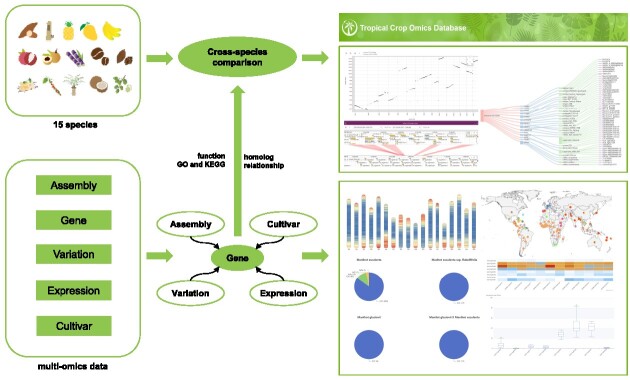
Abstract
Tropical crops are vital for tropical agriculture, with resource scarcity, functional diversity and extensive market demand, providing considerable economic benefits for the world's tropical agriculture-producing countries. The rapid development of sequencing technology has promoted a milestone in tropical crop research, resulting in the generation of massive amount of data, which urgently needs an effective platform for data integration and sharing. However, the existing databases cannot fully satisfy researchers’ requirements due to the relatively limited integration level and untimely update. Here, we present the Tropical Crop Omics Database (TCOD, https://ngdc.cncb.ac.cn/tcod), a comprehensive multi-omics data platform for tropical crops. TCOD integrates diverse omics data from 15 species, encompassing 34 chromosome-level de novo assemblies, 1 255 004 genes with functional annotations, 282 436 992 unique variants from 2048 WGS samples, 88 transcriptomic profiles from 1997 RNA-Seq samples and 13 381 germplasm items. Additionally, TCOD not only employs genes as a bridge to interconnect multi-omics data, enabling cross-species comparisons based on homology relationships, but also offers user-friendly online tools for efficient data mining and visualization. In short, TCOD integrates multi-species, multi-omics data and online tools, which will facilitate the research on genomic selective breeding and trait biology of tropical crops.
Graphical Abstract
Graphical Abstract.
Introduction
Tropical crops, cultivated in tropical regions, encompass a wide range of varieties and can be classified into different categories based on their uses, including rubber crops, tropical food crops, tropical fruit trees, tropical oil crops, tropical spice beverages, tropical medicinal plants and more (1). The functional diversity of these crops gives rise to a broad market demand, making a significant positive impact on the economic growth of tropical agricultural producing countries worldwide. Meanwhile, the rapid advancement of next-generation sequencing technologies has facilitated the generation and accumulation of vast amounts of multi-omics data in tropical crops, enabling the comprehensive elucidation of gene functions and networks under diverse physiological and environmental stress conditions (2).
In the field of genomics, the genomes of diverse tropical crops have been successfully deciphered, with the utilization of advanced technologies such as Hi-C, BioNano optical mapping and Telomere-to-Telomere (T2T), leading to enhanced precision and integrity of the genome sequences (3–10). Based on population genome studies, variation maps have been generated for several tropical crops, including pineapple, mango, cassava and longan, revealing valuable insights into their domestication history (11–14). In the field of transcriptomics, extensive identification of key genes and signalling pathways has provided crucial clues for increasing yields (e.g. rubber tree latex production, sugarcane sugar content) and enhancing tolerance to biotic and abiotic stresses (e.g. pests, diseases, drought, cold and high salinity) (15–17). Additionally, some multi-omics analyses have facilitated a comprehensive exploration of complex physiological processes and polygenic traits, leading to a deeper understanding of the interactions and intricate regulatory mechanisms among different molecules (such as mRNAs, proteins and metabolites) underlying phenotypic traits. (18–21). However, the raw sequencing data and genome sequences of these studies are scattered in different databases, which poses great challenges for data reuse and integrated analysis. Therefore, it is necessary to establish a centralized data sharing platform to provide convenient data sharing services for researchers.
Optimal data integration needs to cover a broad range of species and omics levels while ensuring high-quality datasets. At present, several internationally developed tropical crop databases, such as CassavaBase (22), HeveaDB (23), PGD (24), Sapbase (25), ArecaceaeMDB (26) and TropGeneDB (27), have emerged. CassavaBase, HeveaDB and PGD integrate multi-omics data, but they are only designed for a single species. Similarly, Sapbase and ArecaceaeMDB incorporate multi-omics data from multiple species, but they serve the family Sapindaceae and Arecaceae, respectively. TropGeneDB, is an information system to manage genetic, molecular and phenotypic data for 11 tropical crops, but it lacks multi-omics data and is not up to date. A comprehensive compilation of data encompassing diverse species not only facilitates cross-species research, but also helps to break bottlenecks caused by insufficient data for some species (28). While discoveries of new gene functions and transcriptome conservation levels in Arabidopsis, rice and barley through homologous genes (29,30), cross-species studies on tropical crops remain limited. Therefore, integrating multi-omics data from multiple tropical crops, refining homologous gene relationships among species and creating an inclusive database for data archiving, analysis, and visualization will significantly advance tropical crop research.
Here, we present TCOD (https://ngdc.cncb.ac.cn/tcod), a specialized, integrated and open-access resource for tropical crops. Currently, TCOD includes whole-genome sequencing (WGS) data, RNA sequencing (RNA-Seq) data, genomes, gene functional annotations, homolog relationships and germplasm information for 15 tropical crops. Multiple WGS and RNAseq datasets are analyzed using standardized workflows, resulting in the generation of comprehensive variation maps and expression profiles for each species. In addition, TCOD is well-equipped with a diverse set of online analysis tools for data mining, further bolstering its role in providing free public service for scientific research.
Materials and methods
Data collection
High-throughput sequencing data, genome sequences, annotation information and germplasm resource entries for 15 tropical crops were compiled from major databases, with detailed volumes and sources are shown in Table 1.
Table 1.
