Abstract
Transcription factors (TFs), transcription co-factors (TcoFs) and their target genes perform essential functions in diseases and biological processes. KnockTF 2.0 (http://www.licpathway.net/KnockTF/index.html) aims to provide comprehensive gene expression profile datasets before/after T(co)F knockdown/knockout across multiple tissue/cell types of different species. Compared with KnockTF 1.0, KnockTF 2.0 has the following improvements: (i) Newly added T(co)F knockdown/knockout datasets in mice, Arabidopsis thaliana and Zea mays and also an expanded scale of datasets in humans. Currently, KnockTF 2.0 stores 1468 manually curated RNA-seq and microarray datasets associated with 612 TFs and 172 TcoFs disrupted by different knockdown/knockout techniques, which are 2.5 times larger than those of KnockTF 1.0. (ii) Newly added (epi)genetic annotations for T(co)F target genes in humans and mice, such as super-enhancers, common SNPs, methylation sites and chromatin interactions. (iii) Newly embedded and updated search and analysis tools, including T(co)F Enrichment (GSEA), Pathway Downstream Analysis and Search by Target Gene (BLAST). KnockTF 2.0 is a comprehensive update of KnockTF 1.0, which provides more T(co)F knockdown/knockout datasets and (epi)genetic annotations across multiple species than KnockTF 1.0. KnockTF 2.0 facilitates not only the identification of functional T(co)Fs and target genes but also the investigation of their roles in the physiological and pathological processes.
Graphical Abstract
Graphical Abstract.
Introduction
The complex transcriptional regulation is a fundamental process in establishing and maintaining biological systems (1). The major regulators of the transcriptional regulation programs are transcription factors (TFs) and transcription co-factors (TcoFs) (2). TFs are DNA-binding proteins that recognize their TF binding sites (TFBSs) located in cis-regulatory regions (promoters, enhancers and super-enhancers) of target genes (3). The binding of TFs to these regulatory regions directly controls the regulation of their target gene expression, thereby affecting almost all biological processes (4). TcoFs are proteins that are not DNA-binding in the context of transcriptional regulation, but are involved in interacting with TFs (5). Meanwhile, the chromatin features within T(co)F-bound cis-regulatory regions, such as SNP, expression quantitative trait locus (eQTL), chromatin interaction, chromatin accessibility and DNA methylation are synergistic to supervise gene transcription (6). Thus, effective identification and comprehensive (epi)genetic annotation for TFs, TcoFs and their target genes are crucial for investigating T(co)F functions and gene expression regulation.
Chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) technique and gene expression profile analysis technique before and after T(co)F knockdown or knockout have been used for experimentally identifying T(co)Fs bound to specific locations in the genome and exploring T(co)F functions. Based on these techniques, an increasing number of databases have been developed to systematically identify and annotate TFs or TcoFs. For example, AnimalTFDB is an animal TF and TcoF database that classifies and annotates genome-wide TFs and TcoFs (7). JASPAR is an open-access database containing manually curated, nonredundant TF-binding profiles (8). PlantTFDB is a plant TF database including well-annotated TFs for 165 plant species (9). In addition, GenomeCRISPR collects CRISPR/Cas9 screening datasets in human cell lines and enables users to explore the behavior of genes (10). CRISP-view is a database of CRISPR- or RNAi-based genetic screening spanning various phenotypes, including in vitro and in vivo cell proliferation or viability, immunotherapy response, virus infection and protein expression (11). We developed KnockTF 1.0 in 2019 to provide a large number of available resources for human gene expression profile datasets associated with TF knockdown and knockout and annotate TFs and their target genes in a tissue/cell type-specific manner (12).
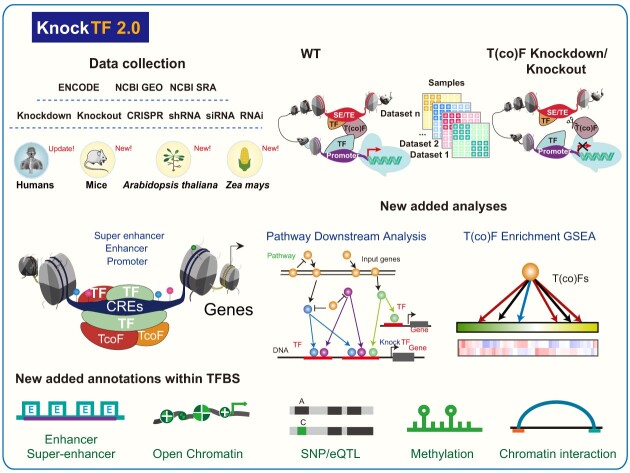
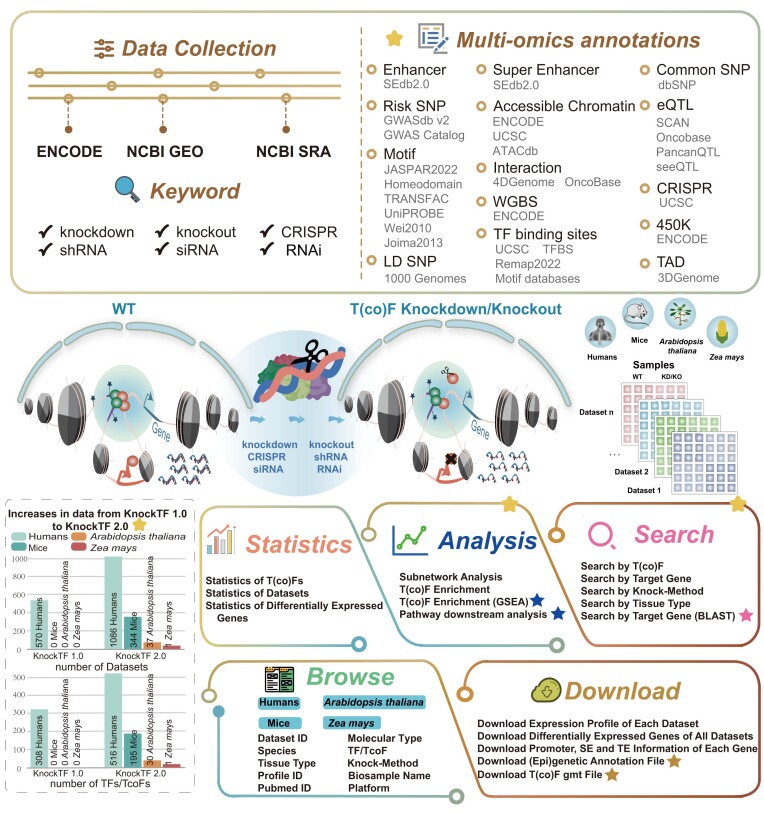
In the recent 3 years, abundant ChIP-seq data and RNA-seq/microarray data before/after T(co)F knockdown/knockout were newly released to decode specific functional properties and gene regulatory networks. Moreover, T(co)F knockdown/knockout experiments in mice can reveal the gene regulatory mechanism in various tissues such as the heart and brain (13). The TF knockdown/knockout data in Arabidopsis thaliana and Zea mays have been generated to model plant disease resistance and contribute to exploring and understanding of plant gene function, disease onset and drug target discovery (14,15). KnockTF 1.0 is insufficient to meet the needs of T(co)F investigation in multiple species. A central challenge is storing, processing and evaluating T(co)F knockdown/knockout datasets spanning multiple tissue/cell types of different species and in a standardized manner, where users can gain new biological insights through data mining. Thus, we developed KnockTF 2.0 (http://www.licpathway.net/KnockTF/index.html), an updated and significantly expanded database, to provide more T(co)F knockdown/knockout datasets and analysis functions for dissecting the transcriptional regulatory cues in humans, mice, A. thaliana and Z. mays (Figure 1). Specifically, KnockTF 2.0 stores 1468 manually curated RNA-seq and microarray datasets associated with 612 TFs and 172 TcoFs disrupted by different knockdown/knockout techniques and across multiple tissue/cell types of different species (∼2.5 times larger than that of KnockTF 1.0). Compared with KnockTF 1.0, KnockTF 2.0 newly supported 304 TFs and 172 TcoFs (∼2 times larger than that of KnockTF 1.0), greatly enhancing the ability and coverage for investigating T(co)F-related transcription regulation. Besides the expanded T(co)F knockdown/knockout datasets, KnockTF 2.0 added the detailed and abundant (epi)genetic annotation information for T(co)F target genes, including super-enhancers, enhancers, TFBSs, common SNPs, risk SNPs, linkage disequilibrium (LD) SNPs, eQTLs, methylation sites, DNase I hypersensitivity sites (DHSs), chromatin interactions, chromatin accessibility regions, CRISPR/Cas9 target sites and topologically associating domains (TADs), in humans and mice. These annotations deepen our understanding of the multidimensional regulatory mechanism of the binding of T(co)Fs to their target genes.
