Abstract
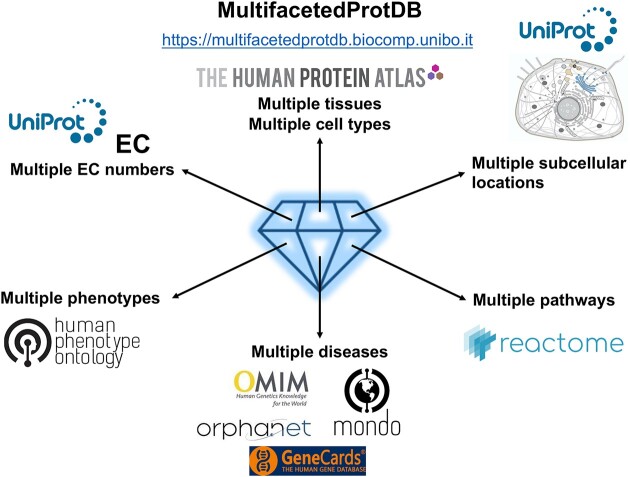
MultifacetedProtDB is a database of multifunctional human proteins deriving information from other databases, including UniProt, GeneCards, Human Protein Atlas (HPA), Human Phenotype Ontology (HPO) and MONDO. It collects under the label ‘multifaceted’ multitasking proteins addressed in literature as pleiotropic, multidomain, promiscuous (in relation to enzymes catalysing multiple substrates) and moonlighting (with two or more molecular functions), and difficult to be retrieved with a direct search in existing non-specific databases. The study of multifunctional proteins is an expanding research area aiming to elucidate the complexities of biological processes, particularly in humans, where multifunctional proteins play roles in various processes, including signal transduction, metabolism, gene regulation and cellular communication, and are often involved in disease insurgence and progression. The webserver allows searching by gene, protein and any associated structural and functional information, like available structures from PDB, structural models and interactors, using multiple filters. Protein entries are supplemented with comprehensive annotations including EC number, GO terms (biological pathways, molecular functions, and cellular components), pathways from Reactome, subcellular localization from UniProt, tissue and cell type expression from HPA, and associated diseases following MONDO, Orphanet and OMIM classification. MultiFacetedProtDB is freely available as a web server at: https://multifacetedprotdb.biocomp.unibo.it/.
Graphical Abstract
Graphical Abstract.
Introduction
Characterising multifunctional proteins is an expanding research area aiming to elucidate the complexities of biological processes (1–5). Proteins endowed with remarkable versatility play roles in various processes, including signal transduction, metabolism, gene regulation, and cellular communication, and are often involved in disease insurgence and progression. Advances in structural biology techniques facilitate the understanding of multiple molecular mechanisms and recent research identified and characterized new multifunctional proteins, uncovering their roles in disease development and progression, which in turn is the key to understand their potential as therapeutic targets and eventually to delve into the outcomes related to diseases comorbidity (6–12).
Originally lens crystallins were recognised as previously known metabolic enzymes (13,14). The term ‘moonlighting’ was put forward by Constance Jeffery (15) while Joran Piatigorsky proposed ‘gene sharing’ (16). Since then, different types of ‘moonlighting’ have been described (17–20) and functionally/structurally characterized in relation to their multiple functions (21). Multitasking proteins can perform different functions, mostly related to cell type and location, oligomeric state, concentration of cellular ligands, substrates, and cofactors (22,23). In literature (see (7), among others) multifunctional proteins are addressed as pleiotropic, multidomain, promiscuous (in relation to enzymes catalysing multiple substrates) and moonlighting (with two or more molecular functions). For this reason, we introduce here the term ‘multifaceted’, hoping to comprise the different categories, and focus on the human reference proteome, its link to diseases and known drugs, when available.
Databases like UniProt (https://uniprot.org, (24)), NCBI Protein (https://www.ncbi.nlm.nih.gov/protein/), and PDB (https://www.rcsb.org/, (25)) lack explicit labelling of multifunctional proteins. Specific resources are then needed to focus on this important and increasing subset of proteins, whose characterisation will help unravelling the mechanisms of cell complexity. To date, three resources collecting experimental data are available: MoonDB (http://moondb.hb.univ-amu.fr/, (26,27)), MoonProt (http://www.moonlightingproteins.org/, (28)) and MultitaskProtDB II (http://wallace.uab.es/multitaskII/, (29)). When restricted to humans, these databases collect 47, 103 and 185 proteins, respectively. Noticeably, the overlap among the three databases is limited (only 15 human proteins are shared among them) and, when merged, they provide information on 241 human proteins.