Statistics and sources of data collected
| Species | WGS projects | WGS samples | RNASeq projects | RNASeq samples | Genomes | Cultivars |
|---|---|---|---|---|---|---|
| areca | − | − | − | − | 1 from CNGB CNSA (31) | 42 from GRIN (32) |
| banana | 5 from NCBI SRA (33) 3 from EBI ENA (34) | 65 from NCBI SRA 165 from EBI ENA | 12 from NCBI SRA 1 from EBI ENA | 388 from NCBI SRA 21 from EBI ENA | 3 from Banana Genome Hub (35) | 209 from GRIN |
| cassava | 8 from NCBI SRA 1 from CassavaBase (22) | 399 from NCBI SRA 174 from CassavaBase | 13 from NCBI SRA | 356 from NCBI SRA | 2 from ITBB-CATAS 1 from HNU 1 from NCBI Genome (36) 1 from Phytozome (37) | 6,074 from CIAT 4,359 from IITA |
| cocoa | 2 from NCBI SRA | 207 from NCBI SRA | 4 from NCBI SRA | 130 from NCBI SRA | 2 from NCBI Genome | 208 from GRIN |
| coconut | - | - | 1 from NCBI SRA | 9 from NCBI SRA | 1 from NCBI Genome | 60 from GRIN |
| coffee | 1 from NCBI SRA | 93 from NCBI SRA | 5 from NCBI SRA 4 from EBI ENA | 123 from NCBI SRA 57 from EBI ENA | 4 from NCBI Genome | 500 from GRIN |
| litchi | 1 from NCBI SRA | 72 from NCBI SRA | 10 from NCBI SRA | 228 from NCBI SRA | 1 from NCBI Genome | 91 from GRIN |
| longan | 1 from NCBI SRA | 95 from NCBI SRA | 3 from NCBI SRA | 90 from NCBI SRA | 1 from NCBI Genome 1 from NGDC GWH (38) | 68 from GRIN |
| mango | 1 from NCBI SRA | 48 from NCBI SRA | 4 from NCBI SRA | 49 from NCBI SRA | 2 from NCBI Genome 1 from NGDC GWH | 310 from GRIN |
| oil palm | 1 from NCBI SRA 1 from DDBJ DRA (39) | 26 from NCBI SRA 72 from DDBJ DRA | 9 from NCBI SRA 1 from EBI ENA | 126 from NCBI SRA 16 from EBI ENA | 1 from NCBI Genome | 91 from GRIN |
| pepper | − | − | − | − | 1 from HZAU | 43 from GRIN |
| pineapple | 1 from NCBI SRA 1 from DDBJ DRA | 86 from NCBI SRA 1 from DDBJ DRA | 8 from NCBI SRA | 208 from NCBI SRA | 2 from NCBI Genome | 362 from GRIN |
| rubber tree | 1 from NGDC GSA (40) | 545 from NGDC GSA | 6 from NCBI SRA | 102 from NCBI SRA | 1 from RRI-CATAS 1 from NCBI Genome | 110 from GRIN |
| sugarcane | − | − | 6 from NCBI SRA | 82 from NCBI SRA | 1 from NCBI Genome 1 from NGDC GWH | 682 from GRIN |
| vanilla | − | − | 1 from NCBI SRA | 12 from NCBI SRA | 4 from NCBI Genome | 172 from GRIN |
| Total | 28 | 2,048 | 88 | 1,997 | 34 | 13,381 |
Note: CIAT: The International Center for Tropical Agriculture (https://ciat.cgiar.org); IITA: International Institute of Tropical Agriculture (https://my.iita.org/accession2/); HNU: Hainan University; HZAU: Huazhong Agricultural University; ITBB-CATAS: Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Science; RRI-CATAS: Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences.
Processing of WGS-Seq data
The collected WGS data were processed using the standard analysis pipelines provided by the Genome Variation Map (GVM) (41). After performing quality control on raw sequencing reads using Trimmomatic v0.36 (42), clean reads were mapped to the reference genome using BWA-MEM (43). Then the mapping results were converted and sorted using Samtools v1.13 (44), and duplicates were marked using MarkDuplicates in GATK v4.1.2.0 (45). For lack of known variations, the high-quality variants called by GATK HaplotypeCaller and Bcftools v1.13 (44) were merged and then input to Basic Quality Score Recalibration (BQSR). Intermediate GVCF files were generated for each sample by HaplotypeCaller, which then were pooled together to generate a VCF file containing all raw variants using CombineGVCFs and GenotypeGVCFs in GATK. These raw variants were further filtered using SelectVariants and VariantFiltration in GATK with recommended parameters. The functional effects of the variants were annotated using VEP v8.4 (46). The minor allele frequency (MAF) of the variants were calculated using vcftools v.0.1.13 (47).
Processing of RNA-Seq data
The collected RNA-Seq data were processed using the standard analysis pipelines provided by the Gene Expression Nebula (GEN) (48). After quality control with Fastp v0.20.0 (49), the reads were aligned to the reference genome using Hisat2 v2.0.5 (50) to evaluate data quality. Projects with an average alignment rate above 50% were selected for subsequent analysis. RseQC v2.6.4 (51) was used to determine strand-specific library sequencing. The high-quality reads were aligned to the genome using the STAR v2.7.1a (52), generating BAM results, which were subsequently processed by the RSEM v1.3.1 (53) for expression quantification, resulting in the generation of gene expression and transcript expression matrices. Additionally, Limma (54) was utilized to perform differential expression analysis under various sample comparison scenarios.
Functional annotation of genomes
To address the issue of lacking information or inconsistent descriptions in genome annotation, we conducted a unified functional annotation on each gene while preserving the original annotation. This involved utilizing the Nr (55) and Uniprot (56) databases to identify similar functional proteins, the Pfam (57) and Interpro (58) databases to identify conserved domains, and the eggnog-mapper webserver (59) to obtain the GO (60) terms and KEGG (61) pathways associated with the genes.
Identification of homologous genes
The protein sequences of each gene are collected and inputted into the OrthoFinder v2.5.4 (62) to identify their homologous genes in other species. The parsing and tabulation of homologous gene results are guided by the structural framework of the Homologous Gene Database (HGD) (63).
Genome synteny analysis
Collinearity analysis includes comparisons between reference genomes of different species and comparisons between genomes of different subspecies within the same species. Genome-wide synteny analysis was performed using MUMmer v4.0.0.rc1 (64). Initially, the ‘nucmer’ program was utilized to generate comprehensive comparisons of nucleotide sequences, employing the parameters -g 1000 -c 100 for interspecies comparisons and default parameters for intraspecies comparisons. Subsequently, the intraspecies comparison results were filtered using the ‘delta-filter’ program with the parameters -m -i 90 -l 10000.