Figure 1.
Database content and construction. KnockTF 2.0 provides a large number of manually curated T(co)F knockdown/knockout datasets in humans, mice, A. thaliana and Z. mays. Detailed and abundant (epi)genetic annotation information for TF target genes can be collected, including super-enhancers, enhancers, TFBSs, common SNPs, risk SNPs, CRISPR/Cas9 target sites and so forth. KnockTF 2.0 contains a variety of functions to browse, search, analyze, and download T(co)F knockdown/knockout datasets.
TFs, TcoFs and their target genes can constitute a complex transcriptional regulatory network, which is worthy of in-depth discussion and research. In recent years, the regulatory network model mediated by TFs or TcoFs has been largely revealed by enrichment analysis methods or algorithmic techniques. Several popular prediction algorithms have been developed, such as MARGE (16), BART (17), Lisa (18) and ANANSE (19), which perform precise TF enrichment in different dimensions by collecting TF target genes, constructing gene reverse engineering networks and integrating epigenetic data. These algorithms and ideas of integrating multi-omics data to accurately predict TFs prompted us to perform the enrichment analysis of T(co)F target genes and transcriptional regulatory network analysis. Therefore, we added a new analysis function ‘T(co)F Enrichment (GSEA)’ to the KnockTF 2.0 database. We also added new analysis and search functions ‘Pathway Downstream Analysis’ and ‘Search by Target Gene (BLAST)’ for better use of the database (Table 1). Overall, KnockTF 2.0 is a user-friendly platform for exploring T(co)F functions and the expression and (epi)genetic annotations of their target genes, which may help detect gene expression and transcriptional regulation in complex diseases and biological processes and provide valuable data resources for scientific research.
Table 1.
KnockTF 2.0 data content compared with KnockTF 1.0
| Function type | Data type/specific function | KnockTF 1.0 | KnockTF 2.0 | Fold increase |
|---|---|---|---|---|
| Interaction table | Species | Humans | Humans, Mice, Arabidopsis thaliana, Zea mays | 4 |
| Dataset | 570 | 1468 | ∼2.5 | |
| TF | 308 | 612 | ∼2 | |
| TcoF | 0 | 172 | New | |
| Annotation | Super-enhancer | 331 601 | 1 717 744 | ∼5 |
| Enhancer | 14 867 092 | 79 709 120 | ∼5 | |
| Common SNP | No | Yes | New | |
| Risk SNP | No | Yes | New | |
| LD SNP | No | Yes | New | |
| eQTL | No | Yes | New | |
| Methylation site | No | Yes | New | |
| CRISPR | No | Yes | New | |
| Chromatin accessibility | No | Yes | New | |
| Chromatin interaction region | No | Yes | New | |
| DHS | No | Yes | New | |
| TAD | No | Yes | New | |
| TF ChIP-seq | Yes | Yes | – | |
| TF motif | Yes | Yes | – | |
| Analysis function | T(co)F Enrichment (GSEA) | No | Yes | New |
| Pathway Downstream Analysis | No | Yes | New | |
| Subnetwork Analysis | Yes | Yes | – | |
| T(co)F Enrichment | Yes | Yes | – | |
| Search function | Search by Target Gene (BLAST) | No | Yes | New |
| Search by T(co)F | Yes | Yes | – | |
| Search by Target Gene | Yes | Yes | – | |
| Search by Knock-Method | Yes | Yes | – | |
| Search by Tissue Type | Yes | Yes | – |
Data expansion and preprocessing
T(co)F knockdown/knockout datasets
A significant data improvement from KnockTF 1.0 to KnockTF 2.0 is the increase in multispecies data. Currently, KnockTF 2.0 contains 344 T(co)F knockdown/knockout datasets in mice, 37 datasets in A. thaliana and 1 dataset in Z. mays, and also extended 1086 T(co)F knockdown/knockout datasets in humans. The T(co)F knockdown/knockout datasets in KnockTF 2.0 are 2.5 times larger than those in KnockTF 1.0. The datasets correspond to 612 TFs and 172 TcoFs, which are about 2 times larger than those in KnockTF 1.0. Specifically, raw knockdown and knockout data were collected from NCBI GEO/SRA (20) using keywords ‘knockdown’, ‘knockout’, ‘shRNA’, ‘siRNA’ and ‘CRISPR’. The collected results were manually reviewed by reading the title, summary and overall design of each GSE series and the detailed protocol of each GSM sample. After a careful manual review, the identified knockdown and knockout data were matched against the TF/TcoF lists from AnimalTFDB (7), TcoFBase (21), TcoF-DB (22) and PlantTFDB (9). The matched T(co)F knockdown and knockout data were manually checked to confirm whether they met the following criteria: (i) Each dataset contained the same GSE series and GPL platform. (ii) The same TF or TcoF was perturbed using a certain knockdown or knockout technique for each dataset. (iii) Each dataset was processed in a particular kind of tissue/cell type of the same species. (iv) Each dataset contained both before and after knockdown/knockout samples. Data that met these criteria were filtered and organized into one T(co)F knockdown/knockout dataset. The whole process was manually checked by at least two researchers to ensure high quality. As a result, 821 new T(co)F knockdown/knockout datasets were collected from the NCBI GEO/SRA database (20), containing 487 T(co)Fs from 626 series and 109 platforms. Some of the data were collected from the ENCODE database (23). By screening for knockdown/knockout techniques and target categories, 77 new T(co)F knockdown/knockout datasets were collected, each subject to one knockdown or knockout technique for a particular human tissue/cell type. Gene expression profiles and gene quantification files were downloaded from the GEO and ENCODE databases, respectively. Statistical significances for differential expression were calculated using fold change (FC) and limma (24), as for KnockTF 1.0.
