Abstract
Purpose
Gain of chromosome 8q has been associated with poor prognosis in uveal melanoma (UM), and an increase in the absolute number of 8q-copies correlated with an even shorter survival. Splicing factor 3b subunit 1 (SF3B1)-mutated (SF3B1MUT) tumors display structural chromosomal anomalies and frequently show a partial gain of chromosome 8qter. A recent subset of SF3B1MUT UM with early-onset metastases has been identified, prompting the investigation of the relationship between survival, 8q gain, and SF3B1MUT UM.
Design
Retrospective cohort study.
Subjects
Patients diagnosed with UM who underwent enucleation or received a biopsy at the Erasmus MC Cancer Institute or the Rotterdam Eye Hospital, The Netherlands were included.
Methods
Fifty-nine patients with SF3B1MUT tumors and 211 patients with BRCA1 associated protein 1 (BAP1)-mutated (BAP1MUT) tumors were included in this study. Copy number status and gene expression were assessed using either a single nucleotide polymorphism array, fluorescence in situ hybridization, and karyotyping, or a combination of these techniques. Disease-free survival was determined and a cut-off of 60 months was used to define early-onset metastatic disease.
Main Outcome Measures
Disease-free survival.
Results
Forty-eight patients with SF3B1MUT UM (81%) had chromosome 8q gain (3 copies, 78%; 4 copies, 22%). Kaplan–Meier analysis of SF3B1MUT UM did not indicate a difference in survival in patients with or without gain of 8q (P = 0.99). Furthermore, the number of 8q copies was not associated with survival when comparing early (P = 0.97) versus late (P = 0.23) metastases group. In contrast, the presence of 8q gain (86%) was correlated with a decreased survival in BAP1MUT UM (P = 0.013).
Conclusions
We did not find a correlation between 8q gain and early-onset metastasis in SF3B1MUT tumors. Gain of 8q has no additional predictive value in SF3B1MUT tumors. In contrast, 8q gain is predictive of a worse prognosis in patients with BAP1MUT tumors. Thus, gain of chromosome 8q has additional predictive value for BAP1MUT tumors, but not for SF3B1MUT tumors.
Financial Disclosure(s)
The author(s) have no proprietary or commercial interest in any materials discussed in this article.
Keywords: Aberration, Chromosome 8q gain, Copy number variation, Prognosis
Uveal melanoma (UM) is the most common primary intraocular malignancy in adults.1 Recurrent mutations in secondary driver genes BRCA1 associated protein 1 (BAP1), splicing factor 3b subunit 1 (SF3B1), and eukaryotic translation initiation factor 1A X-linked (EIF1AX),2 as well as characteristic copy number variations (CNV)3 and gene expression profiles4 are used in prognostication. Previously, studies using fluorescence in situ hybridization (FISH) probes and array comparative genomic hybridization revealed that gain of chromosome 8q (> 2 copies) in UM has a strong correlation to a short survival compared with normal or low copies (< 2 copies) of chromosome 8q.5,6 An increased number of 8q copies was also associated with a worse prognosis (the more copies of 8q, the worse the prognosis).5 Furthermore, chromosome 8q status in UM patients with different iris colors was assessed in a large cohort study.7 Authors describe the correlation of chromosome 8q gain to worse survival in patients with light iris color, but not in those with dark iris color.7 Previously, we reported that CNV profiles are mutation-specific and BAP1-mutated (BAP1MUT) UM are characterized by gains or losses of entire chromosomes, chromosome arms, and recurrent isochromosomes.2,3 Splicing factor 3b subunit 1-mutated (SF3B1MUT) UM are characterized by multiple structural chromosomal aberrations at the distal ends of chromosomes (gain of chromosomes 6p and 8q as well as loss of chromosomes 6q and 11q). The EIF1AX-mutated UM are characterized by gain of chromosome 6p. In BAP1MUT UM, gain of 8q is the result of the gain of an entire chromosome or isochromosome formation. However, in SF3B1MUT UM, 8q gain is the outcome of structural chromosome changes resulting in a gain of only the distal end of 8q.3 Both BAP1MUTand some of the SF3B1MUT UM patients develop early-onset metastatic disease,8 prompting us to investigate the relationship between gain of 8q, survival, and BAP1MUT or SF3B1MUT UM.
