Abstract
Objective: The aim of this study was to identify the active components of Shengxian Decoction (SXT) and to elucidate the multi-component, multi-target, and multi-pathway regulatory mechanisms underlying the efficacy of SXT in treating lung adenocarcinoma (LUAD). Methods: The effects of SXT extract on proliferation, migration, and invasion capabilities of human LUAD cells were determined through 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), wound healing, and Transwell assays. High-Performance Liquid Chromatography (HPLC) was employed to pinpoint the primary active constituents of SXT. The SXT-active component-target-pathway network and protein-protein interaction (PPI) network were constructed based on network pharmacology. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using DAVID. The clinical significance of key targets was assessed using several external databases, and molecular docking confirmed the binding affinities between key targets and SXT active components. Results: SXT significantly inhibited the proliferation, migration and invasion of human LUAD cells. HPLC identified and quantified seven active SXT components. Network pharmacology yielded 197 targets, 128 signaling pathways, and 448 GO terms. The PPI network and external validation underscored 13 key targets significantly associated with the influence of SXT on LUAD progression. Molecular docking demonstrated strong interactions between SXT active components and key targets. Conclusion: SXT treats LUAD through a multifaceted approach involving various components, targets, and pathways. This research offers novel insights into the constituents and molecular mechanisms of SXT in LUAD therapy.
Keywords: Shengxian Decoction (SXT), lung adenocarcinoma (LUAD), active components, network pharmacology, molecular docking
Introduction
Epidemiological data indicate that lung cancer is the leading cause of cancer-related mortality globally, with five-year survival rates ranging from 4% to 17% [1,2]. According to China’s 2021 statistics, lung cancer was the most prevalent and lethal cancer in 2015, with non-small cell lung cancer (NSCLC) accounting for nearly 85% of cases [1,3]. Among NSCLC histological subtypes, lung adenocarcinoma (LUAD) is the most common.
Currently, LUAD patients may undergo surgical resection, thoracic radiotherapy, cytotoxic chemotherapy, immunotherapy, targeted therapy, or combinations thereof. However, these treatments can have adverse effects that restrict their application [4]. Traditional Chinese Medicine (TCM) has emerged as a viable alternative for LUAD treatment due to its efficacy and lower side effects [5]. Nevertheless, the complexity of TCM, involving multiple components and targets, poses challenges in elucidating its mechanisms of action. Advances in computational technology have facilitated network pharmacology and molecular docking to predict active components, key targets, and mechanisms of TCM action, significantly reducing drug development costs and offering new insights into the material basis and mechanisms of TCM [6-8].
Shengxian Decoction (SXT), a classic TCM formula commonly used for the syndrome of pectoral Qi sinking, is comprised of five herbs including Astragali Radix (A. radix), Anemarrhenae Rhizoma (A. rhizoma), Bupleuri Radix (B. radix), Platycodonis Radix (P. radix) and Cimicifugae Rhizoma (C. rhizoma) [9]. The constituent herbs, and phytochemicals they contain, have also been reported to be effective in cancer prevention and treatment, including LUAD. A. radix and B. radix, for example, have been used alongside radiotherapy to improve survival rates and lessen chemotherapy toxicity in lung cancer [10,11]. A. rhizoma and C. rhizoma extracts are rich in various active compounds with antitumor activity against the LUAD cell line A549 [12,13], and P. radix is a component of various TCM formulas for lung tumor treatment [14]. Furthermore, our earlier findings revealed that SXT serum inhibit A549 cell proliferation in vitro and that SXT administration significantly reduced tumor growth in A549 xenografts in nude mice [15]. However, the chemical composition of SXT and the possible mechanisms associated with its effect remain unclear.
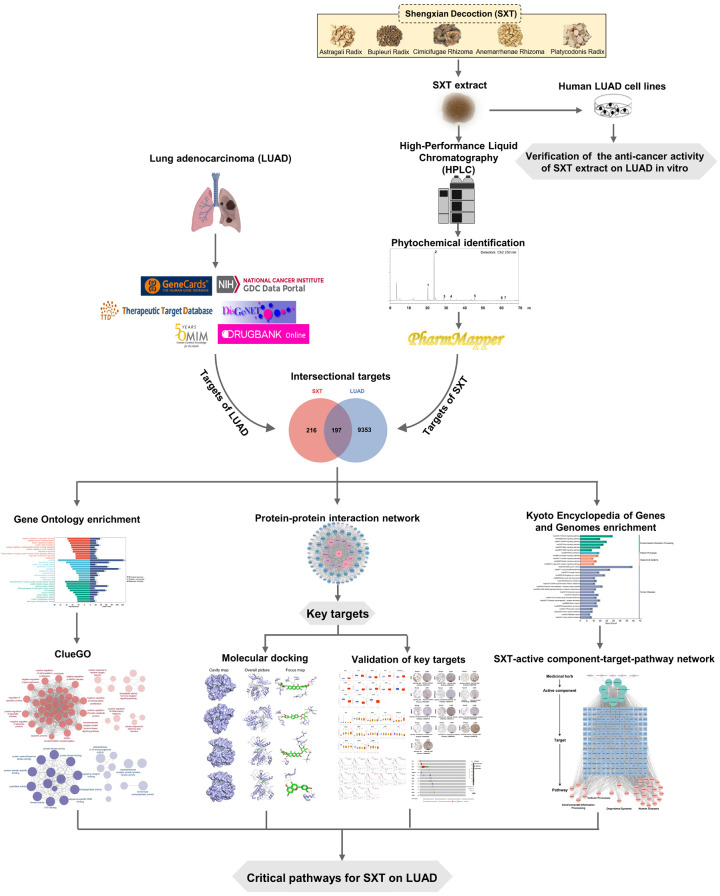
This study first assessed the inhibitory effects of SXT extract on human LUAD cells through in vitro assays. Subsequently, the primary active components of SXT were identified using High-Performance Liquid Chromatography (HPLC). The key targets of SXT active components for LUAD treatment were predicted using network pharmacology and validated with external databases. Lastly, the interactions between the active components of SXT and key targets in LUAD were determined through molecular docking. The methodologies employed in this study are depicted in Figure 1.
Figure 1.
The schematic diagram of the network pharmacology of Shengxian Decoction (SXT) for lung adenocarcinoma (LUAD).
Materials and methods
Preparation of SXT extract
The botanical drug mixture of A. radix, A. rhizoma, B. radix, P. radix and C. rhizoma (12:6:3:3:2) was soaked in distilled water (v:v = 1:12) for 2 h and then boiled for 30 min. The decoction was filtrated and the resulting residues were subsequently collected for two additional decoctions, each with 8 volumes of distilled water and boiled for 30 min. The sum of the decoctions was condensed using reduced pressure (-0.07~-0.08 MPa) at 70°C. The resultant concentrate was freeze-dried and labeled as SXT extract (extraction rate: 38.95%). The obtained SXT extract was stored at -20°C before chemical and pharmacological studies.
Cell line and culture
Human LUAD cell lines (A549 and NCl-H1299) and human embryonic lung fibroblasts WI-38 were obtained from the American Type Culture Collection (ATCC). The cells were cultured in Ham’s F-12K medium (A549 cells), RPMI-1640 medium (NCl-H1299 cells), and MEM medium (WI-38 cells), respectively. The aforementioned medium items were acquired from the vendor Procell, and all of the mediums were supplemented with 10% fetal bovine serum (Biosharp, China) and 1% penicillin-streptomycin solution (Procell, China). The cells were subjected to incubation in a controlled environment consisting of 5% CO2/95% air at 37°C within a humidified incubator.
Cell viability assay and morphology observations
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to assess the cell viability. The cells in logarithmic growth phase were inoculated in 96-well plates at a density of 2 × 103 cells well-1 and allowed to adhere overnight. Cells were then treated with culture medium containing 0, 2, 4, 6, 8, and 10 mg/mL SXT extract. After 24 h or 48 h of treatment, cells were treated with 5 mg/mL MTT solution, incubated at 37°C for 4 h, and then the MTT solution was replaced with 150 μL of DMSO for complete dissolution of formazan crystals. The absorbance (OD) at 490 nm was measured by SpectraMax 190 absorbance plate reader (Molecular Devices, USA).
For morphological observation, cells (1 × 105 cells well-1) were seeded in 6-well plates and allowed to adhere overnight. The cells were subsequently treated with fresh medium containing 0, 2, 4, 6, 8, and 10 mg/mL SXT for 48 h. Cell morphology was recorded under a ×100 microscope using DM IL LED inverted laboratory microscope (Leica Microsystems, Germany).
Wound healing assay
A wound healing assay was performed to evaluate the effect of SXT extract on A549 and NCl-H1299 cell motility. Cells were seeded in 6-well plates at a density of 1 × 105 cells well-1 and cultured overnight. The cell monolayer was scraped with 200 μL pipette tips at 90% confluency, and cell debris was removed by washing with PBS twice. Cells were then cultured in serum-reduced media (2% FBS) with 5 mg/mL SXT extract. Cell migration into the scratched region was examined and recorded at 0, 24, and 48 h and wound closure speed was assessed using ImageJ software.
Cell invasion assay
The Transwell invasion assay was used to assess the effect of SXT extract on the invasiveness of A549 and NCl-H1299 cells. A Transwell chamber insert (BD Corporation, Franklin Lakes, NJ, United States) with 300 μg/mL Matrigel was used in this study. 2 × 104 cells suspended in 100 μL of serum-free culture medium containing 5 mg/mL SXT extract were added into the top invasion insert, while 600 μL of culture medium with 10% FBS was loaded into the well below. After 24 h of incubation, invasive cells that passed through the filter were fixed with methanol and stained with 1% crystal violet. The inserts were rinsed, air-dried, viewed, and photographed. Invasion activity was assessed by counting at least five random fields in each chamber at ×200 magnification.
Preparation of SXT extract sample and mixed-standard solutions
SXT extract (6 mg/mL) and the stock solutions of seven standard substances (neomangiferin, mangiferin, calycosin 7-O-glucoside, isoferulic acid, luteolin, formononetin and saikosaponin A) were carefully prepared in specific concentrations with methanol. Mixed-standard solutions were prepared by mixing the individual reference stock solutions and the concentrations for each standard substance are shown in Table 1. All samples were filtered through 0.22 μm microporous membrane prior to HPLC injection.
Table 1.