Database implementation
TCOD was implemented by SpringBoot (https://spring.io/projects/spring-boot; a free and powerful framework for developing standalone java applications) and Mybatis (https://mybatis.org/mybatis-3; a first-class persistence framework with support for custom SQL, stored procedures and advanced mappings), referring to the framework of iDog and iSheep database (65,66). Data storage and management were realized using MySQL (https://dev.mysql.com; the world's most popular relational database management system). Web user interfaces were developed using JSP (Jakarta Server Pages, a template engine for web applications), HTML (HyperText Markup Language), CSS (Cascading Style Sheets), Bootstrap (https://getbootstrap.com; a powerful, feature-packed frontend toolkit), AJAX (Asynchronous JavaScript and XML; a technique for creating fast and dynamic web pages, allowing partial updates of web pages without reloading the whole page.) and Jquery (https://jquery.com; a fast, small and feature-rich JavaScript library). For dynamic data visualization, Echart (https://echarts.apache.org/en/index.html; a declarative framework for rapid construction of web-based visualization), Highcharts (https://www.highcharts.com; a JavaScript plug-in to create interactive charts) and DataTables (https://datatables.net; a plug-in for the jQuery JavaScript library to render HTML tables) were incorporated to generate charts and tables. Furthermore, third-party software, including NCBI BLAST+ (55), JBrowser2 (67), Primer3Web (68) and ClusterProfiler (69), are invoked for the secondary development of online tools.
Database content and usage
Overview of TCOD
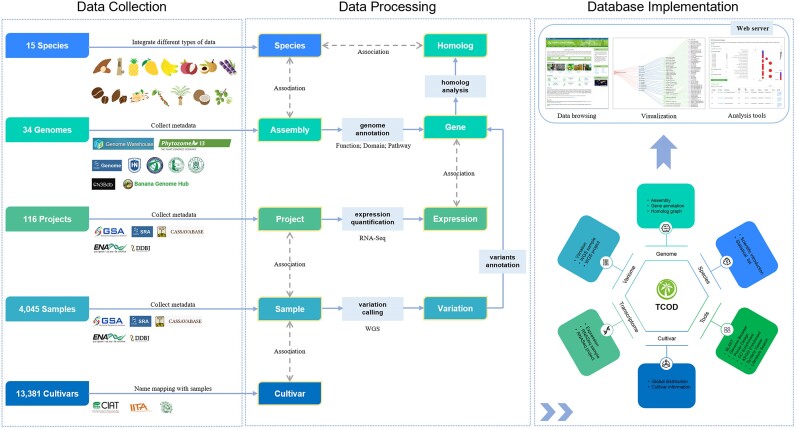
TCOD is dedicated to becoming a comprehensive multi-omics data platform that serves the field of tropical crop research, offering users a one-stop solution for data acquisition and online analysis. By integrating the genome, variome, transcriptome and cultivar data from 15 typical tropical crops, TCOD has achieved the integration of multi-omics data within individual species, utilizing genes as a bridge to connect various dimensions of omics data. Furthermore, leveraging homologous genes as entry points enables the comparison of omics characteristics across different species. As of 1 August 2023, TCOD houses 34 chromosome-level de novo assemblies, 1 255 004 genes with functional annotations, 282 436 992 unique variants from 2048 WGS samples, 88 transcriptomic profiles from 1997 RNA-Seq samples and 13 381 germplasm items. Additionally, TCOD incorporates a range of online tools to facilitate data mining within the database and visualization of analysis results (Figure 1).
Figure 1.
The construction pipelines of TCOD, including data collection, data processing and database implementation.
Genome
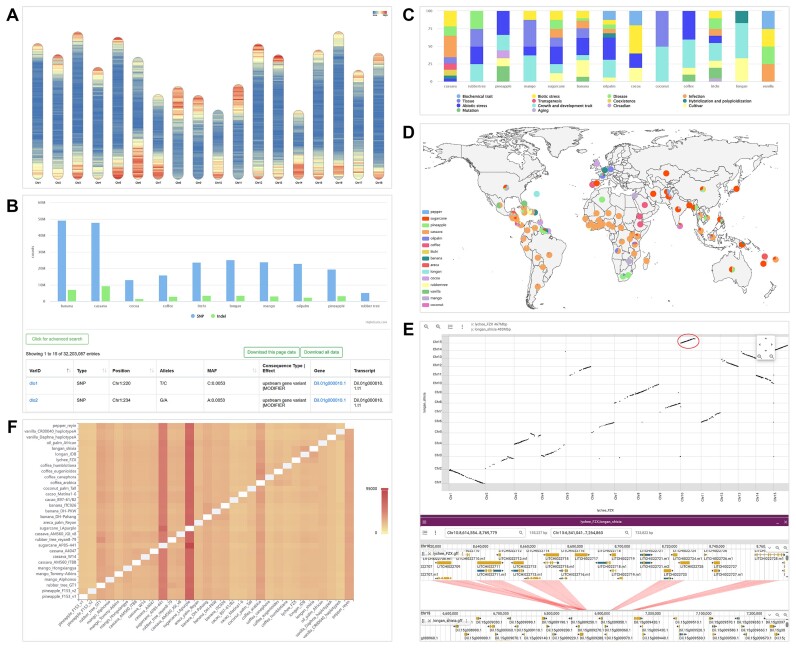
The genome in TCOD includes whole genome sequences and genes, and the genome sequences comprise the ‘Assembly’ module in the system. By integrating assemblies released by multiple public platforms and provided by partners, 34 chromosome-level de novo genomes from 15 species have been collected, covering 30 different varieties. Focusing on each genome, users can browse basic information such as sequencing technologies, coverage, total length, scaffold number, N50 value, GC content and published literature. In addition, We calculated the number of genes on each chromosome and used the Rideogram package (70) to create a heatmap of gene density distribution, visualizing the gene-rich regions in each genome (Figure 2A).
Figure 2.
Screenshots of TCOD. (A) Gene density distribution in the genome of rubber_tree_reyan8-79. (B) Genome-wide variation maps for 10 species. (C) Classification of RNA-Seq studies. (D) Global distribution map of multiple tropical crops. (E) Visualizing collinearity between the genomes lychee_FZX and longan_shixia, the red circle indicates the selected region for a more detailed linear synteny view. (F) Heatmap of homologous genes between genome pairs.