Methods
This is a retrospective cohort study of patients diagnosed with UM between 1994 and 2022 and included in the Rotterdam Ocular Melanoma Study Group (ROMS) database. All patients underwent enucleation or received a biopsy at the Erasmus MC Cancer Institute or the Rotterdam Eye Hospital, The Netherlands. Copy number variations were assessed with ≥ 1 of the following methods: single nucleotide polymorphism (SNP) array, karyotyping, or FISH. Uveal melanoma with concomitant mutations in BAP1, SF3B1, or EIF1AX were excluded. In total, 59 patients with a confirmed SF3B1MUT tumor were included. In 211 samples, BAP1 status was confirmed with either a BAP1 mutation and negative BAP1 immunohistochemistry or both. Because tumors with an EIF1AX mutation typically do not metastasize,9 we excluded these tumors in this study. The disease-free survival (DFS) (determined as the interval from treatment until metastasis or metastasis and subsequent death due to UM or until the last follow-up) was determined in all patients and a cut-off of 60 months was used to identify early-onset metastatic disease in UM patients.8 Informed consent was obtained from all patients. The study adhered to the guidelines of the Declaration of Helsinki and was approved by the Medical Ethics Committee of the Erasmus Medical Centre (OZR nr 2009-17, MEC-2009-375, 12 November 2009).
Copy Number Analysis
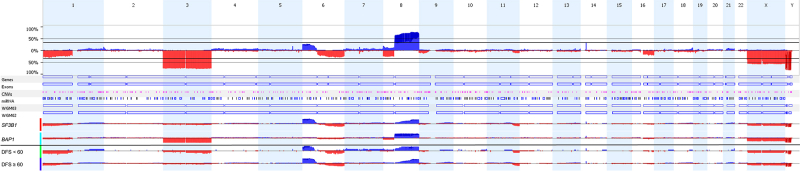
For the SNP array, 200 ng of tumor DNA was used for whole-genome copy number analyses by SNP arrays. For the entire set, 6 types of Illumina Human SNP array platforms were used according to the manufacturers protocol (CytoSNP-12 v2.0 BeadChip [n = 12], CytoSNP-12 v2.1 [n = 35], OmniExpress-12 v1 [n = 29], CytoSNP-850K BeadChips [n = 67] and GSAMD_V1 [n = 34] and GSAMD_V2 [n = 23] [Bead Studio Illumina]). The resolution to detect CNV of all chip types is ≥ 30 kb. Nexus Copy Number 10.0 (BioDiscovery, Inc) was used to calculate the total percentage aneuploidy of the entire genome, the total copy number events, and to visualize the whole-genome SNP array data. A summary plot showing the SNP-array-derived chromosomal patterns of primary UM can be found in the supplementary material Figure S1 (available at www.ophthalmologyscience.org). We used cytogenetic and FISH data from our ROMS cohort, which have been previously described by Van den Bosch et al5 and Yavuzyigitoglu et al.3 Because techniques have changed over the years when including patients in the ROMS cohort, the different techniques were mostly used alternatively in patients, e.g., the earliest patients included only have cytogenetic/FISH data (from 1992 until 2010), and the latest patients included all only have SNP array data (from 2010 onward), though almost all tumors from the earliest patients were later analyzed with the SNP array. Gain of the distal ends of chromosome 8q was defined at location 8q23qter, which was used as the cut-off point by Yavuzyigitoglu et al.3
Survival Analysis
The DFS was determined from the date of diagnosis until the date of metastasis or last follow-up. Patients were censored when they were lost to follow-up, when death from a cause other than UM occurred, or when death without any cause was reported. Kaplan–Meier survival analyses were performed to estimate survival probabilities with either metastasis or death due to metastases. Cox proportional hazard regression analysis was performed to identify the value of the prognostic factors, adjusted for age and sex. Differences between groups were assessed using the t test. The analyses were performed using the statistical software GraphPad Prism 9.4.0 (GraphPad Software, Inc). A P value of < 0.05 was considered statistically significant.