Quantitative analysis of SXT extract by HPLC
| Active component | Content (mg/g) | Regression equation | Correlation coefficient (R2) | Linear range (μg/mL) |
|---|---|---|---|---|
| Neomangiferin | 1.0270 ± 0.0222 | Y = 35038X-916.22 | 1 | 0.3125-40 |
| Mangiferin | 3.5320 ± 0.0775 | Y = 73591X+58.217 | 0.9999 | 0.3125-40 |
| Calycosin 7-O-Glucoside | 0.4264 ± 0.0143 | Y = 42342X+3521.4 | 1 | 0.3125-40 |
| Isoferulic acid | 0.2700 ± 0.0180 | Y = 59103X-2313.6 | 1 | 0.3125-40 |
| Luteolin | 0.2958 ± 0.0188 | Y = 66345X-5221.5 | 1 | 0.3125-40 |
| Formononetin | 0.0454 ± 0.0012 | Y = 112880X+2208.6 | 1 | 0.1-40 |
| Saikosaponin A | 6.4690 ± 0.1189 | Y = 585.38X+265.33 | 0.9997 | 2.5-320 |
Note: mean ± SD; n = 6; SXT, Shengxian Decoction; HPLC, High-Performance Liquid Chromatography.
Quantification of main active components in SXT by HPLC
The chromatographic study of SXT extract was performed using an LC-2030C HPLC (Shimadzu, Japan). Separation was accomplished by using an Agilent ZORBAX SB-C18 (4.6 × 250 mm, 5 μm) column (Agilent Technologies, USA). The mobile phase was composed of acetonitrile (solvent A) and 0.2% formic acid aqueous solution (solvent B) with gradient elution: 0~5 min, 99% B; 5~20 min, 99%~83% B; 20~25 min, 83%~80% B; 25~33 min, 80%~77% B; 33~40 min, 77%~70% B; 40~50 min, 70%~67% B; 50~65 min, 67%~50% B; 65~70 min, 50%~30% B. The flow rate was maintained at 0.8 mL/min, while the column temperature was set to 30°C and the injection volume was 10 μL. The discovery of the peak occurred at a wavelength of 250 nm.
The experiment involved generating a standard curve using a concentration gradient. This was done to evaluate the linear correlation between the concentration of the analyte (X) and the corresponding peak areas obtained (Y). An external standard method was used to determine the amount of each detected compound. The results were expressed as mg/g dry weight.
Statistical analysis
In the quantitative identification of SXT extract by HPLC, six replicate determinations of SXT extract were performed, and the other experiments were performed in triplicates. All of the results were presented as the mean ± standard deviation (SD). One-way ANOVA was used for multiple comparisons, and the Tukey method was used to compare any two groups of data. Significant differences were determined at *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. All data were analyzed using GraphPad Prism 9 (San Diego, CA, United States).
Target prediction of SXT active components
PharmMapper [16-18] (http://lilab-ecust.cn/pharmmapper/index.html) matches active component with the internal pharmacophores model database for reverse pharmacophores to identify potential drug targets to obtain the corresponding targets of the active components. The Structure-Data File of SXT active components were searched in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) then entered the PharmMapper to obtain the predicted genes for SXT active components. The gene symbol names were converted by UniProt (https://www.uniprot.org/) database, and duplicates were removed.
LUAD-related targets prediction
From the transcriptome expression profiles of LUAD retrieved from The Cancer Genome Atlas Program (TCGA) database (http://cancergenome.nih.gov/), samples of RNA expression data were extracted, which included 59 normal samples and 539 LUAD tumor samples. Later, the data were processed using the edge R package, and **P ≤ 0.01 and |log2FC| > 2 were used as screening criteria to analyze the above differentially expressed genes. To improve the comprehensiveness and reliability of data collection on targets relevant to this disease, genes data from five online platforms were supplemented. LUAD-related targets were retrieved from GeneCards (https://www.genecards.org/), DisGeNET (https://www.disgenet.org/), Online Mendelian Inheritance in Man (OMIM, https://omim.org/), Therapeutic Target Database (TTD, http://db.idrblab.net/ttd/) and DrugBank (https://www.drugbank.ca/) with “LUAD” and “lung cancer” as the keyword for searching. These disease genes were then compared with LUAD differentially expressed genes obtained through the TCGA database so that all genes obtained were associated with LUAD progression. All the genes in each database were combined together and then duplicates were removed. SXT active component targets and LUAD targets were inputted to draw Venn diagram, and the common targets were collected.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
GO and KEGG enrichment analysis of common targets of SXT active components for LUAD was performed using the DAVID (https://david.ncifcrf.gov/) database. The top 30 terms were plotted at Bioinformatics online tools (https://www.bioinformatics.com.cn). ClueGO is used as a Cytoscape plug-in to analyze the interrelationship of terms and functional groups in biological networks and to present them as a network diagram. The GO enrichment results obtained from the DAVID database were imported into ClueGO for classification and grouping by function, where Biological Processes (BPs) and Molecular Functions (MFs) of GO analysis were highlighted.
Constructed SXT-active component-target-pathway network
SXT-active component-target-pathway network was built with Cytoscape 3.9.0.
Construction of network
A protein-protein interaction (PPI) network was constructed by importing the common targets to the STRING database (https://www.string-db.org/) with a confidence score set to 0.4, the species restricted to “Homo sapiens”, and concealing free points. Cytoscape 3.9.0 was used to visualize the PPI network and topological analysis was performed by Cytoscape-CytoNCA [19]. Degree centrality (DC), Closeness centrality (CC), and Betweenness centrality (BC) are used to measure the importance of the protein in the PPI network. The top 13 targets with the largest DC value were recognized as key targets for SXT active components in the treatment of LUAD.
External validation of key targets
In this study, we use UALCAN (http://ualcan.path.uab.edu) [20,21] to explore the association of the key target mRNA expression in sample type and cancer stage. Significant difference was considered at *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. To investigate the expression of key targets in LUAD tissues, we analyzed the key targets in the Human Protein Atlas database (HPA, https://www.proteinatlas.org/) [22]. Overall survival (OS) analysis was carried out on the Kaplan-Meier mapper database (http://kmplot.com/analysis/) [23]. The hazard ratio (HR) and log rank P value were calculated and displayed in the graph, and a log rank *P ≤ 0.05 and **P ≤ 0.01 were set as a significant difference. The LUAD (TCGA, PanCancer Atlas) dataset containing 566 samples was selected for analysis in cBioPortal (https://www.cbioportal.org/). Information on the genetic alterations of the key targets was obtained.
Molecular docking validation of SXT active components with key targets
The Structure-Data Files of SXT active components were converted to the mol2 format using Chem3D software. The crystal structures of key targets were obtained from the RCSB protein databank (PDB, http://www.rcsb.org/). Water molecules, ligands and other hetero atoms were removed using PyMOL. AutoDock Vina was used to dock SXT active components on LUAD molecules to explore the binding pose and interactions between the drug and the target protein.
Results
Anti-proliferative effect of SXT extract on LUAD cells (A549 and NCl-H1299) and human embryonic lung fibroblasts (WI-38)
MTT assays were performed to investigate the anti-cancer potential of SXT extract against human LUAD cells (A549 and NCl-H1299) and human embryonic lung fibroblasts WI-38. As shown in Figure 2A, compared to control, SXT extract exhibited significant cytotoxic effects on both LUAD cell lines in a dose- and time-dependent manner, while the killing effect on WI-38 cells was relatively light. The IC50 values of SXT extract against A549/NCl-H1299/WI-38 cells were determined to be 7.19/11.76/19.27 mg/mL and 4.33/3.81/9.22 mg/mL after 24 h and 48 h of treatment, respectively. Microscopic examination of cell morphology (Figure 2B) highlighted the selective cytotoxic activity of SXT extract on LUAD cells juxtaposed with normal, non-cancerous cells within a concentration of 6 mg/mL over 24 h of treatment.
Figure 2.
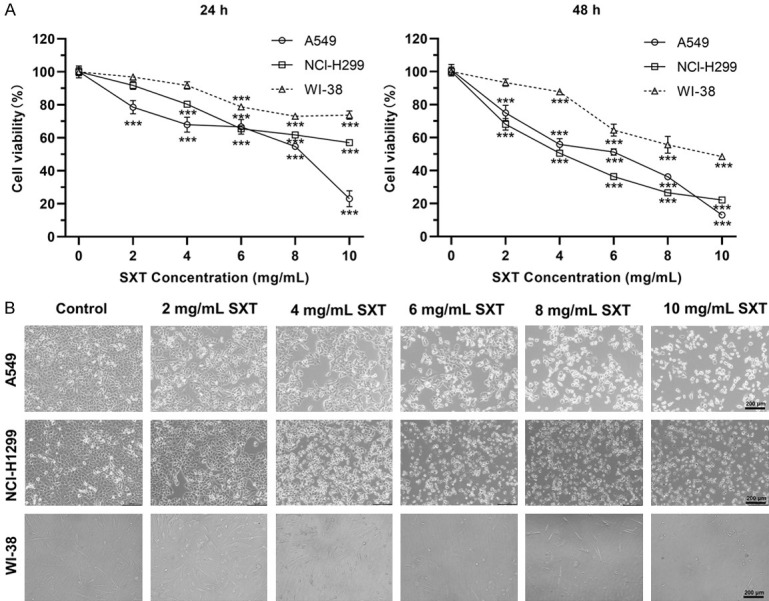
Cytotoxicity effects of Shengxian Decoction (SXT) extract on human lung adenocarcinoma cells and non-carcinoma cells. A. Cell viability assays of control and 24 h/48 h SXT extract-treated cells showing a dose-dependent cytotoxicity in A549 and NCI-H1299, while the killing effect on WI-38 cells was relatively light (mean ± SD; n = 3). ***P ≤ 0.001; B. Morphological changes in each group of cells after 48 h SXT extract treatment was observed under ×100 microscope (scale bar, 200 μm).
SXT extract inhibits migration and invasion of LUAD cells
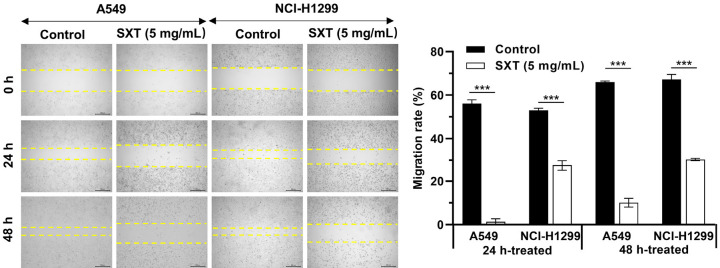
We next examined the effect of SXT extract on LUAD cell migration and invasion as these cells are known for their high metastatic potential. Figure 3 illustrates that 48 h of SXT extract (5 mg/mL) treatment significantly reduced A549 and NCl-H1299 cell migration by 55.87% and 36.77%, respectively. The effect was evident that SXT extract significantly (***P ≤ 0.001) inhibited the motility of A549 and NCI-H1299 cells migrating into the wound area compared to control cells. Figure 4 demonstrates that following 24 h of SXT extract (5 mg/mL) treatment, A549 and NCI-H1299 invasiveness significantly (***P ≤ 0.001) reduced by more than 90%. These findings indicate the potential for SXT extract to inhibit LAUD metastasis in vitro.