The ‘Gene’ module was committed to facilitate researchers to find functional genes. By collating gene structure and function descriptions extracted from each genome annotation, 1 255 004 gene entries were captured to build this module. It supports dynamic conditional retrieval and target gene set downloads by selecting genome version, chromosome coordinate, gene name and gene function. Gene acts as bridges connecting multiple omics data in TCOD. The detailed gene information includes ‘basic information’, ‘genome and sequence’, ‘homolog information’, ‘variant information’, ‘expression information’ and ‘visualization’, from which users can infer the potential biological function of the gene.
Variation
The ‘Variation’ module was designed to provide researchers with an overall and reliable genome-wide variation dataset. By collecting WGS data from different samples and using a standard variation analysis pipeline, this module provides genome-wide variation maps for 10 species (Figure 2B). To facilitate the search and download of interested variants, a unique identifier was assigned to each variant and a multi-conditional retrieval method was supported. For each variant, we provide not only basic information such as variant coordinate, reference and alternative allele, minor allele frequency, etc., but also detailed functional annotation information such as consequence type, variant effect and genotype distribution in the population, which supplies pointcuts for population diversity research.
Expression
The ‘Expression’ module is dedicated to providing evidence to explore the diverse regulatory mechanisms involved in genes. Through the unified analysis of high-quality RNA-Seq datasets from different projects, this module offers transcriptome profiles of 13 species under various experimental conditions. Moreover, to cater to specific research interests, we have included category tags for each project, such as biotic stress, abiotic stress, disease and infection, allowing users to efficiently access datasets of their interest categories (Figure 2C). For each dataset, we support the visualization and download of gene expression profiles, and provide a list of differentially expressed genes under different comparison conditions.
Cultivar
The ‘Cultivar’ module was intended to provide germplasm data to support research on crop breeding and domestication. By integrating varieties from CIAT, IITA and GRIN, a total of 13 381 non-redundant entries have been collected and the number of varieties in specific geographic regions was plotted on a world map, offering a visual representation of the global distribution of germplasm resources for 15 tropical crops (Figure 2D). Taking the cassava accession as an example, researchers can view copious descriptive information for a more comprehensive assessment of the sample. The description consists of three parts, including passport data (such as accession name, synonyms, DOI, origin country), botanical characteristics (such as plant height, stem color, developed leaf color, petiole color) and agronomic characteristics (such as storage root form, root color, dry matter content, hydrocyanic acid content).
Species
The ‘Species’ module organizes multi-species and multi-omics data collected in the database on a species basis, helping researchers to rapidly access the data resources of concerned species. By aggregating data information from different sections, it not only provides concise scientific overview of each species (including geographical distribution, applications and genome sequencing), but also grants direct access to genomes, genes, variants, expressions, projects, samples and cultivars.
Tools
To facilitate data mining and analysis on TCOD, we have set up several commonly easy-to-use tools in the ‘Tools’ module, including BLAST, Genome Browser, Primer Design, Literature Search, GO Enrichment, KEGG Enrichment, Synteny Viewer and Homolog Finder. The BLAST tool, built on NCBI BLAST+, is specifically designed for sequence searching against the genome, coding sequence (CDS) and protein sequences of 15 tropical crops. The Genome Browser provides smooth visualization of genomic sequences, genes and variants in selected region by selecting a reference genome, and supports exporting the visualization results of selected regions to images. The Primer Design is developed based on Primer3web, aimed at assisting users in designing primers for downstream experiments. The Literature Search utilizes the interface offered by the NGDC OpenLB database (71) to facilitate rapid retrieval of relevant literature in the field of tropical crops. The GO Enrichment and KEGG Enrichment tools facilitate enrichment analysis of GO and KEGG pathways for target gene sets, providing downloadable results and visualizing enrichment pathway bubble diagrams. The Synteny Viewer allows users to visualize collinear relationships between genomes, offering inter-species and intra-species collinearity views with dot plots, and the ability to zoom in on specific regions to explore the genes within the collinear regions (Figure 2E). The Homology Finder offers a heat map to illustrate the number of homologous gene between genome pairs (Figure 2F) and supports user searches based on gene ID, gene function and custom species combinations.
Metadata and download
To ensure data traceability and reusability, we included metadata information for all used projects and samples, categorizing them into their corresponding omics categories. Raw sequencing data can be downloaded from the FTP or HTTP addresses provided via the ‘runs’ link within each module, and variant files (GVCF/VCF) for each WGS sample can also be downloaded. Additionally, the download page provides access to genomes and annotation files, variant results, expression matrices and germplasm resources for local data mining.
An example of using TCOD
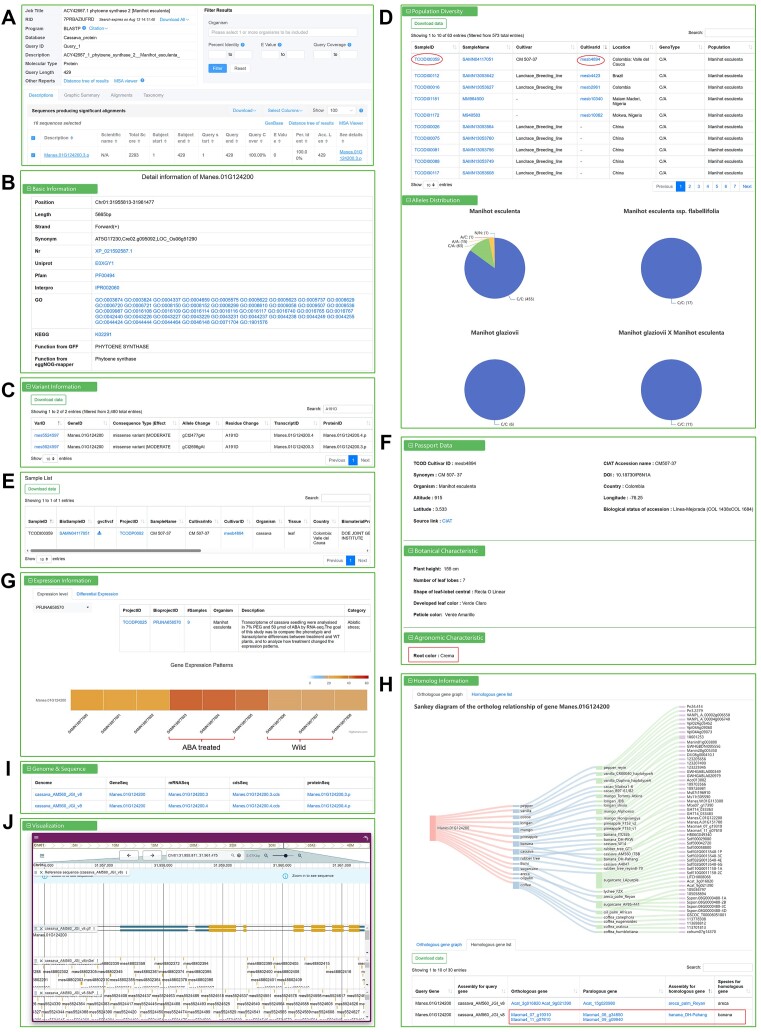
Carotenoid production is limited by the expression level of phytoene synthase (PSY). A non-conservative amino acid exchange (A191D) triggered by a single nucleotide polymorphism (C > A) in PSY2 was reported to lead to a marked increase of carotenoid synthesis in cassava storage roots (72). Here, we use PSY2 as an example to demonstrate how to access data in TCOD.
