Abstract
Objective: Ribonuclease P RNA component H1 (RPPH1) is a long non-coding RNA (lncRNA) associated with cancer progression. Higher RPPH1 expression in breast and cervical cancer samples than that in normal tissues were observed through the lncRNASNP2 database; therefore, silencing RPPH1 expression might be a potential strategy for cancer treatments, even though RPPH1 is also an RNA subunit of ribonuclease P involved in processing transfer RNA (tRNA) precursors and the effect of RPPH1 knockdown is not yet fully understood. Methods: Differentially expressed genes (DEGs) were identified through RNA sequencing in each shRNA-transfected RPPH1 knockdown MDA-MB-231, RPPH1 knockdown HeLa cell, and respective control cells, then the gene ontology enrichment analysis was performed by IPA and MetaCore database according to these DEGs, with further in vitro experiments validating the effect of RPPH1 silencing in MDA-MB-231 and HeLa cells. Results: Hundreds of down-regulated DEGs were identified in RPPH1 knockdown MDA-MB-231 and HeLa cells while bioinformatics analysis revealed that these genes were involved in pathways related to immune response and cancerogenesis. Compared to mock- and vector-transfected cells, the production of mature tRNAs, cell proliferation and migration capacity were inhibited in RPPH1-silenced HeLa and MDA-MB-231 cells. Additionally, RPPH1 knockdown promoted G1 cell cycle arrest mainly through the down-regulation of cyclin D1, although glycolytic pathways were only affected in RPPH1 knockdown HeLa cells but not MDA-MB-231 cells. Conclusion: This study demonstrated that knockdown RPPH1 affected tRNA production, cell proliferation and metabolism. Our findings might provide insight into the role of RPPH1 in tumor development.
Keywords: Long noncoding RNA, ribonuclease P RNA component H1, transfer RNA, next-generation sequencing, tumor progression
Introduction
Most cancer cells have a high level of energy metabolism, so the dysregulation of tRNAs is seen as an inevitable result [1,2]. Several studies have indicated that up-regulation of tRNAs is involved in the proliferation and migration of cancer cells [3-5]. tRNAs are a group of highly conserved RNA molecules that convert genetic code into amino acids to assist protein synthesis, and under normal circumstances, the expression levels of tRNAs depend on the tissue type, cell state and environment [6,7]. However, the expression levels of tRNAs encoded by the nucleus and mitochondria in breast cancer cells are up to 10-fold higher than those in normal breast tissue [8]. Another study showed that the up-regulation of specific tRNAs (tRNAGluUUC and tRNAArgCCG) promotes breast cancer metastasis by directly increasing EXOSC2 expression [9] while an up-regulated tRNAiMet level also enhances the migration and invasion of melanoma cells [10]. These studies indicated that the up-regulation of specific tRNAs increases the malignancy of tumor cells.
Ribonuclease P (RNase P) is a ribonucleoprotein endonuclease that cleaves the 5’ end of precursor-tRNAs (pre-tRNAs) to form a mature 5’ terminus of the tRNA molecule [11-13], and besides tRNA processing, the subunits of RNase P have also been found to be involved in DNA damage repair and innate immunity against pathogens [14,15]. This highly conserved enzyme is mostly composed of one RNA molecule and one or more protein subunits in bacteria, archaea, yeast and human cells [13]. In human cells, RNase P includes ten different proteins and one RNA subunit, namely ribonuclease P RNA component H1 (RPPH1) [12]. RPPH1, the only RNA component of RNase P, provides the catalytic activity of pre-tRNA procession in human cells [16,17] with recent studies indicating that the expression level of RPPH1 as a long non-coding RNA (lncRNA), is closely related to cancer progression [18-20]. For instance, RPPH1 has been found to be up-regulated in gastric and breast cancers [19,21] and for the latter, it regulates CDC42 expression by binding to miR-330-5p thereby promoting the progression of breast cancer by directly targeting miR-122 [19,20]. LncRNA RPPH1 presented in exosome promotes cancer metastasis through the interaction of β-III tubulin while promoting macrophage M2 polarization in the colorectal cancer model [18]; furthermore, circRPPH1 as derived from the RPPH1 gene has been reported to promote breast cancer progression through miR-328-3p/HMGA2, miR-146b-3p/E2F2 or miR-542-3p/ARHGAP1 axes, respectively [22-24]. These results indicate that the RPPH1 gene plays an important role in cell proliferation and metastasis through interaction with specific miRNAs.
Targeting of oncogenic lncRNA is a novel approach to cancer treatment [25]. RPPH1 is not only an lncRNA but a component of RNase P as well, with the RPPH1 expression level being closely related to the protein synthesis of a cell and the molecular function of RPPH1 being closely related to pre-tRNAs processing [26], and therefore RPPH1 is speculated to be related to the protein synthesis of a cell.
Since RPPH1 may play a crucial role in tumor progression, the effect of RPPH1 silencing in cancer cells is an issue that should be investigated; however, comprehensive analysis in RPPH1 knockdown tumor cells has not been elucidated. In this study, two cancer cell lines with high RPPH1 expression, namely HeLa (cervical cancer) and MDA-MB-231 (breast cancer) cells, were investigated using a short hairpin RNA inhibition strategy. The differences in gene expression between RPPH1 knockdown cells and mock cells and enriched pathways were analyzed using next-generation sequencing and bioinformatics tools. The results provide a more comprehensive understanding of the effect of RPPH1 inhibition on tumors.
Materials and methods
Cell culture
Human cervical cancer (HeLa) and breast cancer (MDA-MB-231) were purchased from the American Type Culture Collection (ATCC; Rockville, MD, USA). All cells were cultured in high Glucose of Dulbecco’s modified Eagle’s medium (GIBCO; Grand Island, NY, USA) supplemented with 44 mM NaHCO3, 10% Fetal Bovine Serum (FBS) at 37°C in 5% CO2. The stable shRPPH1 expression clones were maintained in growth medium containing 50 μg/mL hygromycin B.
Cell transfection
Cells were cultured in 6-well plates at a density of 1×105 per well. After 16 h, the cells were transfected with expression constructs (2 μg) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s guidelines. In order to obtain a stable clone, the transfected cells were treated by 300 µg/mL hygromycin B. The selected hygromycin-B-resistant colonies were analyzed by RT-PCR to identify RPPH1 expression.
Design and construction of the shRNA expression vector
Plasmid vectors and constructs were constructed through standard molecular cloning techniques. The oligonucleotides used in this study were purchased from commercial suppliers. The stable shRNA expression vector pSUPER/Hygr was constructed by subcloning the hygromycin resistance gene expression cassette (Hygr) isolated from pDsRed2-N1 into the pSUPER. The shRNA expression vectors were constructed by inserting an annealed oligonucleotide duplex into the restriction enzyme sites (BglII/HindIII) in the pSUPER/Hygr vector. The oligonucleotides were used for this study: shRPPH1-1-F, 5’-gatcccc-GCCAGCGAAGTGAGTTCAA-ttcaagaga-TTGAACTCACTTCGCTGGC-tttttggaaa-3’; shRPPH1-1-R, agcttttccaaaaa-GCCAGCGAAGTGAGTTCAA-tctcttgaa-TTGAACTCACTTCGCTGGC-ggg.
Next-generation sequencing
Four cancer cell samples including HeLa and HeLa-shRPPH1-1-52 (HeLa RPPH1 knockdown clone), MDA-MB-231 and MDA-MB-231-shRPPH1-1-72 (MDA-MB-231 RPPH1 knockdown clone) were collected. The extracted total RNA of four samples were analyzed through genomics Biotechnology Company (New Taipei City, Taiwan), using poly-T oligo-attached beads to purify mRNA, while the samples were also fragmented and primed for cDNA synthesis. The library was identified by Agilent 2100 Bio-analyzer and Real-Time PCR System. In this study, genes that matched both fold change (FC) < 0.65 and false discovery rate (FDR) < 0.05 were considered down-regulated genes and further analyzed through bioinformatics tools.