Results
Characteristics
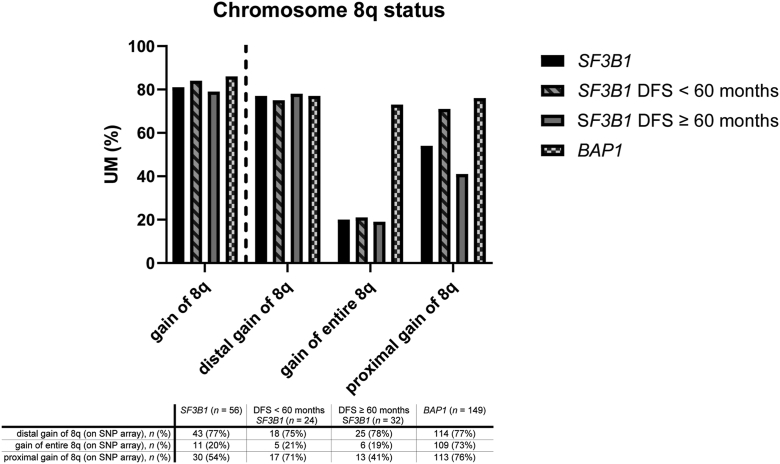
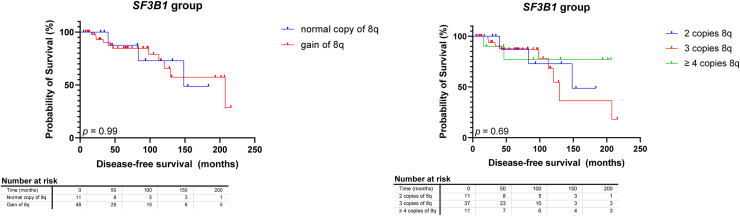
Fifty-nine tumors were included in the SF3B1 group. Gain of chromosome 8q (> 2 copies of 8q) was present in 48 tumors (81%) (Fig 2, Table 1). One SF3B1MUT UM had proximal 8q gain without a gain in the distal ends. Cox proportional hazard analysis could not confirm the reported independent5 prognostic value of gain of chromosome 8q in general (> 2 copies) in SF3B1MUT UM (hazard ratio [HR]: 0.9366 [95% confidence interval {CI}: 0.2809–4.231]), nor for the number of additional 8q copies (3 copies of 8q, HR: 1.145 [95% CI: 0.3174–5.412]; ≥ 4 copies of 8q, HR: 0.5687 [95% CI: 0.07347–3.494]) in SF3B1MUT tumors. Kaplan–Meier survival analysis also indicated no difference in survival in patients with or without 8q gain (P = 0.99). Furthermore, no difference was found in survival between 2, 3, or ≥ 4 copies (P = 0.69) (Fig 3).
Figure 2.
Bar chart of 8q status in splicing factor 3b subunit 1-mutated (SF3B1MUT) and BRCA1 associated protein 1-mutated (BAP1MUT) tumors, as well as in SF3B1MUT tumors with disease-free survival (DFS) < 60 months and DFS ≥ 60 months. SNP = single nucleotide polymorphism; UM = uveal melanoma.
Table 1.
