Abstract
Enhancer RNAs (eRNAs) transcribed from distal active enhancers serve as key regulators in gene transcriptional regulation. The accumulation of eRNAs from multiple sequencing assays has led to an urgent need to comprehensively collect and process these data to illustrate the regulatory landscape of eRNAs. To address this need, we developed the eRNAbase (http://bio.liclab.net/eRNAbase/index.php) to store the massive available resources of human and mouse eRNAs and provide comprehensive annotation and analyses for eRNAs. The current version of eRNAbase cataloged 10 399 928 eRNAs from 1012 samples, including 858 human samples and 154 mouse samples. These eRNAs were first identified and uniformly processed from 14 eRNA-related experiment types manually collected from GEO/SRA and ENCODE. Importantly, the eRNAbase provides detailed and abundant (epi)genetic annotations in eRNA regions, such as super enhancers, enhancers, common single nucleotide polymorphisms, expression quantitative trait loci, transcription factor binding sites, CRISPR/Cas9 target sites, DNase I hypersensitivity sites, chromatin accessibility regions, methylation sites, chromatin interactions regions, topologically associating domains and RNA spatial interactions. Furthermore, the eRNAbase provides users with three novel analyses including eRNA-mediated pathway regulatory analysis, eRNA-based variation interpretation analysis and eRNA-mediated TF–target gene analysis. Hence, eRNAbase is a powerful platform to query, browse and visualize regulatory cues associated with eRNAs.
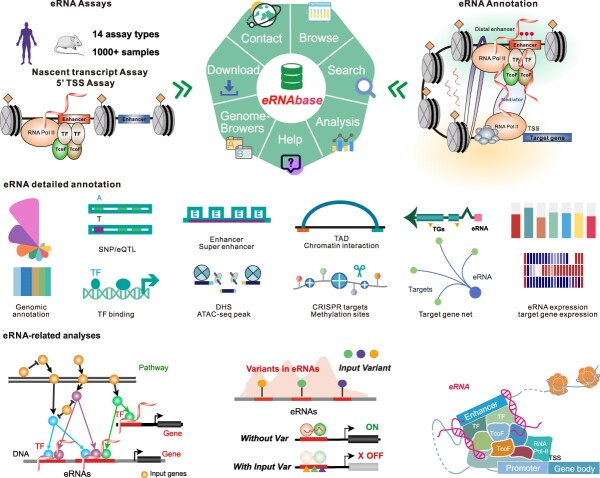
Graphical Abstract
Graphical Abstract.
Introduction
Enhancers are distal cis-regulatory element that couples with promoters to form the enhancer-promoter regulation circuit to trigger cell-specific gene transcription patterns (1,2). Emerging evidence has revealed that active enhancers, especially super enhancers, possess transcription potentials, generating bi-directional non-coding transcripts defined as enhancer RNAs (eRNAs) (3–5). Previous studies reported that eRNAs with cell-specific expression manner played crucial regulatory roles in complex diseases (6). For instance, CCAT1 eRNA within the oncogenic super-enhancer can coordinate the trafficking of the MYC gene to promote colon cancer cell proliferation (7). The overexpression of NET1e in cancer cells can promote tumor growth and induce resistance to anticancer compounds in breast cancer (8). Mechanistically, increasing evidence suggests that eRNAs mediate gene transcription in conjunction with transcription regulators and genetic variants. For instance, selective gene regulation programs of the transcriptional activator PAX3-FOXO1 were determined by enhancer architecture and eRNA transcription from the super enhancer region (9). eRNA can act as a scaffold to activate YAP/TEAD ER-bound enhancers in breast cancer (10). eRNAs transcribed from enhancers are enriched for context-specific genetic variants, which can alter enhancer activities and contribute to pathogenesis (11–13). Moreover, studies demonstrated the regulatory roles of eRNAs in histone modification, chromatin accessibility and gene expression by constructing a chromatin loop, thereby communicating regulatory cues from enhancers to their target gene (14,15). These studies highlight the crucial role and significance of eRNA-mediated regulation, especially in cell-specific pathological processes, cancer cell proliferation and oncogenesis.
Numerous eRNA-related databases have been developed recently to investigate the biological significance of eRNAs. For instance, HeRA and Animal-eRNAdb focus on detecting eRNAs expressed from well-annotated enhancer regions (16,17). Based on this strategy, Prof. Han’s group also elaborated on the pan-cancer eRNA regulatory atlas in eRic and GPIeR (8,18). However, eRNAs show low-abundance characteristics in cells. Typical RNA-seq captures eRNAs with extremely low efficiency. Two categories of assays (TSS and nascent transcript assays) have been developed to fill this gap (19). HACER, PINTS and FANTOM5 provide a list of human eRNAs from these genome-wide RNA-seq assays, such as CAGE and GRO/PRO-seq (20,21). While these advanced databases have promoted eRNA research, they also face limitations, especially concerning the eRNA identification method and scale. More importantly, only a small subset of annotation information was provided in above databases, the comprehensive genetic and epigenetic annotation information of eRNAs is lacking. Because of the rapid accumulation of eRNA-associated high-throughput data from multiple experiments (e.g. Bru-seq, BruChase-seq, mNET-seq, NET-CAGE and PRO/GRO-cap), there is an urgent need to comprehensively collect and effectively process these data (22–26). Numerous studies showed that eRNAs as well as the related variants and TFs, have a strong influence on human diseases and biological processes (27–29). Integrating abundant (epi)genetic annotation and regulatory analyses will be extremely useful for elucidating the mechanism of action of eRNAs on transcriptional regulation. Therefore, a resource for eRNAs that provides comprehensive eRNA annotations and eRNA-associated regulatory analyses is highly desirable.
In this study, we developed the eRNAbase (http://bio.liclab.net/eRNAbase/index.php) to document a large number of available resources of human and mouse eRNAs and to provide comprehensive annotation and analyses for eRNAs. The current version of eRNAbase cataloged 10399928 eRNAs from 1012 samples including 858 human samples and 154 mouse samples. These eRNAs were first identified from 14 eRNA-related experiment types manually collected from GEO/SRA and ENCODE, including Bru-seq, CAGE, BruChase-seq, PRO/GRO-seq, PRO/GRO-cap, mNET-seq, NET-CAGE, csRNA-seq, BruUV-seq, Start-seq, CoPRO and RAMPAGE. Importantly, eRNAbase provides detailed and abundant (epi)genetic annotations in eRNA regions, such as super enhancers, enhancers, common single nucleotide polymorphisms (SNPs), expression quantitative trait loci (eQTL), risk SNPs, transcription factor binding sites (TFBSs), CRISPR/Cas9 target sites, DNase I hypersensitivity sites (DHSs), chromatin accessibility regions, methylation sites, chromatin interactions regions, topologically associating domains (TADs) and RNA spatial interactions. Furthermore, eRNAbase provides users with three types of eRNA regulatory analyses, including eRNA-mediated pathway regulatory analysis, eRNA-based variation interpretation analysis and eRNA-mediated TF–target gene analysis. eRNAbase is a user-friendly database to query, browse and visualize information associated with eRNAs (Figure 1).