MTT cell growth assay
Cells were seeded on 96-well plates at a density of 1×103 cells per well in 100 μL growth medium. The cells were treated with 10 μL MTT solution (5 mg/mL) for 5 h, disrupted in 200 μL dimethyl sulfoxide (DMSO), and measured at 590 nm using an ELISA reader (VERSA tunable microplate reader, Molecular Dynamics, Sunnyvale, CA, USA).
Colony formation assay
Cells were loaded into 6-well plates at a density of 2×103 cells per well. After 7-10 days, the colonies were stained with crystal violet for 10 min and rinsed with deionized distilled water, photographed using a Nikon D80 digital camera (10 Mega-pixel; Nikon Corp., Tokyo, Japan), and scored using AlphaEase FC software (Alpha Innotech Inc., San Leandro, CA, USA).
Wound healing migration assay
Cells were plated into 6-well until confluency and scratched using a sterile plastic 1 mL micropipette tip, then photographed using an inverted phase-contrast microscope every 12 h until complete healing.
Boyden chamber migration assay
Cells were seeded on 10 cm Petri dishes to 70-80% confluency and harvested with 0.1% trypsin (Cambrex, East Rutherford, NJ, USA). A total of 2.5×104 cells were loaded into 8-μm pore-size polycarbonated filters (Nucleopore Crop., Pleasanton, CA, USA) in a 48-well Boyden chamber (Neuro Probe Inc., Gaithersburg, MD, USA). The chemotactic migration of cells was induced by 10% fetal calf serum (FCS) in the lower chamber. After 12 h of incubation, the migrated cells were fixed with 100% ethanol, stained with Liu’s staining solution, then photographed and analyzed using an inverted phase-contrast microscope. Experiments were repeated in triplicate.
Cell synchronization by double thymidine block
Cells were seeded on 6-well plates at a density of 1×105 cells per well. After 16 h, the cells were treated with thymidine to a final concentration of 2 mM and then cultured for 18 h in tissue culture incubator. After 18 h, thymidine was removed by 1X PBS and then cell samples were incubated for 9 h with fresh media in tissue culture incubator. After 9 h, they were then treated with thymidine to a final concentration of 2 mM and incubated for 15 h in tissue culture incubator. Finally, thymidine was removed by 1X PBS, the samples cultured for 10 h in fresh media, then analyzed quantitatively by flow cytometry using a FACS CaliburTM flow cytometer (Beckton Dickinson, San Jose, CA, USA).
Western blot analysis
Total protein from cultured cells was harvested by 1X Trypsin-EDTA Solution and lysed by RIPA lysis buffer (0.1% SDS, 0.5% sodium deoxycholate, 150 mM NaCl, 50 mM Tris, pH 8.0, 1% NP-40). The protein extracts (25 µg) were separated by 10% SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane. The antibodies used were mouse monoclonal anti-HK1 (G-1, Santa Cruz Biotechnology, Santa Cruz, CA, USA), goat polyclonal anti-HK2 (C-14, Santa Cruz Biotechnology), sheep polyclonal anti-LDH5 (ab9002, Abcam), rabbit polyclonal anti-cyclin A (H-432, Santa Cruz Biotechnology), rabbit polyclonal anti-cyclin B1 (H-433, Santa Cruz Biotechnology), rabbit monoclonal anti-cyclin D1 (ab134175, Abcam), rabbit polyclonal anti-cyclin E (M-20, Santa Cruz Biotechnology), rabbit polyclonal anti-IL-6 (ab6672, Abcam), rabbit polyclonal anti-GAPDH (ab9485, Abcam).
pH measurement in conditioned medium
Cells were plated into 6-well plates at a density of 2×105 cells per well. The cells were cultured until confluency and then incubated in fresh culture medium for 24 h. The colors and pH values of the conditioned media were photographed using a Nikon D80 digital camera (10 Mega-pixel; Nikon Corp., Tokyo, Japan) and measured using a FE20-FiveEasyTM pH meter (Mettler Toledo, Zchwerzenbach, Switzerland).
Analysis of survival rate
The correlations between the RPPH1 expression and overall survival rates of breast and cervical cancer samples were evaluated by gene chip and RNA sequencing data in Kaplan-Meier (KM) plotter (http://kmplot.com) database [27]. The high and low RPPH1 expression groups were divided according to the “auto select best cutoff” methodology in the website.
Statistical analysis
Data were expressed as mean ± standard deviation. Student’s t-test was used for analysis of difference between each experimental and control group. P-value of < 0.05 was considered to be significant.
Results
RPPH1 expression in various cancers and effect on prognosis
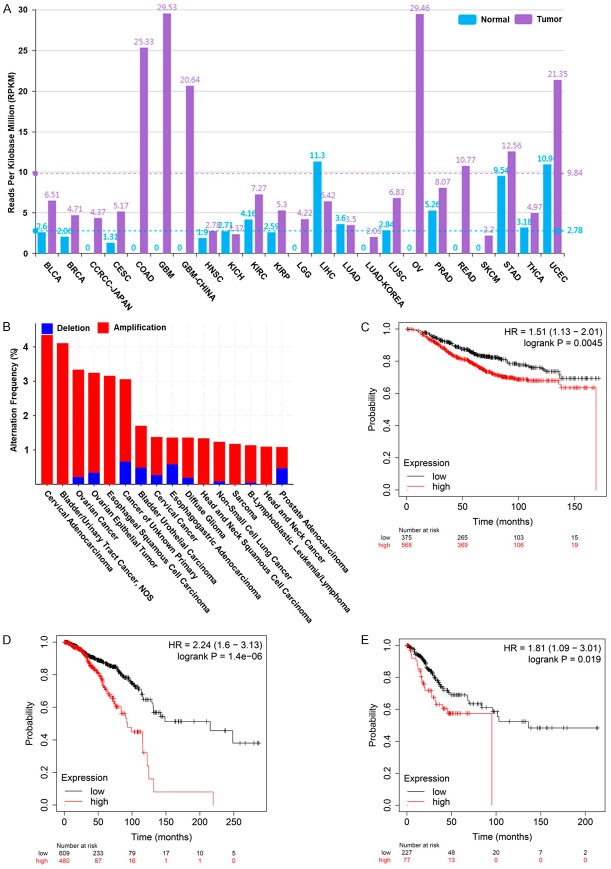
To investigate the expression of RPPH1 in human cancer cells, the level of RPPH1 in various human cancers was analyzed through the lncRNASNP2 website (http://bioinfo.life.hust.edu.cn/lncRNASNP). The data indicated that RPPH1 is more highly expressed in most human cancers, including BRCA (breast invasive carcinoma) (4.71 RPKM) and CESC (cervical squamous cell carcinoma) (5.17 RPKM), than in normal tissue types (Figure 1A); additionally, the RPPH1 gene is amplified in several cancers, especially cervical cancer, with a 4.35% alteration frequency, as determined from a cBioPortal website (http://www.cbioportal.org) analysis (Figure 1B). To understand the correlation between the level of RPPH1 expression and the prognosis of patients, we analyzed the prognosis of patients with breast cancer through the Kaplan-Meier Plotter website (https://kmplot.com/analysis/). The results indicated that breast cancer patients with a high RPPH1 expression level had poor prognosis according to the data from gene chip and RNA sequencing (Figure 1C and 1D). Similar results were observed when analyzing patients with cervical cancer (Figure 1E).
Figure 1.