Genetic and epigenetic annotations
We emphatically expanded the detailed and abundant (epi)genetic annotation information for TF target genes to better decipher gene expression programs regulated by TFs or TcoFs in the T(co)F knockdown/knockout datasets (Supplementary Table S1). All these genetic and epigenetic annotations could help better understand the regulatory mechanism of TFs and TcoFs.
Enhancers and super-enhancers
KnockTF 2.0 significantly added mouse enhancer and super-enhancer sets and extended the scale of human enhancer and super-enhancer sets. Specifically, 1 167 518 super-enhancers from 1739 human H3K27ac ChIP-seq samples and 550 226 super-enhancers from 931 mouse H3K27ac ChIP-seq samples were collected from the SEdb 2.0 database, which was developed by our group in a previous study (25). Similarly, the number of enhancers was also dramatically increased, containing 79 664 341 human enhancers and 44 779 mouse enhancers collected from SEdb 2.0. Currently, the ChIP-seq samples related to enhancers and super-enhancers in KnockTF 2.0 are more than five times larger than those in KnockTF 1.0.
TF binding data
KnockTF 2.0 largely extended TF ChIP-seq data and motif data for identifying TFs that bound to cis-regulatory regions of their target genes. First, 51 616 973 nonredundant binding regions of 817 human TFs and 32 985 444 nonredundant binding regions of 648 mouse TFs were collected from ReMap 2022 (26) across multiple tissue or cell types. BEDTools (27) was used for identifying TF-binding peaks that overlapped with the promoters, enhancers, or super-enhancers of genes in each T(co)F knockdown/knockout dataset. Second, we collected position weight matrices of 869 vertebrate TF motifs from JASPAR 2022 (8), UniPROBE (28), Homeodomains (29), Jolma2013 (30) and Wei2010 (31). A total of 665 plant TF motifs were downloaded from JASPAR 2022 (8). The Find Individual Motif Occurrences program (32) with a threshold of P < 1.0E-06 was used for scanning motif occurrences and further identifying TF motifs within the promoters, enhancers, or super-enhancers of target genes.
Common SNP/LD SNP/risk SNP/Eqtl
We downloaded 38 063 729 human common SNPs from dbSNP (33) and used VCFTools (v0.1.13) to screen SNPs with a minimum allelic frequency >0.05. LD SNPs of five super-populations (African, Ad Mixed American, East Asian, European and South Asian) were calculated using phased genotype information accompanying the 1000 Genomes Project phase 3 (34). Meanwhile, we obtained 264 514 human risk SNPs from GWAS Catalog and GWASdb v2 (35) and collected 2886133 human eQTLs from PancanQTL (36), seeQTL (37), SCAN (38) and OncoBase (39).
Methylation/CRISPR
We obtained DNA methylation states of 30 392 523 methylation sites of 450k array and 166 855 665 methylation sites of whole-genome shotgun bisulfite sequencing from ENCODE (23). We also downloaded CRISPR/Cas9 target sites from UCSC (40), which were annotated with predicted specificity (off-target effects) and predicted efficiency (on-target cleavage) using the CRISPOR tool (41).
Chromatin interaction/DHS/chromatin accessibility region/ TAD
We downloaded the chromatin interaction data from 4DGenome (42) and OncoBase (39), including data from ChIA-PET 3C, 4C, 5C and Hi-C. DHS annotation data of cis-regulatory regions were downloaded from UCSC (40) and ENCODE (23). A total of 69 860 705 human DHSs of 293 samples and 9 802 229 mouse DHSs of 56 samples were obtained. We collected more than 130 000 000 chromatin accessibility regions from ATACdb, which was developed by our group in a previous study (43). In addition, 72 019 human TADs covering 21 tissues or cell lines were obtained from the 3D Genome Browser (44).
Database improved user interface
New effective analysis tools in the ‘Analysis’ page
KnockTF 1.0 provided two analytical tools to explore transcriptional regulatory networks in depth, including Subnetwork Analysis and TF Enrichment. These analytical functions depended on a TF–differentially expressed gene (DEG) network. This network was constructed by combining all nonredundant TF–DEG pairs of the TF knockdown/knockout datasets, with TFs and their DEGs as nodes and TF–DEG pairs as edges. Currently, in KnockTF 2.0, we relied on the new datasets to reconstruct human and mouse T(co)F–DEG networks separately for species-specific network analysis. More importantly, we added two new analytical tools named ‘T(co)F Enrichment (GSEA)’ and ‘Pathway Downstream Analysis’.