Characteristics of All SF3B1MUT and BAP1MUT Tumors in the Rotterdam Ocular Melanoma Study Group Cohort
| Characteristics | SF3B1 (n = 59) | SF3B1 8q Gain (n = 48) | SF3B1 No 8q Gain (n = 11) | 3 copies 8q (n = 37) | ≥ 4 copies 8q (n = 11) | BAP1(n = 211) | BAP1 8q Gain (n = 181) | BAP1 No 8q Gain (n = 30) | 3 Copies 8q (n = 97) | ≥ 4 Copies 8q (n = 84) |
|---|---|---|---|---|---|---|---|---|---|---|
| Male, n (%) | 32 (54) | 26 (54) | 6 (55) | 20 (54) | 6 (55) | 105 (50) | 94 (52) | 11 (37) | 53 (55) | 41 (49) |
| Age, yrs (mean ± SD) | 55.3 ± 15.3 | 55.9 ± 15.3 | 52.7 ± 15.4 | 54.5 ± 14.2 | 60.9 ± 18.6 | 65.2 ± 12.8 | 65.3 ± 12.7 | 65.0 ± 14.0 | 65.0 ± 13.1 | 65.7 ± 12.2 |
| DFS, mos (mean ± SD) | 80.3 ± 57.5 | 78.2 ± 57.6 | 89.6 ± 58.9 | 71.8 ± 50.1 | 99.7 ± 76.8 | 41.7 ± 45.0 | 39.9 ± 44.4 | 53.0 ± 47.7 | 43.3 ± 47.4 | 35.9 ± 40.5 |
| 8q gain, n (%) | 48 (81) | - | - | - | - | 181 (86) | - | - | - | - |
BAP1 = BRCA1 associated protein 1; BAP1MUT = BAP1 mutated; DFS = disease-free survival; SD = standard deviation; SF3B1 = splicing factor 3b subunit 1; SF3B1MUT = SF3B1 mutated.
Figure 3.
Kaplan–Meier survival curves in splicing factor 3b subunit 1-mutated (SF3B1MUT) tumors stratified to chromosome 8q status and chromosome 8q copy numbers. No differences were found between chromosome 8q status (P = 0.99) and chromosome 8q copy numbers (P = 0.69).
Chromosome 8q in BAP1MUT Tumors
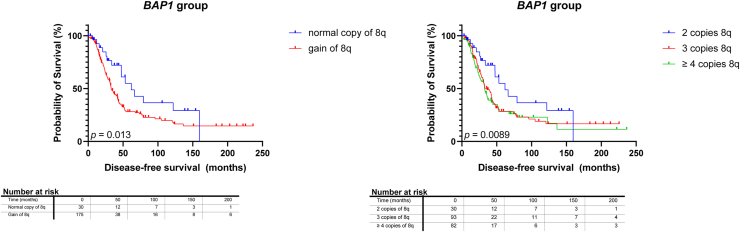
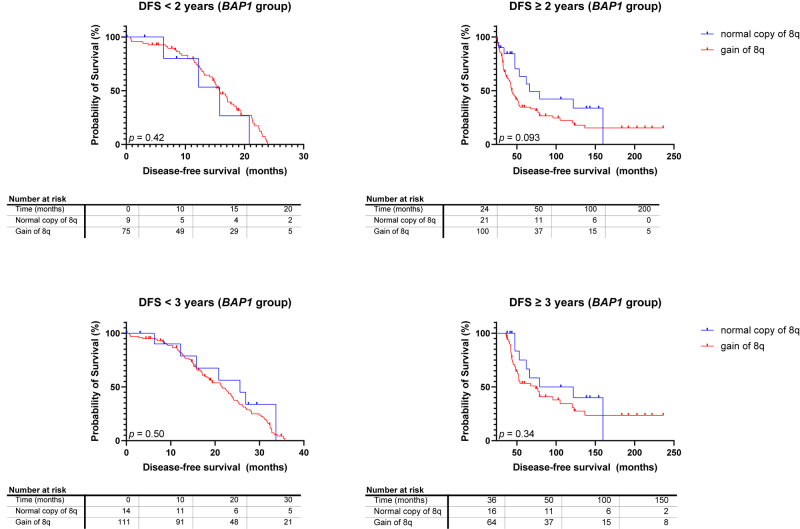
Although 8q gain is seen often in SF3B1MUT UM, it also frequently occurs in BAP1MUT UM. We assessed the survival of 211 UM patients with either immunohistochemically BAP1-negative tumors and tumors with pathogenic BAP1 mutations or both. One hundred thirty patients with BAP1MUT UM developed metastases. Gain of chromosome 8q was present in 181 tumors (86%) (Fig 2, Table 1), including 6 BAP1MUT UM with only gain in the proximal 8q without distal gain, and this correlated with worse survival in BAP1MUT UM (P = 0.013) (Fig 4). However, when the analysis was repeated with a cut-off DFS of 2 and 3 years, no significant differences were found Figure S5 (available at