The status of RPPH1 in clinical samples. A. RPPH1 expression in multiple human cancers and normal tissues (data obtained from the lncRNASNP2 website http://bioinfo.life.hust.edu.cn). The blue and purple bars represent normal tissues and cancers respectively. RPPH1 data were obtained from the BLCA: bladder urothelial carcinoma, BRCA: breast invasive carcinoma, CCRCC: clear cell renal cell carcinoma, CESC: cervical squamous cell carcinoma, COAD: colon adenocarcinoma, GBM: glioblastoma multiforme, HNSC: head and neck squamous cell carcinoma, KICH: kidney chromophobe, KIRC: kidney renal clear cell carcinoma, KIRP: kidney renal papillary cell carcinoma, LGG: brain lower grade glioma, LIHC: liver hepatocellular carcinoma, LUAD: lung adenocarcinoma, LUSC: lung squamous cell carcinoma, OV: ovarian serous cystadenocarcinoma, PRAD: prostate adenocarcinoma, READ: rectum adenocarcinoma, SKCM: skin cutaneous melanoma, STAD: stomach adenocarcinoma, THCA: thyroid carcinoma, UCEC: uterine corpus endometrial carcinoma. B. The genomic mutation rate of the RPPH1 gene in various cancers. Different cancer types are distinguished according to the status of RPPH1 genomic loci. The red and blue bars represent the amplification and deletion of genomic loci respectively. The alteration frequency was analyzed from the cBioportal website (http://www.cbioportal.org). C. Kaplan-Meier analysis estimates of the overall survival plots for RPPH1 expression of gene chip data in breast cancer. D. Kaplan-Meier analysis estimates of the overall survival plots for RPPH1 expression of RNA sequencing data in breast cancer. E. Kaplan-Meier analysis estimates of the overall survival plots for RPPH1 expression of gene chip data in cervical cancer. The survival estimation was analyzed from the Kaplan-Meier Plotter website (https://kmplot.com/analysis/).
Construction and evaluation of differentially expressed genes in RPPH1 knockdown cancer cells
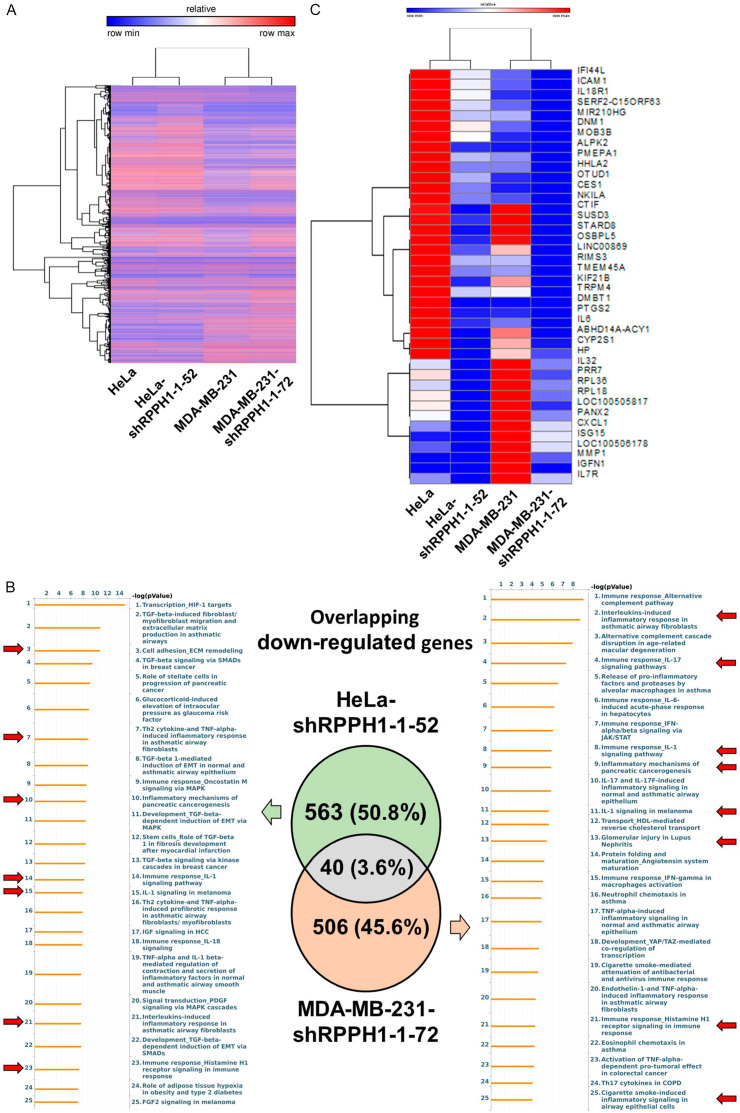
Breast cancer cell line MDA-MB-231 and cervical cancer cell line HeLa were chosen to investigate the molecular mechanism of RPPH1. RPPH1 knockdown cancer cells were generated using the RNAi-mediated gene silencing technique. The expression levels of RPPH1 in RPPH1 knockdown clones including HeLa-shRPPH1-1-39 and HeLa-shRPPH1-1-52 as well as MDA-MB-231-shRPPH1-1-72 and MDA-MB-231-shRPPH1-1-78 were significantly inhibited compared with those in mock- and vector-transfected cells, as determined via RT-PCR analysis (Figure 2A). To further investigate the difference in transcriptional profiles between RPPH1 knockdown cells and mock cells, HeLa-shRPPH1-1-52 cells and MDA-MB-231-shRPPH1-1-72 cells were selected and analyzed using RNA sequencing. The study flowchart is shown in Figure 2B.
Figure 2.
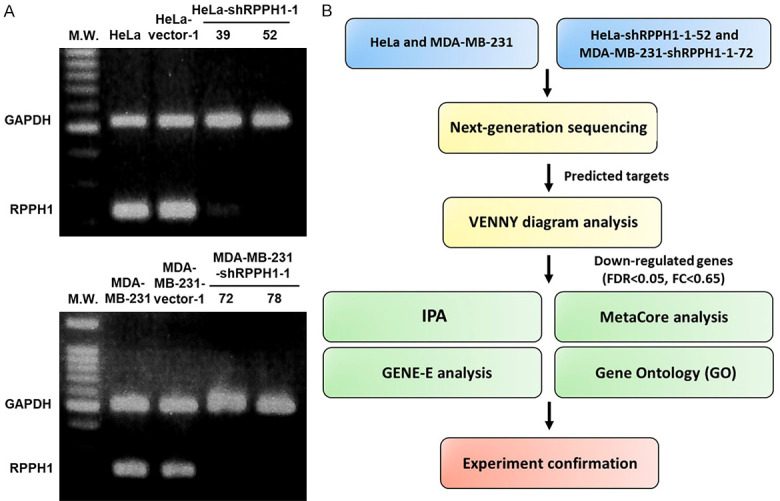
The effect of RPPH1 knockdown in MDA-MB-231 and HeLa cells. A. Detection of RPPH1 expression in untransfected mock cells, vector control, and RPPH1 knockdown clones HeLa-shRPPH1-1-39, HeLa-shRPPH1-1-52, MDA-MB-231-shRPPH1-1-72 and MDA-MB-231-shRPPH1-1-78. One-step RT-PCR was performed with 0.5 μg of total RNAs isolated from cells as indicated. B. Flowchart of study design. HeLa-shRPPH1-1-52, MDA-MB-231-shRPPH1-1-72 and mock cells were deep-sequenced for RNA expression and analyzed using bioinformatics databases. Significantly down-regulated genes with false discovery rate < 0.05 and fold change < 0.65 were further analyzed using bioinformatics tools and confirmed using experimental evidence.
Identification of potential affected pathways in two RPPH1 knockdown cancer cells through bioinformatic analysis
The sequencing results of the four cancer cells (HeLa, HeLa-shRPPH1-1-52, MDA-MB-231, and MDA-MB-231-shRPPH1-1-72) were analyzed using CLC Genomics Workbench, with a total of 15,972 expressed genes being identified and clustered using GENE-E software (Figure 3A). Based on the criteria of a fold change (FC) of < 0.65 and a false discovery rate (FDR) of < 0.05, 603 and 546 down-regulated genes with differential expression were identified in HeLa-shRPPH1-1-52 and MDA-MB-231-shRPPH1-1-72 respectively. These differentially expressed genes (DEGs) were input into the MetaCore and IPA databases for gene ontology enrichment analysis. Various enriched pathways involved in the immune response and cancerogenesis in RPPH1 knockdown cells were observed. Four identical enriched pathways including “Immune responses_IL-1 signaling pathway”; “IL-1 signaling in melanoma”; “Interleukins-induced inflammatory responses in asthmatic airway fibroblast”; and “Immune response_Histamine H1 receptor signaling in immune response” in both cell lines were observed (Figure 3B). In addition, only 40 overlapping DEGs in two RPPH1 knockdown cells were found and then clustered using GENE-E software (Figure 3C). According to metabolic pathways analysis of MetaCore database, the results showed that the (L)-valine, methionine, (L)-threonine, (L)-alanine and (L)-proline pathways were affected in RPPH1 knockdown cells through the down-regulation of TRPM4, ICAM1 and ISG15 (Figure 4A), suggesting that these metabolic pathways were common pathways involved in different RPPH1 knockdown cancer cells.