We hereby introduce MultifacetedProtDB, an integrated and manually curated database providing a comprehensive collection of multifunctional human proteins. MultifacetedProtDB has been built by: (i) merging the above-mentioned datasets; (ii) searching new multifunctional proteins reported in the recent literature; (iii) collecting enzymes endowed with multiple EC codes, emphasizing the differences at the digit-level codes.
The final dataset contains 1103 multifaceted proteins, of which 812 are enzymes. Therefore, MultifacetedProtDB increases by more than 4 times the number of multifunctional proteins reported in currently available resources.
MultifacetedProtDB links directly to GeneCards (https://www.genecards.org/, (30)) and to UniProt/Swiss-Prot for functional and structural features, providing access to protein structures from PDB, and AlphaFold models (AlphaFold Protein Structure Database, https://alphafold.ebi.ac.uk/, (31)), when available. Furthermore, evidence of protein family classification is provided by InterPro and Pfam (InterPro https://www.ebi.ac.uk/interpro/), (32). Catalytic reactions and EC numbers, when it is the case, GO terms (33), including biological pathways, molecular functions, and cellular components, subcellular location and protein-protein interactions are derived from UniProt. Biological pathways link directly to Reactome (https://reactome.org/, (34)). The database enables easy access to protein variants (UniProt variant viewer, https://www.uniprot.org/help/disease_phenotypes_variants_section). Entries in MultifacetedProtDB are associated to diseases, deriving from UniProt, Humsavar (https://ftp.uniprot.org/pub/databases/uniprot/knowledgebase/complete/docs/humsavar.txt), Monarch (https://monarchinitiative.org/, (35)) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, (36)). Diseases are categorized according to MONDO (https://mondo.monarchinitiative.org/, (37)), ICD10 (https://www.cdc.gov/nchs/icd/icd10.htm), Orphanet (https://www.orpha.net/consor/cgi-bin/index.php) and OMIM (https://www.omim.org/, (38)). Links are provided to HPO phenotypes (https://hpo.jax.org/, (39)) along with valuable insights into tissue and cell type expression patterns, sourced from the Human Protein Atlas (https://www.proteinatlas.org/). We also include information, when available, on specific drugs, by linking to GeneCards. Links to quoted literature is preserved and/or included (publications, https://www.uniprot.org/help/return_fields_databases).
We offer an advanced search option with which the user can browse the dataset including up to four conditions, which can be selected from a drop-down menu, and are connected by logical operators (AND, OR, NOT). The information available in the database offers a comprehensive, richly annotated resource for investigating the role of multifunctional proteins within the complex realm of cellular processes, in particular in relation to the development of diseases.
Web server implementation
The database is stored on a PostgreSQL DBMS (v.13; https://www.postgresql.org). For the backend, we adopted the Django Python application server (v.4.1, https://www.djangoproject.com). The look-and-feel of the website graphical interface has been implemented focusing on simplicity, ease of use and optimization of the interaction with the user. The frontend web pages are designed using the Bootstrap framework (v5.3.0, https://getbootstrap.com). JQuery (v3.7, https://jquery.com) implements interactivity. DataTables (v1.13.4, https://datatables.net) was adopted for visualizing tabular data. To improve web server interoperability and cross-referencing, all the identifiers of external databases were linked to the original resources.
Content of MultifacetedProtDB
The overall content of MultifacetedProtDB is summarized in Table 1. The database contains information on 1103 multifaceted proteins, 812 of which are endowed with at least one EC number. For the remaining 291, no catalytic function has been reported so far. Proteins cluster into 913 groups when their sequences are aligned with MMSeq2 (40) at 50% sequence identity with a sequence coverage higher than 80%.
Table 1.
