Abstract
Biomarkers play an important role in various area such as personalized medicine, drug development, clinical care, and molecule breeding. However, existing animals’ biomarker resources predominantly focus on human diseases, leaving a significant gap in non-human animal disease understanding and breeding research. To address this limitation, we present BioKA (Biomarker Knowledgebase for Animals, https://ngdc.cncb.ac.cn/bioka), a curated and integrated knowledgebase encompassing multiple animal species, diseases/traits, and annotated resources. Currently, BioKA houses 16 296 biomarkers associated with 951 mapped diseases/traits across 31 species from 4747 references, including 11 925 gene/protein biomarkers, 1784 miRNA biomarkers, 1043 mutation biomarkers, 773 metabolic biomarkers, 357 circRNA biomarkers and 127 lncRNA biomarkers. Furthermore, BioKA integrates various annotations such as GOs, protein structures, protein–protein interaction networks, miRNA targets and so on, and constructs an interactive knowledge network of biomarkers including circRNA–miRNA–mRNA associations, lncRNA–miRNA associations and protein–protein associations, which is convenient for efficient data exploration. Moreover, BioKA provides detailed information on 308 breeds/strains of 13 species, and homologous annotations for 8784 biomarkers across 16 species, and offers three online application tools. The comprehensive knowledge provided by BioKA not only advances human disease research but also contributes to a deeper understanding of animal diseases and supports livestock breeding.
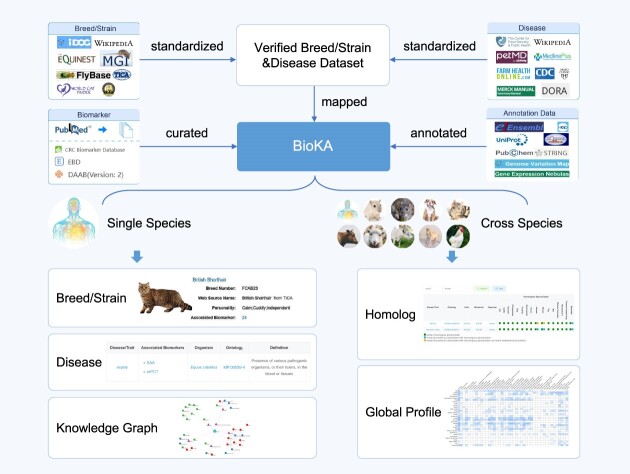
Graphical Abstract
Graphical Abstract.
Introduction
Biological markers, commonly referred to markers or biomarkers, serve as quantifiable and measurable indicators of certain biological states in normal and pathogenic processes, as well as potential pharmacologic responses to therapeutics (1–3). Biomarkers have always been used in clinical medicines and can help in earlier disease diagnoses and efficient therapy of several disorders (4). And biomarkers can also help farmers, veterinarians, livestock researchers and the livestock industry to diagnose and therapy of animal disease, evaluate dietary responses to different feeds, fertility and ascertain other important economic or breeding traits associated with livestock (5).
In recent years, there has been a remarkable surge in biomarker research driven by significant advancements in high-throughput sequencing techniques (6), omics technologies (7) and computer-aided biomarker discovery methods (8). Biomarkers have been reported for various conditions in human disease research, including cancer (9,10), aging (11), depression (12), neurodegenerative diseases (13) and osteoarthritis (14,15). Veterinary disease research (16) has also identified biomarkers for conditions such as avian leukosis virus subgroup J infection in chickens (17), kidney disease in cats (18) and horses (19). To promote the implementation of discovered biomarkers in medicine or industry (20), various model animals are utilized to validate biomarker during different phases of drug development (21). For instance, laboratory mice are employed as xenograft animal models in breast cancer treatment research (22), while flies are used as disease models in therapeutic drug discovery (23). Additionally, an abundant of biomarkers associated with economic traits have been discovered in animal breeding, such as meat quality (24), and fertility (25). However, these biomarkers are mainly scattered across public literatures, posing challenges in retrieving related research information. Therefore, it is necessary to curate high-quality biomarkers from existing literature and establish public data resources that facilitate widespread sharing of biomarkers. These initiatives are paramount as they promote both the clinical translation of biomarkers and advancements in agricultural economics.
At present, several publicly accessible biomarker databases are available, including BCSCdb (26) for human cancer stem cells, CBD (27) for human colorectal cancers, CellMarker (28) for human and mouse cell markers, DAAB (29) for allergy and asthma, EBD (30) for human eye diseases, ExoBCD (31) for human breast cancer, HFBD (32) for human heart failure, MarkerDB (33) for various human disease, and ResMarkerDB (34) for antibody therapy response in human breast and colorectal cancer. These databases offer online services for browsing, searching, and downloading biomarker information, greatly benefiting biomarker research. However, limitations still persist. Firstly, most of these databases primarily focus on specific singleton human disease and lack critical information about laboratory animals, such as breed/strain, age, weight and experiment results. This limitation hampers the retrieval and research of cross-species information. For instance, laboratory mice are commonly used as animal models to validate findings in various human disease research, with specific strains like BALB/c nude for hepatocellular carcinoma (35) and C57BL/6J for obesity and type 2 diabetes (36). The strain, sex and age variations of mice can significantly impact the composition of body fluids during biological processes (37). Therefore, it is crucial to provide this information for rigorous evaluation and efficient retrieval once a biomarker has been identified. Secondly, the annotated information provided by most of these databases is not well integrated, for example, regulatory relation, homologs and multi-omics are not available, which is not conducive to a comprehensive understanding of biomarkers. It is worth noting that circRNA–miRNA–mRNA regulatory networks play pivotal roles in diseases such as Alzheimer's disease, diabetes, and cancers (38), while lncRNAs promote tumor progression as miRNAs sponges (39). And orthologs (40) are essential for cancer biotherapy, particularly immunotherapy and gene therapy (41). Therefore, integrating regulatory network information, homologs, and multi-omics data annotation (42) will greatly advance biomarker discovery and functional research. Furthermore, these databases mainly focus on human biomarkers, disregarding non-human animals. There is a considerable gap in the availability of a comprehensive biomarker database covering a wide range of domestic animal diseases and livestock breeding traits. To advance both human disease research and animal disease/breeding research, it is imperative to construct a comprehensive biomarker knowledgebase that encompasses biomarkers with disease/trait associations and breed/strain information for a wide range of animals, while also integrating various annotations.