First, the PSY2 protein sequence (NCBI accession number: ACY42667) is aligned to the database using the ‘BLAST’ tool, revealing 100% sequence similarity with Manes.01G124200 (Figure 3A). Clicking the ID link in the ‘See details’ column redirects to view detailed gene information, the ‘Basic Information’ reveals its function as phytoene synthase and provides its functional domains, as well as GO and KEGG annotations (Figure 3B). When searching for the keyword ‘A191D’ in the ‘variant information’, we can quickly find the consistent variant as mes5524597 (Figure 3C). Clicking on it leads to the ‘Variation’ module, where we discover that among the 573 cassava samples, 63 have this variation, all of which belong to the Manihot esculenta population (Figure 3D). For samples of interest, such as TCODI00359, representing the CM 507-37 variety from Colombia, we can either click on the sampleID to access the ‘Sample’ module for metadata information (Figure 3E) and download the variant GVCF file, or click on the cultivarID mesb4894 to enter the ‘Cultivar’ module and view corresponding phenotype data, such as the root color being crema (Figure 3F). The ‘Expression information’ displays Manes.01G124200’s expression level (TPM value) in each sample of the project. In the project TCODP0025, the TPM value of wild two-month-old cassava seedling samples was lower than that of cassava seedling samples treated with abscisic acid (ABA). The differential expression analysis indicated its significance, suggesting that the gene may be involved in the response to ABA stress (Figure 3G).
Figure 3.
Using TCOD to obtain data for each section associated with the PSY2. (A) The gene Manes.01G124200 was identified with 100% sequence similarity to PSY2 in the tropical crop protein database by blastp. (B) The function annotation of the gene Manes.01G124200 in TCOD. (C) The variant ID mes5524597 was obtained by filtering with the keyword ‘A191D’ among the 2480 variants in the gene Manes.01G124200. (D) Genotype distribution of variant mes5524597 in 573 cassava samples. (E) Sample metadata corresponding to the sampleID TCODI00359. (F) Cultivar information corresponding to the cultivarID mesb4894. (G) The expression level of gene Manes.01G124200 in different samples of project TCODP0025. (H) Homologous relationship of gene Manes.01G124200 in other species. (I) The corresponding genome and sequence of the gene Manes.01G124200. (J) Genome browser for visualizing different tracks in the gene Manes.01G124200.
In addition to supporting published studies with larger-scale sample datasets, the ‘homologous information’ offers inspiration for cross-species functional studies. By utilizing the visualized orthologous gene relationship graph, we can make preliminary predictions regarding which tropical crops may possess similar gene functions (Figure 3H). In light of this, our emphasis lies on banana species, as elevating the vitamin A content in banana fruit can effectively combat vitamin A deficiency in developing countries across Africa and Southeast Asia (73). The ‘Homologous gene list’ shows that the gene Manes.01G124200 has two orthologous genes and two paralogous genes in the banana genome (banana_DH-Pahang), suggesting potential similar roles in banana as the gene Manes.01G124200 in cassava and functional analysis can be further conducted based on the GO pathway, KEGG pathway, variation and expression information of the homologous gene. Notably, the expression of Macma4_09_g09940 (with 95% similarity to MtPsy2a protein sequence) has been confirmed to lead to an increase in vitamin A content (74), implying the PSY2 gene plays universally applicable function in both cassava and banana. Moreover, TCOD supports gene sequence downloads (Figure 3I) and genomic region visualization (Figure 3J), facilitating users in conducting subsequent analyses conveniently.
Discussion and future plans
Tropical crops have significant economic value and incomparable biological status, and the rapid development of multi-omics research has highlighted an urgent need for the integration and sharing of massive data. Internationally, existing tropical crop databases either lack multi-omics data or have a limited scope, focusing solely on specific species or families. Presently, TCOD stands as the most comprehensive multi-omics database, encompassing the largest diversity of tropical crop species.
When compared to other established databases, TCOD exhibits the following key characteristics: (i) multi-species and multi-omics data integration: TCOD aggregates diverse data resources covering 15 tropical crop species, including genome sequences, genome variations, gene functions, gene expressions and germplasm information. By correlating multi datasets in units of genes, TCOD provides a user-friendly platform with efficient data browsing, retrieval and downloading capabilities, making it a one-stop resource for researchers seeking valuable information on tropical crops. (ii) High-quality datasets: Genomes incorporated into TCOD reach a level of chromosomal integrity and are accompanied by uniform and comprehensive gene annotation. Additionally, by collecting raw sequencing data from diverse sources and implementing a standardized analysis pipeline, TCOD generates relatively complete variation maps for each species, along with expression profiles under different physiological conditions, providing a valuable dataset for the artificial intelligence breeding of tropical crops. (iii) Diverse online tools: To enhance effective data mining, TCOD offers a series of online tools for sequence similarity comparison, downstream primer design, literature searches, genes pathway enrichment and pairwise genome consensus linear views. (iv) Cross-species analysis: TCOD furnishes gene homology relationships across various species, allowing cross-species comparisons of gene functions and multi-omics characteristics, which facilitates in-depth exploration into the shared biological attributes among different organisms.