T(co)F Enrichment (GSEA)
The enrichment analysis of TF target genes is an important issue in studying transcription regulation. In the ‘T(co)F Enrichment (GSEA)’ function, KnockTF 2.0 embedded the popular gene set enrichment analysis (GSEA) algorithm (45) to perform the enrichment analysis of T(co)F target genes. Based on the T(co)F target gene pairs identified from T(co)F knockdown/knockout profiles and TF ChIP-seq/motif data as the background gene sets, users could input the differential gene rank list and select relevant parameters, including gene set size, permutation times, P-value/adjusted P-value cutoffs, and species to perform T(co)F enrichment analysis and visualize and download analysis results. Specifically, we considered the top 100 genes with the most significant differential expression changes in the gene expression profile of each T(co)F knockdown/knockout dataset as a gene set S, so that each gene set corresponded to a TF or TcoF. The disease-related differential gene rank list entered by the users was referred to as the rank list L of GSEA. The objective of GSEA was to determine whether the members of a gene set S were mainly located at the top or bottom of L. If the members are significantly located at the top or bottom of L under the threshold of P < 0.01, it indicated that the corresponding TF or TcoF regulated the user-entered disease-related DEGs. Thus, all the potential T(co)Fs that played key roles in regulating disease occurrence and progression were identified.
Pathway Downstream Analysis
TFs are usually located in the terminal of signaling pathways. They can strongly control the expression of cell identity-specific genes by binding to DNA regulatory elements. However, the downstream information of TFs is absent in signaling pathways. To address this issue, KnockTF 2.0 provides T(co)F-related pathway downstream analysis. In the ‘Pathway Downstream Analysis’ function, we manually collected 2881 pathways from 10 pathway databases and innovatively reconstructed traditional signaling pathways by integrating T(co)F target gene relationships from T(co)F knockdown/knockout datasets. Entering a list of genes of interest, users can select relevant parameters. KnockTF 2.0 can map them into the reconstructed signaling pathway map model. A hypergeometric test was used to calculate the statistical significance of the intersection between the input gene nodes and the TFs in the terminal of each pathway. KnockTF 2.0 can also identify the significant pathways and label the terminal downstream T(co)Fs of pathways. In the detail page, users can find target genes of the terminal T(co)Fs. Hence, users can obtain the regulatory axes of the genes of interest, including pathway genes, T(co)Fs and their downstream target genes.
A new ‘Search’ interface for conveniently retrieving
KnockTF 2.0 has a more user-friendly inquiry mode. Currently, it provides five kinds of query methods, including ‘Search by TF’, ‘Search by Target Gene’, ‘Search by Knock-Method’, ‘Search by Tissue Type’ and ‘Search by Target Gene (BLAST)’. Among these, ‘Search by Target Gene (BLAST)’ was newly added in KnockTF 2.0. When the user enters a gene sequence of interest and selects species and the degree of differential expression, KnockTF 2.0 can align the corresponding gene name related to the sequence by running the BLAST program (46,47). Then, T(co)F knockdown/knockout datasets in which the gene acted as a T(co)F target are presented as an interactive table on the result page. In every row of the table, the information including Dataset ID, Target Gene, T(co)F, Molecular Type, Knock-Method, Tissue Type, Biosample Name, Profile ID, Platform, Mean expression of control samples, Mean expression of knockdown/knockout samples, FC and log2FC are described. Users can click ‘Dataset ID’ to view details of each T(co)F knockdown or knockout dataset and click ‘Target Gene’ to view detailed descriptions of each target gene.
More user-friendly updates for quick retrieval
We rewrote the websites and analysis code and updated new website servers in KnockTF 2.0 to improve the access speed and user experience. KnockTF 2.0 provides a newly designed and user-friendly browsing interface. Users can browse through the T(co)F knockdown/knockout datasets of different species by category, including humans, mice, A. thaliana and Z. mays.
Case study
Case study of T(co)F Enrichment (GSEA)
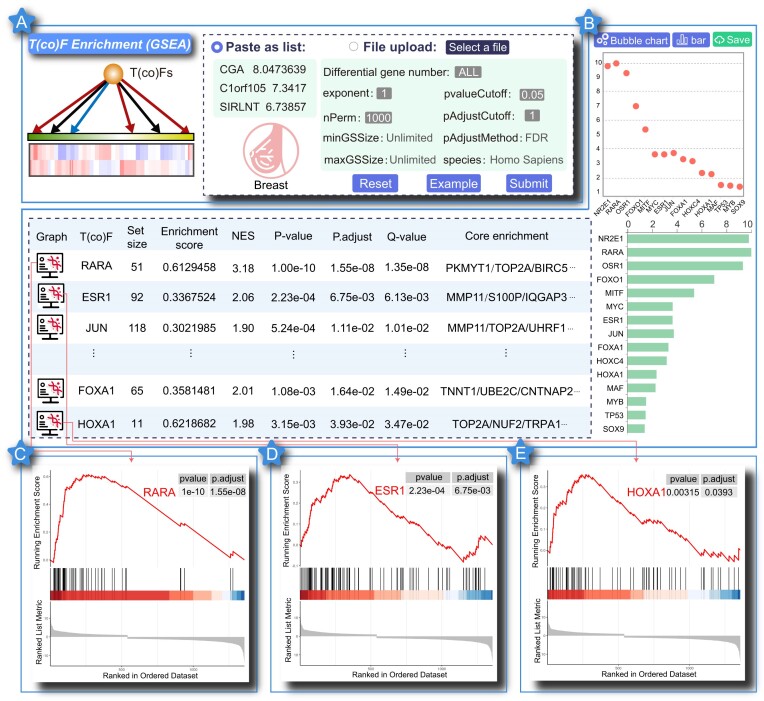
We performed the ‘T(co)F Enrichment (GSEA)’ function by inputting the differentially expressed gene rank list of breast cancer from the TCGA project (http://cancergenome.nih.gov/abouttcga) to demonstrate the new use and potential applications of GSEA-based T(co)F enrichment analysis in human complex diseases (Figure 2A). Thus, in the GSEA program, the breast cancer-related differential gene rank list entered was rank list L and the top 100 differentially expressed genes of each T(co)F knockdown/knockout dataset comprised the gene set S. We selected recommended category options including ‘species: Human, pvalueCutoff: 0.05 and pAdjustCutoff: 1’ (Figure 2A). The analysis results and charts showed that 32 gene sets (S) were significantly enriched at the top or bottom of L with P < 0.01. Each gene set was labeled by the corresponding T(co)F name (Figure 2B). Most of these T(co)Fs were validated to be associated with breast cancer. For example, TF RARA represented the most significantly enriched gene set (P = 1.00e-10, FDR = 1.55e-08), and the enrichment score (ES) was 0.6129 (Figure 2C). The result indicated that TF RARA regulated the differential expression of breast cancer genes and might play a crucial role in the molecular regulation