Figure 3.
Effect of SXT extract on the migration capacity of A549 and NCI-H1299 cells. Wound healing assay and its quantitation showing strong inhibition with 5 mg/mL of SXT extract treatment (magnification, ×40; scale bar, 200 μm). Quantitative data are represented as mean ± SD (n = 3). ***P ≤ 0.001. SXT, Shengxian Decoction.
Figure 4.
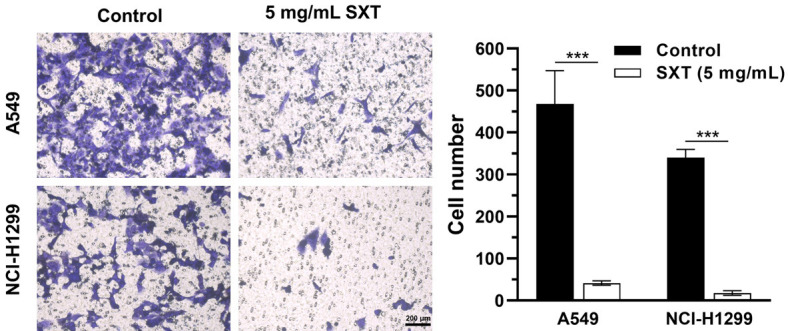
Effect of SXT extract on the invasion capacity of A549 and H1299 cells. Invasion assays and its quantitation showing strong inhibition with 24 h treatment of SXT extract (5 mg/mL) (magnification, ×200; scale bar, 200 μm). Quantitative data are represented as mean ± SD (n = 3). ***P ≤ 0.001. SXT, Shengxian Decoction.
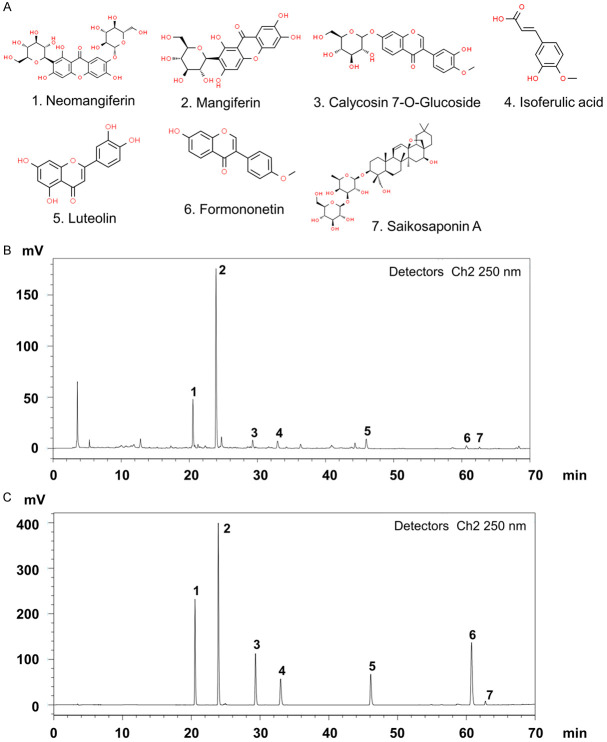
Identification of active components in SXT extract
The chemical fingerprint of the SXT extract was established using HPLC. Seven chemicals were used as standard samples for comparison with the SXT extract (Figure 5A). Analysis revealed several peaks (Figure 5B), indicating the presence of multiple substances. The active components of SXT were identified by comparing the retention times of major peaks with those of the standards. Peaks 1-7 in the SXT extract corresponded to neomangiferin, mangiferin, calycosin 7-O-glucoside, isoferulic acid, luteolin, formononetin, and saikosaponin A, respectively (Figure 5C). Table 1 lists the concentrations of these compounds in the SXT extract, with saikosaponin A (6.47 mg/g) and mangiferin (3.53 mg/g) being the most abundant. These results suggest that the prepared SXT extract contains various active components, whose mechanism of action in LUAD treatment will be further explored using a network pharmacology approach.
Figure 5.
Identification of anti-cancer phytochemicals in SXT extract by HPLC analysis. A. Potential chemical compounds in SXT extract responsible for anti-cancer effect; B. HPLC chromatogram of SXT extract; C. HPLC chromatogram of mixed-standard substance; Peak assignment as follows: 1. Neomangiferin, 2. Mangiferin, 3. Calycosin 7-O-glucoside, 4. Isoferulic acid, 5. Luteolin, 6. Formononetin, 7. Saikosaponin A. SXT, Shengxian Decoction; HPLC, High-Performance Liquid Chromatography.
Common targets of SXT active components and LUAD
The PharmMapper database provided 289 targets for neomangiferin, 288 for mangiferin, 286 for calycosin 7-O-glucoside, 260 for isoferulic acid, 288 for luteolin, 242 for formononetin, and 285 for saikosaponin A. After removing duplicates, 413 targets related to SXT active components were identified.
The TCGA database yielded 598 samples, comprising 539 tumor and 59 normal samples. Analysis revealed 7365 differentially expressed genes, visualized in volcano plots with genes of increased expression in red and decreased expression in blue in LUAD tumor samples (Figure 6A). From the GeneCards, DisGeNET, OMIM, TTD, and DrugBank databases, 2007, 58, 1060, 227, and 82 LUAD-related targets were identified, respectively. After eliminating duplicates, 9550 targets related to LUAD were compiled. Ultimately, 197 common targets were derived from the intersection of drug- and disease-related targets (Figure 6B), suggesting that SXT active components modulates LUAD progression through these targets.
Figure 6.
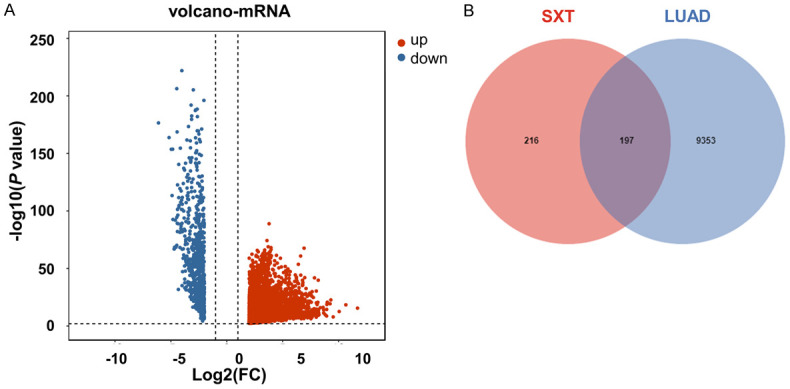
Target selection. A. Volcano maps of LUAD differential expressed genes. Red, upward; Blue, downward; B. Venn diagram of active component targets in SXT and LUAD targets. SXT, Shengxian Decoction; LUAD, lung adenocarcinoma.
Biological process and pathway enrichment analysis
GO enrichment analysis of 197 common targets were assessed by DAVID to explore the dynamic activity of SXT active components in LUAD using a filtering threshold of **P ≤ 0.01 (Supplementary Table 1). Analysis revealed 320 BPs including negative regulation of the apoptotic process and positive regulation of phosphoinositide 3-kinase (PI3K) signaling, 39 Cellular Components (CCs) involving the cytosol and extracellular region, and 89 MFs including transmembrane receptor protein tyrosine kinase activity. The top 10 related entries for BPs, MFs, and CCs are shown in a category summary chart (Figure 7A). To further analyze the interrelationship of terms and functional groups in biological networks, 320 BPs and 89 MFs were analyzed separately using ClueGO to enrich for entries with similar groups, displayed as a network graph. As illustrated in Figure 8, the analysis of the most populated group in the functional enrichment network map of MFs highlighted the inclusion of kinase-related molecular functions, such as protein kinase binding and protein serine/threonine kinase activity. Additionally, apoptosis-related biological processes constituted a significant portion of the enrichment network graph, including the regulation of apoptotic processes. This suggests that the SXT active components might regulate cellular behavior, especially apoptosis, by regulating protein kinases.
Figure 7.
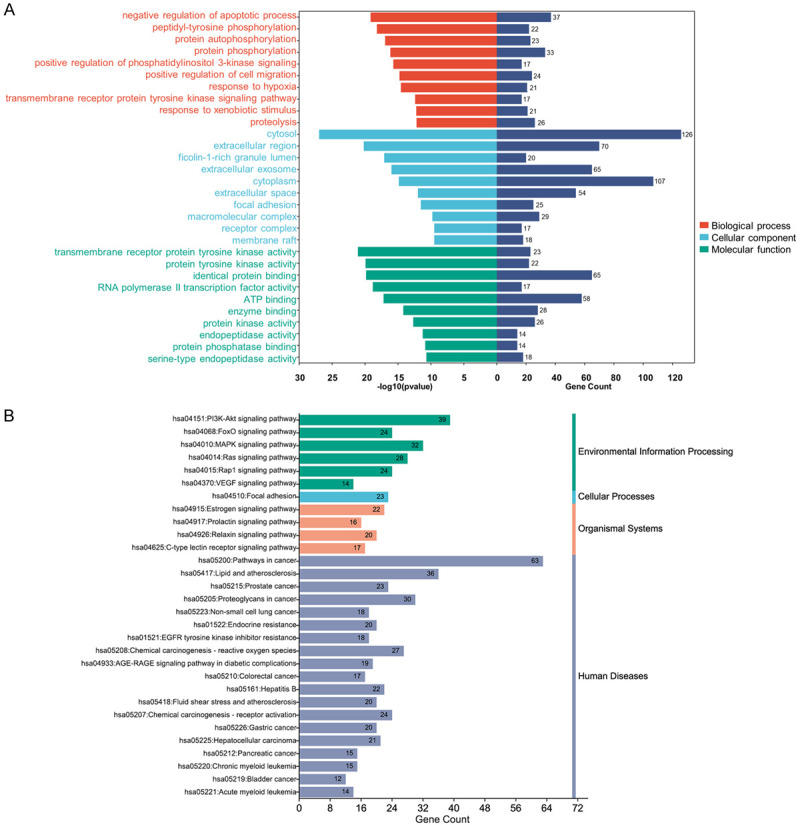
GO functional enrichment analysis and KEGG analysis of 197 common targets. A. The Bar chart of the top 10 terms of biological processes, cellular components and molecular functions extracted according to the P value based on GO enrichment analysis. In order to be able to show it in the figure, RNA polymerase II transcription factor activity and ligand-activated sequence-specific DNA binding to RNA polymerase II transcription factor activity were deleted in the GO analysis; B. The Bar chart of the top 30 terms extracted according to the P value based on KEGG enrichment analysis. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 8.