In the future, Multi-omics data of tropical crops with extremely rich species diversity will be generated and collected in TCOD. We also plan to add additional types of omics data (e.g. metabolome, proteome) and establish Variant-Gene-Trait associations through manual collection and curation of GWAS related literature, providing a comprehensive prior knowledge module for tropical crop trait research. Moreover, the advancement in machine learning and artificial intelligence technology holds the promise of integrating biological knowledge and omics data to achieve precise breeding (75). At present, it has been realized in some staple crops such as rice, maize and wheat (76–78), but it is still relatively behind in the field of tropical crops. To bridge this gap, TCOD will continue its unwavering commitment to extensively explore multi-omics data using these cutting-edge methods and technologies, providing vital data support for tropical crop breeding to enter the generation of the ‘5Gs’ - (genome, germplasm, gene, genomic breeding and gene editing) (79). We also enthusiastically welcome comments and suggestions from researchers worldwide to enhance and improve TCOD.
Acknowledgements
We thank Tingting Chen and Xu Chen for their help in the transmission and storage of raw sequencing data, thank Cuiping Li and Dongmei Tian for their help in variation analysis, thank Yuansheng Zhang and Tongtong Zhu for their help in transcriptome analysis, thank Zhuojing Fan for her help in image design, thank Dong Zou for providing the data interface of the OpenLB database and thank Yingke Ma for her technical support in the development of BLAST tools. Meanwhile, we are grateful to Prof. Yiming Bao, Prof. Zhang Zhang, Prof. Shuhui Song and Prof. Yuan Gao for their constructive suggestions.
Contributor Information
Hailong Kang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Tianhao Huang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Guangya Duan, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yuyan Meng, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Xiaoning Chen, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Shuang He, Sanya Nanfan Research Institute, Hainan University, Sanya 572025, China.
Zhiqiang Xia, Sanya Nanfan Research Institute, Hainan University, Sanya 572025, China.
Xincheng Zhou, Institute of Tropical Biosciences and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Haikou 571101, China.
Jinquan Chao, Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou 571101, China.
Bixia Tang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Zhonghuang Wang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Junwei Zhu, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Zhenglin Du, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Yanlin Sun, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Sisi Zhang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China.
Jingfa Xiao, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Weimin Tian, Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou 571101, China.
Wenquan Wang, Sanya Nanfan Research Institute, Hainan University, Sanya 572025, China.
Wenming Zhao, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences and China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Data availability
TCOD is freely available online at https://ngdc.cncb.ac.cn/tcod and does not require user to register.
Funding
National Key R&D Program of China [2018YFD1000505 to W.Z., 2018YFD1000500 to W.W.]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB38050300]; National Natural Science Foundation of China [32100506, 32100511, 32170678]; Genomics Data Center Operation and Maintenance of Chinese Academy of Sciences [CAS-WX2022SDC-XK05]; Developing Bioinformatics Platform in Hainan Yazhou Bay Seed Lab [B21HJ0001]. Funding for open access charge: National Key R&D Program of China [2018YFD1000505].
Conflict of interest statement. None declared.
References
- 1. Smith N.J., Williams J.T., Plucknett D.L., Talbot J.P.. Tropical Forests and their Crops. 2018; Cornell University Press. [Google Scholar]
- 2. Yang Y., Saand M.A., Huang L., Abdelaal W.B., Zhang J., Wu Y., Li J., Sirohi M.H., Wang F.. Applications of multi-Omics technologies for crop improvement. Front. Plant Sci. 2021; 12:563953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu J., Shi C., Shi C.C., Li W., Zhang Q.J., Zhang Y., Li K., Lu H.F., Shi C., Zhu S.T.et al.. The chromosome-based rubber tree genome provides new insights into spurge genome evolution and rubber biosynthesis. Mol. Plant. 2020; 13:336–350. [DOI] [PubMed] [Google Scholar]
- 4. Hu W., Ji C., Shi H., Liang Z., Ding Z., Ye J., Ou W., Zhou G., Tie W., Yan Y.et al.. Allele-defined genome reveals biallelic differentiation during cassava evolution. Mol. Plant. 2021; 14:851–854. [DOI] [PubMed] [Google Scholar]
- 5. Zhang Q., Qi Y., Pan H., Tang H., Wang G., Hua X., Wang Y., Lin L., Li Z., Li Y.et al.. Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nat. Genet. 2022; 54:885–896. [DOI] [PubMed] [Google Scholar]
- 6. Yang Y., Huang L., Xu C., Qi L., Wu Z., Li J., Chen H., Wu Y., Fu T., Zhu H.et al.. Chromosome-scale genome assembly of areca palm (Areca catechu). Mol. Ecol. Resour. 2021; 21:2504–2519. [DOI] [PubMed] [Google Scholar]
- 7. Belser C., Baurens F.C., Noel B., Martin G., Cruaud C., Istace B., Yahiaoui N., Labadie K., Hribova E., Dolezel J.et al.. Telomere-to-telomere gapless chromosomes of banana using nanopore sequencing. Commun. Biol. 2021; 4:1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu G., Feng J., Xiang X., Wang J., Salojarvi J., Liu C., Wu Z., Zhang J., Liang X., Jiang Z.et al.. Two divergent haplotypes from a highly heterozygous lychee genome suggest independent domestication events for early and late-maturing cultivars. Nat. Genet. 2022; 54:73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Piet Q., Droc G., Marande W., Sarah G., Bocs S., Klopp C., Bourge M., Siljak-Yakovlev S., Bouchez O., Lopez-Roques C.et al.. A chromosome-level, haplotype-phased Vanilla planifolia genome highlights the challenge of partial endoreplication for accurate whole-genome assembly. Plant Commun. 2022; 3:100330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu L., Xu Z., Wang M., Fan R., Yuan D., Wu B., Wu H., Qin X., Yan L., Tan L.et al.. The chromosome-scale reference genome of black pepper provides insight into piperine biosynthesis. Nat. Commun. 2019; 10:4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang P., Luo Y., Huang J., Gao S., Zhu G., Dang Z., Gai J., Yang M., Zhu M., Zhang H.. The genome evolution and domestication of tropical fruit mango. Genome Biol. 2020; 21:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen L.Y., VanBuren R., Paris M., Zhou H., Zhang X., Wai C.M., Yan H., Chen S., Alonge M., Ramakrishnan S.et al.. The bracteatus pineapple genome and domestication of clonally propagated crops. Nat. Genet. 2019; 51:1549–1558. [DOI] [PubMed] [Google Scholar]