Here, we present BioKA (https://ngdc.cncb.ac.cn/bioka), a comprehensive and high-quality disease/trait biomarker knowledge base for animals, including model animals, domestic animals and human. BioKA meticulously curate biomarkers and integrates various annotations, such as GOs, protein structures, protein–protein interaction networks, miRNA targets, metabolism details, expression, variations, and homologous genes, into a single web platform. It provides a one-step service for cross-species research, along with free public data services for browsing, retrieval, comparison and downloading.
Materials and methods
Data collection
To provide reliable information on biomarkers across multiple animal species, a comprehensive literature review was conducted. Firstly, literature published before 10 July 2023, was searched using pre-defined keywords such as ‘biomarker’, ‘marker’, ‘indicator’ and ‘predictor’ for each species. A total of 64 935 literatures were obtained with other basic information (e.g. title, journal, paper type, abstract) through NCBI E-utilities and utilized as the original data. Secondly, reviews, comments, letters, editorials and non-English literature were excluded from the dataset. Abstracts were further analyzed using the Natural Language Toolkit (https://www.nltk.org/) to identify sentences containing both an entity term (e.g. ‘biomarker’, ‘marker’, ‘indicator’ or ‘target’) and a qualifying word (e.g. ‘diagnostic’, ‘prognostic’, ‘therapeutic’ or ‘valuable’) using regular expression operations in Python (https://www.python.org/). This meticulous filtering process resulted in a refined collection of 19297 corresponding articles, serving as the data source for BioKA. Finally, the entire collection of papers underwent thorough filtration and curation. Papers with a specific focus on a particular disease/trait, concise conclusions, and detailed experimental information (e.g. in vitro, in vivo, clinical tests, behavioral tests) were selected for the curation of essential biomarker information (name, associated disease/trait, usage, conclusion), sample and experiment details (e.g. numbers, breed/strain, age, sex, technology, tissue, significance), as well as regulatory relation (e.g. circRNA–miRNA). Additionally, in order to enhance the comprehensiveness of BioKA, three curated resources focusing on human biomarkers CBD (27), DAAB (29) and EBD (30), were integrated into the BioKA according to the curation data standardization.
Breed/strain and disease information were extracted from various websites (Table 1). Functional annotations for biomarkers were collected from multiple resources. Gene or mutation biomarker annotations were gathered from Ensembl (43), protein annotations from UniProt (44) and protein–protein interaction networks from STRING (45). miRNA annotations were sourced from miRBase (46), with predicted miRNA targets from TargetScan (47) and curated miRNA targets obtained from miRTarBase (48). Metabolite annotations were collected from PubChem (49), while multi-omics and across species annotations were collected from CNCB-NGDC (50), including variation annotations were gathered from GVM (51), expression annotations from GEN (52) and homologs information from HGD (53).
Table 1.
Data source for breed/strain and disease
| Meta-data type | Data source |
|---|---|
| Breed/strain | TICA (https://tica.org/) |
| CFA (https://cfa.org/) | |
| Equinest (http://www.theequinest.com/) | |
| MGI (63) | |
| FlyBase (64) | |
| World Cat Finder (https://worldcatfinder.com/) | |
| Wikipedia | |
| iDog (54) | |
| Non-human animal disease | CFSPH (https://www.cfsph.iastate.edu/) |
| petMD (https://www.petmd.com/) | |
| Farm Health Online (https://www.farmhealthonline.com/US/) | |
| DORA (http://dora.missouri.edu/) | |
| Merck Vet Manual (https://www.merckvetmanual.com/) | |
| Human disease | Mayo Clinic (https://www.mayoclinic.org/diseases-conditions) |
| CDC (https://www.cdc.gov/biomonitoring) | |
| MedlinePlus (https://medlineplus.gov/) | |
| Wikipedia (https://www.wikipedia.org/) |
Data processing
The entire data processing procedure involved the preprocessing of breed/strain and disease information, the standardization of biomarkers and the integration of annotations. For breed/strain and disease data, the original information was collected from various public websites mentioned above and parsed as structured data. The extracted breed names and disease names were then subjected to preprocessing, which included removing abbreviations, punctuation, and adjusting word order. Breed names were standardized following the naming rule from iDog (54), while disease names were mapped to the Mondo Disease Ontology (MONDO) (https://obofoundry.org/ontology/mondo.html). Based on the standardized names, the parsed information was integrated and manually verified to create a high-quality dataset of breed/strain and disease. In total, 2410 breeds and 177638 strains for 15 species, along with 4823 disease records for 14 species, were obtained.
The standardization of disease/trait terms for biomarkers in BioKA was achieved by mapping them to a merged and customized ontology as Animal Disease and Trait Ontology (ADTO). This ontology incorporates MONDO, Vertebrate trait ontology (https://obofoundry.org/ontology/vt.html), Animal Phenotype and Trait Ontology (51), Mammalian Phenotype Ontology (55) and Environmental conditions, treatments, and exposures ontology (https://obofoundry.org/ontology/ecto.html). The breed/strain names were also standardized using the same procedure as mentioned for breed/strain standardization. To obtain a unique set of biomarkers for each species, various biomarker names, including full names, abbreviations, and ID mapping names (such as Ensembl gene ID, UniProt ID, Entrez ID), were utilized to eliminate redundancy.
