes D.P., Salathé M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016;7 doi: 10.3389/fpls.2016.01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X., Qiao Y., Meng F., Fan C., Zhang M. Identification of maize leaf diseases using improved deep convolutional neural networks. IEEE Access. 2018;6:30370–30377. doi: 10.1109/ACCESS.2018.2844405. [DOI] [Google Scholar]
- 16.Agarwal M., Gupta S.K., Biswas K.K. Development of Efficient CNN model for Tomato crop disease identification. Sustainable Computing: Informatics and Systems. 2020;28 doi: 10.1016/j.suscom.2020.100407. [DOI] [Google Scholar]
- 17.Abdollahi J. Proceedings - 2022 27th International Computer Conference. Computer Society of Iran; 2022. Identification of medicinal plants in ardabil using deep learning: identification of medicinal plants using deep learning. [DOI] [Google Scholar]
- 18.Amuthalingeswaran C., Sivakumar M., Renuga P., Alexpandi S., Elamathi J., Hari S.S. Proceedings of the International Conference on Trends in Electronics and Informatics, 2019. Institute of Electrical and Electronics Engineers; 2019. Identification of medicinal plant's and their usage by using deep learning; pp. 886–890. [DOI] [Google Scholar]
- 19.Gao L., Lin X. Fully automatic segmentation method for medicinal plant leaf images in complex background. Comput. Electron. Agric. 2019;164 doi: 10.1016/j.compag.2019.104924. [DOI] [Google Scholar]
- 20.Kumar A., Kumar D.B. Automatic recognition of medicinal plants using machine learning techniques. Gedrag & Organisatie Review. 2020;33:166–175. doi: 10.37896/gor33.01/012. [DOI] [Google Scholar]
- 21.Nguyen Quoc T., Truong Hoang V. 2020 International Conference on Information and Communication Technology Convergence. 2020. Medicinal Plant identification in the wild by using CNN; pp. 25–29. [DOI] [Google Scholar]
- 22.Patil S., Sasikala M. 2022. Segmentation and Identification of Medicinal Plant through Weighted KNN, Multimedia Tools and Applications. [DOI] [Google Scholar]
- 23.Sivaranjani C., Kalinathan L., Amutha R., Kathavarayan R.S., Jegadish Kumar K.J. 2019 International Conference on Computational Intelligence in Data Science. Institute of Electrical and Electronics Engineers; 2019. Real-time identification of medicinal plants using machine learning techniques; pp. 1–4. [DOI] [Google Scholar]
- 24.Malik O.A., Ismail N., Hussein B.R., Yahya U. Automated real-time identification of medicinal plants species in natural environment using deep learning models-A case study from borneo region. Plants. 2022;11 doi: 10.3390/plants11151952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le T.-L., Tran D.-T., Hoang V.-N. Proceedings of the 5th Symposium on Information and Communication Technology. Association for Computing Machinery; New York, NY, USA: 2014. Fully automatic leaf-based plant identification, application for Vietnamese medicinal plant search; pp. 146–154. [DOI] [Google Scholar]
- 26.Raghukumar A.M., Narayanan G. vol. 2020. 2020. Comparison of machine learning algorithms for detection of medicinal plants; pp. 56–60. (Proceedings of the 4th International Conference on Computing Methodologies and Communication). [DOI] [Google Scholar]
- 27.Pacifico L.D.S., Britto L.F.S., Oliveira E.G., Ludermir T.B. 2019 8th Brazilian Conference on Intelligent Systems. Institute of Electrical and Electronics Engineers; 2019. Automatic classification of medicinal plant species based on color and texture features; pp. 741–746. [DOI] [Google Scholar]
- 28.Duong-Trung N., Da Quach L., Nguyen M.H., Nguyen C.N. Association for Computing Machinery International Conference Proceeding Series; 2019. A Combination of Transfer Learning and Deep Learning for Medicinal Plant Classification; pp. 83–90. Part F1479. [DOI] [Google Scholar]
- 29.Pushpa B.R., Rani N.S. Ayur-PlantNet: an unbiased light weight deep convolutional neural network for Indian Ayurvedic plant species classification. Journal of Applied Research on Medicinal and Aromatic Plants. 2023 doi: 10.1016/j.jarmap.2023.100459. [DOI] [Google Scholar]
- 30.Muneer A., Fati S.M. Efficient and automated herbs classification approach based on shape and texture features using deep learning. IEEE Access. 2020;8:196747–196764. doi: 10.1109/ACCESS.2020.3034033. [DOI] [Google Scholar]
- 31.Pushpanathan K., Hanafi M., Masohor S., Ilahi W.F.F. MYLPHerb-1: a dataset of Malaysian local perennial herbs for the study of plant images classification under uncontrolled environment. Pertanika Journal of Science and Technology. 2022;30:413–431. doi: 10.47836/PJST.30.1.23. [DOI] [Google Scholar]
- 32.Kan H.X., Jin L., Zhou F.L. Classification of medicinal plant leaf image based on multi-feature extraction. Pattern Recogn. Image Anal. 2017;27:581–587. doi: 10.1134/S105466181703018X. [DOI] [Google Scholar]