- 13. Wang J., Li J., Li Z., Liu B., Zhang L., Guo D., Huang S., Qian W., Guo L. Genomic insights into longan evolution from a chromosome-level genome assembly and population genomics of longan accessions. Hortic. Res. 2022; 9:uhac021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu W., Ji C., Liang Z., Ye J., Ou W., Ding Z., Zhou G., Tie W., Yan Y., Yang J.. Resequencing of 388 cassava accessions identifies valuable loci and selection for variation in heterozygosity. Genome Biol. 2021; 22:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Men X., Wang F., Chen G.-Q., Zhang H.-B., Xian M.. Biosynthesis of natural rubber: current state and perspectives. Int. J. Mol. Sci. 2018; 20:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ali A., Khan M., Sharif R., Mujtaba M., Gao S.-J.. Sugarcane Omics: an update on the current status of research and crop improvement. Plants. 2019; 8:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ning Y., Yang D.-d., Yu X.-c., Cao X.. Multi-omics-driven development of alternative crops for natural rubber production. J. Integr. Agric. 2023; 22:959–971. [Google Scholar]
- 18. Ding Z., Fu L., Tie W., Yan Y., Wu C., Dai J., Zhang J., Hu W.. Highly dynamic, coordinated, and stage-specific profiles are revealed by a multi-omics integrative analysis during tuberous root development in cassava. J. Exp. Bot. 2020; 71:7003–7017. [DOI] [PubMed] [Google Scholar]
- 19. Bittencourt C.B., Carvalho da Silva T.L., Rodrigues Neto J.C., Vieira L.R., Leão A.P., de Aquino Ribeiro J.A., Abdelnur P.V., de Sousa C.A.F., Souza M.T. Jr. Insights from a Multi-Omics Integration (MOI) Study in Oil Palm (Elaeis guineensis Jacq.) Response to Abiotic Stresses: part one—salinity. Plants. 2022; 11:1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leão A.P., Bittencourt C.B., Carvalho da Silva T.L., Rodrigues Neto J.C., Braga Í.d.O., Vieira L.R., de Aquino Ribeiro J.A., Abdelnur P.V., de Sousa C.A.F., Souza Júnior M.T.. Insights from a Multi-Omics Integration (MOI) study in oil palm (Elaeis guineensis Jacq.) response to abiotic stresses: part two—drought. Plants. 2022; 11:2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takahashi S., Saito K., Jia H., Kato H.. An integrated multi-omics study revealed metabolic alterations underlying the effects of coffee consumption. PLoS One. 2014; 9:e91134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fernandez-Pozo N., Menda N., Edwards J.D., Saha S., Tecle I.Y., Strickler S.R., Bombarely A., Fisher-York T., Pujar A., Foerster H.. The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res. 2015; 43:D1036–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng H. HeveaDB: a hub for rubber tree genetic and genomic resources. The Rubber Tree Genome. 2020; 137–152. [Google Scholar]
- 24. Xu H., Yu Q., Shi Y., Hua X., Tang H., Yang L., Ming R., Zhang J.. PGD: pineapple Genomics Database. Hortic. Res. 2018; 5:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li J., Chen C., Zeng Z., Wu F., Feng J., Liu B., Mai Y., Chu X., Wei W., Li X.. SapBase (Sapinaceae Genomic DataBase): a central portal for functional and comparative genomics of Sapindaceae species. 2022; bioRxiv doi:29 November 2022, preprint: not peer reviewed 10.1101/2022.11.25.517904. [DOI] [PubMed]
- 26. Yang Z., Liu Z., Xu H., Li Y., Huang S., Cao G., Shi M., Zhu J., Zhou J., Li R.et al.. ArecaceaeMDB: a comprehensive multi-omics database for Arecaceae breeding and functional genomics studies. Plant Biotechnol. J. 2023; 21:11–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamelin C., Sempere G., Jouffe V., Ruiz M.. TropGeneDB, the multi-tropical crop information system updated and extended. Nucleic Acids Res. 2013; 41:D1172–D1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fu Y., Liu H., Dou J., Wang Y., Liao Y., Huang X., Tang Z., Xu J., Yin D., Zhu S.et al.. IAnimal: a cross-species omics knowledgebase for animals. Nucleic Acids Res. 2023; 51:D1312–D1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Armstead I., Donnison I., Aubry S., Harper J., Hörtensteiner S., James C., Mani J., Moffet M., Ougham H., Roberts L.. Cross-species identification of Mendel's I locus. Science. 2007; 315:73–73. [DOI] [PubMed] [Google Scholar]
- 30. Hartmann A., Berkowitz O., Whelan J., Narsai R.. Cross-species transcriptomic analyses reveals common and opposite responses in Arabidopsis, rice and barley following oxidative stress and hormone treatment. BMC Plant Biol. 2022; 22:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo X., Chen F., Gao F., Li L., Liu K., You L., Hua C., Yang F., Liu W., Peng C.. CNSA: a data repository for archiving omics data. Database. 2020; 2020:baaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Volk G.M., Richards C.M.. Availability of genotypic data for USDA-ARS National Plant Germplasm System accessions using the genetic resources information network (GRIN) database. HortScience. 2008; 43:1365–1366. [Google Scholar]
- 33. Leinonen R., Sugawara H., Shumway M.. The sequence read archive. Nucleic Acids Res. 2011; 39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burgin J., Ahamed A., Cummins C., Devraj R., Gueye K., Gupta D., Gupta V., Haseeb M., Ihsan M., Ivanov E.. The european nucleotide archive in 2022. Nucleic Acids Res. 2023; 51:D121–D125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Droc G., Martin G., Guignon V., Summo M., Sempéré G., Durant E., Soriano A., Baurens F.-C., Cenci A., Breton C.. The banana genome hub: a community database for genomics in the Musaceae. Hortic. Res. 2022; 9:uhac221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sayers E.W., Beck J., Bolton E.E., Bourexis D., Brister J.R., Canese K., Comeau D.C., Funk K., Kim S., Klimke W.et al.. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021; 49:D10–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N.et al.. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012; 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen M., Ma Y., Wu S., Zheng X., Kang H., Sang J., Xu X., Hao L., Li Z., Gong Z.et al.. Genome Warehouse: a Public Repository Housing Genome-scale Data. Genomics Proteomics Bioinformatics. 