www.ophthalmologyscience.org). Based on the Cox regression model, tumors with 3 copies of 8q were not associated with a decreased survival when compared with 2 copies of 8q (HR: 1.645 [95% CI: 0.9486–3.034]), though ≥ 4 copies of 8q showed a significant decrease in survival when compared with normal copies of 8q (HR: 2.172 [95% CI: 1.246–4.023]).
Figure 4.
Kaplan–Meier survival curves in BRCA1 associated protein 1-mutated (BAP1MUT) tumors stratified to chromosome 8q status and chromosome 8q copy numbers. Gain of chromosome 8q was correlated with a worse prognosis (P = 0.013) with increased copy numbers of 8q correlating with shorter disease-free survival (P = 0.0089).
Chromosome 8q in SF3B1MUT Tumors with DFS < 60 and DFS ≥ 60 Months
Next, SF3B1MUT tumors were classified based on DFS (DFS < 60 months [n = 25] and DFS ≥ 60 months [n = 34]) (Table 2). The distal gains of chromosome 8q (defined as 8q23qter3) were similar in patients with a DFS < 60 months (74%) and DFS ≥ 60 months (78%) based on SNP array data (Fig 2). Gain of the entire chromosome 8q was similar in patients with a DFS < 60 months (21%) and DFS ≥ 60 months (19%) (Fig 2). Proximal gain of chromosome 8q (defined as gain at least between 8q11-8q23) was more prominent in patients with a DFS < 60 months (71%) compared with patients with a DFS ≥ 60 months (41%), though most of these also had distal gains (therefore, these can also be seen as an incomplete gain of the entire chromosome). Fifteen patients with SF3B1 mutation have metastatic disease, including 8 patients with a DFS < 60 months (88% with 8q gain) and 7 patients with a DFS ≥ 60 months (71.4% with 8q gain).8 Six patients in the DFS < 60 months group died of other causes.
Table 2.
Characteristics of 8q Status of All SF3B1MUT Grouped Based on DFS in the Rotterdam Ocular Melanoma Study Group Cohort
| SF3B1MUT Tumors | DFS < 60 mos (n = 25) | DFS ≥ 60 mos (n = 34) |
|---|---|---|
| 2 copies of 8q, n (%) | 4 (16) | 7 (21) |
| 3 copies of 8q, n (%) | 16 (64) | 21 (61) |
| ≥ 4 copies of 8q, n (%) | 5 (20) | 6 (18) |
| 8q gain, n (%) | 21 (84) | 27 (79) |
DFS = disease-free survival; SF3B1MUT = splicing factor 3b subunit 1-mutated.