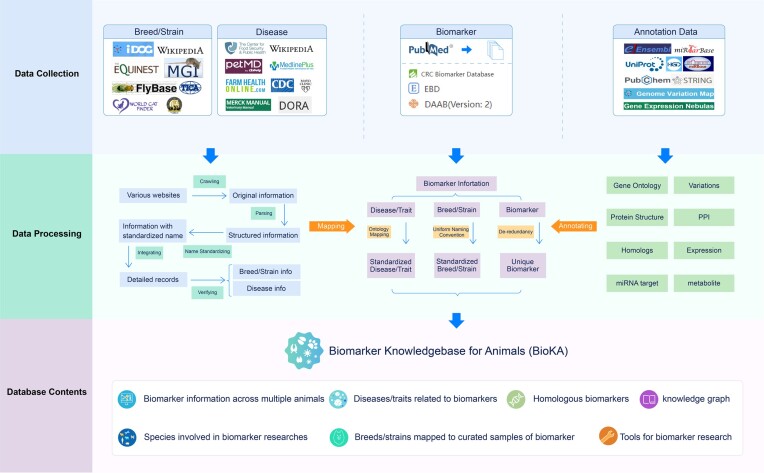
Finally, the breed/strain and disease names of biomarkers were mapped to the verified breed/strain and disease dataset mentioned earlier. Subsequently, comprehensive annotated information, including GOs, protein structures, protein–protein interaction networks, miRNA targets, metabolism, expression, variations and homologs, was obtained by annotating the biomarkers using the various resources mentioned above. This process resulted in the extraction of 16 296 distinct biomarkers for 31 species, with 951 diseases/traits and 308 curated breeds/strains. Additionally, we identified 8784 biomarkers associated with homologs. The entire procedure described above is depicted in Figure 1.
Figure 1.
Overview of data collection, data processing and database contents of BioKA.
Database implementation
BioKA was developed utilizing SpringBoot (https://spring.io/projects/spring-boot;a versatile back-end framework for creating standalone Java applications). The data storage engine employed was MySQL (https://www.mysql.com/;a widely-used and free relational database management system). For the front-end service, Vue3 (https://vuejs.org/; a highly performant and adaptable framework for building web user interfaces) was utilized. To enhance the usability of the web interface, Element Plus (https://element-plus.org/; a UI library based on Vue 3) was integrated. Additionally, data visualization capabilities were implemented using D3.js (https://d3js.org/; an open-source JavaScript library renowned for its ability to create visually appealing and interactive visualizations).
Database contents and usage
Data summarize
BioKA manually curates a diverse range of biomarkers for various animals and integrates three curated biomarker databases based on curated data criteria. Currently, BioKA houses 16296 biomarkers associated with 951 diseases/traits from a comprehensive compilation of 4747 literature references across 31 species, including 9 model animals, 16 domestic animals, 5 other animals and human. Meticulously curated biomarkers include 11 925 gene/protein biomarkers, 1784 miRNA biomarkers, 1043 mutation biomarkers, 773 metabolic biomarkers, 357 circRNA biomarkers and 127 lncRNA biomarkers. To facilitate convenient retrieval, BioKA provides integrated annotation information such as GOs, protein structures, protein–protein interaction networks, miRNA targets, metabolism, expression, variations, and homologs. Additionally, BioKA constructs an interactive knowledge network of biomarkers, which includes 7320 entities and 401 208 links across 10 species. Moreover, BioKA offers detailed information on 308 breeds/strains of 13 species and provides homologous annotations for 8784 biomarkers across 16 species. Table 2 presents the statistical summary of the data, while Figure 2A provides an illustrative overview of a colored global profile normalized by the number of biomarkers.
Table 2.
Biomarker statistics for each species
| Organism | Common name | NCBI taxon ID | #Breeds/strains | #Diseases/traits | #Biomarkers |
|---|---|---|---|---|---|
| Felis catus | cat | 9685 | 28 | 45 | 121 |
| Bos taurus | cattle | 9913 | 30 | 66 | 987 |
| Gallus gallus | chicken | 9031 | 2 | 56 | 485 |
| Pan troglodytes | chimpanzee | 9598 | − | 3 | 14 |
| Macaca fascicularis | cynomolgus macaque | 9541 | − | 18 | 42 |
| Canis lupus familiaris | dog | 9615 | 143 | 72 | 839 |
| Equus asinus | donkey | 9793 | 1 | 6 | 32 |
| Anas platyrhynchos | duck | 8839 | − | 6 | 25 |
| Drosophila melanogaster | Fruit fly | 7227 | 6 | 4 | 92 |
| Meriones unguiculatus | gerbil | 10047 | − | 6 | 8 |
| Ailuropoda melanoleuca | giant panda | 9646 | − | 2 | 5 |
| Capra hircus | goat | 9925 | 11 | 35 | 275 |
| Mesocricetus auratus | golden hamster | 10036 | − | 4 | 13 |
| Anser anser | goose | 8843 | − | 6 | 23 |
| Cavia porcellus | guinea pig | 10141 | − | 13 | 53 |
| Equus caballus | horse | 9796 | 31 | 82 | 693 |
| Homo sapiens | human | 9606 | − | 178 | 6960 |
| Callithrix jacchus | marmoset | 9483 | − | 9 | 12 |
| Mus musculus | mouse | 10090 | 31 | 84 | 3408 |
| Sus scrofa | pig | 9823 | 13 | 75 | 930 |
| Oryctolagus cuniculus | rabbit | 9986 | 3 | 13 | 33 |
| Rattus norvegicus | rat | 10116 | 4 | 53 | 422 |
| Macaca mulatta | rhesus macaque | 9544 | − | 16 | 37 |
| Caenorhabditis elegans | Roundworm | 6239 | − | 1 | 1 |
| Ovis aries | sheep | 9940 | 5 | 49 | 146 |