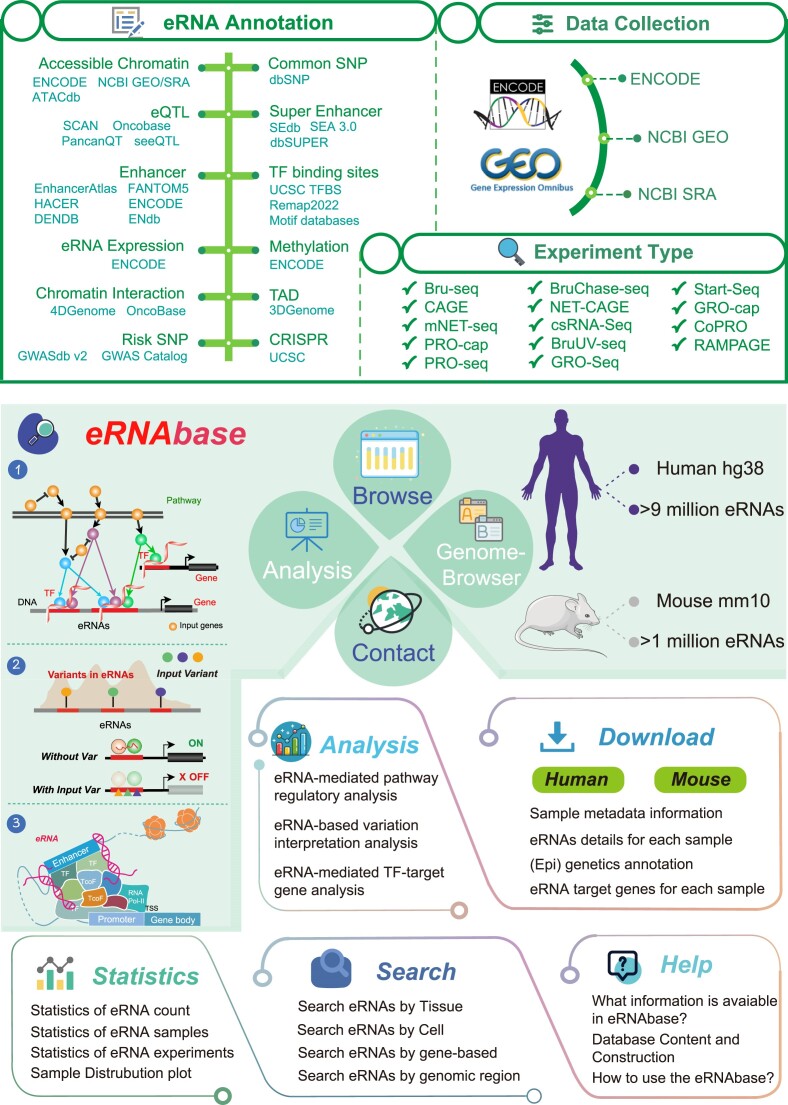
Figure 1.
Data collection and construction of eRNAbase. eRNAs of eRNAbase were identified from 14 experiment types manually collected from GEO/SRA and ENCODE. Abundant (epi) genetic annotations, such as super enhancers, enhancers, open chromatins, common SNPs, eQTLs, risk SNPs, TFBS, CRISPR target sites, methylation sites, chromatin interactions regions and TADs. Moreover, eRNAbase contains analysis tools and functions to analyze, query, browse and visualize information associated with eRNAs.
Materials and methods
Data processing and eRNA identification
In the current eRNAbase, we manually collected 1012 eRNA sequencing samples, including 858 human samples and 154 mouse samples. These samples were firstly collected from 14 eRNA-related experiments from GEO/SRA and ENCODE with the keywords, including ‘Bru-seq, CAGE, BruChase-seq, PRO/GRO-seq, PRO/GRO-cap, mNET-seq, NET-CAGE, csRNA-seq, BruUV-seq, Start-seq, CoPRO and RAMPAGE’ (30–32). To make these datasets more available, eRNAbase performed quality control for sequencing data and used a unified pipeline, named PINTS, to identify eRNAs (19). Briefly, we first used fastp (version: 0.12.4) to evaluate the sequencing quality and filter the low-quality reads with the parameters (-adapter_sequence -low_complexity_filter -l 14 -w 16) (33). Second, STAR (version: 2.7.10b) was used to align the human genome (Hg38) or mouse genome (Mm10) with the following parameters (STAR -runThreadN 16 -outSAMtype BAM SortedByCoordinate) (34). Third, pints_caller (version: 1.1.8) was used to identify bidirectional eRNA peaks from BAM files with the parameters (pints_caller –bam –exp-type -thread 16). Finally, distal eRNA peaks were filtered out using Bedtools (version: 2.30.0) with the parameters (bedtools intersect -a bidirectional_peaks.bed -b promoters.bed -v > bidirectional_peaks.distalTREs.bed). A total of 10399928 eRNAs were identified based on the aforementioned protocols.
Identification of TF binding sites of eRNA genomic loci
Identification of TFs that are recruited to eRNA genomic loci can provide valuable insights into the regulatory roles of eRNAs in transcriptional regulation (35). This study used two strategies to map potential TFBSs on eRNA genome loci via integrating TF ChIP-seq data and motif scanning methods (36). For the first strategy, 51 616 973 nonredundant TF binding peaks from 817 human TF ChIP-seq datasets and 32 985 444 nonredundant TF binding peaks from 648 mouse TF ChIP-seq datasets from multiple cells and tissues were collected from ReMap 2022 (37). Then, we used the genome intersection command of Bedtools to find the overlap regions between eRNAs and TF ChIP-seq peaks. Each overlapped region represented the potential TFBSs on eRNAs. For the second strategy of the motif scanning method, we first integrated the position weight matrices of ∼700 TF motifs from multiple sources, including JASPAR CORE 2020 vertebrates, Homeodomains, Jolma2013, UniPROBE and Wei2010. Then, we used Find Individual Motif Occurrences (FIMO) (version: 5.4.0), which was the constituent of the MEME Suite toolkit, to scan for the motif binding activities within each eRNA region (38). Finally, we identified potential TFs binding to eRNA genomic loci with a strict experiential P-value threshold of 1.0E-06 (39).