Statistics of MultifacetedProtDB
| Proteins (#) | PDB (#, # proteins) | Reactome paths (# leaves, # internal nodes*, # proteins)a | Reactome roots (#)a | InterPro (#, # proteins) | Pfam (#, # proteins) | Diseases (# MONDO, # ICD10 categories and chapters, # proteins) | |
|---|---|---|---|---|---|---|---|
| Enzymes | 812 | 10 469 entries | 662 leaf nodes | 27 | 1943 entries | 761 entries | 619 MONDO diseases |
| 520 proteins | 163 internal nodes | 812 proteins | 807 proteins | 172 ICD10 categories | |||
| 698 proteins | 16 ICD10 chapters | ||||||
| 321 proteins | |||||||
| Non-enzymes | 291 | 2840 entries | 465 leaf nodes | 25 | 784 entries | 314 entries | 309 MONDO diseases |
| 223 proteins | 106 internal nodes | 291 proteins | 287 proteins | 117 ICD10 categories | |||
| 252 proteins | 13 ICD10 chapters | ||||||
| 110 proteins | |||||||
| All proteins | 1103 | 13 147 entries | 849 leaf nodes | 27 | 2626 entries | 1045 entries | 895 MONDO diseases |
| 743 proteins | 198 internal nodes | 1103 proteins | 1094 proteins | 213 ICD10 categories | |||
| 950 proteins | 17 ICD10 chapters | ||||||
| 431 proteins |
#: number of
*: “internal nodes” represent the intermediate nodes within the Reactome hierarchy.
a: Reactome entries related to “Disease” and “Drug ADME” are not considered when counting the number of Reactome pathways and roots.
The three-dimensional structures of 743 proteins have been resolved, at least partially, and are reported in 13 147 PDB entries. 630 proteins are endowed with more than one resolved structure; among them, carbonic anhydrase 2 (UniProt: P00918) is the most extreme case, being associated with 1031 different PDB entries.
Some 86% of all proteins (698 enzymes and 252 non enzymes) participate in Reactome pathways that are part of all the 27 main Reactome roots (excluding ‘Disease’ and ‘Drug ADME’). All proteins are endowed with InterPro annotations and only 9 (of which, 5 enzymes) lack annotations in Pfam. Collectively, proteins in MultifacetedProtDB are covered by 2626 and 1045 InterPro and Pfam domains, respectively.
The most represented Pfam domains are: PF00782 (Dual specificity phosphatase, catalytic domain, 33 proteins), PF00069 (Protein kinase domain, 32 proteins), PF00067 (Cytochrome P450, 21 proteins), PF00029 (Connexin, 21 proteins), PF00501 (AMP-binding enzyme, 19 proteins), PF00106 (short chain dehydrogenase, 17 proteins), PF13193 (AMP-binding enzyme C-terminal domain, 15 proteins), PF00856 (SET domain, 14 proteins), PF00005 (ABC transporter, 14 proteins), PF02798 (Glutathione S-transferase, N-terminal domain, 13 proteins) and PF00030 (Beta/Gamma crystallin, 13 proteins).
We collected associations among proteins and diseases merging the information reported in UniProt, Humsavar, Monarch initiative and ClinVar and reported the disease nomenclatures provided by the MONDO ontology, the OMIM and Orphanet catalogs, and the ICD10 classification scheme.
Some 30% of proteins in our database (321 enzymes and 110 non enzymes) are associated with 895 MONDO diseases classified into 213 ICD10 categories and into 17 (out of the 19) ICD10 main chapters, after excluding chapters not describing diseases with a genetic component (namely, XX: ‘External causes of morbidity and mortality’, XXI: ‘Factors influencing health status and contact with health services’, and XXII: ‘Codes for special purposes’). The most represented chapter is ‘XVII: Congenital malformations, deformations and chromosomal abnormalities’ accounting for 226 diseases associated with 135 multifaceted proteins.
Of the 895 diseases, 323 are included in the Orphanet catalog of rare diseases. Overall, when considering the phenotypic characterization of diseases, 430 proteins are linked to terms out of the Human Phenotype Ontology (HPO).