| Anser cygnoides | Swan goose | 8845 | − | 1 | 1 |
| Xenopus tropicalis | tropical clawed frog | 8364 | − | 1 | 2 |
| Meleagris gallopavo | turkey | 9103 | − | 2 | 2 |
| Bubalus bubalis | water buffalo | 89462 | − | 6 | 22 |
| Bos grunniens | yak | 30521 | − | 7 | 18 |
| Danio rerio | zebrafish | 7955 | − | 32 | 595 |
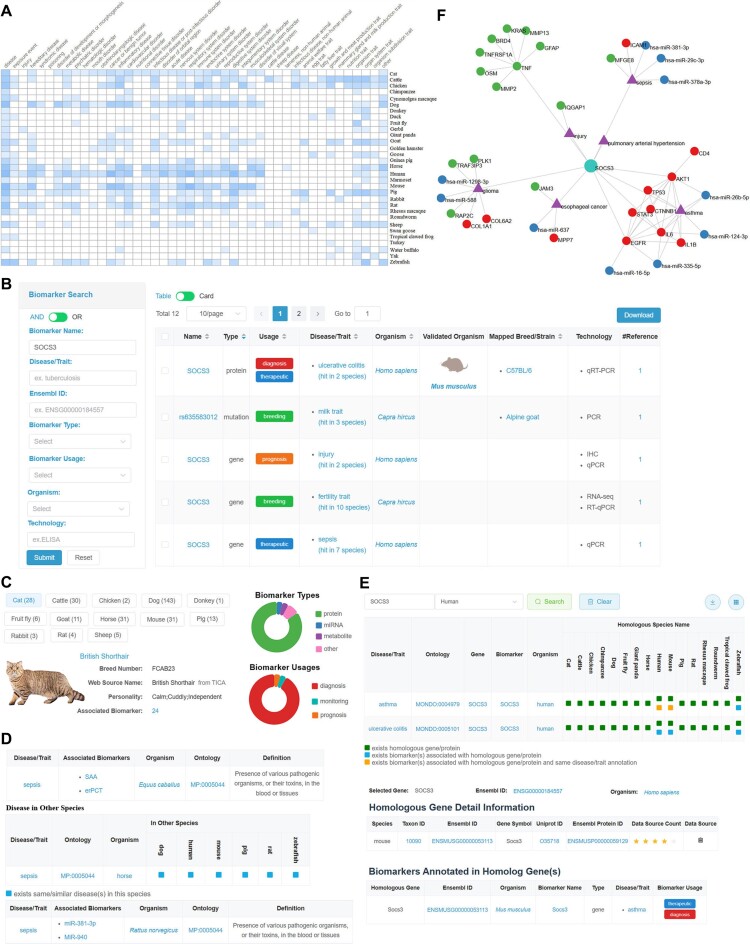
Figure 2.
Screenshots of BioKA. (A) A global profile presenting biomarkers across 31 species. (B) The biomarker list for SOCS3 is sorted by type in descending order, and a filter panel for results filtering. (C) The breed/strain list, and a statistical pie chart for biomarker type and usage respectively. (D) The disease/trait table list, and a summarized panel showed in sepsis detailed information page for associated biomarkers discovered in seven species. (E) Homologs annotated with biomarker disease/trait information. (F) A knowledge graph is constructed based on the annotated information of biomarkers and integrated associations in BioKA.
Browsing, retrieval and download biomarkers across multiple animals
BioKA offers a user-friendly panel for efficient exploration of diverse biomarkers across multiple animals (Figure 2B). The website provides two view modes: table view and card view, enabling users to access basic information on each biomarker including name, type, disease/trait, usage, mapped breed/strain, organism and validated organism. Notably, the ‘Validated Organism’ feature focuses on human studies and highlights model animals used for validated biomarkers through various experimental approaches (e.g. in vivo, in vitro, clinical tests). Clicking on the validated organism name provides comprehensive details of the corresponding sample and experimental information. Additionally, BioKA incorporates an intuitive multi-condition filtering module conveniently positioned in the left panel. Users can easily toggle between ‘and/or’ filtering modes and apply multiple conditions to efficiently filter biomarkers of interest.
To efficiently query contents of interest, BioKA is equipped with several search channels: (i) a basic search function on the home page is provided for quick fuzzier retrieval of results by various conditions for biomarker (e.g. name, synonyms, organism, Ensembl ID), disease (e.g. disease name, synonyms, abbreviation, organism, definition) and organism (e.g. latin name, common name, taxon ID), respectively; (ii) an advanced search function on the ‘Search’ page with multiple conditions is available for directly accessing BioKA by a list of terms by biomarker name and disease/trait name; (iii) a search mode for an intuitive graph by biomarker name and disease/trait is available on the ‘Knowledge Graph’ page; (iv) a search mode for using gene and protein sequence is available on the ‘Tools’ page by integrating NCBI BLAST (56) tools. All query results can be downloaded freely, and a summarized list of biomarkers organized by species, disease/trait and type can be accessible on the ‘Download’ page.
To facilitate the retrieval of disease or trait information associated with biomarkers of interest, biomarker tools for disease/trait annotation are also available. Users can select an organism, breed/strain, and input a list of biomarker names. Subsequently, a heatmap with annotated disease/trait information is generated, ensuring convenient access to relevant data.
Mapped breeds/strains for biomarkers
BioKA provides meticulously curated mapped breed/strain information, facilitating comprehensive descriptions of curated samples (Figure 2C). By precisely mapping breed/strain names associated with biomarker samples to a verified breed/strain dataset, BioKA compiles an extensive collection of 308 mapped breeds/strains across 13 species. Users can explore available species by selecting tagged buttons, revealing a list with essential details such as breed/strain name, synonyms, and personality traits. Convenient filtering criteria like name, size, weight, height, temperament allow users to efficiently narrow down their search within the breed/strain list. Clicking on a specific breed/strain name directs users to a dedicated page that presents crucial information, such as height, weight, life expectancy, origin and historical background. This page also provides statistics in the form of visualized pie charts and a detailed list of associated biomarkers for further exploration.