Moreover, we embedded 5 547 656 human TFBSs and 2 858 356 mouse TFBSs from UCSC genome browser and used the genome intersection command of Bedtools to identify the intersected TFBSs within eRNA regions (40).
Epigenetic and genetic annotation for eRNAs
Genome mutation events and epigenetic state changes of eRNAs were considered key issues in investigating the regulatory mechanisms of diseases from the transcription regulation facet. In this study, we provided abundant epigenetic and genetic annotations for unveiling eRNAs, such as super enhancers, enhancers, accessible chromatins, DHS, 3D chromatin interactions, TFBS, DNA methylation sites, common SNPs, risk SNPs and eQTLs. Furthermore, we also provided the eRNA-related gene editing sites of CRISPR for low-throughput knockout experiments. All the detailed information of (epi)genetic annotations was provided in Supplementary Table S1.
Super enhancer and enhancer
eRNAs are increasingly considered as a critical marker for active enhancers and super enhancers genome wide. Notably, cell-specific gene transcription patterns driver by distal enhancers, especially super enhancers, were demonstrated to play significant roles in maintaining cell identity (6,41). We manually collected and processed H3K27ac ChIP-seq data from GEO/SRA, ENCODE, Roadmap and GGR databases for the super enhancer category annotation. Briefly, we first used Bowtie software to align human (Hg38) or mouse (Mm10) genomes for H3K27ac ChIP-seq data and then used MACS2 software to call H3K27ac binding peaks. ROSE software was used to identify super enhancers by merging H3K27ac-binding peaks. We also collected human and mouse super enhancers from SEA and dbSuper (42,43). As a result, 2 678 273 human super enhancers and 11609 mouse super enhancers were integrated into the current eRNAbase. For enhancer category annotation, we collected the archived enhancer data from EnhancerAtlas, HACER, ENCODE, FANTOM5, DENDB and ENdb, including 14 797 266 human enhancers and 439 092 mouse enhancers (20,21,32,44,45).
Chromatin accessibility regions
Emerging evidence has proved that eRNA regions have high chromatin accessibility, which can recruit transcription regulators to mediate the activation of downstream genes. ATAC-seq and DNase-seq are considered as the golden methods to identify accessible chromatins or DHSs. We collected more than 130 000 000 chromatin accessibility regions from ATACdb, which was developed by our lab, to annotate accessible chromatins for eRNAs. In brief, we manually processed 2723 human and mouse ATAC-seq datasets from GEO/SRA and performed Bowtie2 and MACS2 to call chromatin accessibility peaks. We also collected 69 860 705 human DHSs from 293 samples and 9 802 229 mouse DHSs from 56 samples from ENCODE DNase-seq datasets.
Common SNP/risk SNP/eQTL
Genetic variants, especially SNPs within eRNAs determined eRNA activity and TF binding events, prominently affecting gene regulatory circuit states. eRNAbase provides multiple SNP annotation information eRNAs from diverse sources. In detail, 37 302 978 human common SNPs from dbSNP were downloaded and filtered SNPs with a minimum allelic frequency (MAF) > 0.05 out via VCFTools (v0.1.13) (46). A total of 351 728 human risk SNPs from GWAS Catalog and GWASdb v2 were also curated (47). We also collected 11 995 221 human eQTLs from PancanQTL, seeQTL, SCAN, Oncobase and GTEx (48–52). Besides, 81432271 mouse common SNPs from dbSNP were also released in the current eRNAbase. In the analysis panel, we identified the linkage disequilibrium SNPs (r2 = 0.8) for five super-populations (African, Ad Mixed American, East Asian, European and South Asian) via Plink (v1.9).
Chromatin interaction/methylation/RNA spatial interaction
Chromatin loops link the regulatory cues from distal and proximal elements, directing the cell-specific gene transcription (53). In eRNAbase, we collected 34 342 926 human chromatin interactions from 4DGenome and Oncobase, including data from ChIA-PET 3C, 4C, 5C and Hi-C. Furthermore, 72019 human TADs from 21 tissues or cell lines from 3D Genome Browser were embedded (54,55). Dynamic DNA methylation states, which are highly related to chromatin interaction conformation, also have the regulatory effects on eRNA activity (56). The eRNAbase recorded DNA methylation states from 166 855 665 methylation sites of whole-genome shotgun bisulfite sequencing and 30 392 523 methylation sites of the 450k array from ENCODE. Emerging RNA spatial interaction experiment, such as RIC-seq, enables the identification of the specific molecules that interact with eRNAs, shedding new light on the molecular mechanisms of enhancer function and gene regulation. We also collected the human RIC-seq derived eRNA interaction data from published datasets of GSE190214 and GSE221551. Totally, 1 026 628 pairs of RNA interactions from 9 tissues/cells were provided in eRNAbase.
eRNA expression/CRISPR
Cell-specific eRNA expression patterns could help find key regulators in pathological processes. To provide global normalized eRNA expression in tissues, we downloaded the same-batch BAM files of human and mouse total RNA-seq from ENCODE project. Then, we used SAMtools (version: 1.13) and Bedtools to call the read count number and calculate the log-normalized CPM (counts per million) in eRNA regions. The eRNAbase provided eRNA expression in 30 human tissues/cells and 11 mouse tissues. Knockout experiments were the efficient method to investigate the biological functions of RNAs or DNA regulatory elements. We also provided CRISPR/Cas9 target sites. These sites were annotated for predicted specificity (off-target effects) and predicted efficiency (on-target cleavage) using various algorithms through the tool CRISPOR from UCSC.
Assigning target genes of eRNAs
Identifying eRNA downstream target genes is important to reveal the biological function and regulatory mechanisms of eRNAs. We used two strategies (geneMapper and BETA (version: 1.0.7)) based on genome distances to comprehensively annotate the potential target genes (57,58). For the geneMapper method, we input eRNA regions in the bed format file and used a Python script (ROSE geneMapper.py) to assign downstream target genes. The target genes were classified into three strategies including overlap, proximal and closest. For the BETA method, we also input eRNA regions into BETA script and obtained the target genes with distance and score thresholds. The target genes from the aforementioned two methods were integrated and used for further analysis. Furthermore, we provided the target gene expression in humans and mice from GTEx, ENCODE, TCGA, CCLE and FANTOM5.