The proteomic distribution of multifaceted proteins in our database is characterized in terms of subcellular location, tissues and tissue cell types. The subcellular location has been extracted from the Swiss-Prot curated annotation: 1005 proteins are distributed among 168 locations, out of the 561 defined in the UniProt controlled vocabulary (https://www.uniprot.org/locations). Of the 1005 proteins, 411 are annotated in only one location, while 594 are endowed with multiple locations. The most multilocalised protein is Annexin A1 (P04083), endowed with 16 different locations.
As to the distribution among tissues and tissues cell types, data have been downloaded from the Human Protein Atlas (HPA): 804 proteins are distributed among 58 tissues and 127 cell types, out of the 64 tissues and 145 cell types defined in HPA. Only 27 proteins are specific for a single tissue. The remaining proteins are expressed in at least two tissues and, in most cases, they have a very low specificity. 549 proteins are found in at least 30 different tissues; the extreme case is represented by 120 proteins detected in 49 tissues.
Enzymes in MultifacetedProtDB
Over 73% of our database includes enzymes. Enzymes can be multifunctional when their catalytic activity is performed independently of other activities in the cell, or when, depending on the environment, they change their catalytic activity at different extents (22). For enzymes, the Enzyme Commission (EC) number is a four-level traditional code, referring to the catalysed biochemical reaction and following the recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB) (https://iubmb.qmul.ac.uk/enzyme/). EC numbers describe the catalysed biochemical reaction (1st digit), and the relationship among protein activity (2nd digit), substrates and products (3rd and 4th digits). Presently EC codes include seven major classes: (i) oxidoreductases, (ii) transferases, (iii) hydrolases, (iv) lyases, (v) isomerases, (vi) ligases, (vii) translocases (28).
For sake of clarity, Table 2 groups multifunctional enzymes into three sets. The first set includes enzymes that are annotated only with one EC number in UniProt, and can perform other functions (Table 2, second row); the second set includes enzymes that are characterized by EC numbers belonging to different classes as indicated by the change of the first digit (Table 2, third row). The third group includes all the enzymes in which the first digit is conserved while the remaining three may change at the second, third and/or fourth level (Table 2, fourth row). In each group, enzymes can be associated to diseases (Table 2, rightmost column). It is evident that 39.5% of the multifunctional enzymes are disease associated. A recent estimate reports that some 1494 enzymes in humans are involved in diseases (41,42). Therefore, it turns out that presently about 21% of enzymes involved in diseases are multifunctional.
Table 2.
Statistics of enzymes in MultifacetedProtDB
| Enzymes (#) | PDB (#, # enzymes) | Reactome paths (# leaves, # internal nodes*, # enzymes)a | Reactome roots (#)a | InterPro (#, # enzymes) | Pfam (#, # enzymes) | Diseases (# MONDO, # ICD10 categories and chapters, # enzymes) | |
|---|---|---|---|---|---|---|---|
| Enzymes | 812 | 10 469 entries | 662 leaf nodes | 27 | 1943 entries | 761 entries | 619 MONDO diseases |
| 520 enzymes | 163 internal nodes | 812 enzymes | 807 enzymes | 172 ICD10 categories | |||
| 698 enzymes | 16 ICD10 chapters | ||||||
| 321 enzymes | |||||||
| Enzymes with only 1 EC | 144 | 3315 entries | 287 leaf nodes | 23 | 623 entries | 213 entries | 173 MONDO diseases |
| 120 enzymes | 82 int. nodes | 144 enzymes | 142 enzymes | 89 ICD10 categories | |||
| 135 enzymes | 13 ICD10 chapters | ||||||
| 82 enzymes | |||||||
| Enzymes with Multiple ECs with different 1st levels | 136 | 1472 entries | 149 leaf nodes | 18 | 527 entries | 200 entries | 123 MONDO diseases |
| 88 proteins | 27 int. nodes | 136 enzymes | 136 enzymes | 74 ICD10 categories | |||
| 112 enzymes | 12 ICD10 chapters | ||||||
| 56 enzymes | |||||||
| Enzymes with Multiple ECs with different figures other than the 1st | 532 | 5750 entries | 488 leaf nodes | 27 | 1110 entries | 440 entries | 349 MONDO diseases |
| 312 enzymes | 107 int. nodes | 532 enzymes | 529 enzymes | 161 ICD10 categories | |||
| 451 enzymes | 15 ICD10 chapters | ||||||
| 183 enzymes |
#: number of
*: ‘internal nodes’ indicates the intermediate nodes within the Reactome hierarchy.
a: Reactome entries related to ‘Disease’ and ‘Drug ADME’ are not considered when counting the number of Reactome pathways and roots.