Discussion
Many studies identified gain of chromosome 8q as one of the prognostic factors in UM.10, 11, 12, 13 In a previous study, we have already assessed 220 UM with the FISH technique and concluded that chromosome 8q gain is an important independent prognostic factor in UM.5 Furthermore, a higher percentage of gain of 8q in tumor cells was significantly correlated to a worse prognosis. This was later confirmed by Cassoux et al6 with multiplex ligation-dependent probe amplification assay in 338 UM, and by Versluis et al14 with digital polymerase chain reaction in 66 enucleated UM, where they found that an increased 8q copy number was correlated with metastatic risk. Wierenga et al recently showed that gain of chromosome 8q has a significant role in survival in UM patients with light iris color compared with those with dark iris color.7
One of the most important commercial methods to prognosticate UM is the gene expression profiling test,4 which uses the expression of 15 genes to classify UM into 2 groups, a low metastatic risk group (reported as class 1) and a high metastatic risk group (reported as class 2). Class 2 tumors frequently had either BAP1 mutations and monosomy 3 (M3) or both, whereas most of class 1 tumors had 2 copies of chromosome 3.15 Additionally, Robertson et al2 divided UM into 4 molecular and clinical clusters. Uveal melanoma with M3 was associated with a poor prognosis, whereas UM with disomy 3 (D3) showed a more favorable prognosis. Disomy 3 tumors have the least chromosomal aberrations, where gain of 8q can be present, but most commonly partial or total gain of chromosome 6p.2 Alternatively, M3 tumors showed gain of 6p and partial gain of 8q, often a result of isochromosome formation.3 Therefore, chromosome 8q gain is associated with M3 and correlated with poor prognosis. Furthermore, although not always done in the past, UM can also be prognosticated by mutation status. As 8q gain often coexists with M3, gain of 8q is often seen in BAP1MUT tumors. Even though both SF3B1MUT and BAP1MUT tumors display 8q gain,2,3 the correlation between the amount of 8q copies and mutation status in UM has yet to be assessed.
In this study, no association was found between 8q gain and SF3B1MUT tumors in relation to survival. Furthermore, we show that 8q gain is not discriminating for the survival of early versus late metastasizing SF3B1MUT tumors. Uveal melanoma with gain of 8q in combination with D3 is correlated to a lower metastatic risk compared with those with gain of 8q and M3.6 A possible explanation could be because these tumors with 8q gain in combination with D3 could be classified as SF3B1MUT tumors (as SF3B1MUT tumors have D3 in general). However, although rare, BAP1MUT tumors (with positive BAP1 immunohistochemistry) can also have D3, making it more difficult to correlate 8q to prognosis based only on gene expression profiling classification.2,16 Furthermore, as BAP1MUT tumors have a peak in metastatic disease (large peak at approximately 3 years) within the first 5 years after diagnosis, we repeated the analysis at 2 and 3 years, though no significant differences were found (Fig S5). Additionally, chromosome 8q status in this study was assessed with various methods over the years. Where karyotyping and FISH were used in the past, the SNP array has become the first line of choice to detect CNV, as it offers relatively fast and objective results as well as a minimal required amount of tissue. Single nucleotide polymorphism array can detect submicroscopic alterations which can be missed with either karyotyping and FISH or both. Therefore, the lack of 8q gain in some of the patients in this study could be explained due to the difference in techniques used over the years, though most tumors have had SNP array analysis. Furthermore, in the ROMS cohort, only 1 SF3B1MUT tumor with 8q gain had loss of chromosome 8p whereas 40% of BAP1MUT tumors with 8q gain had loss of chromosome 8p, strengthening the findings of small chromosomal aberrations in the SF3B1 group. The current study is limited by its small sample size, and consequently the small number of events. Conducting further studies with a larger cohort would enhance the validity and robustness of these findings.
Therefore, in the current study, we have shown that the prognostic relevance of chromosome 8q status is only observed in BAP1MUT tumors, and not in SF3B1MUTtumors. In addition, BAP1MUT tumors showed a trend toward decreased survival with increased numbers of 8q copies compared with normal copies of 8q. Furthermore, BAP1MUT tumors contribute to > 50% of all UM and are characterized by gains and losses of entire chromosomes or chromosome arms. In contrast, SF3B1MUT UM karyotypes are more complex and CNV often have recurrent distal breakpoints on chromosomes 6 and 8. This difference in CNV patterns could indicate a separate tumorigenesis mechanism in both groups. In this study, the correlation between 8q gain and survival was observed only in BAP1MUT tumors, not SF3B1MUT tumors. Therefore, we conclude that the reduced survival reported in UM with chromosome 8q gain is primarily influenced by BAP1MUT tumors, which have not been specifically identified in this context.