of breast cancer development. In fact, the epigenetic functional plasticity of the RARA mechanism in mammary epithelial cells is necessary for normal morphogenetic processes, especially for preventing the development of breast cancer due to the potential effects of physiological retinoic acid (48). Factors that hinder the epigenetic function of RARA enable physiological retinoic acid to drive abnormal morphogenesis through nontranscriptional RARA, resulting in cell transformation (49,50). The gene set corresponding to TF ESR1 was significantly enriched at the top of the differential gene rank list with P = 2.23e-04 and FDR = 6.75e-03 (ES = 0.3368, NES = 2.06) (Figure 2D). Preclinical and clinical studies showed that ESR1 mutations preexisted in primary tumors and were enriched during metastasis (51). In addition, ESR1 mutations expressed a unique transcriptional profile that favored tumor progression, suggesting that selected ESR1 mutations might influence metastasis (52). Some research groups employed sensitive assays using patient fluid biopsies to track ESR1 or trunk cell mutations to predict tumor progression and treatment effectiveness; some of these techniques could eventually be used to guide sequential treatment regimens for patients (53,54). The gene set corresponding to TF HOXA1 was significantly enriched at the top of the breast cancer differential gene rank list with P = 3.15e-03 and FDR = 3.93e-02 (ES = 0.6219, NES = 1.9804) (Figure 2E). The HOXA1 expression in human breast cancer was documented as early as 25 years ago. A few years later, HOXA1 was identified as a suitable proto-oncogene in breast tissue (55). Over the next 20 years, the molecular data on how the HOXA1 protein acts, the factors that promote activation and maintenance of HOXA1 gene expression, and the identity of its target genes have accumulated and provided a broader perspective on the relationship between TFs and breast tumorigenesis (56,57). The experimental data showed that HOXA1 overexpression alone was sufficient to promote the carcinogenic transformation of breast epithelial cells. Moreover, the HOXA1 overexpression was systematically associated with cancer progression and poor prognosis (58). The aforementioned case analysis indicated that using GSEA-based T(co)F enrichment analysis, KnockTF 2.0 could locate all the potential T(co)Fs that play important roles in the regulation of diseases, providing a theoretical basis for the study of the molecular mechanisms of transcription regulation in human complex diseases.
Figure 2.
Results of ‘T(co)F Enrichment (GSEA)’ analysis function. (A) User input the breast cancer–related differential gene rank list. (B) Results overview for ‘T(co)F Enrichment (GSEA)’ analysis function. (C) GSEA plot of TF RARA. (D) GSEA plot of TF ESR1. (E) GSEA plot of TF HOXA1.
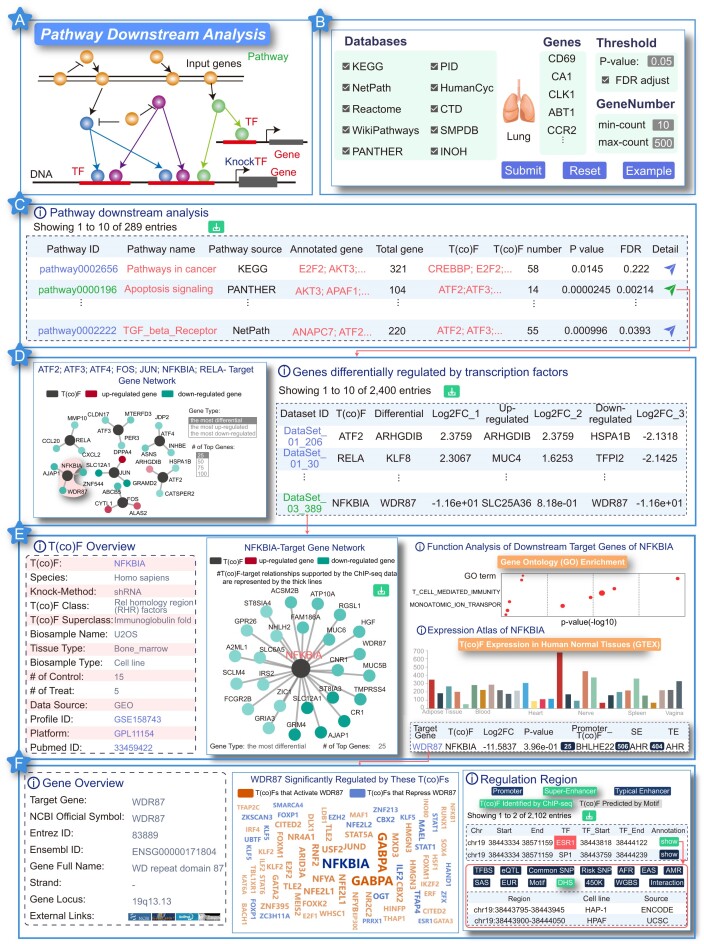
Case study of Pathway Downstream Analysis
We selected 2833 differentially expressed genes associated with pulmonary hypertension as the input to highlight the use of ‘Pathway Downstream Analysis’ (Figure 3A). Parameter options included ‘Databases: All, Threshold: P-value = 0.05 and GeneNumber: [10–500]’ (Figure 3B). On the page of analysis results, 289 significantly enriched signaling pathways were identified, including DNA methylation, apoptosis signaling pathway, T cell receptor signaling pathway, interleukin signaling pathway and so forth. Meanwhile, KnockTF 2.0 labeled terminal downstream TFs of pathways. For example, 32 pulmonary hypertension–related DEGs were mapped to the ‘Apoptosis signaling pathway’ and 7 terminal downstream TFs of this pathway were significantly annotated by hypergeometric test P = 0.0000245, including ATF2, ATF3, ATF4, FOS, JUN, NFKBIA and RELA (Figure 3C). These TFs were associated with pulmonary hypertension. For example, TF NFKBIA, with considerable diagnostic value, was identified as the diagnostic biomarker and actionable target to expand treatment options for patients with pulmonary hypertension (59). When clicking on the ‘detail’ button of ‘Apoptosis signaling pathway’, we found that the target genes of the terminal TFs were regulated by these TFs (Figure 3D). Therefore, we obtained the regulatory axes of the genes of interest, including apoptosis signaling pathway, seven terminal downstream TFs (ATF2, ATF3, ATF4, FOS, JUN, NFKBIA and RELA), and their downstream target genes. We could then view the details of these TFs and their target genes in the corresponding T(co)F knockdown/knockout datasets by clicking on ‘Dataset ID’. For example, when clicking on ‘DataSet_03_389’, the detailed descriptions including TF overview, NFKBIA-target gene network, target gene information, function analysis, upstream pathway analysis and expression analysis were displayed (Figure 3E). Furthermore, by clicking on ‘Target Gene’ (for example, WDR87), we could obtain target gene overview and the abundant (epi)genetic annotation information (Figure 3F). Similarly, for 289 significantly enriched signaling pathways identified, we could obtain 289 regulatory axes including signaling pathways, terminal downstream TFs and their downstream target genes, which provided a good theoretical basis and biological explanation for the study of pulmonary hypertension at the level of biological pathways and transcriptional regulation. Thus, the function of ‘Pathway Downstream Analysis’ could be used for unlocking the full range of disease molecular mechanisms in genomic studies, improving researchers' understanding of biological processes at the molecular level. Normative biological pathways can help us systematically understand higher biological functions and inherent interdependencies.