User interface
Search interface for eRNAs
eRNAbase is a comprehensive platform for users to search, browse, visualize and analyze eRNAs of interest. In the ‘Search’ interface, eRNAbase provides four query methods for searching eRNAs based on eRNA characteristics and sample information: ‘Search by Genomic Region’ (input genomic position and select the optional sample metadata), ‘Search by Target Genes’ (input gene symbols of interest and select species and methods), ‘Search by Tissue Type’ (input tissue of interest and select species) and ‘Search by Cell Type’ (input cell type of interest and select species) (Figure 2A and C). Brief information of the search results is displayed in a table on the result page. For instance, for the methods based on eRNA regions or target genes, the result table returned the eRNA information including eRNA ID, eRNA region, sample ID, tissue/cell type and experiment type. For methods based on tissue or cell type, the result table described the basic information of samples such as ‘Browse’ page, including sample ID, species, tissue type, biosample type, cell type and experiment type (Figure 2B). Users can click the sample ID to obtain the summarized regulatory information for searching the eRNA sample. In the sample page, eRNAbase provides information on sample overview, eRNA genomic annotation, eRNA detail, annotation data source and software parameters (Figure 2D). In the genomic annotation section, eRNAbase provides genomic proportions and peak distribution statistics calculated using ChIPseeker. The eRNA ‘Details’ section provides comprehensive annotation for eRNAs, including eRNA ID, genome location, size, bidirectional genome location and detailed (epi)genetic information in eRNA regions (Figure 2D). Users can click the eRNA ID to obtain details regarding the eRNA (Figure 2E).
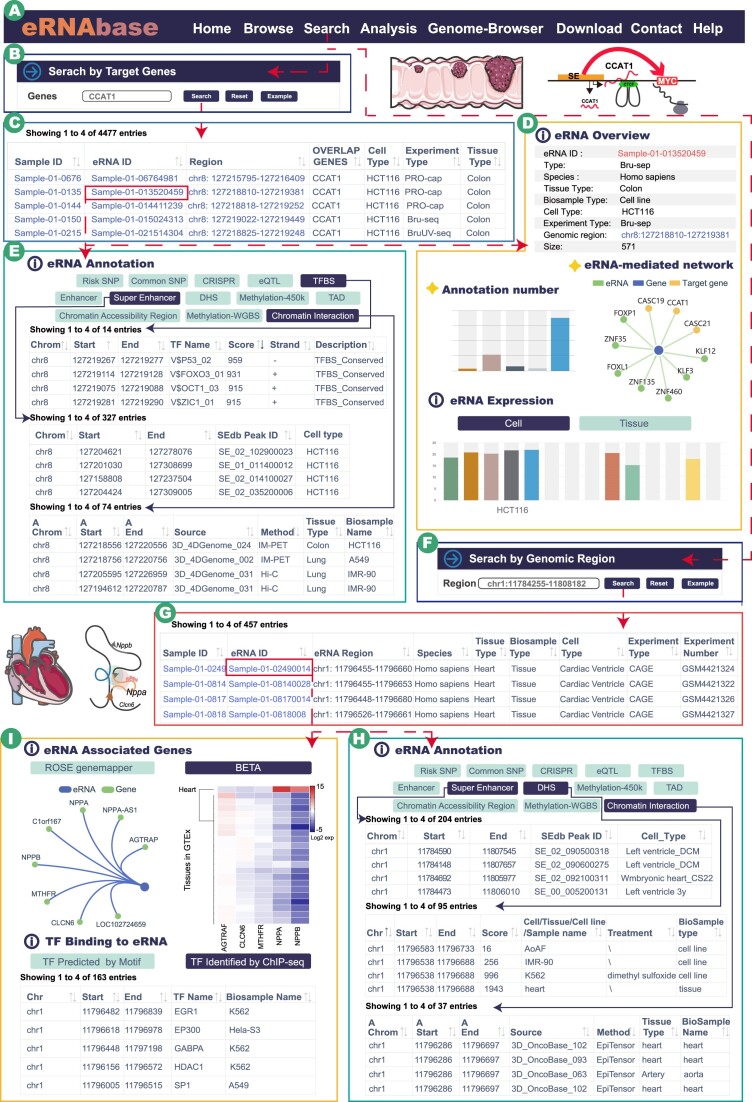
Figure 2.
Main interfaces and usage of eRNAbase. (A) The navigation bar of eRNAbase. (B) Data browse page of eRNAbase. (C) Users can search eRNAs of interest via four queries. (D) Sample interface of eRNA sets from human K562 cell line, including sample overview, eRNA detail, genome proportion plot, peak distribution plot and chromosome distribution plot. (E) Detailed annotation information of eRNAs of interest, including eRNA overview information, annotation statistic, eRNA-mediated network, (epi)genetic annotation interactive table, eRNA associated genes, target gene expression, eRNA expression and TF binding information. (F) The eRNAbase provides three online analysis tools to help dissect eRNA-related regulation. (G) Genome browser interface. (H) Download interface.
On the ‘Details’ page, eRNAbase provides six sections to list the comprehensive annotation for eRNAs: eRNA overview, eRNA annotation, eRNA-associated genes, TFs binding to eRNAs, overlap with other eRNAs and eRNA expression (Figure 2E). In the section of eRNA annotation, eRNAbase provides a large amount of (epi)genetic annotation information, including super enhancers, enhancers, common SNPs, risk SNPs, CRISPRs, eQTLs, TFBS, DHS, DNA methylation sites, TAD, 3D chromatin interactions and RNA spatial interactions. Alternatively, for the results of searches by eRNA regions or target genes, users can obtain the eRNAs of interest directly.