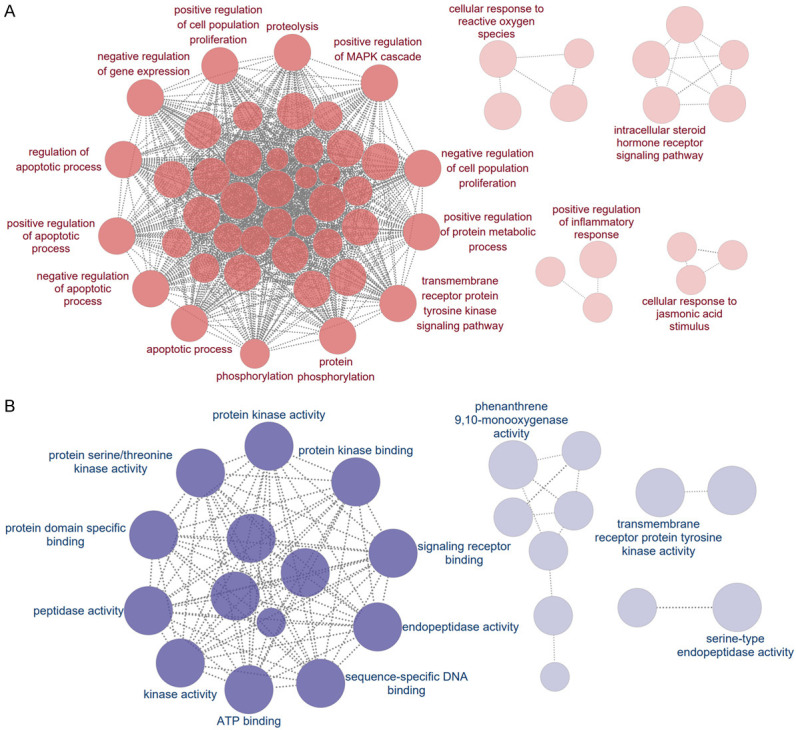
The interaction network of GO terms generated by the Cytoscape plug-in ClueGO. A. The functional networks with the most terms enriched by biological processes are shown; B. The functional networks with the most terms enriched by molecular functions are shown. The GO terms are rendered as nodes, with the size of the node representing importance. GO, Gene Ontology.
Significantly relevant signaling pathways of 197 common targets were explored with KEGG enrichment analyses using DAVID. This analysis identified 128 pathways showing significant differences (**P ≤ 0.01, Supplementary Table 2). The top 30 pathways with the highest -log10 (P value) were selected for visual analysis bioinformatics platform (Figure 7B). The results of the KEGG enrichment analysis included: 1) environmental information processing pathways such as the PI3K-protein kinase (Akt) signaling pathway, forkhead box protein O (FoxO) signaling pathway, and mitogen-activated protein kinase (MAPK) signaling pathway; 2) cellular processes involving focal adhesion; 3) organismal systems involving the estrogen signaling pathway; and 4) human diseases pathways including those in cancer, NSCLC, and epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor resistance. These findings suggest that SXT active components may play a significant role in intervening these pathways in LUAD.
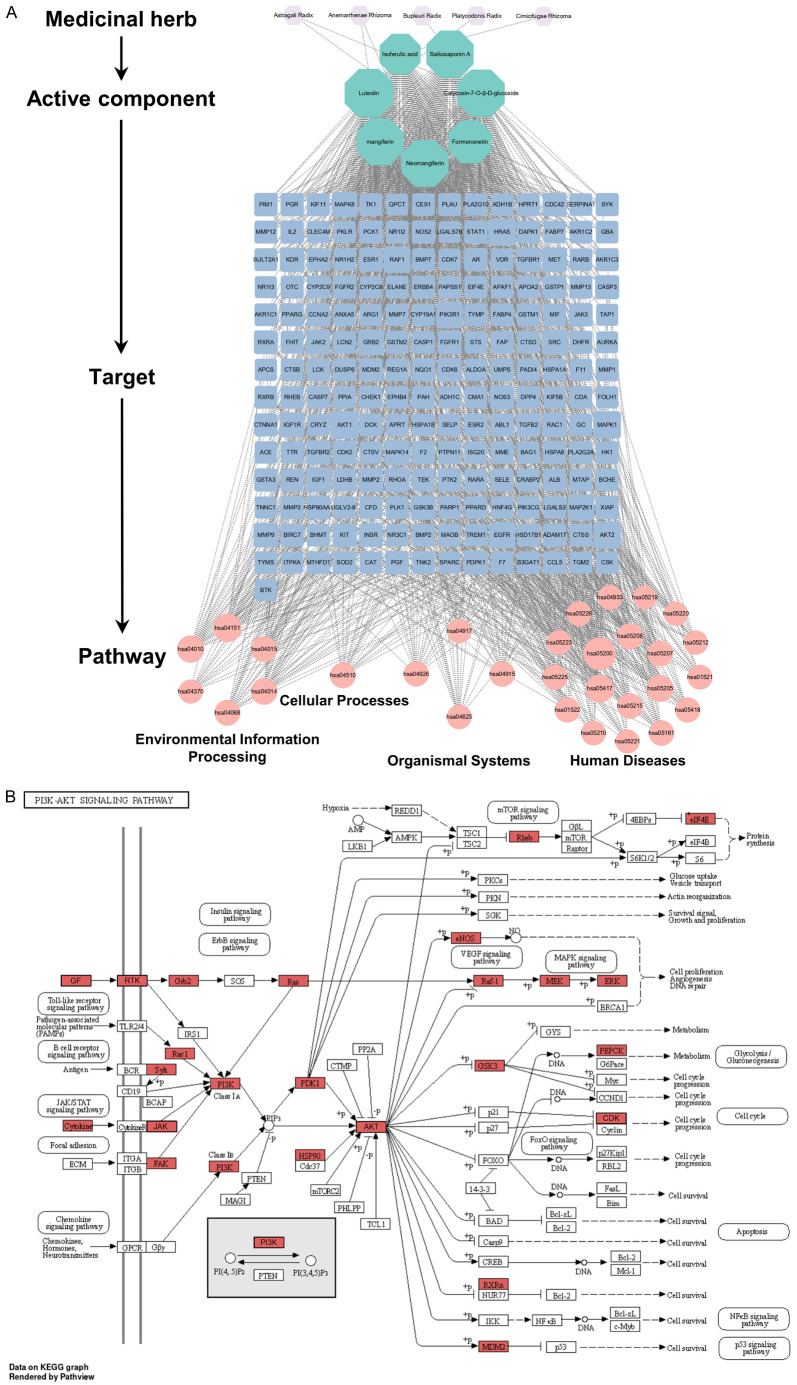
SXT-active component-target-pathway network
The SXT-active component-target-pathway network, depicted in Figure 9A, comprised 239 nodes, including 5 herbs, 7 active components, 197 common targets, and 30 KEGG enrichment pathway. Analysis of this network suggested that the therapeutic effect of SXT on LUAD is mediated through a complex interplay of multiple components, targets, and pathways. Notably, the PI3K-Akt signaling pathway emerged as the second most significantly enriched pathway after the generic cancer pathway, indicating its potential critical role in LUAD treatment (Figure 9B).
Figure 9.
Construction and pathway analysis of the SXT-active component-target-pathway. A. SXT-active component-target-pathway. The nodes with different colors and shapes represent the herbs, components, targets, and pathways, and an edge is an association between the nodes; B. PI3K-Akt signaling pathway. The red nodes represent potential targets of SXT active component for the treatment of LUAD, arrows represent the activation effect, T arrows represent the inhibition effect and segments show the activation effect or inhibition effect. SXT, Shengxian Decoction; PI3K, phosphoinositide 3-kinase; Akt, protein kinase; LUAD, lung adenocarcinoma.
PPI network construction and analysis of important targets
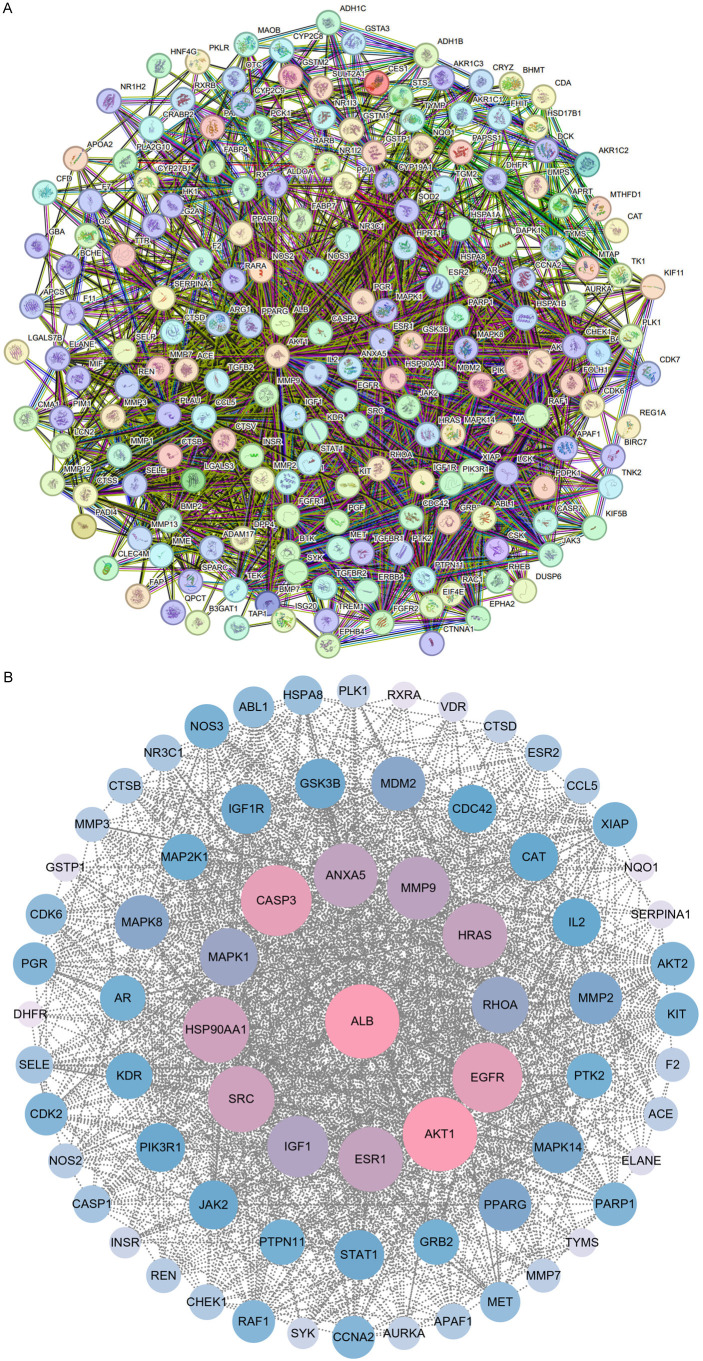
The PPI network, constructed using STRING for the 197 common targets, is shown in Figure 10A. It includes 193 nodes and 2372 edges, with 4 isolated nodes removed. The Cytoscape 3.9.0 platform was used to visualize the PPI networks. Significant targets were identified using CytoNCA based on their DC, CC, and BC values, with thresholds set at DC ≥ 19, CC ≥ 0.48, and BC ≥ 58.36, resulting in a total of 72 important targets. The DC value, indicative of the number of connections to a node, highlights the importance of a node within the network. The top 13 targets, having the highest degrees, were identified as key targets for SXT in LUAD treatment, underlining their critical role in the network (Figure 10B; Supplementary Table 3). Table 2 shows the 13 key targets and their specific values for DC, CC, and BC were also presented.