Figure 3.
Results of ‘Pathway Downstream Analysis’ function. (A) Model diagram of pathway analysis. (B) User input 2833 differentially expressed genes associated with pulmonary hypertension. (C) Results overview for ‘Pathway Downstream Analysis’ function. (D) When clicking on the ‘detail’ button, target genes of the terminal TFs could be found. (E) Details of the TF and their target genes in the corresponding T(co)F knockdown/knockout datasets could be viewed by clicking on ‘Dataset ID’. (F) Detailed information on the target gene overview and the abundant (epi)genetic annotations could be viewed by clicking on ‘Target Gene’.
Discussion and future directions
The universe of transcription regulation has developed rapidly and has become one of the most widely studied fields (60). Identifying TFs, TcoFs and their target genes is key to understanding transcriptional regulatory mechanisms in disease development and biological processes (61). Gene expression profile datasets of multispecies T(co)F knockdown/knockout have accumulated rapidly, providing a basis for identifying T(co)F target genes and clarifying the potential biological functions of T(co)Fs (62). Therefore, we developed the updated version of KnockTF that significantly extended T(co)F knockdown/knockout data for new tissue/cell types in humans. To date, KnockTF is still the first human gene expression profile database of T(co)F knockdown/knockout with the largest number of human T(co)F knockdown/knockout expression data. We also newly added multispecies T(co)F knockdown/knockout datasets to expand the dataset scale. All the datasets span multiple different species, from plants (A. thaliana and Z. mays) to mice to humans, providing an important basis for dissecting the transcriptional regulatory cues across multiple species.
Characterizing the multifaceted contribution of (epi)genetic annotations to T(co)F target genes is a major challenge to investigate genetic and epigenetic influences on transcription mediated by T(co)Fs. KnockTF 2.0 showed the most comprehensive (epi)genetic annotations for T(co)F target genes in each T(co)F knockdown/knockout dataset, including super-enhancers, enhancers, TFBSs, common SNPs, risk SNPs, LD SNPs, eQTLs, methylation sites, DHSs, chromatin interactions, chromatin accessibility regions, CRISPR/Cas9 target sites and TADs. Recent studies demonstrated that the epigenomic landscape of gene regulation, including DNA methylation and chromatin accessibility, can vary considerably, contributing to distinct gene expression programs and biological functions (63). Our previous study showed that some core TFs could form core transcription regulatory circuitry by binding to super-enhancers and further activating the transcription of specific genes in cancer development. The recurrent structural differences at TAD boundaries and significant alterations in intra-TAD chromatin interactions have been identified to reflect differences in gene expression (64). Overall, the annotation results deepen our understanding of genetic and epigenetic regulation of the transcriptional machinery across multiple tissue/cell types of different species, providing a multidimensional and in-depth perspective to gain insight into the regulatory mechanism of T(co)Fs.
KnockTF 2.0 provides particularly useful query and analysis functions to interpret knockdown/knockout technique-mediated transcriptional regulation. Among these, the function of ‘Search by Target Gene (BLAST)’ provides a more friendly way to conveniently retrieve T(co)F knockdown/knockout datasets. The GSEA-based T(co)F enrichment analysis tool can help researchers identify all potential T(co)Fs that play key roles in regulating genes of interest. Also, the functional pathway downstream analysis tool can help researchers identify the significant pathways, label terminal downstream T(co)Fs and obtain the regulatory axes, including pathway genes, T(co)Fs and their downstream target genes. In summary, the updated KnockTF was developed to include major improvements and significantly enhance its utility for a broader community of foundation researchers, cell/molecular biologists, geneticists and data scientists. We believe these improvements can make KnockTF more comprehensive and useful. The gene expression profile data before/after T(co)F knockdown/knockout for various species will continue to grow. We will regularly update the KnockTF database to make it a core resource for T(co)F functions and gene expression regulation.
Supplementary Material
Contributor Information
Chenchen Feng, National Health Commission Key Laboratory of Birth Defect Research and Prevention & School of Computer, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, University of South China, Hengyang, Hunan, 421001, China; School of Medical Informatics, Daqing Campus, Harbin Medical University, Daqing, 163319, China.
Chao Song, The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Institute of Cardiovascular Disease, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Department of Cardiology, Hengyang Medical School, University of South China, Hengyang, China.
Shuang Song, The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, University of South China, Hengyang, Hunan, 421001, China; Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Cell Biology and Genetics, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Guorui Zhang, The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, University of South China, Hengyang, Hunan, 421001, China; Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Cell Biology and Genetics, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Mingxue Yin, The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, University of South China, Hengyang, Hunan, 421001, China; Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Cell Biology and Genetics, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Yuexin Zhang, The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Institute of Cardiovascular Disease, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Department of Cardiology, Hengyang Medical School, University of South China, Hengyang, China.
Fengcui Qian, The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Institute of Cardiovascular Disease, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Department of Cardiology, Hengyang Medical School, University of South China, Hengyang, China.
Qiuyu Wang, National Health Commission Key Laboratory of Birth Defect Research and Prevention & School of Computer, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, University of South China, Hengyang, Hunan, 421001, China; Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Cell Biology and Genetics, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Maozu Guo, School of Electrical and Information Engineering, Beijing University of Civil Engineering and Architecture, Beijing 100044, China.