Figure 3.
The results of RNA sequencing and bioinformatic analysis. A. The heat maps of 15,972 gene expression levels in RPPH1 knockdown cells and mock cells were analyzed using CLC Genomics Workbench and visualized using GENE-E software. Red color indicates up-regulated gene; blue color indicates down-regulated gene. Genes and samples were clustered using the one minus Pearson correlation method. B. Differentially expressed genes (DEGs) were identified. A total of 603 and 546 genes in the orange circle and the green circle indicate down-regulated DEGs in Hela-shRPPH1-1-52 and MDA-MB-231-shRPPH1-1-72 respectively. Potential enriched pathways were analyzed using MetaCore databases. Red arrow indicates the pathway involving tumor progression and immune responses. C. Gene expression using heatmap visualization of 40 overlapping down-regulated DEGs in RPPH1 knockdown cells by GENE-E software. Red color indicates up-regulated gene; blue color indicates down-regulated gene. Genes and samples were clustered using the one minus Spearman’s rank correlation method.
Figure 4.
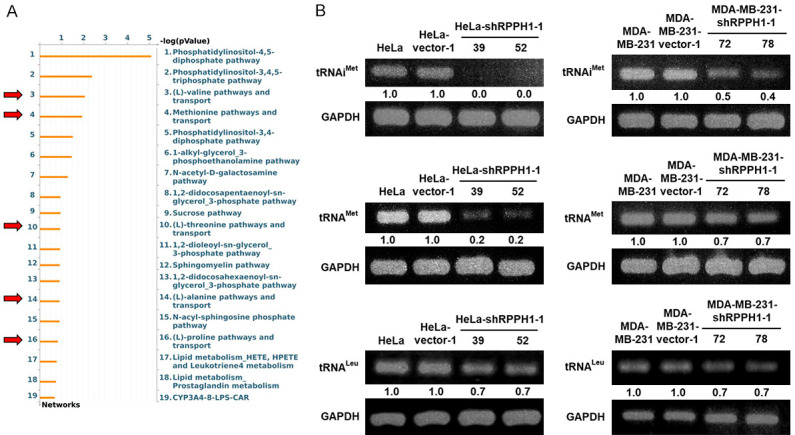
Several amino acid pathways and tRNA production are down-regulated in RPPH1 knockdown cells. A. The metabolic pathways of 40 down-regulated DEGs in RPPH1 knockdown cells were analyzed using MetaCore bioinformatics tools. Red arrow indicates the pathway associated with amino acid metabolism. B. RPPH1 knockdown cells of tRNA expression in HeLa and MDA-MB-231 cells. One-step RT-PCR was performed with 0.5 μg of total RNAs isolated from cells as indicated.
Inhibition of RPPH1 expression decreases mature tRNA generation
RNase P is a ribonuclease that cleaves the 5’ sequence of pre-tRNAs to assist tRNA maturation [11,28]. Among its subunits, RPPH1 plays an important role in catalytic activity [16,17]. To elucidate whether RPPH1 inhibition affects the activity of RNase P in cervical cancer and breast cancer, several tRNA products were measured using RT-PCR. The RPPH1 knockdown cells showed a large reduction in tRNAiMet, tRNAMet, and tRNALeu generation compared to that in the mock- and vector-transfected cells, especially tRNAiMet (Figure 4B). These results confirmed that the Cancer inhibition of RPPH1 expression decreases the production of mature tRNAs and several amino acid pathways in cervical and breast cancer cells.
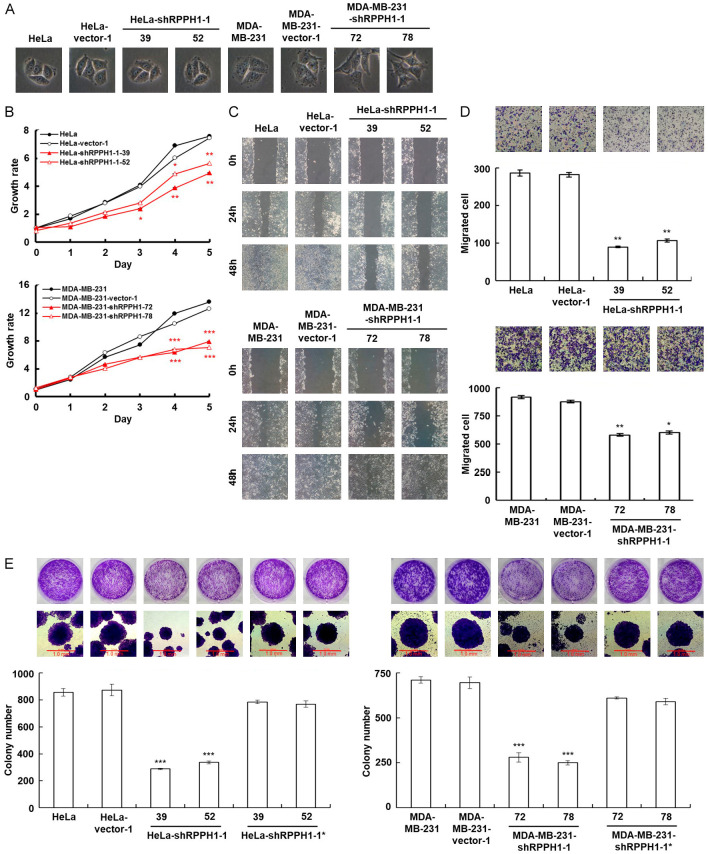
Down-regulation of RPPH1 expression decreases tumor cell proliferation and migration
The cell morphology of RPPH1-knockdown cancer cells is shown in Figure 5A. Because several down-regulated genes, including GRO-1, GRO-2, MMP1, COX-2, IL-6, and ICAM-1, are involved in tumor-related pathways (Table 1), the tumor progression-associated properties were further determined. To investigate the effect of RPPH1 knockdown on tumor proliferation, the RPPH1 knockdown cells were examined using the MTT cell growth assay. The RPPH1 knockdown cells showed a slower growth rate than that of the mock- and vector-transfected cells (Figure 5B). Moreover, slower cell motility was observed in RPPH1 knockdown cells compared to the mock- and vector-transfected cells in wound-healing migration assay (Figure 5C). The number of migrated RPPH1 knockdown cells decreased in the short term of the Boyden chamber migration assay compared to that of the mock- and vector-transfected cells (Figure 5D). Colony formation assay is a technique to evaluate the capability of clonal expansion. The number and size of colonies greatly decreased in RPPH1 knockdown cells compared to the mock- and vector-transfected cells (Figure 5E). These results showed that the presence of RPPH1 in both cancer cell lines contributed to proliferation and migration capabilities.
Figure 5.
Inhibition of RPPH1 decreases cancer cell proliferation and migration. A. The cell morphology of RPPH1 knockdown HeLa and MDA-MB-231 cells (600× magnification). B. MTT cell growth assay of RPPH1 knockdown HeLa and MDA-MB-231 cells. Cells (as indicated) were cultured in 96-well plates and then subjected to the MTT assay. *P < 0.05, **P < 0.01, ***P < 0.001. C. Wound healing migration assay of RPPH1 knockdown cells. Cells (as indicated) were cultured until reaching confluency and then subjected to the scratched wound-healing migration assay; wounds were then imaged after incubation for various time periods (as labeled, 100× magnification). D. Boyden chamber migration assay of RPPH1 knockdown cells. Cells (as indicated) were loaded into Boyden chambers and incubated for 6 h. The migrated cells were stained, imaged, and enumerated (100× magnification). *P < 0.05, **P < 0.01. E. Colony formation assay of RPPH1 knockdown HeLa and MDA-MB-231 cells. Cells (as indicated) were cultured in 6-well plates for 6 days. The grown colonies were stained and scored (100× magnification). ***P < 0.001.