Diseases/traits related to biomarkers
BioKA provides comprehensive and standardized disease/trait information, delivering curated descriptions alongside disease ontology, definitions, and associated biomarkers (Figure 2D). Through meticulous mapping of disease/trait names to the merged ADTO ontology, BioKA compiles an extensive collection encompassing 951 diseases/traits across 31 species. Users can explore the disease/trait list by selecting specific names from the ontology tree, ensuring efficient filtering and navigation. Clicking on a disease name redirects users to a page with four sections: basic information (e.g. symptoms, treatments, prevention advice), biomarker statistical for type and usage, disease occurrence in other species, and disease ontology. In ‘disease occurrence in other species’ section, clicking on the blue colored block explores the detailed list of associated biomarkers for further exploration.
Homolog annotated with disease/trait for biomarkers
BioKA integrates HGD homologs and annotates them with disease/trait information linked to biomarkers, enhancing understanding of gene effects on diseases/traits (Figure 2E). Disease/trait terms of biomarkers are mapped to the merged ADTO ontology and annotated to homologs using gene/protein symbols. This process filters 276 disease/trait terms for 8784 biomarkers, resulting in annotations assigned to 7905 homologs across 16 species. Users can select a disease/trait term of interest to view different annotations for multi-species homologs, indicated by colored icons. The green icon signifies that the queried disease/trait only has homologs within the corresponding species. Clicking on an orange icon presents a concise table list featuring homologs and a biomarker list associated with the same queried disease/trait, facilitating further research into biomarker function.
Highly integrated knowledge graph with interactive visualization
To better integrate and visualize the knowledge embraced in BioKA, various regulatory relationships such as circRNA–miRNA–mRNA associations, lncRNA–miRNA associations, protein–protein associations, SNP–gene–trait associations and gene/protein/circRNA/lncRNA/miRNA–trait associations are systematically combined and visualized as a knowledge graph (Figure 2F). This integration involves collecting circRNA–miRNA associations from curated biomarker information, integrating miRNA–mRNA associations from miRTarBase, mapping gene names from Ensembl to obtain lncRNA–miRNA associations, and incorporating protein–protein associations from STRING. After mapping all biomarkers to these associations, BioKA has successfully constructed a comprehensive knowledge graph that includes 7320 entities (such as 17 circRNAs, 331 disease/traits, 301 mutations, 519 miRNAs, 26 lncRNAs, 5434 genes and 692 proteins) and 401 208 links across 10 species. Users can conveniently search for specific biomarker names and diseases/traits related to their organism of interest on the ‘Knowledge Graph’ page. This will provide them with an overview of a centralized knowledge graph tailored to their research needs. By default, BioKA displays 10 associations for each node based on the number of degrees of associated nodes/traits. However, users have the flexibility to adjust the display settings to show the entire knowledge graph, including predicted nodes and associations, or limit it to nodes and associations mapped within BioKA. Additionally, clicking on a node within the knowledge graph allows users to access a detailed biomarker page for further research exploration.
An example using BioKA
Suppressor of cytokine signaling (SOCS) proteins act as negative feedback regulators within the JAK-STAT signalling pathway. SOCS3, a member of the SOCS family, negatively regulates cytokines and their associated pathways, influencing cell proliferation, apoptosis, and other biological processes (57). Several studies have explored the therapeutic potential and diagnostic biomarker value of SOCS proteins (58). A basic search on SOCS3 reveals its involvement in 10 diseases/traits across four species: goat, mouse, zebrafish and human (Figure 2B). Clicking on a protein biomarker of SOCS3 associated with ulcerative colitis in human opens a new page with seven sections providing general information, summary, curated information, GO annotation, structure, interaction and homologs. The summary section (Figure 3A) highlights SOCS3 as a potential therapeutic and diagnostic biomarker for human diseases. A colored map summarizes various diseases/traits associated with SOCS3, including hereditary diseases, inflammatory diseases, immune system disorders, and digestive system disorders. Clicking on a colored block presents a table with detailed disease/trait information. The curated information section (Figure 3B) provides literature-based details, including animal group information, experiment details, and conclusions. Notably, the curated information includes a validated animal model using the C57BL/6 mouse strain to support the conclusions. The GO Annotation section (Figure 3C) presents a table illustrating SOCS3’s gene function in the negative regulation of receptor signaling pathway via JAK-STAT, in line with its known function. The Structure section (Figure 3D) displays a predicted protein structure of SOCS3 generated by AlphaFold, along with zoom and download options. The Interaction section (Figure 3E) showcases an interaction network, highlighting proteins such as JAK2, STAT3 and IL6ST that interact with SOCS3. The Homolog section (Figure 3F) identifies five homologous biomarkers in human, one in mouse, two in zebrafish (socs3a and socs3b). Clicking on a colored respiratory system disorder term for Socs3 in mouse reveals its association with asthma, suggesting potential therapeutic and diagnostic applications.
Figure 3.
BioKA usage example. (A) Summarized disease/trait panel for human-specific SOCS3. (B) Curated information from literature sources for SOCS3. (C) GO Annotation data integrated from UniProt for SOCS3. (D) Protein structure information integrated from UniProt for SOCS3. (E) Protein–protein interaction network integrated from STRING for SOCS3. (F) Homologous information integrated from HGD for SOCS3. (G) Variation information integrated from GVM for human-specific SOCS3. (H) Expression information for SOCS3 in human integrated from GEN. (I) Predicted miRNA list integrated from TargetScan for human-specific SOCS3.
Clicking on a gene biomarker associated with SOCS3 with ashma in human opens a new page featuring 8 sections. The Variation section (Figure 3G) provides a summarized variation profile for SOCS3, highlighting 49 variants in the primer UTR, downstream gene, upstream gene, and intron regions. Clicking on a colored block reveals a detailed variation list, including alleles and protein residue changes for further investigation. In the Gene Expression section (Figure 3H), a summarized expression profile showing SOCS3 expression in 38 biological contexts. Clicking on the respiratory system term reveals a detailed list of differential expression, highlighting the significance of SOCS3 expression in 5 high-quality RNA-seq datasets, specifically in human trachea-bronchial tissues (59). The Targeted miRNAs section (Figure 3I) presents a list of predicted miRNAs targeting SOCS3, including has-miR-455–5p which have been identified by Zhongzhong Tu et al. (60). Notably, miR-455-5p (61) is also a biomarker in BioKA. Clicking on the Biomarker ID will open a detail page for further exploration.