Figure 10.
PPI network to demonstrate SXT active component targets for LUAD treatment. A. PPI network diagram of common targets; B. The PPI network diagram of important targets above the median of degree centrality, closeness centrality and betweenness centrality, with the center and first circle representing the key targets. PPI, protein-protein interaction; SXT, Shengxian Decoction; LUAD, lung adenocarcinoma.
Table 2.
Ranking of key targets degree
| No. | Target | DC | CC | BC |
|---|---|---|---|---|
| 1 | ALB | 118 | 0.72 | 5615.69 |
| 2 | AKT1 | 111 | 0.70 | 2916.85 |
| 3 | EGFR | 98 | 0.66 | 2002.56 |
| 4 | SRC | 87 | 0.63 | 1008.92 |
| 5 | HSP90AA1 | 85 | 0.63 | 1505.92 |
| 6 | CASP3 | 85 | 0.64 | 1347.02 |
| 7 | MMP9 | 83 | 0.63 | 1548.34 |
| 8 | HRAS | 81 | 0.62 | 1040.10 |
| 9 | ESR1 | 79 | 0.61 | 1281.94 |
| 10 | IGF1 | 77 | 0.61 | 721.80 |
| 11 | RHOA | 68 | 0.58 | 593.45 |
| 12 | MAPK1 | 67 | 0.58 | 817.36 |
| 13 | ANXA5 | 61 | 0.58 | 250.16 |
Note: Degree centrality (DC) represents the number of links to the node; closeness centrality (CC) represents the distance between the individuals and all other peers in a network; betweenness centrality (BC) is used to measure how often a node lies on the shortest path between nodes in the network. ALB, albumin; AKT1, serine/threonine protein kinase 1; EGFR, epidermal growth factor receptor; SRC, steroid receptor coactivator; HSP90AA1, heat shock protein 90 alpha family class A member 1; CASP3, caspase-3; MMP9, matrix metalloproteinase-9; HRAS, harvey rat sarcoma viral oncogene homolog; ESR1, estrogen receptor 1; IGF1, insulin like growth factor 1; RHOA, ras homolog gene family member A; MAPK1, mitogen-activated protein kinase 1; ANXA5, annexin A5.
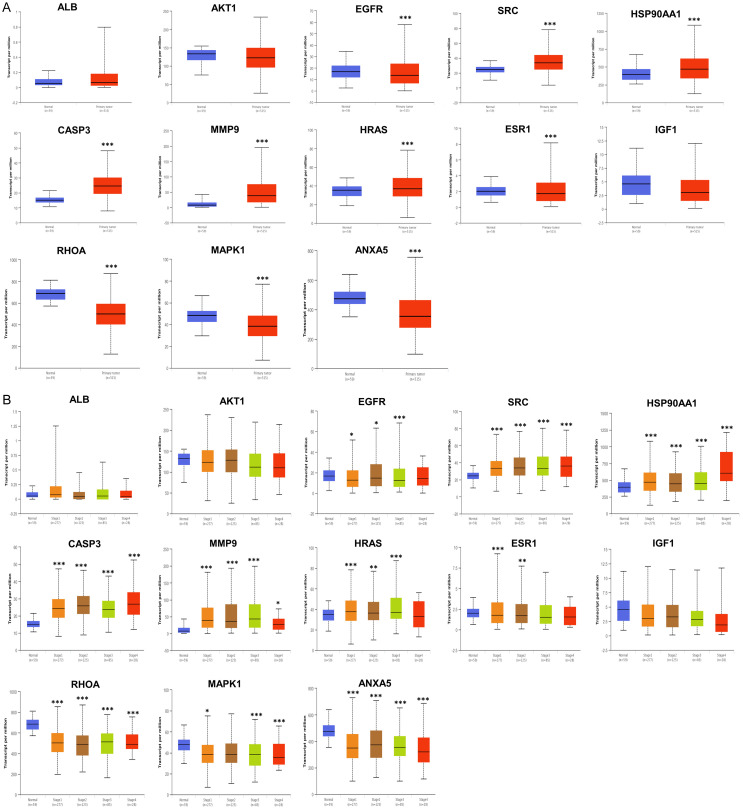
Validation of key targets in different databases
To elucidate the possible therapeutic significance and prognostic value of these 13 key targets, their mRNA expression was analysed. The results showed that the mRNA levels of steroid receptor coactivator (SRC), heat shock protein 90 alpha family class A member 1 (HSP90AA1), caspase-3 (CASP3), matrix metalloproteinase-9 (MMP9) and harvey rat sarcoma viral oncogene homolog (HRAS) were significantly higher in LUAD than in normal lung tissues. In contrast, EGFR, estrogen receptor 1 (ESR1), ras homolog gene family, member A (RHOA), MAPK1 and annexin A5 (ANXA5) mRNA expression levels were significantly decreased (Figure 11A). Next, we investigated the correlation between the mRNA levels of 13 key targets and different cancer stages of LUAD in the UALCAN database. As shown in Figure 11B, when compared with normal lung tissues, the mRNA levels of SRC, HSP90AA1, CASP3 and MMP9 were increased significantly in stages one to four LUAD tissues, while the expression levels of RHOA and ANXA5 were decreased significantly. For EGFR levels, there was a significant decrease in stage one to three LUAD tissue, but no significant change in stage four. Furthermore, the mRNA expression of serine/threonine protein kinase 1 (AKT1) and insulin like growth factor 1 (IGF1) in different tumor stages had different degrees of expression than that in normal lung tissues, but the difference was not significant.
Figure 11.
Validation of the mRNA expression of key targets in UALCAN. A. The mRNA levels of 13 key targets in LUAD tissues and normal lung tissues. B. The mRNA levels of 13 key targets in different tumor stages of LUAD (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). LUAD, lung adenocarcinoma; ALB, albumin; AKT1, serine/threonine protein kinase 1; EGFR, epidermal growth factor receptor; SRC, steroid receptor coactivator; HSP90AA1, heat shock protein 90 alpha family class A member 1; CASP3, caspase-3; MMP9, matrix metalloproteinase-9; HRAS, harvey rat sarcoma viral oncogene homolog; ESR1, estrogen receptor 1; IGF1, insulin like growth factor 1; RHOA, ras homolog gene family member A; MAPK1, mitogen-activated protein kinase 1; ANXA5, annexin A5.
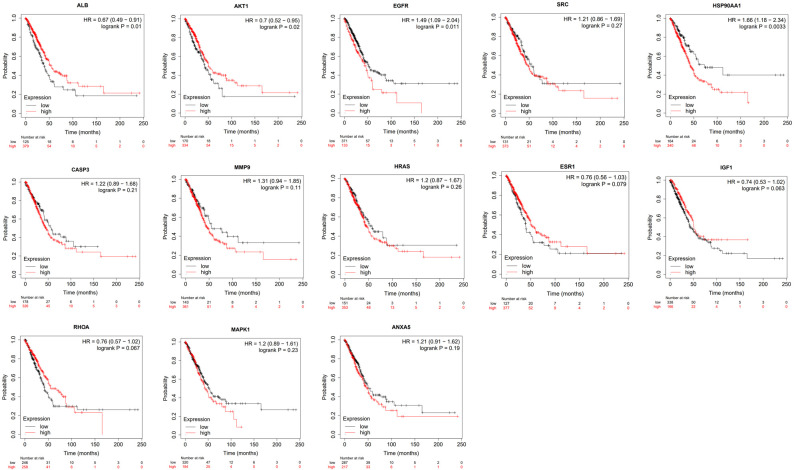
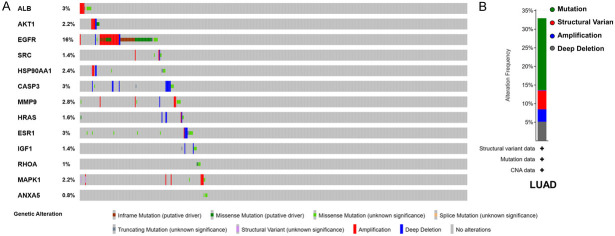
We further studied the expression levels of 13 key target proteins in LUAD. As shown in Figure 12, compared with normal lung tissues, the expression levels of EGFR, SRC, HSP90AA1, CASP3, HRAS, ESR1, IGF1, MAPK1 and ANXA5 were increased in LUAD tissues, while the expression levels of albumin (ALB) and RHOA were decreased in LUAD tissues. These results demonstrated that multiple key targets were abnormally expressed at the protein level in LUAD patients. Figure 13 shows the impact of 13 key target expressions on the OS rate of LUAD patients. Results indicated that low expression of ALB and AKT1 and high expression of EGFR and HSP90AA1 were closely connected with a poor survival prognosis. We next analyzed the frequency and types of gene changes in 13 key targets in 507 LUAD patients. As shown in Figure 14, the overall alteration rate of 13 key targets was 32.94% (167/507) including gene mutation, gene amplification, deep deletion and multiple alterations. The rate of genetic alteration rate of individual genes varied was highest for EGFR (16%) and lowest for ANXA5 (0.8%).