Table 1.
Enrichment by pathway maps
| Maps | Gene Ratio | p-value | FDR | Gene |
|---|---|---|---|---|
| IL-1 signaling in melanoma | 6/42 | 1.097E-10 | 4.092E-08 | GRO-1, GRO-2, MMP-1, COX-2 (PTGS2), IL-6, ICAM1 |
| Interleukins-induced inflammatory response in asthmatic airway fibroblasts | 5/35 | 4.636E-09 | 8.647E-07 | GRO-1, GRO-2, COX-2 (PTGS2), IL-6, ICAM1 |
| Immune response_IL-1 signaling pathway | 6/82 | 6.981E-09 | 8.680E-07 | GRO-1, MMP-1, COX-2 (PTGS2), IL-6, ICAM1, MMP-13 |
| Immune response_IL-17 signaling pathways | 5/60 | 7.564E-08 | 7.053E-06 | GRO-1, MMP-1, COX-2 (PTGS2), IL-6, ICAM1 |
| Inflammatory mechanisms of pancreatic cancerogenesis | 5/67 | 1.326E-07 | 9.892E-06 | GRO-1, COX-2 (PTGS2), IL-6, ICAM1, IL-32 (NK4) |
| Cigarette smoke-induced inflammatory signaling in airway epithelial cells | 4/36 | 5.394E-07 | 3.353E-05 | COX-2 (PTGS2), IL-6, ICAM1, sICAM1 |
| Glomerular injury in Lupus Nephritis | 5/92 | 6.547E-07 | 3.489E-05 | GRO-1, GRO-2, MMP-1, IL-6, ICAM1 |
| Release of pro-inflammatory factors and proteases by alveolar macrophages in asthma | 4/44 | 1.231E-06 | 5.738E-05 | GRO-1, GRO-2, MMP-1, IL-6 |
| Immune response_Histamine H1 receptor signaling in immune response | 4/47 | 1.611E-06 | 6.677E-05 | MMP-1, IL-6, ICAM1, MMP-13 |
| Activation of TNF-alpha-dependent pro-tumoral effect in colorectal cancer | 4/49 | 1.909E-06 | 7.120E-05 | GRO-1, COX-2 (PTGS2), IL-6, ICAM1 |
| Immune response_IL-18 signaling | 4/63 | 5.274E-06 | 1.788E-04 | IL-18R1, COX-2 (PTGS2), IL-6, ICAM1 |
| TNF-alpha and IL-1 beta-mediated regulation of contraction and secretion of inflammatory factors in normal and asthmatic airway smooth muscle | 4/65 | 5.979E-06 | 1.859E-04 | GRO-1, GRO-2, COX-2 (PTGS2), IL-6 |
RPPH1 knockdown cells promote G1 arrest through down-regulation of cyclin D1
In the MTT cell growth assay, the RPPH1 knockdown cells showed a slower growth rate than that of the mock- and vector-transfected cells; additionally, the RPPH1 knockdown cells took more than 48 hours longer to reach confluency in the wound-healing migration assay compared to the control group. To investigate the relevance of a slow growth rate and cell cycle progression in RPPH1 knockdown cells, the cells were measured at different time points using a flow cytometer, with the RPPH1 knockdown cells exhibiting a significant delay in the G1 phase (Figure 6A). The cell-cycle-related proteins were down-regulated in RPPH1 knockdown cells compared to the mock- and vector-transfected cells, especially the cyclin D1 proteins (Figure 6B). These results indicated that the knockdown of RPPH1 expression affects cell cycle progression mainly through the down-regulation of cyclin D1.
Figure 6.
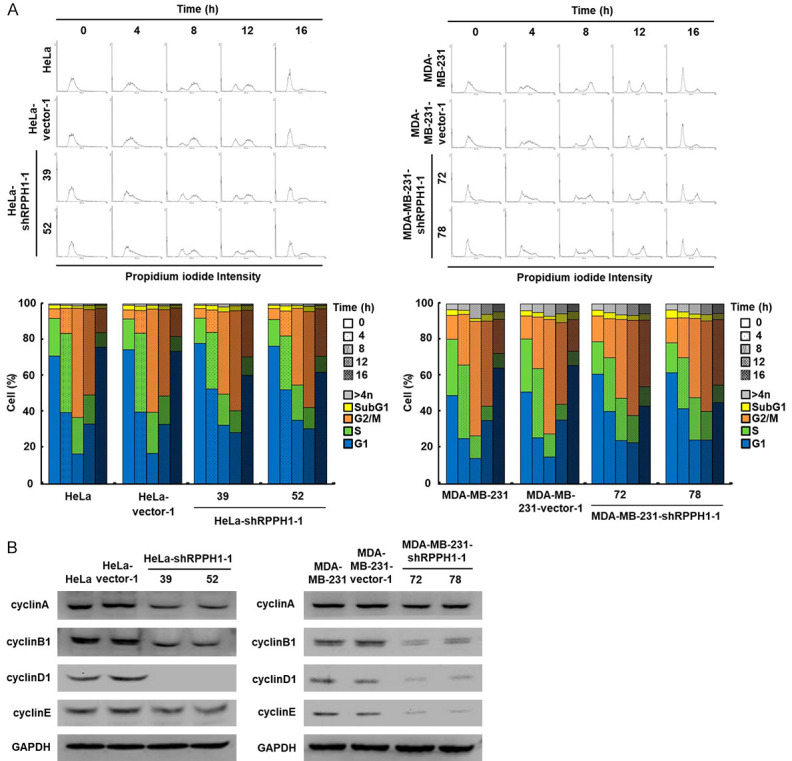
Knockdown of RPPH1 decreases cell cycle progression in HeLa cells. A. Cell cycle progression of RPPH1 knockdown HeLa and MDA-MB-231 cells. Cells (as indicated) were treated with thymidine and then fixed in ethanol and stained with PI. The fluorescence intensities of cells in each cell cycle were examined and analyzed using a flow cytometer. B. Western blotting of proteins involved in cell cycle progression in RPPH1 knockdown HeLa and MDA-MB-231 cells. Total proteins isolated from cells were hybridized with antibodies against cell-cycle-related proteins. The level of GAPDH served as a control for protein loading.