Discussion and future plans
Biomarkers play crucial roles in clinical medicine and drug development (2) and are also vital in animal breeding (62). Comparing to existing biomarker resources (Table 3), BioKA offers several advanced features. Firstly, BioKA provides disease/trait associated biomarkers for human and 30 animals including model animals, domestic, and livestock animals, facilitating comprehensive cross-species biomarker research. Secondly, BioKA providing clear information on breed/strain details obtained from diverse public web resources, which is meticulously standardized and integrated into curated samples of biomarkers. Furthermore, BioKA offers comprehensive functional annotation, interaction, structure, regulatory and multi-omics annotations for a wide range of biomarkers, creating a comprehensive knowledge base along with a visualized knowledge graph for further research. Its unique browsing feature allows simultaneous exploration of homologs that are annotated in biomarkers across multiple species. Additionally, BioKA provide four efficient approaches for retrieving biomarkers, including basic keyword search, advanced batch search, gene/protein sequence search and knowledge graph search. And query results can be freely downloaded. The comprehensive knowledge provided by BioKA not only advances human disease research but also enhances our understanding of animal diseases and supports livestock breeding endeavors.
Table 3.
The features of existed biomarker resources
| Database | Species | Disease/trait | Breed/strain | Regulatory | Homologs | Multi-omics | Knowledge graph |
|---|---|---|---|---|---|---|---|
| BCSCdb | Human | Various human cancer | × | √ | × | × | × |
| CBD | Human | Colorectal cancer | × | × | × | × | × |
| CellMarker | Human, mouse | Various cancer | × | × | × | × | × |
| DAAB-v2 | Human, mouse, rat | Allergy, asthma | √ | √ | × | × | × |
| EBD | Human | Eye related disease | × | × | × | × | × |
| HFBD | Human | Heart failure | × | × | × | × | × |
| MarkerDB | Human | Various human disease | × | × | × | × | × |
| ResMarkerDB | Human | Breast, colorectal cancer | × | × | × | × | × |
| BioKA | Human and 30 animals | Various disease/trait | √ | √ | √ | √ | √ |
In the future, we will continuously maintain and update BioKA by curating and integrating more biomarker findings. Our plans include curating additional types of biomarkers, such as image and cell biomarkers, and integrating high-quality biomarkers sourced from reputable public resources like CellMarker (28) to further enrich the biomarker repertoire. Moreover, we aim to expand the organism coverage by incorporating reptiles such as snakes and turtles, as well as aquatic animals like shrimp, to meet the diverse research requirements. Additionally, we intend to develop a user-friendly biomarker submission feature to facilitate the timely release and sharing of newly discovered biomarkers by researchers.
Acknowledgements
We thank G.V.M., G.E.N., H.G.D. team in National Genomics Data Center (NGDC) for providing data retrieve interface, and thank the high-performance computing platform of NGDC for providing the powerful computational resources.
Contributor Information
Yibo Wang, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yihao Lin, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Sicheng Wu, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jiani Sun, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yuyan Meng, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Enhui Jin, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Demian Kong, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Guangya Duan, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Shaoqi Bei, Qilu University of Technology (Shandong Academy of Sciences), Shandong 250353, China.
Zhuojing Fan, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Gangao Wu, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Lili Hao, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Shuhui Song, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Bixia Tang, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Wenming Zhao, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Data availability
BioKA is available online for free at https://ngdc.cncb.ac.cn/bioka and does not require user registration.
Funding
National Natural Science Foundation of China [32100506, 32170678, 32100511]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
References
- 1. Zhao X., Modur V., Carayannopoulos L.N., Laterza O.F.. Biomarkers in pharmaceutical research. Clin. Chem. 2015; 61:1343–1353. [DOI] [PubMed] [Google Scholar]
- 2. Califf R.M. Biomarker definitions and their applications. Exp. Biol. Med. (Maywood). 2018; 243:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lippi G., Mattiuzzi C.. The biomarker paradigm: between diagnostic efficiency and clinical efficacy. Pol. Arch. Med. Wewn. 2015; 125:282–288. [DOI] [PubMed] [Google Scholar]
- 4. Ahmad A., Imran M., Ahsan H.. Biomarkers as biomedical bioindicators: approaches and techniques for the detection, analysis, and validation of novel Biomarkers of diseases. Pharmaceutics. 2023; 15:1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goldansaz S.A., Guo A.C., Sajed T., Steele M.A., Plastow G.S., Wishart D.S.. Livestock metabolomics and the livestock metabolome: a systematic review. PLoS One. 2017; 12:e0177675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khalilpour A., Kilic T., Khalilpour S., Alvarez M.M., Yazdi I.K.. Proteomic-based biomarker discovery for development of next generation diagnostics. Appl. Microbiol. Biotechnol. 2017; 101:475–491. [DOI] [PubMed] [Google Scholar]
- 7. Wheelock C.E., Goss V.M., Balgoma D., Nicholas B., Brandsma J., Skipp P.J., Snowden S., Burg D., D’Amico A., Horvath I.et al.. Application of 'omics technologies to biomarker discovery in inflammatory lung diseases. Eur. Respir. J. 2013; 42:802–825. [DOI] [PubMed] [Google Scholar]