User-friendly interface for browsing eRNA
The ‘Data-Browse’ page is organized as an interactive and alphanumerically sortable table that allows users to quickly browse samples and customize filters through ‘Species,’ ‘Tissue type,’ ‘Biosample Type,’ ‘Cell Type’ and ‘Experiment type.’ Users can use the ‘Show entries’ drop-down menu to change the number of the displayed records per page (Figure 2A and B). Then, users can click on ‘Sample ID’ to view the eRNAs for a given sample.
eRNA-related online analysis tool
The eRNAbase provides three analysis tools to help biologists comprehensively explore the biological function and mechanisms of eRNAs (Figure 2F). Pathway analysis is an effective strategy to identify abnormal biological processes of diseases. While TFs are frequently located at the end of pathways, their downstream regulatory events remain unknown. In eRNA-mediated pathway regulatory analysis, users can submit a list of genes of interest to obtain enriched pathways through a hypergeometric test. Within each pathway, the eRNAbase locates the terminal TFs and terminal TF-bound eRNAs, forming genes/pathways/TFs/eRNA regulatory axes. The genetic variants located at eRNAs determine eRNA activity and TF binding affinity, inducing aberrant gene expression programs. In eRNA-based variation interpretation analysis, users can submit a variant ID (rsID) or genomic region (bed format) to eRNAbase to locate the eRNAs that overlapped with the variant or variants within the submitted genomic region. The results from this analysis also displayed the annotation information of submitted variant(s), including genomic coordinates, variant type, linkage disequilibrium SNP and disease information. Moreover, TF binding activity is highly related to enhancer activity and eRNA biogenesis. The eRNAs can also serve as the scaffolds or intermediates for TF–target gene regulation. In eRNA-mediated TF–target gene analysis, users can input paired lists of TFs and genes of interest and then select species. The eRNAbase can identify eRNA-mediated TF-gene pair(s) in different tissue types. Meanwhile, users can set different TF identification methods through the ‘Type’ option, including motif and ChIP-seq methods.
Genome browser and data download
The eRNAbase uses the JBrowse genome browser to visualize the regulatory information of human and mouse eRNAs by customizing tracks of interest, such as eRNA regions, genes, super-enhancer and variants (Figure 2G). The eRNA regions, eRNA downstream target genes and all annotation data are available for individual or batch download on the ‘Download’ page. Additionally, we also provided the merged eRNA sets of human and mouse on the ‘Download’ page. Users can quickly download data of interest (Figure 2H).
Case study
Search for CCAT1 eRNA
Colon cancer-associated transcript 1 (CCAT1) is a widely studied eRNA transcribed from the super enhancer region of the 8q24 locus (59,60). Increasing evidence has suggested that CCAT1 participates in various biological processes, such as cell proliferation, invasion, metastasis and apoptosis via functioning as a regulator in transcription regulation in multiple cancers. We performed a search case to illustrate the regulatory information for CCAT1 eRNA in eRNAbase. Briefly, we first input CCAT1 and selected the ‘ROSE genemapper’ method in the target gene search (Figure 3A and B). The results showed that >4000 eRNA records were searched out, indicating that CCAT1 was a hot region to produce eRNA transcript (Figure 3C). CCAT1 was first identified from colon cancers, and then we selected the ‘Sample-01–013520459’ eRNA in HCT-116 cell lines of colon cancer, which was overlapped with the target gene CCAT1 (Figure 3C).
Figure 3.
Case study panels of eRNAbase. (A) The navigation bar of eRNAbase. (B) Case study of CCAT1 query in target gene search. CCAT1 is a well-investigated eRNAs that plays key roles in cancers, especially in colon cancer. (C) Interactive result table of CCAT1 search, including the sample ID, eRNA ID, regions, genes and sample metadata information. (D-E) eRNA detailed information of the selected eRNA ‘Sample-01–013520459″, including eRNA overview, eRNA-mediated network, (epi)genetic annotation interactive table (TFBS, Super enhancer and Chromatin interaction), eRNA associated genes and eRNA expression. (F) Case study of NPPA super enhancer query in genomic region search. (G) Interactive result table of genomic region search. (H–I) eRNA detailed annotation information of the selected eRNA ‘Sample-01–02490014’.
The eRNAbase provides comprehensive epigenetic and genetic annotations for eRNAs. Users can click the eRNA ID to explore the potential regulatory information. For example, in the ‘Details’ page of eRNA ‘Sample-01–013520459’, the results showed that the 571-bp-long eRNA covered multiple annotations and was high-expressed in HCT-116 cell line (Figure 3D), suggesting that this eRNA might play crucial roles in transcription regulation. Some colon cancer-related TF binding events were also detected within this eRNA, such as TP53, FOXO3s1 and OCT1 (Figure 3E, upper) (61,62). CCAT1 was demonstrated as a key super enhancer-driven transcript. The results showed that 327 super enhancers overlapped with this eRNA (Figure 3E, middle). Notably, previous studies reported that CCAT1 maintained the chromosome loop of MYC, promoting the expression of MYC (60). In the annotation result of chromatin interaction, some colon-specific validated interactions within MYC promoters were identified (Figure 3E, bottom). These results indicated that the eRNAbase has a strong ability to explore the potential regulatory mechanisms for eRNAs.
Search for NPPA super enhancer
Super enhancers-mediated chromatin circuits play crucial roles in cell identity and diseases, whereas eRNA signals within super enhancer region can be used to infer its activities. Heart-specific expressed NPPA is controlled by a shared super-enhancer (63). We obtained the super-enhancer region of NPPA from SEdb 2.0 and searched eRNAs based on the method of genomic region search (Figure 3F). As a result, four eRNAs in diverse samples overlapped with the input super enhancer (Figure 3G). We selected the eRNAs with eRNA ID ‘Sample-01-08170014’ as the example. In the eRNA detail page, the results showed that a lot of heart tissue-specific annotation information was enriched in this eRNA region, such as DHS and chromatin interactions, indicating that the eRNAbase could support tissue-specific research (Figure 3H). In addition, NPPA were identified by the eRNA-associated gene network. The expression heatmap showed that NPPA was highly expressed in heart tissue, which coincided with the biomarker feature of NPPA. TF binding to the eRNA module listed the potential binding TFs in eRNA regions, including the validated transcription regulators of EGR1, EP300, HDAC1 and SP1 (Figure 3I) (64–66). The aforementioned results also illustrate the availability and biological value of the eRNAbase.
Discussion
eRNA is a type of noncoding RNA transcribed from distal enhancers, which mediates cell-specific gene expression (67). Numerous studies have revealed that aberrant eRNA expression patterns can disrupt the chromatin circuits of disease genes to promote disease development (68). The eRNA data needs to be collected and processed to investigate the regulatory landscape of eRNA. Some advanced databases have been developed to illustrate eRNA regulation. For instance, HACER, FANTOM5 and PINTS cataloged eRNAs derived from nascent transcript sequencing data, including CAGE and GRO/PRO-seq (19–21). Conversely, HeRA and Animal-eRNAdb detected the high-expression transcripts located within enhancer regions from RNA-seq data (16,17). Prof. Han’s group also developed eRIC and GPIeR to reveal cancer-related eRNA transcripts (8,18). These databases have promoted eRNA research, but they have limitations, especially regarding eRNA identification methods, scale and annotation. The eRNAbase was highlighted in eRNA scale and annotation (Supplementary Table S2). Briefly, the eRNAbase extends the eRNA scale to >5 times the existing eRNA databases, recording 10 399 928 eRNAs from 1012 samples including 858 human samples and 154 mouse samples. Moreover, abundant epigenetic and genetic annotations within eRNAs are integrated to help users explore potential functions and the regulation of eRNAs in diseases and biological processes. Overall, eRNAbase serves as a powerful platform for eRNA-related regulation enriched with abundant annotation information.