Figure 12.
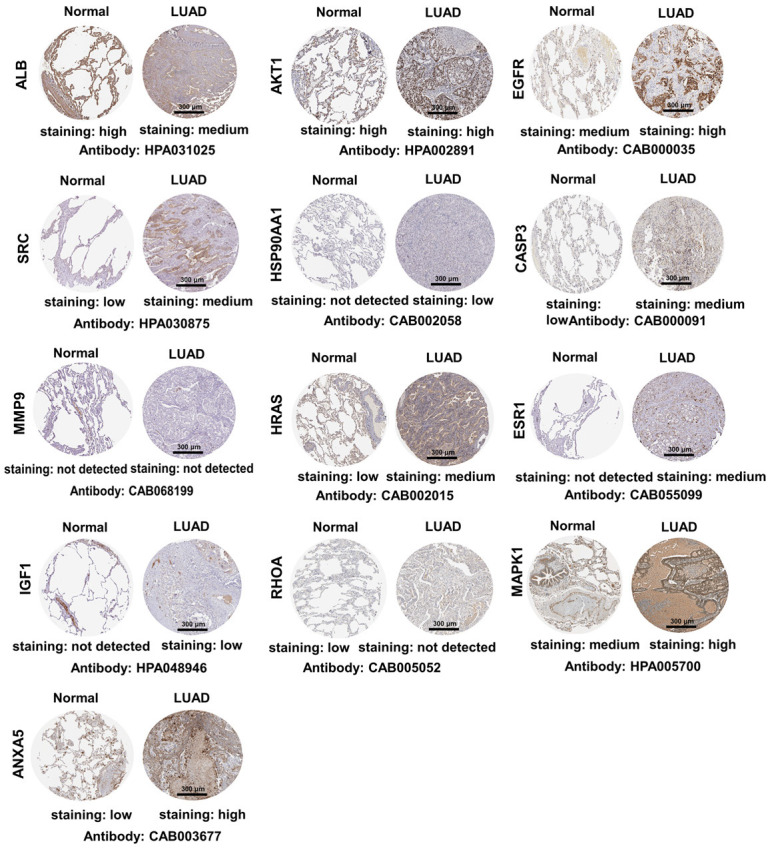
Immunohistochemical images (magnification, ×20) of key target protein expression levels in the Human Protein Atlas database. Scale bar, 300 μm. The abbreviations in the figure are the same as in Figure 11.
Figure 13.
Overall survival analysis of 13 key targets in LUAD in Kaplan-Meier mapper database. *P ≤ 0.05, **P ≤ 0.01. HR, hazard ratio. Other abbreviations in the figure are the same as in Figure 11.
Figure 14.
Genetic alterations in 13 key targets in LUAD patients in cBioPortal. A. OncoPrint visual summary of genetic alterations detected in 13 key targets. B. Summary of alterations in 13 key targets in LUAD. The abbreviations in the figure are the same as in Figure 11.
Molecular docking validation
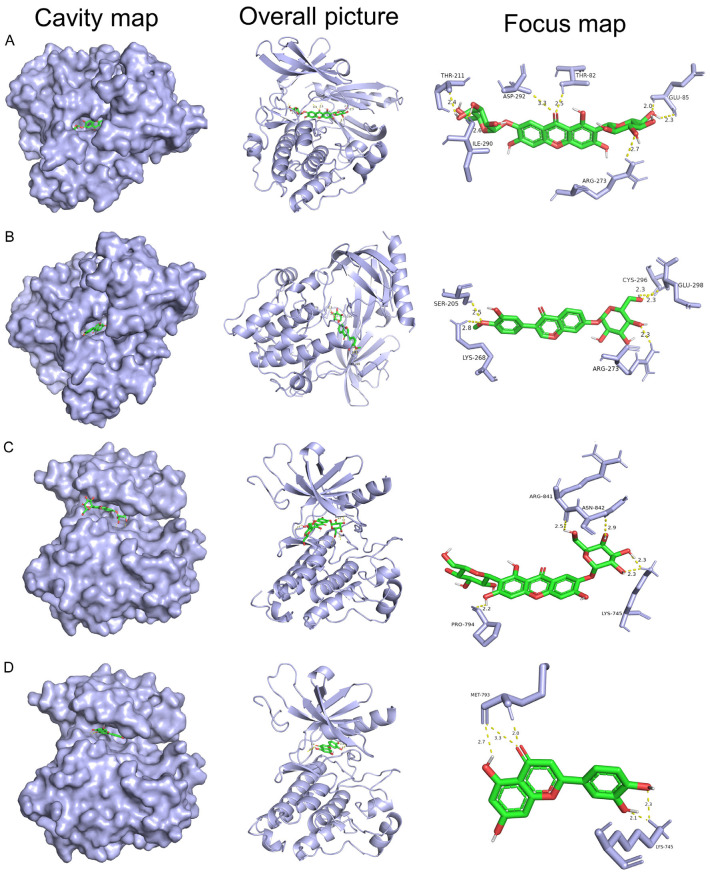
Validation including mRNA and protein expression, survival analysis, and genetic alterations, verified modulation of key targets. Subsequent molecular docking assessed the binding affinities of SXT active components to these key targets. The binding energies between SXT active components and the 13 key targets were all below -5.0 kJ∙mol-1, demonstrating good ligand-receptor interactions (Figure 15; Supplementary Table 4). KEGG pathway analysis revealed that SXT active components inhibits LUAD primarily by modulating the PI3K-Akt signaling pathway, with AKT1 being a pivotal target in apoptosis regulation within this pathway. External database analysis identified EGFR as playing a significant role in LUAD progression and exhibiting the highest degree of variation. These findings align with the PPI results, suggesting that AKT1 and EGFR are critical targets of SXT active components in LUAD treatment. We visualized the top two targets with the lowest binding energy to AKT1 and EGFR from SXT active components by PyMol, respectively. As shown in Table 3 and Figure 16, neomangiferin and calycosin 7-O-glucoside bind well to the AKT1 active pocket, showing high binding affinities, both with a binding energy of -10.7 kJ∙mol-1. Neomangiferin binds well to the AKT1 active pocket by binding to amino acid residues glutamic acid (GLU)-85, arginine (ARG)-273, threonine (THR)-82, aspartic acid (ASP)-292, isoleucine (ILE)-290 and THR-211. Calycosin 7-O-glucoside was bound to ATK1 by 5 hydrogen bonds to amino acid residues serine (SER)-205, lysine (LYS)-268, ARG-273, cysteine (CYS)-296 and GLU-298. Neomangiferin and luteolin bind well to EGFR with binding energies of -9.1 kJ∙mol-1 and -8.7 kJ∙mol-1, respectively. Neomangiferin binds to EGFR through binding to amino acid residues ARG-841, asparagine (ASN)-842, LYS-745 and proline (PRO)-794. Luteolin binds to EGFR through binding to amino acid residues methionine (MET)-793 and LYS-745. The docking junction results suggest that the SXT active components may be a potential inhibitor for the treatment of LUAD and that the AKT1 and EGFR play a critical role.
Figure 15.
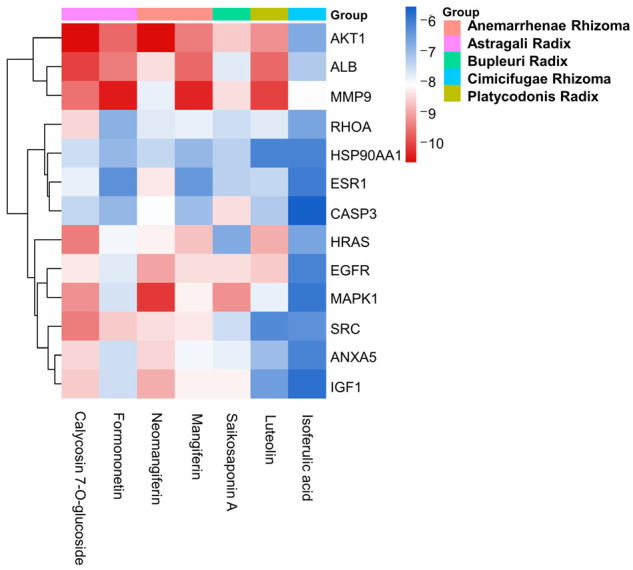
Molecular docking heat map. The abbreviations in the figure are shown in Figure 11.
Table 3.
Protein, ligand, hydrogen bond and binding energy in molecular docking
| Protein | PDB ID | Compounds | H-bond | Binding energy/(kJ∙mol-1) |
|---|---|---|---|---|
| AKT1 | 3O96 | Neomangiferin | GLU-85, ARG-273, THR-82, ASP-292, ILE-290, THR-211 | -10.7 |
| AKT1 | 3O96 | Calycosin 7-O-glucoside | SER-205, LYS-268, ARG-273, CYS-296, GLU-298 | -10.7 |
| EGFR | 7KXZ | Neomangiferin | ARG-841, ASN-842, LYS-745, PRO-794 | -9.1 |
| EGFR | 7KXZ | Luteolin | MET-793, LYS-745 | -8.7 |
Note: AKT1, serine/threonine protein kinase 1; EGFR, epidermal growth factor receptor; GLU, glutamic acid; ARG, arginine; THR, threonine; ASP, aspartic acid; ILE, isoleucine; SER, serine; LYS, lysine; CYS, cysteine; ASN, asparagine; PRO, proline; MET, methionine.
Figure 16.
The picture of molecule docking model of (A) Neomangiferin with AKT1, (B) Calycosin 7-O-glucoside with AKT1, (C) Neomangiferin with EGFR, and (D) Luteolin with EGFR. The green model represents the SXT active components, and the grey model indicates the key target proteins. The hydrogen bonds were represented by yellow dotted lines, and the length was marked around the lines. AKT1, serine/threonine protein kinase 1; EGFR, epidermal growth factor receptor; SXT, Shengxian Decoction; GLU, glutamic acid; ARG, arginine; THR, threonine; ASP, aspartic acid; ILE, isoleucine; SER, serine; LYS, lysine; CYS, cysteine; ASN, asparagine; PRO, proline; MET, methionine.