- 8. Lin Y., Qian F., Shen L., Chen F., Chen J., Shen B.. Computer-aided biomarker discovery for precision medicine: data resources, models and applications. Brief Bioinform. 2019; 20:952–975. [DOI] [PubMed] [Google Scholar]
- 9. Hristova V.A., Chan D.W.. Cancer biomarker discovery and translation: proteomics and beyond. Expert Rev. Proteomics. 2019; 16:93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu D., Zhang P., Ma J., Xu J., Yang L., Xu W., Que H., Chen M., Xu H.. Serum biomarker panels for the diagnosis of gastric cancer. Cancer Med. 2019; 8:1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Picca A., Calvani R., Coelho-Junior H.J., Landi F., Marzetti E.. Anorexia of aging: metabolic changes and biomarker discovery. Clin. Interv. Aging. 2022; 17:1761–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang C.L., Li Y.J., Lu S., Zhang T., Xiao R., Luo H.R.. Fluoxetine ameliorates depressive symptoms by regulating lncRNA expression in the mouse hippocampus. Zool Res. 2021; 42:28–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeromin A., Bowser R.. Biomarkers in neurodegenerative diseases. Adv. Neurobiol. 2015; 15:491–528. [DOI] [PubMed] [Google Scholar]
- 14. Mobasheri A. Osteoarthritis year 2012 in review: biomarkers. Osteoarthritis Cartilage. 2012; 20:1451–1464. [DOI] [PubMed] [Google Scholar]
- 15. Munjal A., Bapat S., Hubbard D., Hunter M., Kolhe R., Fulzele S.. Advances in molecular biomarker for early diagnosis of osteoarthritis. Biomol. Concepts. 2019; 10:111–119. [DOI] [PubMed] [Google Scholar]
- 16. Myers M.J., Smith E.R., Turfle P.G.. Biomarkers in Veterinary medicine. Annu. Rev. Anim. Biosci. 2017; 5:65–87. [DOI] [PubMed] [Google Scholar]
- 17. Yan Y., Zhang H., Gao S., Zhang H., Zhang X., Chen W., Lin W., Xie Q.. Differential DNA methylation and gene expression between ALV-J-positive and ALV-J-negative chickens. Front Vet Sci. 2021; 8:659840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ichii O., Ohta H., Horino T., Nakamura T., Hosotani M., Mizoguchi T., Morishita K., Nakamura K., Sasaki N., Takiguchi M.et al.. Urinary exosome-derived microRNAs reflecting the changes in renal function in cats. Front. Vet. Sci. 2018; 5:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Galen G.V., Olsen E., Siwinska N.. Biomarkers of kidney disease in horses: a review of the current literature. Animals (Basel). 2022; 12:2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Twomey J.D., Brahme N.N., Zhang B.. Drug-biomarker co-development in oncology - 20 years and counting. Drug Resist. Updat. 2017; 30:48–62. [DOI] [PubMed] [Google Scholar]
- 21. Aigner B., Renner S., Kessler B., Klymiuk N., Kurome M., Wunsch A., Wolf E.. Transgenic pigs as models for translational biomedical research. J. Mol. Med. (Berl.). 2010; 88:653–664. [DOI] [PubMed] [Google Scholar]
- 22. Quan M., Oh Y., Cho S.Y., Kim J.H., Moon H.G.. Polo-like Kinase 1 regulates chromosomal instability and paclitaxel resistance in breast cancer cells. J. Breast Cancer. 2022; 25:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pandey U.B., Nichols C.D.. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 2011; 63:411–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. T X., X Z., X X., J L., L Z., F G.. Physiochemical properties, protein and metabolite profiles of muscle exudate of chicken meat affected by wooden breast myopathy. Food Chem. 2020; 316:126271. [DOI] [PubMed] [Google Scholar]
- 25. Chapinal N., Carson M.E., LeBlanc S.J., Leslie K.E., Godden S., Capel M., Santos J.E., Overton M.W., Duffield T.F.. The association of serum metabolites in the transition period with milk production and early-lactation reproductive performance. J. Dairy Sci. 2012; 95:1301–1309. [DOI] [PubMed] [Google Scholar]
- 26. Firdous S., Ghosh A., Saha S.. BCSCdb: a database of biomarkers of cancer stem cells. Database (Oxford). 2022; 2022:baac082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang X., Sun X.F., Cao Y., Ye B., Peng Q., Liu X., Shen B., Zhang H.. CBD: a biomarker database for colorectal cancer. Database (Oxford). 2018; 2018:bay046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu C., Li T., Xu Y., Zhang X., Li F., Bai J., Chen J., Jiang W., Yang K., Ou Q.et al.. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2023; 51:D870–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sircar G., Saha B., Jana T., Dasgupta A., Gupta Bhattacharya S., Saha S.. DAAB: a manually curated database of allergy and asthma biomarkers. Clin. Exp. Allergy. 2015; 45:1259–1261. [DOI] [PubMed] [Google Scholar]
- 30. Zhang X., Kong L., Liu S., Zhang X., Shang X., Zhu Z., Huang Y., Ma S., Jason H., Kiburg K.V.et al.. EBD: an eye biomarker database. Bioinformatics. 2023; 39:btad194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang X., Chai Z., Pan G., Hao Y., Li B., Ye T., Li Y., Long F., Xia L., Liu M.. ExoBCD: a comprehensive database for exosomal biomarker discovery in breast cancer. Brief. Bioinform. 2021; 22:bbaa088. [DOI] [PubMed] [Google Scholar]
- 32. He H., Shi M., Lin Y., Zhan C., Wu R., Bi C., Liu X., Ren S., Shen B.. HFBD: a biomarker knowledge database for heart failure heterogeneity and personalized applications. Bioinformatics. 2021; 37:4534–4539. [DOI] [PubMed] [Google Scholar]
- 33. Wishart D.S., Bartok B., Oler E., Liang K.Y.H., Budinski Z., Berjanskii M., Guo A., Cao X., Wilson M.. MarkerDB: an online database of molecular biomarkers. Nucleic Acids Res. 2021; 49:D1259–D1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perez-Granado J., Pinero J., Furlong L.I.. ResMarkerDB: a database of biomarkers of response to antibody therapy in breast and colorectal cancer. Database (Oxford). 2019; 2019:baz060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sheng P., Zhu H., Zhang W., Xu Y., Peng W., Sun J., Gu M., Jiang H.. The immunoglobulin superfamily member 3 (IGSF3) promotes hepatocellular carcinoma progression through activation of the NF-kappaB pathway. Ann. Transl. Med. 2020; 8:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng J., Ma H., Huang Y., Li J., Li W.. Ruminococcaceae_UCG-013 promotes obesity resistance in mice. Biomedicines. 2022; 10:3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. RE M., J K., MK D., PD W.. Biomarker discovery in animal health and disease: the application of post-genomic technologies. Biomark. Insights. 2007; 2:185–196. [PMC free article] [PubMed] [Google Scholar]