The eRNAbase is a user-friendly database to query, browse and visualize information associated with eRNAs, which has multiple highlights and advantages compared with the existing databases as follows: (i) eRNAs were first identified from 14 eRNA-related experiment types manually collected from GEO/SRA and ENCODE, including Bru-seq, CAGE, BruChase-seq, PRO/GRO-seq, PRO/GRO-cap, mNET-seq, NET-CAGE, csRNA-seq, BruUV-seq, Start-seq, CoPRO and RAMPAGE; (ii) comprehensive genetic and epigenetic annotations within eRNA regions include TF bindings, super-enhancers, enhancers, common SNPs, eQTLs, CRISPR editing sites, risk SNPs, DHS, DNA methylation sites, 3D chromatin interactions, RNA spatial interactions and TADs; (iii) quality control and unified software parameters are used to process sequencing data; (iv) the visualization function is used to annotate eRNA regions; (v) novel and useful online analysis tools are available to investigate eRNA-mediated pathways, variations and TF regulation; (vi) a customized genome browser provides users with an intuitive view of the regulatory information of eRNA adjacent regions and embeds several annotation tracks and (vii) the user-friendly database displays eRNA regions and associated annotation information with interactive tables.
In summary, the eRNAbase is a comprehensive resource for eRNAs, with the largest scale and the most comprehensive annotation details for both human and mouse eRNAs. The current version of the eRNAbase also provides online analysis tools, allowing researchers to explore regulatory patterns of eRNAs across multi-omics facets. In forthcoming versions of the eRNAbase, we aim to update this useful resource by extending eRNA scales from more experiments (e.g. RNA-seq and single-cell data) and by adding more epigenetic and genetic annotations (e.g. functional variants and somatic mutations). Moreover, the current version of eRNAbase used PINTS to identify eRNAs in all datasets, which was designed for detecting eRNA TSSs and candidate enhancers. This pipeline may lose the information of the entire eRNA transcription unit. We will use more powerful pipelines to identify eRNAs in the next updated versions. We are confident that eRNAbase will emerge as a useful and effective platform for exploring potential functions and the regulation of eRNAs in diseases and various biological processes.
Supplementary Material
Contributor Information
Chao Song, The First Affiliated Hospital & Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Guorui Zhang, The First Affiliated Hospital & Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences & MOE Key Lab of Rare Pediatric Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Cell Biology and Genetics, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Xinxin Mu, The First Affiliated Hospital & Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Chenchen Feng, School of Computer, University of South China, Hengyang, Hunan, 421001, China.
Qinyi Zhang, The First Affiliated Hospital & Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences & MOE Key Lab of Rare Pediatric Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Cell Biology and Genetics, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Shuang Song, The First Affiliated Hospital & Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences & MOE Key Lab of Rare Pediatric Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Cell Biology and Genetics, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Yuexin Zhang, The First Affiliated Hospital & Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Mingxue Yin, The First Affiliated Hospital & Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences & MOE Key Lab of Rare Pediatric Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Cell Biology and Genetics, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Hang Zhang, The First Affiliated Hospital & Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; School of Computer, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China.
Huifang Tang, The First Affiliated Hospital & Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Department of Cardiology, Hengyang Medical School, University of South China, Hengyang, China; The First Affiliated Hospital, Cardiovascular Lab of Big Data and Imaging Artificial Intelligence, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Institute of Cardiovascular Disease, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Clinical Research Center for Myocardial Injury in Hunan Province, Hengyang, Hunan, 421001, China.
Chunquan Li, The First Affiliated Hospital & Hunan Provincial Key Laboratory of Multi-omics And Artificial Intelligence of Cardiovascular Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences & MOE Key Lab of Rare Pediatric Diseases, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; Hunan Provincial Maternal and Child Health Care Hospital, National Health Commission Key Laboratory of Birth Defect Research and Prevention, Hengyang Medical School, University of South China, Hengyang, Hunan, 421001, China; School of Computer, University of South China, Hengyang, Hunan, 421001, China; The First Affiliated Hospital, Department of Cardiology, Hengyang Medical School, University of South China, Hengyang, China.
Data availability
The research community can access information freely in the eRNAbase without registration or logging in. The URL of eRNAbase is http://bio.liclab.net/eRNAbase/index.php.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Natural Science Foundation of China [62171166, 62001145, 62302206]; Research Foundation of the First Affiliated Hospital of University of South China for Advanced Talents [20210002-1005 USCAT-2021-01]; China Postdoctoral Science Foundation [2019M661311]; Natural Science Foundation of Hunan Province [2023JJ30547, 2023JJ40594, 2023JJ30536]; Scientific Research Fund Project of Hunan Provincial Health Commission [20201920]; Special Funds for the Construction of Innovative Provinces in Hunan [2020SK4008]; Key Project of Hunan Provincial Science and Technology Innovation [No. 2020SK1013-2]; Clinical Research 4310 Program of the University of South China [20214310NHYCG03]. Funding for open access charge: Research Foundation of the First Affiliated Hospital of University of South China.
Conflict of interest statement. None declared.