Discussion
Current treatment strategies for LUAD typically involve surgery, chemotherapy, radiotherapy, targeted therapy, immunotherapy, or a combination thereof [24]. However, these conventional therapies often result in severe side effects, which can hinder the successful completion of effective radiotherapy and chemotherapy cycles for most patients. Consequently, the search for effective drugs with fewer side effects for LUAD treatment has become a critical area of research. SXT, a commonly used TCM formula, has been clinically applied to treat various diseases, including chronic heart failure and industrial pneumoconiosis [25,26]. Our prior research demonstrated the effectiveness of SXT in inhibiting tumor growth in A549-xenograft nude mice, with low toxicity to visceral tissues. Additionally, a serum pharmacology assay confirmed the efficacy of SXT-containing serum in inhibiting the growth of three types of LUAD cells (A549, NCl-H1975, and SK-LU-1) in vitro [15]. In this study, we further established that SXT extract significantly inhibited proliferation, migration, and invasion in human LUAD cells (A549 and NCl-H1299) in vitro. These findings encouraged us to further study the active components in SXT extract.
Previous studies have identified mangiferin, neomangiferin, calycosin 7-O-glucoside, formononetin, saikosaponin A and isoferulic acid in SXT extract [15,27]. This study marks the first identification of luteolin in SXT extract. Among these components, neomangiferin can partially convert to mangiferin during the decoction process. Mangiferin has shown anti-tumor properties in A549 xenograft mice and may affect LUAD cells proliferation, cell cycle progression, and apoptosis induction through down-regulating miR-92a and miR-27b expression levels [28]. Calycosin 7-O-glucoside and isoferulic acid also exhibit tumor-suppressive effects by impacting the cell cycle [29,30]. Formononetin inhibits tumor growth by targeting the EGFR-Akt-myeloid cell leukemia-1 (Mcl-1) axis in NSCLC [31]. Saikosaponin A inhibits migration and invasion of triple-negative breast cancer cells by down-regulating C-X-C chemokine receptor type (CXCR4) expression, inactivating the Akt-mammalian target of rapamycin (mTOR) signaling pathway, and suppressing MMP-9 and MMP-2 expression [32]. Luteolin attenuates lung cancer cell migration and invasion by inhibiting the focal adhesion kinase and non-receptor tyrosine kinase signaling pathways [33]. To better understand the therapeutic contribution of these active components to LUAD, the chemical composition of SXT extract was identified and quantified. The results showed that SXT extract is particularly rich in saikosaponin A (6.47 mg/g) and mangiferin (3.53 mg/g), which may contribute significantly to the tumor suppression activity of SXT extract.
By integrating data from various databases, 197 targets with therapeutic potential for LUAD treatment by SXT active components were identified. GO analysis indicated that SXT active components could intervene in LUAD progression through biological processes such as apoptosis and protein phosphorylation. KEGG analysis suggested that SXT active components might exert its therapeutic effects on LUAD by regulating pathways like PI3K-Akt, FOXO, and MAPK, all of which have been implicated in lung cancer [34,35]. The constructed SXT-active components-target-pathway network highlighted the significance of the PI3K-Akt pathway, with key targets (including AKT1, EGFR, HSP90AA1, HRAS, IGF1, MAPK3, and MAPK1) predominantly enriched in this pathway. The PI3K-Akt pathway is crucial for regulating various processes, including cell proliferation, apoptosis, necrosis, and inflammation [36]. The EGFR signaling pathway, critical in tumor progression, activates the PI3K-Akt pathways, leading to cell proliferation, invasion, and migration [37]. The EGFR-MAPK pathway is activated in LUAD to promote tumor growth and metastasis [38], while FOXO signaling pathways are triggered by PI3K-Akt, mediating cellular functions related to proliferation and growth [39]. Considering the inhibition of tumor cell proliferation and promotion of apoptosis as viable cancer treatment pathways, this study preliminarily infers that SXT active components could potentially inhibit LUAD by targeting pathways related to cell proliferation and apoptosis.
The 13 key targets with top-degree values may play a critical role in treating LUAD, which were then validated in different databases. EGFR is a determinant that drives the growth and in vivo therapeutic response of LUAD [40]. AKT1 is a key metastasis regulator in LUAD cells and that inhibition of AKT1 promoted migration and invasion of KRAS or EGFR mutated LUAD cells in vitro [41]. SRC promotes the metastasis of various tumors, including NSCLC. Moreover, SRC can be used as an anti-cancer molecular target, and inhibition of SRC can inhibit multiple signaling pathways, including the EGFR pathway [42]. The knockdown of HSP90AA1 is able to inhibit AKT1 expression, thus inhibiting lung cancer cell proliferation [43]. CASP3 plays a central role in executing apoptosis and is involved in carcinogenesis [44]. In addition, analysis of the UALCAN database with the HPA database showed that the mRNA and protein levels of EGFR, SRC, HSP90AA1 and CASP3 were significantly altered in LUAD tissues, which may contribute to the occurrence of LUAD. Based on the Kaplan-Meier mapper database, high expression of EGFR and HSP90AA1, as well as low expression of AKT1, were associated with significant poor survival. The cBioPortal database shows that the mutation rate of EGFR in LUAD is as high as 16%, suggesting that EGFR has an important role in LUAD progression. The above studies indicate that SXT active components can regulate the expression of key targets and inhibit the malignant biological behaviors such as proliferation, migration, and invasive ability of LUAD cells, and thus can be used for the LUAD treatment.
Finally, we verified the binding affinities of compounds to the targets using molecular docking techniques. For instance, neomangiferin bound to AKT1 by forming hydrogen bonds with amino acid residues THR-82 and THR-211 (-10.7 kJ∙mol-1). It was further speculated whether the active binding of neomangiferin to AKT1 could lead to abnormal phosphorylation at threonine residues in the active pocket of ATP and thus contribute to the treatment of LUAD. Luteolin demonstrated strong binding affinity to the ATP-binding pocket of EGFR (-8.7 kJ∙mol-1), and formed a hydrogen bond with the backbone nitrogen of Met793. This interaction between EGFR and Luteolin may disrupt the interaction between EGFR and ATP, thereby inhibiting the activation of downstream signaling pathways, such as the EGFR-PI3K-Akt pathway, closely related to proliferation and apoptosis. The interaction of multiple active components in SXT extract with this pathway suggests that SXT treats LUAD through a multifaceted approach involving multiple components and targets (Figure 17).
Figure 17.
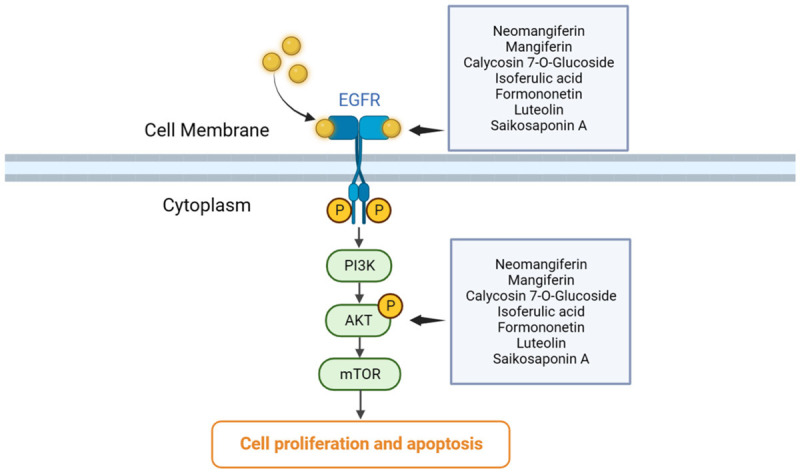
Schematic representation of the multi-component and multi-target of SXT intervening in the development of lung adenocarcinoma. SXT, Shengxian Decoction; EGFR, epidermal growth factor receptor; PI3K, phosphoinositide 3-kinase; AKT, serine/threonine protein kinase; mTOR, mammalian target of rapamycin.
Conclusion
In summary, this study demonstrated the inhibitory activity of SXT extract on LUAD cells in vitro and identified the main bioactive components using HPLC. Furthermore, the potential mechanisms of SXT in LUAD treatment were elucidated through network pharmacology, external validation databases, and molecular docking. It is postulated that SXT may impede LUAD proliferation, migration, and invasion by facilitating interactions between its main active components, such as saikosaponin A and mangiferin, with 13 key targets, including ALB, AKT1, EGFR, and SRC, through EGFR-PI3K-Akt and other signaling pathways. This study provides a theoretical foundation for further investigation and application of SXT in the treatment of LUAD.
Acknowledgements
K. Li thanks the support from the Natural Science Foundation of Sichuan Province (22NSFSC1268) and the National Natural Science Foundation of China (81904081). F. You is grateful to the Science and Technology Major Project of Sichuan Province of China (2022ZDZX0022) and the Central Guidance on Local Science and Technology Development Fund of Sichuan Province of China (2022ZYD0100).
Disclosure of conflict of interest
None.