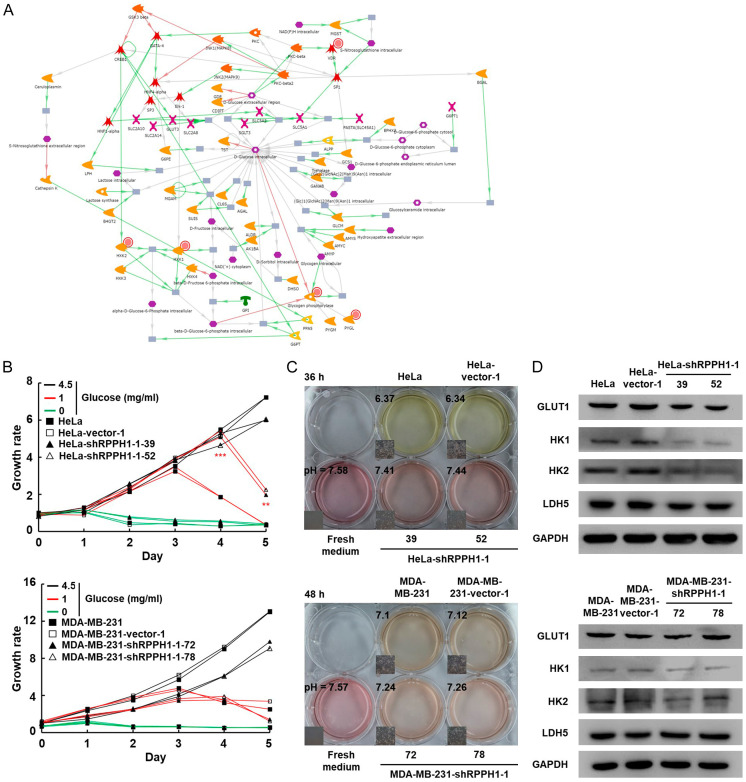
Down-regulated RPPH1 affects glycolytic pathway in cervical cancer
Glucose is an important energy source for cancer cells [29,30]. Malignant cells tend to ferment glucose into lactic acid, even in the presence of abundant oxygen [31,32]. The expression levels of HK1 (hexokinase 1), HK2 (hexokinase 2), and glycogen phosphorylase were significantly down-regulated after silencing RPPH1 in HeLa cell. The network of the glucose pathway in RPPH1 knockdown HeLa cells is shown in Figure 7A. To confirm the bioinformatic analysis, cancer cells were cultured in a medium media containing different concentrations of glucose, and based on cell growth rate, the glucose consumption rate of RPPH1 knockdown HeLa cells was slower than that of the mock- and vector-transfected cells (Figure 7B). In addition, the lactate production decreased compared to that of the mock- and vector-transfected cells, as determined by measuring the pH in the conditioned media of the cells (Figure 7C). To confirm the alteration of the glycolytic pathway in RPPH1 knockdown cells, protein expression was examined using western blotting with the results showing large decreases in GLUT1 (glucose transporter 1), HK1, and HK2 proteins in RPPH1 knockdown cells compared to the mock- and vector-transfected cells (Figure 7D). The reduced LDH5 protein expression in RPPH1 knockdown HeLa cells is consistent with the significantly decreased pH in the conditioned media of the cells (Figure 7C and 7D); in contrast, the glucose consumption rate and lactate production did not reveal significant differences between the RPPH1 knockdown MDA-MB-231 and the mock- and vector-transfected cells (Figure 7B and 7C). Although the protein expressions of HK-1 and HK-2 in RPPH1 knockdown MDA-MB-231 cells were also slightly lower than that in mock- and vector-transfected cells (Figure 7D), the difference in protein expression was not as significant as that in HeLa cells. Taken together, these results show that RPPH1 knockdown affects the glycolytic pathway through the down-regulation of glycolysis-related proteins in the HeLa cells but not in the MDA-MB-231 cells.
Figure 7.
RPPH1 knockdown HeLa cell down-regulates the expression of glucose pathway-related genes. A. Networks of glucose pathway in RPPH1 knockdown HeLa cells determined using MetaCore database. Red circles indicate down-regulated proteins. Green arrows indicate activation of the corresponding proteins; red arrow indicates inhibition; grey arrow indicates unspecified function. B. Glucose dependence assay of RPPH1 knockdown Hela (upper panel) and MDA-MB-231 (lower panel). Cells (as indicated) were cultured in 96-well plates containing different glucose concentrations and examined using the MTT cell growth assay. **P < 0.01, ***P < 0.001. C. pH of conditioned medium cultured with RPPH1 knockdown cells. Cells (as indicated) were cultured until confluency was achieved and then incubated in fresh medium. The color and pH value of the conditioned media were imaged and measured. D. Western blotting of proteins involved in the glycolytic pathway in RPPH1 knockdown cells. Total proteins purified from cells were blotted with antibodies against GLUT1, HK1, HK2, and LDH5.
Discussion
RPPH1 is an RNA unit of RNase P that provides the catalytic activity of tRNA processing [16,17]. According to the data of clinical tumor samples obtained from databases, relatively high RPPH1 expression was found in breast and cervical cancers; additionally, high genomic mutation frequency was observed in cervical cancer, with high expression of RPPH1 being associated with poor prognosis in both breast and cervical cancers. The results of RNA sequencing identified hundreds of genes with differential expression levels in MDA-MB-231 and HeLa cells respectively. Gene ontology analysis showed that various enriched pathways involved in the immune response and cancerogenesis in RPPH1 knockdown cells the two types of cancer cells. Metabolic pathway analysis showed that several pathways were involved in amino acids pathways in RPPH1 knockdown cells based on 40 overlapping DEGs. In MDA-MD-231 and HeLa cells, decreased tRNA production was observed in RPPH1 knockdown cells. Silencing RPPH1 attenuated the capacity of cell proliferation and metastasis and caused G1 phase cell cycle arrest in both cell types. Finally, we demonstrated that RPPH1 knockdown affects the glycolytic pathway in HeLa cells.
tRNA is an indispensable substance in protein synthesis. Dysregulation of tRNA production contributes to several types of human diseases, including cancer [33]. The initiator methionine tRNA (tRNAiMet) shows the most significant decrease among tRNA in RPPH1 knockdown cells. Initiator tRNA is a critical element in the initiation of protein synthesis; it reads the start codon to allow the initiating ribosome to correct the location and initiate translation. Several studies have shown that initiator tRNA is deregulated in specific cancers and is closely related to the development of cancer [10,34,35] as increased tRNAiMet levels drive tumor migration and angiogenesis by enhancing the secretion of type II collagen-rich extracellular matrix from stromal fibroblasts while increasing cell metabolic activity by changing the global tRNA expression profile [34]. These results indicate that RPPH1 could promote tumor progression by regulating the expression level of intracellular tRNA; additionally, RPPH1 knockdown cells were observed to down-regulate gene expressions of the amino acid pathway and transport, including that of TRPM4, ICAM1, and ISG15. These gene are involved in the pathways of valine, methionine, threonine, alanine and proline. In this study, we demonstrated that the inhibition of RPPH1 not only decreases mature tRNA generation but down-regulates several genes involved in the amino acid pathway and transportation as well.
Several studies have shown that chronic inflammation is closely related to the progression of cancer [31,32]. Specific signaling pathways of the immune system are up-regulated to support tumor proliferation, metabolism, and metastasis [36,37]. RPPH1 has been shown to increase the expression of TNF-α and Mcp-1 in diabetic nephropathy (DN) by directly interacting with the DN-related factor galectin-3 (Gal-3) signaling pathway to promote inflammation [38]. In our study, several inflammation-related factors were found to be down-regulated in RPPH1 knockdown cells, including IL-6, GRO-1, ICAM1, and COX-2. IL-6 is a major pro-inflammatory cytokine that transiently responds to tissue injuries and infections. This protein also plays a critical role in the cancer-related inflammatory response and promotes the differentiation and metastasis of cancer cells [39,40]. In four identical enriched pathways in breast and cervical cancer cells, IL-1 might play a critical role in inflammatory pathways of RPPH1 knockdown cells. This IL-1 cytokine family consists of 11 members [41], where IL-1α and IL-1β are associated with pro-tumorigenesis function through promoting angiogenesis, inflammation and induction of myeloid derived suppressor cells [42]. The current study was not able to examine whether RPPH1 knockdown cells affected the behavior of peripheral immune cells or stromal cells in the tumor microenvironment because this was an in vitro study, although it would be worthwhile to study this issue in the future.
The inhibition of RPPH1 has been shown to decrease cell proliferation and affect the cell cycle [19,43]. We confirmed that RPPH1 inhibition promotes cell cycle arrest in the G1 phase mainly through the down-regulation of cyclin D1. Cyclin D1 is a key regulator of cell cycle progression, which drives the G1-to-S phase progression [44,45]. Dysregulation of cyclin D1 has been linked to tumor development and progression [46,47]. Although the other subunits of RNase P Rpp25 and Rpp20 are reported to regulate rRNA gene transcription during G1/S phase [48], the mechanism of how RPPH1 affects the cell cycle through cyclin D1 remains unclear. IL-6 has been shown to induce cell proliferation by the activation of the JAK2/STAT3/cyclin D1 pathway [49-51] while decreased IL-6 expression is observed in RPPH1 knockdown cells. We speculate that IL-6 could serve as a regulator linking RPPH1, cyclin D1 expression and cell cycle G1 arrest.
Dysregulation of the glycolytic pathway is a well-known strategy for cancer cells to obtain energy [52,53]. Most cancer cells increase the expression of specific glycolytic enzymes to promote tumor proliferation and metastasis [54,55]. In our study, we found a unique regulatory mechanism for the glycolytic pathway in RPPH1 knockdown HeLa cells. Several glycolytic enzymes in RPPH1 knockdown HeLa cells are down-regulated, especially hexokinase, which is the first enzyme of the glycolytic pathway, transferring an inorganic phosphate group from ATP to glucose to form glucose-6-phosphate.