2021; 19:584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tanizawa Y., Fujisawa T., Kodama Y., Kosuge T., Mashima J., Tanjo T., Nakamura Y.. DNA Data Bank of Japan (DDBJ) update report 2022. Nucleic Acids Res. 2023; 51:D101–D105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., Dong L., Zhang Z., Yu C., Sun Y.et al.. The Genome Sequence Archive Family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics. 2021; 19:578–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li C., Tian D., Tang B., Liu X., Teng X., Zhao W., Zhang Z., Song S.. Genome Variation Map: a worldwide collection of genome variations across multiple species. Nucleic Acids Res. 2021; 49:D1186–D1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M.. Twelve years of SAMtools and BCFtools. GigaScience. 2021; 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M.et al.. The Genome Analysis Toolkit: a MapReduce framework for analyzing next- generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F.. The ensembl variant effect predictor. Genome Biol. 2016; 17:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T.et al.. The variant call format and VCFtools. Bioinformatics. 2011; 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Zou D., Zhu T., Xu T., Chen M., Niu G., Zong W., Pan R., Jing W., Sang J.et al.. Gene Expression Nebulas (GEN): a comprehensive data portal integrating transcriptomic profiles across multiple species at both bulk and single- cell levels. Nucleic Acids Res. 2022; 50:D1016–D1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen S., Zhou Y., Chen Y., Gu J.. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016; 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang L., Wang S., Li W.. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012; 28:2184–2185. [DOI] [PubMed] [Google Scholar]
- 52. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011; 12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinf. 2009; 10:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. The UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J.et al.. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021; 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Paysan-Lafosse T., Blum M., Chuguransky S., Grego T., Pinto B.L., Salazar G.A., Bileschi M.L., Bork P., Bridge A., Colwell L.et al.. InterPro in 2022. Nucleic Acids Res. 2023; 51:D418–D427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cantalapiedra C.P., Hernandez-Plaza A., Letunic I., Bork P., Huerta-Cepas J.. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021; 38:5825–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. The Gene Ontology Consortium The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021; 49:D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kanehisa M., Goto S.. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Emms D.M., Kelly S.. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019; 20:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Duan G., Wu G., Chen X., Tian D., Li Z., Sun Y., Du Z., Hao L., Song S., Gao Y.. HGD: an integrated homologous gene database across multiple species. Nucleic Acids Res. 2023; 51:D994–D1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Marcais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A.. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 2018; 14:e1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tang B., Zhou Q., Dong L., Li W., Zhang X., Lan L., Zhai S., Xiao J., Zhang Z., Bao Y.. iDog: an integrated resource for domestic dogs and wild canids. Nucleic Acids Res. 2019; 47:D793–D800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Z.-H., Zhu Q.-H., Li X., Zhu J.-W., Tian D.-M., Zhang S.-S., Kang H.-L., Li C.-P., Dong L.-L., Zhao W.-M.. iSheep: an integrated resource for sheep genome, variant and phenotype. Front. Genet. 2021; 12:714852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Diesh C., Stevens G.J., Xie P., De Jesus, Martinez T., Hershberg E.A., Leung A., Guo E., Dider S., Zhang J.et al.. JBrowse 2: a modular genome browser with views of synteny and structural variation. Genome Biol. 2023; 24:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G.. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012; 40:e115–e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L.et al.. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Cambridge (Mass.)). 2021; 2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hao Z., Lv D., Ge Y., Shi J., Weijers D., Yu G., Chen J.. RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ. Computer Science. 2020; 6:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. CNCB-NGDC Members and Partners Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2023. Nucleic Acids Res. 2023; 51:D18–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Welsch R., Arango J., Bar C., Salazar B., Al-Babili S., Beltran J., Chavarriaga P., Ceballos H., Tohme J., Beyer P.. Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell. 2010; 22:3348–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mlalazi B., Welsch R., Namanya P., Khanna H., Geijskes R.J., Harrison M.D., Harding R., Dale J.L., Bateson M.. Isolation and functional characterisation of banana phytoene synthase genes as potential cisgenes. Planta. 2012; 236:1585–1598. [DOI] [PubMed] [Google Scholar]
- 74. Paul J.Y., Khanna H., Kleidon J., Hoang P., Geijskes J., Daniells J., Zaplin E., Rosenberg Y., James A., Mlalazi B.. Golden bananas in the field: elevated fruit pro-vitamin A from the expression of a single banana transgene. Plant Biotechnol. J. 2017; 15:520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yan J., Wang X.. Machine learning bridges omics sciences and plant breeding. Trends Plant Sci. 2023; 28:199–210. [DOI] [PubMed] [Google Scholar]
- 76. Gupta C., Ramegowda V., Basu S., Pereira A.. Using network-based machine learning to predict transcription factors involved in drought resistance. Front. Genet. 2021; 12:652189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liu S., Xu F., Xu Y., Wang Q., Yan J., Wang J., Wang X., Wang X.. MODAS: exploring maize germplasm with multi-omics data association studies. Science Bulletin. 2022; 67:903–906. [DOI] [PubMed] [Google Scholar]
- 78. Ma W., Qiu Z., Song J., Li J., Cheng Q., Zhai J., Ma C.. A deep convolutional neural network approach for predicting phenotypes from genotypes. Planta. 2018; 248:1307–1318. [DOI] [PubMed] [Google Scholar]
- 79. Varshney R.K., Sinha P., Singh V.K., Kumar A., Zhang Q., Bennetzen J.L.. 5Gs for crop genetic improvement. Curr. Opin. Plant Biol. 2020; 56:190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
TCOD is freely available online at https://ngdc.cncb.ac.cn/tcod and does not require user to register.