To conclude, we have evaluated gain of chromosome 8q and its role on DFS in SF3B1MUT and BAP1MUT UM in the ROMS cohort. There is no correlation between 8q gain and early-onset metastasis in SF3B1MUT tumors. We do not see evidence of additional predictive value of gain of 8q in SF3B1MUT tumors. In contrast, 8q gain is only predictive of a worse prognosis in BAP1MUT tumors. To establish and clarify the role of 8q gain in UM in general, a uniform approach toward analyzing CNV, mutation status and transcriptomic data in relation to a DFS over a longer follow-up period in a larger cohort study should be considered. Validation in clinically well-typed patient cohorts with CNV profiles, transcriptome, and mutation data would strengthen these findings.
Manuscript no. XOPS-D-22-00201R4.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s):
The Rotterdam Ocular Melanoma Study Group (ROMS) is a collaborative research group with members from the Rotterdam Eye Hospital, Departments of Ophthalmology, Pathology and Clinical Genetics, of the Erasmus MC, Rotterdam, The Netherlands.
All authors have completed and submitted the ICMJE disclosures form.
The authors have no proprietary or commercial interest in any materials discussed in this article.
Supported by the Henkes foundation (2021-04), Rotterdam, The Netherlands, and the Combined Ophthalmic Research Rotterdam (6.2.0), Rotterdam, The Netherlands. The funding organizations had no role in the design or conduct of this research.
HUMAN SUBJECTS: Human subjects were included in this study. Informed consent was obtained from all patients. The study adhered to the guidelines of the Declaration of Helsinki and was approved by the Medical Ethics Committee of the Erasmus Medical Centre (OZR nr 2009-17, MEC-2009-375, 12 November 2009).
No animal subjects were used in this study.
Author Contributions:
Conception and design: de Klein, Brosens, Kiliç
Data collection: Nguyen, Drabarek, Vaarwater, Yavuzyigitoglu, Verdijk, Paridaens, Naus, de Klein, Brosens, Kiliç
Analysis and interpretation: Nguyen, Vaarwater, de Klein, Brosens, Kiliç
Obtained funding: Kiliç
Overall responsibility: Nguyen, Drabarek, Vaarwater, Yavuzyigitoglu, Verdijk, Paridaens, Naus, de Klein, Brosens, Kiliç
Contributor Information
Emine Kiliç, Email: e.kilic@erasmusmc.nl.
Rotterdam Ocular Melanoma Study group:
Emine Kilic, Annelies de Klein, Erwin Brosens, Nicole C. Naus, Dion Paridaens, Serdar Yavuzyigitoglu, Wojtek Drabarek, Josephine Q.N. Nguyen, Jolanda Vaarwater, and Robert M. Verdijk
Supplementary Data
Figure S1.
A summary plot showing the SNP-array-derived chromosomal patterns of splicing factor 3b subunit 1-mutated (SF3B1MUT) and BRCA1 associated protein 1-mutated (BAP1MUT) tumors, as well as in SF3B1MUT tumors with disease-free survival (DFS) < 60 months and DFS ≥ 60 months. SNP = single nucleotide polymorphism.
Figure S5.
Kaplan–Meier survival curves in BRCA1 associated protein 1-mutated (BAP1MUT) tumors with disease-free survival (DFS) < 2 or 3 and DFS ≥ 2 or 3 years, stratified to chromosome 8q status. Gain of chromosome 8q was correlated with a worse prognosis (P = 0.093) within BAP1MUT tumors with a DFS ≥ 2 years. No differences were found between chromosome 8q status within BAP1MUT tumors with a DFS < 3 or DFS ≥ 3 years, and in BAP1MUT tumors with a DFS < 2 years.