Hexokinase 2 (HK2) has been reported to be up-regulated in various cancers and to enhance the rate of glycolysis [56,57], and importantly, its overexpression might increase the resistance of cancer cells to apoptosis [58]. This enzyme plays a key role in cancer cell survival and development. The current study does not reveal how RPPH1 regulates the expression of hexokinase in HeLa cells. Several studies have shown that IL-6 enhances glycolysis through the up-regulation of HK2 expression [59-61], and this evidence might reveal a possible correlation among RPPH1, IL-6 and the glycolytic pathway in HeLa cells.
This study is the first to comprehensively investigate the inhibitory effect of RPPH1 in cancer cells, although several limitations remain; for instance, the effect of overexpressing RPPH1 or silencing of RPPH1 with different expression levels on tumor growth and cellular transcriptional profiles was not determined, and only one cell line was used in each type of cancer, while detailed regulatory mechanisms between down-regulated RPPH1 and cancerogenesis phenotypes and the impact of RPPH1 inhibition in vivo have also not been investigated. These issues require further investigation.
Conclusions
In this study, we demonstrated that the inhibition of RPPH1 in human cervical (HeLa) and breast (MDA-MB-231) cancer cells decreased mature tRNA generation. Bioinformatics analysis suggested that various cancer progression-related pathways were affected by silencing RPPH1. Compared to the parental MDA-MB-231 and HeLa cells, silencing RPPH1 inhibited tumor progression; additionally, our results showed that RPPH1 affected the glycolytic pathway only in HeLa cells. Current data suggest that the effects of RPPH1 knockdown on cancer cells are multidimensional, and imply that different types of cancer cells could have their own unique regulatory pathways. The results provide novel insight into the role of RPPH1 in tumor development.
Acknowledgements
This study was supported by the Ditmanson Medical Foundation Chia-Yi Christian Hospital (Grant No. R111-074).
Disclosure of conflict of interest
None.
References
- 1.Kim SY. Cancer energy metabolism: shutting power off cancer factory. Biomol Ther (Seoul) 2018;26:39–44. doi: 10.4062/biomolther.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dell’ Antone P. Energy metabolism in cancer cells: how to explain the Warburg and Crabtree effects? Med Hypotheses. 2012;79:388–392. doi: 10.1016/j.mehy.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Santos M, Fidalgo A, Varanda AS, Oliveira C, Santos MAS. tRNA deregulation and its consequences in cancer. Trends Mol Med. 2019;25:853–865. doi: 10.1016/j.molmed.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Hyeon DY, Kim JH, Ahn TJ, Cho Y, Hwang D, Kim S. Evolution of the multi-tRNA synthetase complex and its role in cancer. J Biol Chem. 2019;294:5340–5351. doi: 10.1074/jbc.REV118.002958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou S, Van Bortle K. The Pol III transcriptome: basic features, recurrent patterns, and emerging roles in cancer. Wiley Interdiscip Rev RNA. 2023;14:e1782. doi: 10.1002/wrna.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres AG, Reina O, Stephan-Otto Attolini C, Ribas de Pouplana L. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc Natl Acad Sci U S A. 2019;116:8451–8456. doi: 10.1073/pnas.1821120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018;28:395–404. doi: 10.1038/s41422-018-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165:1416–1427. doi: 10.1016/j.cell.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birch J, Clarke CJ, Campbell AD, Campbell K, Mitchell L, Liko D, Kalna G, Strathdee D, Sansom OJ, Neilson M, Blyth K, Norman JC. The initiator methionine tRNA drives cell migration and invasion leading to increased metastatic potential in melanoma. Biol Open. 2016;5:1371–1379. doi: 10.1242/bio.019075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karasik A, Fierke CA, Koutmos M. Interplay between substrate recognition, 5’ end tRNA processing and methylation activity of human mitochondrial RNase P. RNA. 2019;25:1646–1660. doi: 10.1261/rna.069310.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarrous N, Liu F. Human RNase P: overview of a ribonuclease of interrelated molecular networks and gene-targeting systems. RNA. 2023;29:300–307. doi: 10.1261/rna.079475.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaukat AN, Kaliatsi EG, Skeparnias I, Stathopoulos C. The dynamic network of RNP RNase P subunits. Int J Mol Sci. 2021;22:10307. doi: 10.3390/ijms221910307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abu-Zhayia ER, Khoury-Haddad H, Guttmann-Raviv N, Serruya R, Jarrous N, Ayoub N. A role of human RNase P subunits, Rpp29 and Rpp21, in homology directed-repair of double-strand breaks. Sci Rep. 2017;7:1002. doi: 10.1038/s41598-017-01185-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramanathan A, Weintraub M, Orlovetskie N, Serruya R, Mani D, Marcu O, Stepensky P, Weisblum Y, Djian E, Shaag A, Revel-Vilk S, Fried I, Kotler M, Rouvinski A, Wolf D, Elpeleg O, Jarrous N. A mutation in POLR3E impairs antiviral immune response and RNA polymerase III. Proc Natl Acad Sci U S A. 2020;117:22113–22121. doi: 10.1073/pnas.2009947117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mann H, Ben-Asouli Y, Schein A, Moussa S, Jarrous N. Eukaryotic RNase P: role of RNA and protein subunits of a primordial catalytic ribonucleoprotein in RNA-based catalysis. Mol Cell. 2003;12:925–935. doi: 10.1016/s1097-2765(03)00357-5. [DOI] [PubMed] [Google Scholar]
- 17.Kikovska E, Svard SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Acad Sci U S A. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang ZX, Liu HS, Wang FW, Xiong L, Zhou C, Hu T, He XW, Wu XJ, Xie D, Wu XR, Lan P. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019;10:829. doi: 10.1038/s41419-019-2077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Tang L. Inhibition of breast cancer cell proliferation and tumorigenesis by long non-coding RNA RPPH1 down-regulation of miR-122 expression. Cancer Cell Int. 2017;17:109. doi: 10.1186/s12935-017-0480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Y, Sun Z, Jia H, Luo H, Ye X, Wu Q, Xiong Y, Zhang W, Wan J. Rpph1 upregulates CDC42 expression and promotes hippocampal neuron dendritic spine formation by competing with miR-330-5p. Front Mol Neurosci. 2017;10:27. doi: 10.3389/fnmol.2017.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Li Y, Cheng H. Circ-RPPH1 knockdown retards breast cancer progression via miR-328-3p-mediated suppression of HMGA2. Clin Breast Cancer. 2022;22:e286–e295. doi: 10.1016/j.clbc.2021.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Feng H, Sun SZ, Cheng F, Zhang NQ. Mediation of circ_RPPH1 on miR-146b-3p/E2F2 pathway to hinder the growth and metastasis of breast carcinoma cells. Aging (Albany NY) 2021;13:20552–20568. doi: 10.18632/aging.203439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi L, Sun B, Yang B, Lu S. circRNA RPPH1 facilitates the aggravation of breast cancer development by regulating miR-542-3p/ARHGAP1 pathway. Cancer Biother Radiopharm. 2022;37:708–719. doi: 10.1089/cbr.2020.4381. [DOI] [PubMed] [Google Scholar]