- 33.VijayaLakshmi B., Mohan V. Kernel-based PSO and FRVM: an automatic plant leaf type detection using texture, shape, and color features. Comput. Electron. Agric. 2016;125:99–112. doi: 10.1016/J.COMPAG.2016.04.033. [DOI] [Google Scholar]
- 34.Russel N.S., Selvaraj A. Leaf species and disease classification using multiscale parallel deep CNN architecture. Neural Comput. Appl. 2022;34:19217–19237. doi: 10.1007/S00521-022-07521-W. [DOI] [Google Scholar]
- 35.Sachar S., Kumar A. Deep ensemble learning for automatic medicinal leaf identification. Int. J. Inf. Technol. 2022;14:3089–3097. doi: 10.1007/s41870-022-01055-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dileep M.R., Pournami P.N. AyurLeaf: a deep learning approach for classification of medicinal plants, ieee region 10 annual international conference. Proceedings/TENCON. 2019-Octob. 2019:321–325. doi: 10.1109/TENCON.2019.8929394. [DOI] [Google Scholar]
- 37.Azadnia R., Al-Amidi M.M., Mohammadi H., Cifci M.A., Daryab A., Cavallo E. An AI based approach for medicinal plant identification using deep CNN based on global average pooling. Agronomy. 2022;12 doi: 10.3390/agronomy12112723. [DOI] [Google Scholar]
- 38.Azadnia R., Kheiralipour K. Recognition of leaves of different medicinal plant species using a robust image processing algorithm and artificial neural networks classifier. Journal of Applied Research on Medicinal and Aromatic Plants. 2021;25 doi: 10.1016/j.jarmap.2021.100327. [DOI] [Google Scholar]
- 39.Kaggle 2022. https://www.kaggle.com/
- 40.Das T., Zhong Y., Stoica I., Shenker S. Proceedings of the Association for Computing Machinery Symposium on Cloud Computing. Association for Computing Machinery; New York, NY, USA: 2014. Adaptive stream processing using dynamic batch sizing; pp. 1–13. [DOI] [Google Scholar]
- 41.Jain A.K., Rao P.P., Venkatesh Sharma K. Advanced Analytics and Deep Learning Models. 2022. Optimization techniques in deep learning scenarios: an empirical comparison; pp. 255–282. [DOI] [Google Scholar]
- 42.Dey B., Abir K.A.M., Ahmed R., Salam M.A., Redowan M., Miah M.D., Iqbal M.A. Monitoring groundwater potential dynamics of north-eastern Bengal Basin in Bangladesh using AHP-Machine learning approaches. Ecol. Indicat. 2023;154 doi: 10.1016/j.ecolind.2023.110886. [DOI] [Google Scholar]
- 43.Zhang Y., Wang Y. Recent trends of machine learning applied to multi-source data of medicinal plants. Journal of Pharmaceutical Analysis. 2023 doi: 10.1016/J.JPHA.2023.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mihai S., Yaqoob M., Hung D.V., Davis W., Towakel P., Raza M., Karamanoglu M., Barn B., Shetve D., Prasad R.V., Venkataraman H., Trestian R., Nguyen H.X. Digital twins: a survey on enabling technologies, challenges, trends and future prospects. IEEE Communications Surveys and Tutorials. 2022;24:2255–2291. doi: 10.1109/COMST.2022.3208773. [DOI] [Google Scholar]
- 45.Bahri A., Bourass Y., Badi I., Zouaki H., El moutaouakil K., Satori K. Dynamic CNN combination for Morocco aromatic and medicinal plant classification. International Journal of Computing and Digital Systems. 2022;11:239–249. doi: 10.12785/IJCDS/110120. [DOI] [Google Scholar]
- 46.Akter R., Hosen M.I. ETCCE 2020 - International Conference on Emerging Technology in Computing. Communication and Electronics; 2020. CNN-Based leaf image classification for Bangladeshi medicinal plant recognition. [DOI] [Google Scholar]
- 47.Austen G.E., Bindemann M., Griffiths R.A., Roberts D.L. Species identification by experts and non-experts: comparing images from field guides. Sci. Rep. 2016;6(2016):1–7. doi: 10.1038/srep33634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solow A., Smith W., Burgman M., Rout T., Wintle B., Roberts D. Uncertain sightings and the extinction of the ivory-billed woodpecker. Conserv. Biol. 2012;26:180–184. doi: 10.1111/J.1523-1739.2011.01743.X. [DOI] [PubMed] [Google Scholar]
- 49.Phumthum M., Srithi K., Inta A., Junsongduang A., Tangjitman K., Pongamornkul W., Trisonthi C., Balslev H. Ethnomedicinal plant diversity in Thailand. J. Ethnopharmacol. 2018;214:90–98. doi: 10.1016/J.JEP.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 50.Pärtel J., Pärtel M., Wäldchen J. Plant image identification application demonstrates high accuracy in Northern Europe. Annals of Botany (AoB) Plants. 2021;13 doi: 10.1093/AOBPLA/PLAB050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terry J.C.D., Roy H.E., August T.A. Thinking like a naturalist: enhancing computer vision of citizen science images by harnessing contextual data. Methods Ecol. Evol. 2020;11:303–315. doi: 10.1111/2041-210X.13335. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.