Supplementary Table 1
Supplementary Tables 2-4
References
- 1.Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Sun D, Li H, Cao M, He S, Lei L, Peng J, Chen W. Cancer burden in China: trends, risk factors and prevention. Cancer Biol Med. 2020;17:879–895. doi: 10.20892/j.issn.2095-3941.2020.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou XJ, Li R, Liu X, Qu YQ. Advances in deubiquitinating enzymes in lung adenocarcinoma. J Cancer. 2021;12:5573–5582. doi: 10.7150/jca.56532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li ZH, Yu D, Huang NN, Wu JK, Du XW, Wang XJ. Immunoregulatory mechanism studies of ginseng leaves on lung cancer based on network pharmacology and molecular docking. Sci Rep. 2021;11:18201. doi: 10.1038/s41598-021-97115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W, Tian W, Wang Y, Jin X, Guo H, Wang Y, Tang Y, Yao X. Explore the mechanism and substance basis of Mahuang FuziXixin Decoction for the treatment of lung cancer based on network pharmacology and molecular docking. Comput Biol Med. 2022;151:106293. doi: 10.1016/j.compbiomed.2022.106293. [DOI] [PubMed] [Google Scholar]
- 7.Wang ZY, Li MZ, Li WJ, Ouyang JF, Gou XJ, Huang Y. Mechanism of action of Daqinjiao decoction in treating cerebral small vessel disease explored using network pharmacology and molecular docking technology. Phytomedicine. 2023;108:154538. doi: 10.1016/j.phymed.2022.154538. [DOI] [PubMed] [Google Scholar]
- 8.Li Q, Liu P, Wu C, Bai L, Zhang Z, Bao Z, Zou M, Ren Z, Yuan L, Liao M, Lan Z, Yin S, Chen L. Integrating network pharmacology and pharmacological validation to explore the effect of Shi Wei Ru Xiang powder on suppressing hyperuricemia. J Ethnopharmacol. 2022;298:115679. doi: 10.1016/j.jep.2022.115679. [DOI] [PubMed] [Google Scholar]
- 9.Huang C, Qiu S, Fan X, Jiao G, Zhou X, Sun M, Weng N, Gao S, Tao X, Zhang F, Chen W. Evaluation of the effect of Shengxian Decoction on doxorubicin-induced chronic heart failure model rats and a multicomponent comparative pharmacokinetic study after oral administration in normal and model rats. Biomed Pharmacother. 2021;144:112354. doi: 10.1016/j.biopha.2021.112354. [DOI] [PubMed] [Google Scholar]
- 10.He H, Zhou X, Wang Q, Zhao Y. Does the couse of astragalus-containing Chinese herbal prescriptions and radiotherapy benefit to non-small-cell lung cancer treatment: a meta-analysis of randomized trials. Evid Based Complement Alternat Med. 2013;2013:426207. doi: 10.1155/2013/426207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Zheng XL, Yang L, Shi F, Gao LB, Zhong YJ, Sun H, He F, Lin Y, Wang X. Reactive oxygen species-mediated apoptosis contributes to chemosensitization effect of saikosaponins on cisplatin-induced cytotoxicity in cancer cells. J Exp Clin Cancer Res. 2010;29:159. doi: 10.1186/1756-9966-29-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou C, Yu T, Zhu R, Lu J, Ouyang X, Zhang Z, Chen Q, Li J, Cui J, Jiang F, Jin KY, Sarapultsev A, Li F, Zhang G, Luo S, Hu D. Timosaponin AIII promotes non-small-cell lung cancer ferroptosis through targeting and facilitating HSP90 mediated GPX4 ubiquitination and degradation. Int J Biol Sci. 2023;19:1471–1489. doi: 10.7150/ijbs.77979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong SH, Guo H, Kim HY, Park JB. Ethanol extract of Cimicifugae rhizoma exerted more potent anti-inflammatory and tumor suppressor activities compared with methanol and water extracts. Biomed Res India. 2016;27:1054–1059. [Google Scholar]
- 14.Shen HS, Wen SH. Effect of early use of Chinese herbal products on mortality rate in patients with lung cancer. J Ethnopharmacol. 2018;211:1–8. doi: 10.1016/j.jep.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 15.Li K, You F, Zhang Q, Yuan R, Yuan Q, Fu X, Ren Y, Wang Q, Li X, Zhang Z, Shichiri M, Yu Y. Chemical and biological evidence of the efficacy of Shengxian Decoction for treating human lung adenocarcinoma. Front Oncol. 2022;12:849579. doi: 10.3389/fonc.2022.849579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Ouyang S, Yu B, Liu Y, Huang K, Gong J, Zheng S, Li Z, Li H, Jiang H. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010;38:W609–614. doi: 10.1093/nar/gkq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Pan C, Gong J, Liu X, Li H. Enhancing the enrichment of pharmacophore-based target prediction for the polypharmacological profiles of drugs. J Chem Inf Model. 2016;56:1175–1183. doi: 10.1021/acs.jcim.5b00690. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, Lai L, Pei J, Li H. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45:W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Li Z, Yang M, Wang D, Yu L, Guo C, Guo X, Lin N. Identification of GRB2 and GAB1 coexpression as an unfavorable prognostic factor for hepatocellular carcinoma by a combination of expression profile and network analysis. PLoS One. 2013;8:e85170. doi: 10.1371/journal.pone.0085170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, Creighton CJ, Varambally S. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi: 10.1016/j.neo.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 23.Lánczky A, Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res. 2021;23:e27633. doi: 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim W, Lee WB, Lee JW, Min BI, Baek SK, Lee HS, Cho SH. Traditional herbal medicine as adjunctive therapy for breast cancer: a systematic review. Complement Ther Med. 2015;23:626–632. doi: 10.1016/j.ctim.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Ma Y, Wang BL, Wang L, Huang CY, Sun M, Jiao GY, Zhang F, Chen WS. Effective components of Shengxian Decoction and its mechanism of action in treating chronic heart failure based on UHPLC-Q-TOF-MS integrated with network pharmacology. Zhongguo Zhong Yao Za Zhi. 2021;46:2489–2500. doi: 10.19540/j.cnki.cjcmm.20200915.201. [DOI] [PubMed] [Google Scholar]
- 26.Wang HJ, Ma GQ. Introduction to Professor Ma Guiqin’s experience in treatment of occupational pneumoconiosis. Zhongguo Zhong Yao Za Zhi. 2019;44:2871–2874. doi: 10.19540/j.cnki.cjcmm.20190419.502. [DOI] [PubMed] [Google Scholar]
- 27.Wang M, Li H, Gao Y, Li Y, Sun Y, Liu S, Liu Z. A multidimensional strategy to rapidly identify the chemical constituents in Shengxian Decoction by using ultra-performance liquid chromatography coupled with ion mobility spectrometry quadrupole time-of-flight mass spectrometry. J Sep Sci. 2022;45:3115–3127. doi: 10.1002/jssc.202200267. [DOI] [PubMed] [Google Scholar]
- 28.Chi XJ, Meng JJ, Lin CY, Su QS, Qin YY, Wei RH, Lan D, Huang C. Mangiferin inhibits human lung adenocarcinoma by suppressing miR-27b and miR-92a. Evid Based Complement Alternat Med. 2021;2021:2822950. doi: 10.1155/2021/2822950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bratkov VM, Shkondrov AM, Zdraveva PK, Krasteva IN. Flavonoids from the genus astragalus: phytochemistry and biological activity. Pharmacogn Rev. 2016;10:11–32. doi: 10.4103/0973-7847.176550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long Z, Feng G, Zhao N, Wu L, Zhu H. Isoferulic acid inhibits human leukemia cell growth through induction of G2/M‑phase arrest and inhibition of Akt/mTOR signaling. Mol Med Rep. 2020;21:1035–1042. doi: 10.3892/mmr.2020.10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu X, Gao F, Li W, Zhou L, Liu W, Li M. Formononetin inhibits tumor growth by suppression of EGFR-Akt-Mcl-1 axis in non-small cell lung cancer. J Exp Clin Cancer Res. 2020;39:62. doi: 10.1186/s13046-020-01566-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Zhao L, Han X, Wang Y, Mi J, Wang C, Sun D, Fu Y, Zhao X, Guo H, Wang Q. Saikosaponin A inhibits triple-negative breast cancer growth and metastasis through downregulation of CXCR4. Front Oncol. 2020;9:1487. doi: 10.3389/fonc.2019.01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masraksa W, Tanasawet S, Hutamekalin P, Wongtawatchai T, Sukketsiri W. Luteolin attenuates migration and invasion of lung cancer cells via suppressing focal adhesion kinase and non-receptor tyrosine kinase signaling pathway. Nutr Res Pract. 2020;14:127–133. doi: 10.4162/nrp.2020.14.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao P, Huang Y, Peng J, Yu J, Guo X, Bao F, Dian Z, An S, Xu TR. IRS4 promotes the progression of non-small cell lung cancer and confers resistance to EGFR-TKI through the activation of PI3K/Akt and Ras-MAPK pathways. Exp Cell Res. 2021;403:112615. doi: 10.1016/j.yexcr.2021.112615. [DOI] [PubMed] [Google Scholar]
- 35.Sun T, Zhang J, Deng B, Fan X, Long T, Jin H, Tao S, Kang P, Tan Q. FOXO1 and FOXO3a sensitize non-small-cell lung cancer cells to cisplatin-induced apoptosis independent of Bim. Acta Biochim Biophys Sin (Shanghai) 2020;52:1348–1359. doi: 10.1093/abbs/gmaa129. [DOI] [PubMed] [Google Scholar]
- 36.Iksen, Pothongsrisit S, Pongrakhananon V. Targeting the PI3K/AKT/mTOR signaling pathway in lung cancer: an update regarding potential drugs and natural products. Molecules. 2021;26:4100. doi: 10.3390/molecules26134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang J, Yuan Z, Sun Y, Bu Y, Li W, Fei Z. Ginsenoside Rg3 enhances the anti-proliferative activity of erlotinib in pancreatic cancer cell lines by downregulation of EGFR/PI3K/Akt signaling pathway. Biomed Pharmacother. 2017;96:619–625. doi: 10.1016/j.biopha.2017.10.043. [DOI] [PubMed] [Google Scholar]
- 38.Xu H, Yang X, Xuan X, Wu D, Zhang J, Xu X, Zhao Y, Ma C, Li D. STAMBP promotes lung adenocarcinoma metastasis by regulating the EGFR/MAPK signaling pathway. Neoplasia. 2021;23:607–623. doi: 10.1016/j.neo.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W. FOXO signaling pathways as therapeutic targets in cancer. Int J Biol Sci. 2017;13:815–827. doi: 10.7150/ijbs.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foggetti G, Li C, Cai H, Hellyer JA, Lin WY, Ayeni D, Hastings K, Choi J, Wurtz A, Andrejka L, Maghini DG, Rashleigh N, Levy S, Homer R, Gettinger SN, Diehn M, Wakelee HA, Petrov DA, Winslow MM, Politi K. Genetic determinants of EGFR-driven lung cancer growth and therapeutic response in vivo. Cancer Discov. 2021;11:1736–1753. doi: 10.1158/2159-8290.CD-20-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao G, Pierobon M, Kim IK, Hsu WH, Deng J, Moon YW, Petricoin EF, Zhang YW, Wang Y, Giaccone G. Inhibition of AKT1 signaling promotes invasion and metastasis of non-small cell lung cancer cells with K-RAS or EGFR mutations. Sci Rep. 2017;7:7066. doi: 10.1038/s41598-017-06128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giaccone G, Zucali PA. Src as a potential therapeutic target in non-small-cell lung cancer. Ann Oncol. 2008;19:1219–1223. doi: 10.1093/annonc/mdn048. [DOI] [PubMed] [Google Scholar]
- 43.Niu M, Zhang B, Li L, Su Z, Pu W, Zhao C, Wei L, Lian P, Lu R, Wang R, Wazir J, Gao Q, Song S, Wang H. Targeting HSP90 inhibits proliferation and induces apoptosis through AKT1/ERK pathway in lung cancer. Front Pharmacol. 2022;12:724192. doi: 10.3389/fphar.2021.724192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin J, Zhang Y, Wang H, Chang J, Wei L, Cao L, Zhang Z, Zhang X. Genetic polymorphisms in the apoptosis-associated gene CASP3 and the risk of lung cancer in Chinese population. PLoS One. 2016;11:e0164358. doi: 10.1371/journal.pone.0164358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.