Chunquan Li, National Health Commission Key Laboratory of Birth Defect Research and Prevention & School of Computer, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, University of South China, Hengyang, Hunan, 421001, China; MOE Key Lab of Rare Pediatric Diseases, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Institute of Cardiovascular Disease, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Data availability
The research community can access information freely in the KnockTF 2.0 without registration or logging in. The URL for KnockTF 2.0 is http://www.licpathway.net/KnockTF/index.html.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Natural Science Foundation of China [62171166, 62001145, 62302206, 62272212, 62031003]; Research Foundation of the First Affiliated Hospital of University of South China for Advanced Talents [20210002-1005 USCAT-2021-01]; China Postdoctoral Science Foundation [2019M661311]; Natural Science Foundation of Hunan Province [2023JJ40594, 2023JJ30536]; Scientific Research Fund Project of Hunan Provincial Health Commission [20201920]; Clinical Research 4310 Program of the University of South China [20224310NHYCG05]; and Natural Science Foundation of Heilongjiang Province [LH2021F044]. Funding for open access charge: National Natural Science Foundation of China [62171166, 62001145, 62302206, 62272212, 62031003]; Research Foundation of the First Affiliated Hospital of University of South China for Advanced Talents [20210002-1005 USCAT-2021-01]; China Postdoctoral Science Foundation [2019M661311]; Natural Science Foundation of Hunan Province [2023JJ40594, 2023JJ30536]; Scientific Research Fund Project of Hunan Provincial Health Commission [20201920]; Clinical Research 4310 Program of the University of South China [20224310NHYCG05]; and Natural Science Foundation of Heilongjiang Province [LH2021F044].
Conflict of interest statement. None declared.
References
- 1. Lee T.I., Young R.A.. Transcriptional regulation and its misregulation in disease. Cell. 2013; 152:1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zabidi M.A., Stark A.. Regulatory enhancer-core-promoter communication via transcription factors and cofactors. Trends Genet. 2016; 32:801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T.. The Human transcription factors. Cell. 2018; 172:650–665. [DOI] [PubMed] [Google Scholar]
- 4. Reiter F., Wienerroither S., Stark A.. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 2017; 43:73–81. [DOI] [PubMed] [Google Scholar]
- 5. Gill G. Regulation of the initiation of eukaryotic transcription. Essays Biochem. 2001; 37:33–43. [DOI] [PubMed] [Google Scholar]
- 6. Chen M., Zhu Q., Li C., Kou X., Zhao Y., Li Y., Xu R., Yang L., Yang L., Gu L.et al.. Chromatin architecture reorganization in murine somatic cell nuclear transfer embryos. Nat. Commun. 2020; 11:1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shen W.K., Chen S.Y., Gan Z.Q., Zhang Y.Z., Yue T., Chen M.M., Xue Y., Hu H., Guo A.Y.. AnimalTFDB 4.0: a comprehensive animal transcription factor database updated with variation and expression annotations. Nucleic Acids Res. 2023; 51:D39–D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castro-Mondragon J.A., Riudavets-Puig R., Rauluseviciute I., Lemma R.B., Turchi L., Blanc-Mathieu R., Lucas J., Boddie P., Khan A., Manosalva Perez N.et al.. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022; 50:D165–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jin J., Tian F., Yang D.C., Meng Y.Q., Kong L., Luo J., Gao G.. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017; 45:D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pourcel C., Touchon M., Villeriot N., Vernadet J.P., Couvin D., Toffano-Nioche C., Vergnaud G.. CRISPRCasdb a successor of CRISPRdb containing CRISPR arrays and cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers. Nucleic Acids Res. 2020; 48:D535–D544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cui Y., Cheng X., Chen Q., Song B., Chiu A., Gao Y., Dawson T., Chao L., Zhang W., Li D.et al.. CRISP-view: a database of functional genetic screens spanning multiple phenotypes. Nucleic Acids Res. 2021; 49:D848–D854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feng C., Song C., Liu Y., Qian F., Gao Y., Ning Z., Wang Q., Jiang Y., Li Y., Li M.et al.. KnockTF: a comprehensive human gene expression profile database with knockdown/knockout of transcription factors. Nucleic Acids Res. 2020; 48:D93–D100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miura H., Quadros R.M., Gurumurthy C.B., Ohtsuka M.. Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat. Protoc. 2018; 13:195–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marand A.P., Chen Z., Gallavotti A., Schmitz R.J.. A cis-regulatory atlas in maize at single-cell resolution. Cell. 2021; 184:3041–3055. [DOI] [PubMed] [Google Scholar]
- 15. Tu X., Mejia-Guerra M.K., Valdes Franco J.A., Tzeng D., Chu P.Y., Shen W., Wei Y., Dai X., Li P., Buckler E.S.et al.. Reconstructing the maize leaf regulatory network using ChIP-seq data of 104 transcription factors. Nat. Commun. 2020; 11:5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang S., Zang C., Xiao T., Fan J., Mei S., Qin Q., Wu Q., Li X., Xu K., He H.H.et al.. Modeling cis-regulation with a compendium of genome-wide histone H3K27ac profiles. Genome Res. 2016; 26:1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z., Civelek M., Miller C.L., Sheffield N.C., Guertin M.J., Zang C.. BART: a transcription factor prediction tool with query gene sets or epigenomic profiles. Bioinformatics. 2018; 34:2867–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qin Q., Fan J., Zheng R., Wan C., Mei S., Wu Q., Sun H., Brown M., Zhang J., Meyer C.A.et al.. Lisa: inferring transcriptional regulators through integrative modeling of public chromatin accessibility and ChIP-seq data. Genome Biol. 2020; 21:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu Q., Georgiou G., Frolich S., van der Sande M., Veenstra G.J.C., Zhou H., van Heeringen S.J.. ANANSE: an enhancer network-based computational approach for predicting key transcription factors in cell fate determination. Nucleic Acids Res. 2021; 49:7966–7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Song C., Zhang Y., Wang Y., Feng C., Chen J., Wei L., Pan Q., Shang D., Zhu Y.et al.. TcoFBase: a comprehensive database for decoding the regulatory transcription co-factors in human and mouse. Nucleic Acids Res. 2022; 50:D391–D401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmeier S., Alam T., Essack M., Bajic V.B.. TcoF-DB v2: update of the database of human and mouse transcription co-factors and transcription factor interactions. Nucleic Acids Res. 2017; 45:D145–D150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y., Song C., Zhao J., Zhang Y., Zhao X., Feng C., Zhang G., Zhu J., Wang F., Qian F.et al.. SEdb 2.0: a comprehensive super-enhancer database of human and mouse. Nucleic Acids Res. 2023; 51:D280–D290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hammal F., de Langen P., Bergon A., Lopez F., Ballester B.. ReMap 2022: a database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res. 2022; 50:D316–D325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robasky K., Bulyk M.L.. UniPROBE, update 2011: expanded content and search tools in the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2011; 39:D124–D128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berger M.F., Badis G., Gehrke A.R., Talukder S., Philippakis A.A., Pena-Castillo L., Alleyne T.M., Mnaimneh S., Botvinnik O.B., Chan E.T.et al.. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008; 133:1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G.et al.. DNA-binding specificities of human transcription factors. Cell. 2013; 152:327–339. [DOI] [PubMed] [Google Scholar]