- 38. Khan S., Jha A., Panda A.C., Dixit A.. Cancer-associated circRNA–miRNA–mRNA Regulatory networks: a meta-analysis. Front. Mol. Biosci. 2021; 8:671309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fang Z., Ruan B., Zhong M., Xiong J., Jiang Y., Song Z.. Silencing LINC00491 inhibits pancreatic cancer progression through MiR-188-5p-induced inhibition of ZFP91. J. Cancer. 2022; 13:1808–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. EV. K. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005; 39:309–338. [DOI] [PubMed] [Google Scholar]
- 41. Cafuir L.A., Kempton C.L.. Current and emerging factor VIII replacement products for hemophilia A. Ther. Adv. Hematol. 2017; 8:303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaur H., Kumar R., Lathwal A., Raghava G.P.S.. Computational resources for identification of cancer biomarkers from omics data. Brief. Funct. Genomics. 2021; 20:213–222. [DOI] [PubMed] [Google Scholar]
- 43. Cunningham F., Allen J.E., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Austine-Orimoloye O., Azov A.G., Barnes I., Bennett R.et al.. Ensembl 2022. Nucleic Acids Res. 2022; 50:D988–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. UniProt, C. UniProt: the Universal Protein knowledgebase in 2023. Nucleic Acids Res. 2023; 51:D523–D531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P.et al.. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021; 49:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kozomara A., Birgaoanu M., Griffiths-Jones S.. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019; 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McGeary S.E., Lin K.S., Shi C.Y., Pham T.M., Bisaria N., Kelley G.M., Bartel D.P.. The biochemical basis of microRNA targeting efficacy. Science. 2019; 366:eaav1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang H.Y., Lin Y.C., Cui S., Huang Y., Tang Y., Xu J., Bao J., Li Y., Wen J., Zuo H.et al.. miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2022; 50:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem 2023 update. Nucleic Acids Res. 2023; 51:D1373–D1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. CNCB-NDGC Members and Partners Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2023. Nucleic Acids Res. 2023; 51:D18–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li C., Tian D., Tang B., Liu X., Teng X., Zhao W., Zhang Z., Song S.. Genome variation Map: a worldwide collection of genome variations across multiple species. Nucleic Acids Res. 2021; 49:D1186–D1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y., Zou D., Zhu T., Xu T., Chen M., Niu G., Zong W., Pan R., Jing W., Sang J.et al.. Gene Expression Nebulas (GEN): a comprehensive data portal integrating transcriptomic profiles across multiple species at both bulk and single-cell levels. Nucleic Acids Res. 2022; 50:D1016–D1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Duan G., Wu G., Chen X., Tian D., Li Z., Sun Y., Du Z., Hao L., Song S., Gao Y.et al.. HGD: an integrated homologous gene database across multiple species. Nucleic Acids Res. 2023; 51:D994–D1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tang B., Zhou Q., Dong L., Li W., Zhang X., Lan L., Zhai S., Xiao J., Zhang Z., Bao Y.et al.. iDog: an integrated resource for domestic dogs and wild canids. Nucleic Acids Res. 2019; 47:D793–D800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith C.L., Eppig J.T.. The Mammalian Phenotype ontology as a unifying standard for experimental and high-throughput phenotyping data. Mamm. Genome. 2012; 23:653–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinf. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang X., Tian M., Lin Y., Li L., Sun X., Zhang Z., Kang M., Lin J.. Characterization of the roles of suppressor of cytokine signaling-3 in esophageal carcinoma. Hum. Gene Ther. 2023; 34:495–517. [DOI] [PubMed] [Google Scholar]
- 58. Inagaki-Ohara K., Kondo T., Ito M., Yoshimura A.. SOCS, inflammation, and cancer. JAKSTAT. 2013; 2:e24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bai J., Smock S.L., Jackson G.R. Jr., MacIsaac K.D., Huang Y., Mankus C., Oldach J., Roberts B., Ma Y.L., Klappenbach J.Aet al.. Phenotypic responses of differentiated asthmatic human airway epithelial cultures to rhinovirus. PLoS One. 2015; 10:e0118286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Z T., M X., J Z., Y F., Z H., C T., Y L.. Pentagalloylglucose inhibits the replication of rabies virus via mediation of the miR-455/SOCS3/STAT3/IL-6 pathway. J. Virol. 2019; 93:e00539-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li W., Qi N., Wang S., Jiang W., Liu T.. miR-455-5p regulates atrial fibrillation by targeting suppressor of cytokines signaling 3. J. Physiol. Biochem. 2021; 77:481–490. [DOI] [PubMed] [Google Scholar]
- 62. Wang M., Ibeagha-Awemu E.M.. Impacts of epigenetic processes on the health and productivity of livestock. Front. Genet. 2020; 11:613636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Eppig J.T. Mouse genome informatics (MGI) resource: genetic, genomic, and biological knowledgebase for the laboratory mouse. ILAR J. 2017; 58:17–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thurmond J., Goodman J.L., Strelets V.B., Attrill H., Gramates L.S., Marygold S.J., Matthews B.B., Millburn G., Antonazzo G., Trovisco V.et al.. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019; 47:D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
BioKA is available online for free at https://ngdc.cncb.ac.cn/bioka and does not require user registration.