- 25.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24:257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartkiewicz M, Gold H, Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989;3:488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- 27.Gyorffy B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience. 2023;45:1889–1898. doi: 10.1007/s11357-023-00742-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarrous N. Roles of RNase P and its subunits. Trends Genet. 2017;33:594–603. doi: 10.1016/j.tig.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Cordier-Bussat M, Thibert C, Sujobert P, Genestier L, Fontaine E, Billaud M. Even the Warburg effect can be oxidized: metabolic cooperation and tumor development. Med Sci (Paris) 2018;34:701–708. doi: 10.1051/medsci/20183408017. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz L, Seyfried T, Alfarouk KO, Da Veiga Moreira J, Fais S. Out of Warburg effect: an effective cancer treatment targeting the tumor specific metabolism and dysregulated pH. Semin Cancer Biol. 2017;43:134–138. doi: 10.1016/j.semcancer.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Tekade RK, Sun X. The Warburg effect and glucose-derived cancer theranostics. Drug Discov Today. 2017;22:1637–1653. doi: 10.1016/j.drudis.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orellana EA, Siegal E, Gregory RI. tRNA dysregulation and disease. Nat Rev Genet. 2022;23:651–664. doi: 10.1038/s41576-022-00501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke CJ, Berg TJ, Birch J, Ennis D, Mitchell L, Cloix C, Campbell A, Sumpton D, Nixon C, Campbell K, Bridgeman VL, Vermeulen PB, Foo S, Kostaras E, Jones JL, Haywood L, Pulleine E, Yin H, Strathdee D, Sansom O, Blyth K, McNeish I, Zanivan S, Reynolds AR, Norman JC. The initiator methionine tRNA drives secretion of type II collagen from stromal fibroblasts to promote tumor growth and angiogenesis. Curr Biol. 2016;26:755–765. doi: 10.1016/j.cub.2016.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavon-Eternod M, Gomes S, Rosner MR, Pan T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA. 2013;19:461–466. doi: 10.1261/rna.037507.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murata M. Inflammation and cancer. Environ Health Prev Med. 2018;23:50. doi: 10.1186/s12199-018-0740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P, Sun Y, Peng R, Chen W, Fu X, Zhang L, Peng H, Zhang Z. Long non-coding RNA Rpph1 promotes inflammation and proliferation of mesangial cells in diabetic nephropathy via an interaction with Gal-3. Cell Death Dis. 2019;10:526. doi: 10.1038/s41419-019-1765-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakraborty C, Sharma AR, Sharma G, Lee SS. The interplay among miRNAs, major cytokines, and cancer-related inflammation. Mol Ther Nucleic Acids. 2020;20:606–620. doi: 10.1016/j.omtn.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose-John S. Local and systemic effects of interleukin-6 (IL-6) in inflammation and cancer. FEBS Lett. 2022;596:557–566. doi: 10.1002/1873-3468.14220. [DOI] [PubMed] [Google Scholar]
- 41.Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018;281:8–27. doi: 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun R, Gao DS, Shoush J, Lu B. The IL-1 family in tumorigenesis and antitumor immunity. Semin Cancer Biol. 2022;86:280–295. doi: 10.1016/j.semcancer.2022.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Ham IH, Oh HJ, Jin H, Bae CA, Jeon SM, Choi KS, Son SY, Han SU, Brekken RA, Lee D, Hur H. Targeting interleukin-6 as a strategy to overcome stroma-induced resistance to chemotherapy in gastric cancer. Mol Cancer. 2019;18:68. doi: 10.1186/s12943-019-0972-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Topacio BR, Zatulovskiy E, Cristea S, Xie S, Tambo CS, Rubin SM, Sage J, Koivomagi M, Skotheim JM. Cyclin D-Cdk4,6 drives cell-cycle progression via the retinoblastoma protein’s C-terminal helix. Mol Cell. 2019;74:758–770. e754. doi: 10.1016/j.molcel.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong P, Zhang C, Parker BT, You L, Mathey-Prevot B. Cyclin D/CDK4/6 activity controls G1 length in mammalian cells. PLoS One. 2018;13:e0185637. doi: 10.1371/journal.pone.0185637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med (Berl) 2016;94:1313–1326. doi: 10.1007/s00109-016-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shan YS, Hsu HP, Lai MD, Hung YH, Wang CY, Yen MC, Chen YL. Cyclin D1 overexpression correlates with poor tumor differentiation and prognosis in gastric cancer. Oncol Lett. 2017;14:4517–4526. doi: 10.3892/ol.2017.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reiner R, Krasnov-Yoeli N, Dehtiar Y, Jarrous N. Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I. PLoS One. 2008;3:e4072. doi: 10.1371/journal.pone.0004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo J, Yan R, He X, He J. Constitutive activation of STAT3 and cyclin D1 overexpression contribute to proliferation, migration and invasion in gastric cancer cells. Am J Transl Res. 2017;9:5671–5677. [PMC free article] [PubMed] [Google Scholar]
- 50.Kurosaka M, Machida S. Interleukin-6-induced satellite cell proliferation is regulated by induction of the JAK2/STAT3 signalling pathway through cyclin D1 targeting. Cell Prolif. 2013;46:365–373. doi: 10.1111/cpr.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Won C, Lee CS, Lee JK, Kim TJ, Lee KH, Yang YM, Kim YN, Ye SK, Chung MH. CADPE suppresses cyclin D1 expression in hepatocellular carcinoma by blocking IL-6-induced STAT3 activation. Anticancer Res. 2010;30:481–488. [PubMed] [Google Scholar]
- 52.Lin J, Xia L, Liang J, Han Y, Wang H, Oyang L, Tan S, Tian Y, Rao S, Chen X, Tang Y, Su M, Luo X, Wang Y, Wang H, Zhou Y, Liao Q. The roles of glucose metabolic reprogramming in chemo- and radio-resistance. J Exp Clin Cancer Res. 2019;38:218. doi: 10.1186/s13046-019-1214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bose S, Le A. Glucose metabolism in cancer. Adv Exp Med Biol. 2018;1063:3–12. doi: 10.1007/978-3-319-77736-8_1. [DOI] [PubMed] [Google Scholar]
- 54.Jiang SH, Dong FY, Da LT, Yang XM, Wang XX, Weng JY, Feng L, Zhu LL, Zhang YL, Zhang ZG, Sun YW, Li J, Xu MJ. Ikarugamycin inhibits pancreatic cancer cell glycolysis by targeting hexokinase 2. FASEB J. 2020;34:3943–3955. doi: 10.1096/fj.201901237R. [DOI] [PubMed] [Google Scholar]
- 55.Liu C, Wang X, Zhang Y. The roles of HK2 on tumorigenesis of cervical cancer. Technol Cancer Res Treat. 2019;18:1533033819871306. doi: 10.1177/1533033819871306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nong S, Han X, Xiang Y, Qian Y, Wei Y, Zhang T, Tian K, Shen K, Yang J, Ma X. Metabolic reprogramming in cancer: mechanisms and therapeutics. MedComm (2020) 2023;4:e218. doi: 10.1002/mco2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garcia SN, Guedes RC, Marques MM. Unlocking the potential of HK2 in cancer metabolism and therapeutics. Curr Med Chem. 2019;26:7285–7322. doi: 10.2174/0929867326666181213092652. [DOI] [PubMed] [Google Scholar]
- 58.Zhang XY, Zhang M, Cong Q, Zhang MX, Zhang MY, Lu YY, Xu CJ. Hexokinase 2 confers resistance to cisplatin in ovarian cancer cells by enhancing cisplatin-induced autophagy. Int J Biochem Cell Biol. 2018;95:9–16. doi: 10.1016/j.biocel.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 59.Chen X, Tian J, Su GH, Lin J. Blocking IL-6/GP130 signaling inhibits cell viability/proliferation, glycolysis, and colony forming activity in human pancreatic cancer cells. Curr Cancer Drug Targets. 2019;19:417–427. doi: 10.2174/1568009618666180430123939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ando M, Uehara I, Kogure K, Asano Y, Nakajima W, Abe Y, Kawauchi K, Tanaka N. Interleukin 6 enhances glycolysis through expression of the glycolytic enzymes hexokinase 2 and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3. J Nippon Med Sch. 2010;77:97–105. doi: 10.1272/jnms.77.97. [DOI] [PubMed] [Google Scholar]
- 61.Siu MKY, Jiang YX, Wang JJ, Leung THY, Han CY, Tsang BK, Cheung ANY, Ngan HYS, Chan KKL. Hexokinase 2 regulates ovarian cancer cell migration, invasion and stemness via FAK/ERK1/2/MMP9/NANOG/SOX9 signaling cascades. Cancers (Basel) 2019;11:813. doi: 10.3390/cancers11060813. [DOI] [PMC free article] [PubMed] [Google Scholar]