- 31. Wei G.H., Badis G., Berger M.F., Kivioja T., Palin K., Enge M., Bonke M., Jolma A., Varjosalo M., Gehrke A.R.et al.. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010; 29:2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Genomes Project C., Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012; 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li M.J., Liu Z., Wang P., Wong M.P., Nelson M.R., Kocher J.P., Yeager M., Sham P.C., Chanock S.J., Xia Z.et al.. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016; 44:D869–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gong J., Mei S., Liu C., Xiang Y., Ye Y., Zhang Z., Feng J., Liu R., Diao L., Guo A.Y.et al.. PancanQTL: systematic identification of cis-eQTLs and trans-eQTLs in 33 cancer types. Nucleic Acids Res. 2018; 46:D971–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xia K., Shabalin A.A., Huang S., Madar V., Zhou Y.H., Wang W., Zou F., Sun W., Sullivan P.F., Wright F.A.. seeQTL: a searchable database for human eQTLs. Bioinformatics. 2012; 28:451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gamazon E.R., Zhang W., Konkashbaev A., Duan S., Kistner E.O., Nicolae D.L., Dolan M.E., Cox N.J.. SCAN: SNP and copy number annotation. Bioinformatics. 2010; 26:259–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X., Shi L., Wang Y., Zhong J., Zhao X., Teng H., Shi X., Yang H., Ruan S., Li M.et al.. OncoBase: a platform for decoding regulatory somatic mutations in human cancers. Nucleic Acids Res. 2019; 47:D1044–D1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Karolchik D., Barber G.P., Casper J., Clawson H., Cline M.S., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M.et al.. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014; 42:D764–D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haeussler M., Schonig K., Eckert H., Eschstruth A., Mianne J., Renaud J.B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J.et al.. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016; 17:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Teng L., He B., Wang J., Tan K.. 4DGenome: a comprehensive database of chromatin interactions. Bioinformatics. 2015; 31:2560–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang F., Bai X., Wang Y., Jiang Y., Ai B., Zhang Y., Liu Y., Xu M., Wang Q., Han X.et al.. ATACdb: a comprehensive human chromatin accessibility database. Nucleic Acids Res. 2021; 49:D55–D64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M.N.K., Li Y., Hu M.et al.. The 3D genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018; 19:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 47. Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Salazar M.D., Ratnam M., Patki M., Kisovic I., Trumbly R., Iman M., Ratnam M.. During hormone depletion or tamoxifen treatment of breast cancer cells the estrogen receptor apoprotein supports cell cycling through the retinoic acid receptor alpha1 apoprotein. Breast Cancer Res. 2011; 13:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johansson H.J., Sanchez B.C., Mundt F., Forshed J., Kovacs A., Panizza E., Hultin-Rosenberg L., Lundgren B., Martens U., Mathe G.et al.. Retinoic acid receptor alpha is associated with tamoxifen resistance in breast cancer. Nat. Commun. 2013; 4:2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cunningham T.J., Duester G.. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 2015; 16:110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Herzog S.K., Fuqua S.A.W.. ESR1 mutations and therapeutic resistance in metastatic breast cancer: progress and remaining challenges. Br. J. Cancer. 2022; 126:174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dustin D., Gu G., Fuqua S.A.W.. ESR1 mutations in breast cancer. Cancer. 2019; 125:3714–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jeselsohn R., Bergholz J.S., Pun M., Cornwell M., Liu W., Nardone A., Xiao T., Li W., Qiu X., Buchwalter G.et al.. Allele-specific chromatin recruitment and therapeutic vulnerabilities of ESR1 activating mutations. Cancer Cell. 2018; 33:173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chandarlapaty S., Chen D., He W., Sung P., Samoila A., You D., Bhatt T., Patel P., Voi M., Gnant M.et al.. Prevalence of ESR1 mutations in cell-free DNA and outcomes in metastatic breast cancer: a secondary analysis of the BOLERO-2 clinical trial. JAMA Oncol. 2016; 2:1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu J., Liu J., Lu X.. HOXA1 upregulation is associated with poor prognosis and tumor progression in breast cancer. Exp. Ther. Med. 2019; 17:1896–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang X., Zhu T., Chen Y., Mertani H.C., Lee K.O., Lobie P.E.. Human growth hormone-regulated HOXA1 is a human mammary epithelial oncogene. J. Biol. Chem. 2003; 278:7580–7590. [DOI] [PubMed] [Google Scholar]
- 57. Brock A., Krause S., Li H., Kowalski M., Goldberg M.S., Collins J.J., Ingber D.E.. Silencing HoxA1 by intraductal injection of siRNA lipidoid nanoparticles prevents mammary tumor progression in mice. Sci. Transl. Med. 2014; 6:217ra212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang X., Emerald B.S., Mukhina S., Mohankumar K.M., Kraemer A., Yap A.S., Gluckman P.D., Lee K.O., Lobie P.E.. HOXA1 is required for E-cadherin-dependent anchorage-independent survival of human mammary carcinoma cells. J. Biol. Chem. 2006; 281:6471–6481. [DOI] [PubMed] [Google Scholar]
- 59. Wang L., Zhang W., Li C., Chen X., Huang J.. Identification of biomarkers related to copper metabolism in patients with pulmonary arterial hypertension. BMC Pulm. Med. 2023; 23:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M.. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009; 10:252–263. [DOI] [PubMed] [Google Scholar]
- 61. Ravasi T., Suzuki H., Cannistraci C.V., Katayama S., Bajic V.B., Tan K., Akalin A., Schmeier S., Kanamori-Katayama M., Bertin N.et al.. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010; 140:744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Morin A., Chu E.C., Sharma A., Adrian-Hamazaki A., Pavlidis P.. Characterizing the targets of transcription regulators by aggregating ChIP-seq and perturbation expression data sets. Genome Res. 2023; 33:763–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Roadmap Epigenomics C., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J.et al.. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kloetgen A., Thandapani P., Ntziachristos P., Ghebrechristos Y., Nomikou S., Lazaris C., Chen X., Hu H., Bakogianni S., Wang J.et al.. Three-dimensional chromatin landscapes in T cell acute lymphoblastic leukemia. Nat. Genet. 2020; 52:388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The research community can access information freely in the KnockTF 2.0 without registration or logging in. The URL for KnockTF 2.0 is http://www.licpathway.net/KnockTF/index.html.