References
- 1.Jager M.J., Shields C.L., Cebulla C.M., et al. Uveal melanoma. Nat Rev Dis Primers. 2020;6:24. doi: 10.1038/s41572-020-0158-0. [DOI] [PubMed] [Google Scholar]
- 2.Robertson A.G., Shih J., Yau C., et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204–220 e15. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yavuzyigitoglu S., Drabarek W., Smit K.N., et al. Correlation of gene mutation status with copy number profile in uveal melanoma. Ophthalmology. 2017;124:573–575. doi: 10.1016/j.ophtha.2016.10.039. [DOI] [PubMed] [Google Scholar]
- 4.Harbour J.W., Chen R. The DecisionDx-UM gene expression profile test provides risk stratification and individualized patient care in uveal melanoma. PLOS Curr. 2013;5 doi: 10.1371/currents.eogt.af8ba80fc776c8f1ce8f5dc485d4a618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Bosch T., van Beek J.G., Vaarwater J., et al. Higher percentage of FISH-determined monosomy 3 and 8q amplification in uveal melanoma cells relate to poor patient prognosis. Invest Ophthalmol Vis Sci. 2012;53:2668–2674. doi: 10.1167/iovs.11-8697. [DOI] [PubMed] [Google Scholar]
- 6.Cassoux N., Rodrigues M.J., Plancher C., et al. Genome-wide profiling is a clinically relevant and affordable prognostic test in posterior uveal melanoma. Br J Ophthalmol. 2014;98:769–774. doi: 10.1136/bjophthalmol-2013-303867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wierenga A.P.A., Brouwer N.J., Gelmi M.C., et al. Chromosome 3 and 8q aberrations in uveal melanoma show greater impact on survival in patients with light iris versus dark iris color. Ophthalmology. 2022;129:421–430. doi: 10.1016/j.ophtha.2021.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Drabarek W., van Riet J., Nguyen J.Q.N., et al. Identification of early-onset metastasis in SF3B1 mutated uveal melanoma. Cancers (Basel) 2022;14:846. doi: 10.3390/cancers14030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen J.Q.N., Drabarek W., Yavuzyigitoglu S., et al. Spliceosome mutations in uveal melanoma. Int J Mol Sci. 2020;21:9546. doi: 10.3390/ijms21249546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White V.A., Chambers J.D., Courtright P.D., Chang W.Y., Horsman D.E. Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer. 1998;83:354–359. [PubMed] [Google Scholar]
- 11.Kilic E., Naus N.C., van Gils W., et al. Concurrent loss of chromosome arm 1p and chromosome 3 predicts a decreased disease-free survival in uveal melanoma patients. Invest Ophthalmol Vis Sci. 2005;46:2253–2257. doi: 10.1167/iovs.04-1460. [DOI] [PubMed] [Google Scholar]
- 12.Damato B., Duke C., Coupland S.E., et al. Cytogenetics of uveal melanoma: a 7-year clinical experience. Ophthalmology. 2007;114:1925–1931. doi: 10.1016/j.ophtha.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Sisley K., Rennie I.G., Parsons M.A., et al. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes Cancer. 1997;19:22–28. doi: 10.1002/(sici)1098-2264(199705)19:1<22::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 14.Versluis M., de Lange M.J., van Pelt S.I., et al. Digital PCR validates 8q dosage as prognostic tool in uveal melanoma. PLOS ONE. 2015;10 doi: 10.1371/journal.pone.0116371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harbour J.W. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile. Methods Mol Biol. 2014;1102:427–440. doi: 10.1007/978-1-62703-727-3_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harbour J.W., Onken M.D., Roberson E.D., et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]