References
- 1. Long H.K., Prescott S.L., Wysocka J.. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016; 167:1170–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heinz S., Romanoski C.E., Benner C., Glass C.K.. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015; 16:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li W., Notani D., Rosenfeld M.G.. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 2016; 17:207–223. [DOI] [PubMed] [Google Scholar]
- 4. Rothschild G., Basu U.. Lingering questions about enhancer RNA and enhancer transcription-coupled genomic instability. Trends Genet. 2017; 33:143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sartorelli V., Lauberth S.M.. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 2020; 27:521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen H., Liang H.. A high-resolution map of Human enhancer RNA loci characterizes super-enhancer activities in cancer. Cancer Cell. 2020; 38:701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chachoua I., Tzelepis I., Dai H., Lim J.P., Lewandowska-Ronnegren A., Casagrande F.B., Wu S., Vestlund J., Mallet de Lima C.D., Bhartiya D.et al.. Canonical WNT signaling-dependent gating of MYC requires a noncanonical CTCF function at a distal binding site. Nat. Commun. 2022; 13:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Z., Lee J.H., Ruan H., Ye Y., Krakowiak J., Hu Q., Xiang Y., Gong J., Zhou B., Wang L.et al.. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat. Commun. 2019; 10:4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang S., Wang J., Liu Q., McDonald W.H., Bomber M.L., Layden H.M., Ellis J., Borinstein S.C., Hiebert S.W., Stengel K.R.. PAX3-FOXO1 coordinates enhancer architecture, eRNA transcription, and RNA polymerase pause release at select gene targets. Mol. Cell. 2022; 82:4428–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu C., Li L., Zhang Z., Bi M., Wang H., Su W., Hernandez K., Liu P., Chen J., Chen M.et al.. A non-canonical role of YAP/TEAD is required for activation of estrogen-regulated enhancers in breast cancer. Mol. Cell. 2019; 75:791–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang P., Amarasinghe H.E., Whalley J.P., Tay C., Fang H., Migliorini G., Brown A.C., Allcock A., Scozzafava G., Rath P.et al.. Epigenomic analysis reveals a dynamic and context-specific macrophage enhancer landscape associated with innate immune activation and tolerance. Genome Biol. 2022; 23:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yao P., Lin P., Gokoolparsadh A., Assareh A., Thang M.W., Voineagu I.. Coexpression networks identify brain region-specific enhancer RNAs in the human brain. Nat. Neurosci. 2015; 18:1168–1174. [DOI] [PubMed] [Google Scholar]
- 13. Murakawa Y., Yoshihara M., Kawaji H., Nishikawa M., Zayed H., Suzuki H., Fantom C., Hayashizaki Y.. Enhanced identification of transcriptional enhancers provides mechanistic insights into diseases. Trends Genet. 2016; 32:76–88. [DOI] [PubMed] [Google Scholar]
- 14. Zhou Y., Xu S., Zhang M., Wu Q.. Systematic functional characterization of antisense eRNA of protocadherin alpha composite enhancer. Genes Dev. 2021; 35:1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bose D.A., Donahue G., Reinberg D., Shiekhattar R., Bonasio R., Berger S.L.. RNA binding to CBP stimulates histone acetylation and transcription. Cell. 2017; 168:135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Z., Hong W., Ruan H., Jing Y., Li S., Liu Y., Wang J., Li W., Diao L., Han L.. HeRA: an atlas of enhancer RNAs across human tissues. Nucleic Acids Res. 2021; 49:D932–D938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jin W., Jiang G., Yang Y., Yang J., Yang W., Wang D., Niu X., Zhong R., Zhang Z., Gong J.. Animal-eRNAdb: a comprehensive animal enhancer RNA database. Nucleic Acids Res. 2022; 50:D46–D53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Z., Luo M., Li Q., Liu Y., Lussier C., Zhang J., Ye Y., Guo A.Y., Han L.. Genetic, pharmacogenomic, and immune landscapes of enhancer RNAs across Human cancers. Cancer Res. 2022; 82:785–790. [DOI] [PubMed] [Google Scholar]
- 19. Yao L., Liang J., Ozer A., Leung A.K., Lis J.T., Yu H.. A comparison of experimental assays and analytical methods for genome-wide identification of active enhancers. Nat. Biotechnol. 2022; 40:1056–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T.et al.. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang J., Dai X., Berry L.D., Cogan J.D., Liu Q., Shyr Y.. HACER: an atlas of human active enhancers to interpret regulatory variants. Nucleic Acids Res. 2019; 47:D106–D112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirabayashi S., Bhagat S., Matsuki Y., Takegami Y., Uehata T., Kanemaru A., Itoh M., Shirakawa K., Takaori-Kondo A., Takeuchi O.et al.. NET-CAGE characterizes the dynamics and topology of human transcribed cis-regulatory elements. Nat. Genet. 2019; 51:1369–1379. [DOI] [PubMed] [Google Scholar]
- 23. Paulsen M.T., Veloso A., Prasad J., Bedi K., Ljungman E.A., Magnuson B., Wilson T.E., Ljungman M.. Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods. 2014; 67:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Core L.J., Waterfall J.J., Lis J.T.. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008; 322:1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mahat D.B., Kwak H., Booth G.T., Jonkers I.H., Danko C.G., Patel R.K., Waters C.T., Munson K., Core L.J., Lis J.T.. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat. Protoc. 2016; 11:1455–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nojima T., Gomes T., Grosso A.R.F., Kimura H., Dye M.J., Dhir S., Carmo-Fonseca M., Proudfoot N.J.. Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell. 2015; 161:526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou T., Zhu X., Ye Z., Wang Y.F., Yao C., Xu N., Zhou M., Ma J., Qin Y., Shen Y.et al.. Lupus enhancer risk variant causes dysregulation of IRF8 through cooperative lncRNA and DNA methylation machinery. Nat. Commun. 2022; 13:1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kristjansdottir K., Dziubek A., Kang H.M., Kwak H.. Population-scale study of eRNA transcription reveals bipartite functional enhancer architecture. Nat. Commun. 2020; 11:5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rahnamoun H., Lee J., Sun Z., Lu H., Ramsey K.M., Komives E.A., Lauberth S.M.. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat. Struct. Mol. Biol. 2018; 25:687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katz K., Shutov O., Lapoint R., Kimelman M., Brister J.R., O'Sullivan C. The sequence read Archive: a decade more of explosive growth. Nucleic Acids Res. 2022; 50:D387–D390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luo Y., Hitz B.C., Gabdank I., Hilton J.A., Kagda M.S., Lam B., Myers Z., Sud P., Jou J., Lin K.et al.. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020; 48:D882–D889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen S., Zhou Y., Chen Y., Gu J.. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee J.H., Xiong F., Li W.. Enhancer RNAs in cancer: regulation, mechanisms and therapeutic potential. RNA Biol. 2020; 17:1550–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng C., Song C., Liu Y., Qian F., Gao Y., Ning Z., Wang Q., Jiang Y., Li Y., Li M.et al.. KnockTF: a comprehensive human gene expression profile database with knockdown/knockout of transcription factors. Nucleic Acids Res. 2020; 48:D93–D100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hammal F., de Langen P., Bergon A., Lopez F., Ballester B.. ReMap 2022: a database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res. 2022; 50:D316–D325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y., Song C., Zhao J., Zhang Y., Zhao X., Feng C., Zhang G., Zhu J., Wang F., Qian F.et al.. SEdb 2.0: a comprehensive super-enhancer database of human and mouse. Nucleic Acids Res. 2023; 51:D280–D290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Navarro Gonzalez J., Zweig A.S., Speir M.L., Schmelter D., Rosenbloom K.R., Raney B.J., Powell C.C., Nassar L.R., Maulding N.D., Lee C.M.et al.. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 2021; 49:D1046–D1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gurumurthy A., Yu D.T., Stees J.R., Chamales P., Gavrilova E., Wassel P., Li L., Stribling D., Chen J., Brackett M.et al.. Super-enhancer mediated regulation of adult beta-globin gene expression: the role of eRNA and integrator. Nucleic Acids Res. 2021; 49:1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Khan A., Zhang X.. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016; 44:D164–D171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen C., Zhou D., Gu Y., Wang C., Zhang M., Lin X., Xing J., Wang H., Zhang Y.. SEA version 3.0: a comprehensive extension and update of the Super-Enhancer archive. Nucleic Acids Res. 2020; 48:D198–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao T., Qian J.. EnhancerAtlas 2.0: an updated resource with enhancer annotation in 586 tissue/cell types across nine species. Nucleic Acids Res. 2020; 48:D58–D64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ashoor H., Kleftogiannis D., Radovanovic A., Bajic V.B.. DENdb: database of integrated human enhancers. Database (Oxford). 2015; 2015:bav085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li M.J., Liu Z., Wang P., Wong M.P., Nelson M.R., Kocher J.P., Yeager M., Sham P.C., Chanock S.J., Xia Z.et al.. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2016; 44:D869–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gong J., Mei S., Liu C., Xiang Y., Ye Y., Zhang Z., Feng J., Liu R., Diao L., Guo A.Y.et al.. PancanQTL: systematic identification of cis-eQTLs and trans-eQTLs in 33 cancer types. Nucleic Acids Res. 2018; 46:D971–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xia K., Shabalin A.A., Huang S., Madar V., Zhou Y.H., Wang W., Zou F., Sun W., Sullivan P.F., Wright F.A.. seeQTL: a searchable database for human eQTLs. Bioinformatics. 2012; 28:451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gamazon E.R., Zhang W., Konkashbaev A., Duan S., Kistner E.O., Nicolae D.L., Dolan M.E., Cox N.J.. SCAN: SNP and copy number annotation. Bioinformatics. 2010; 26:259–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li X., Shi L., Wang Y., Zhong J., Zhao X., Teng H., Shi X., Yang H., Ruan S., Li M.et al.. OncoBase: a platform for decoding regulatory somatic mutations in human cancers. Nucleic Acids Res. 2019; 47:D1044–D1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang P., Wu Y., Zhou H., Zhou B., Zhang H., Wu H.. CLNN-loop: a deep learning model to predict CTCF-mediated chromatin loops in the different cell lines and CTCF-binding sites (CBS) pair types. Bioinformatics. 2022; 38:4497–4504. [DOI] [PubMed] [Google Scholar]
- 54. Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M.N.K., Li Y., Hu M.et al.. The 3D genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018; 19:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Teng L., He B., Wang J., Tan K.. 4DGenome: a comprehensive database of chromatin interactions. Bioinformatics. 2015; 31:2560–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pan C.W., Wen S., Chen L., Wei Y., Niu Y., Zhao Y.. Functional roles of antisense enhancer RNA for promoting prostate cancer progression. Theranostics. 2021; 11:1780–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang S., Sun H., Ma J., Zang C., Wang C., Wang J., Tang Q., Meyer C.A., Zhang Y., Liu X.S.. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc. 2013; 8:2502–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xiang J.F., Yin Q.F., Chen T., Zhang Y., Zhang X.O., Wu Z., Zhang S., Wang H.B., Ge J., Lu X.et al.. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014; 24:513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McCleland M.L., Mesh K., Lorenzana E., Chopra V.S., Segal E., Watanabe C., Haley B., Mayba O., Yaylaoglu M., Gnad F.et al.. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J. Clin. Invest. 2016; 126:639–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maddox J., Shakya A., South S., Shelton D., Andersen J.N., Chidester S., Kang J., Gligorich K.M., Jones D.A., Spangrude G.J.et al.. Transcription factor Oct1 is a somatic and cancer stem cell determinant. PLoS Genet. 2012; 8:e1003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu Y., Ao X., Ding W., Ponnusamy M., Wu W., Hao X., Yu W., Wang Y., Li P., Wang J.. Critical role of FOXO3a in carcinogenesis. Mol. Cancer. 2018; 17:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Man J.C.K., van Duijvenboden K., Krijger P.H.L., Hooijkaas I.B., van der Made I., de Gier-de Vries C., Wakker V., Creemers E.E., de Laat W., Boukens B.J.et al.. Genetic dissection of a super enhancer controlling the nppa-nppb cluster in the heart. Circ. Res. 2021; 128:115–129. [DOI] [PubMed] [Google Scholar]
- 64. Marques F.Z., Nelson E., Chu P.Y., Horlock D., Fiedler A., Ziemann M., Tan J.K., Kuruppu S., Rajapakse N.W., El-Osta A.et al.. High-Fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation. 2017; 135:964–977. [DOI] [PubMed] [Google Scholar]
- 65. Rouhi L., Fan S., Cheedipudi S.M., Braza-Boils A., Molina M.S., Yao Y., Robertson M.J., Coarfa C., Gimeno J.R., Molina P.et al.. The EP300/TP53 pathway, a suppressor of the Hippo and canonical WNT pathways, is activated in human hearts with arrhythmogenic cardiomyopathy in the absence of overt heart failure. Cardiovasc. Res. 2022; 118:1466–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li Y., Li Z., Zhang C., Li P., Wu Y., Wang C., Bond Lau W., Ma X.L., Du J.. Cardiac fibroblast-specific activating transcription factor 3 protects against heart failure by suppressing MAP2K3-p38 signaling. Circulation. 2017; 135:2041–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lam M.T., Li W., Rosenfeld M.G., Glass C.K.. Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci. 2014; 39:170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Adhikary S., Roy S., Chacon J., Gadad S.S., Das C.. Implications of enhancer transcription and eRNAs in cancer. Cancer Res. 2021; 81:4174–4182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The research community can access information freely in the eRNAbase without registration or logging in. The URL of eRNAbase is http://bio.liclab.net/eRNAbase/index.php.