One example for each group in Table 2 is described in the following.
An example of a multifaceted protein enzyme associated with a single EC number (row ‘Enzymes with only 1 EC’ in Table 2) is Hexokinase-1 (UniProt: P19367, gene: HK1). Its catalytic activity is described by EC: 2.7.1.1 (Hexokinase, Transferase class); as a second function it recognises bacterial peptidoglycans, playing a role in innate immunity and inflammation. It is a multilocalized protein and it is associated with the mitochondrial outer membrane when acting as an enzyme, while it dissociates to the cytoplasm when interacting with N-acetyl-D-glucosamine, a component of peptidoglycans (3). It is a housekeeping gene, showing a low specific expression pattern, being detected in 90 tissue cell types out of 46 different tissues. Variants of HK1 have been related to 5 different diseases classified in ICD10 chapters III (Diseases of the blood and blood-forming organs and certain disorders involving the immune mechanism), VI (Diseases of the nervous system) and VII (Diseases of the eye and adnexa).
As a second example we select one multiclass enzyme (row ‘Enzymes with Multiple ECs with different 1st levels’ in Table 2). The cytoplasmic C-1-tetrahydrofolate synthase (UniProt: P11586, gene MTHFD1) is a trifunctional enzyme that catalyzes the interconversion of three forms of one-carbon-substituted tetrahydrofolate. The three catalytic activities are described by the following codes: EC: 1.5.1.5 (Methylenetetrahydrofolate dehydrogenase (NADP+), Oxidoreductase class); EC: 3.5.4.9 (Methenyltetrahydrofolate cyclohydrolase, Hydrolase class); and EC: 6.3.4.3 (formate-tetrahydrofolate ligase, Ligase class). The enzyme is monolocalised (cytoplasm) and involved in a single Reactome pathway (Metabolism of folate and pterines, R-HSA-196757). Three-dimensional structures are available for the N-terminal 306-residue long domain, performing both the oxidoreductase and hydrolase activities, while the ligase activity is predicted to be performed in a separate domain, identified by the Pfam domain PF01268 (formate-tetrahydrofolate ligase, residues 319–695). It shows a low specific expression pattern, being detected in 80 tissue cell types out of 49 different tissues (HPA). Variants of MTHFD1 are associated with three diseases: one disease is unclassified by ICD10 and the remaining two belong to ICD10 chapters II (neoplasm) and XVII (congenital malformations, deformations and chromosomal abnormalities) (43).
An example of third group in Table 2 (Enzymes with Multiple ECs with different digits other than the 1st) is the mitochondrial α-aminoadipic semialdehyde synthase (UniProt: Q9UDR5, Gene: AAS), a bifunctional enzyme catalysing the first two steps in lysine degradation, with EC: 1.5.1.8 (saccharopine dehydrogenase (NADP+, l-lysine-forming), oxidoreductase class) and EC: 1.5.1.9 (saccharopine dehydrogenase (NADP+, l-glutamate-forming), oxidoreductase class). It is monolocalised (mitochondrion) and involved in a single Reactome pathway (lysine catabolism, R-HSA-71064). Similarly to the cytoplasmic C-1-tetrahydrofolate synthase, it shows a low specific expression pattern, being detected in 61 tissue cell types out of 46 different tissues. Variants of AAS are associated with three diseases classified in ICD10 chapters IV (Endocrine, nutritional and metabolic diseases) and VI (diseases of the nervous system) (44).
Retrieving from MultifacetedProtein DB
The database can be searched for any of the information fields it includes. ‘ADVANCED SEARCH’ interface can be used to combine different searches, connected with the ‘AND’, ‘OR’, ‘NOT’ boolean operators. The clauses ‘With’ and ‘Multiple’ in the search fields allow to restrict the search to entries endowed with at least one or multiple annotations for the field, respectively.
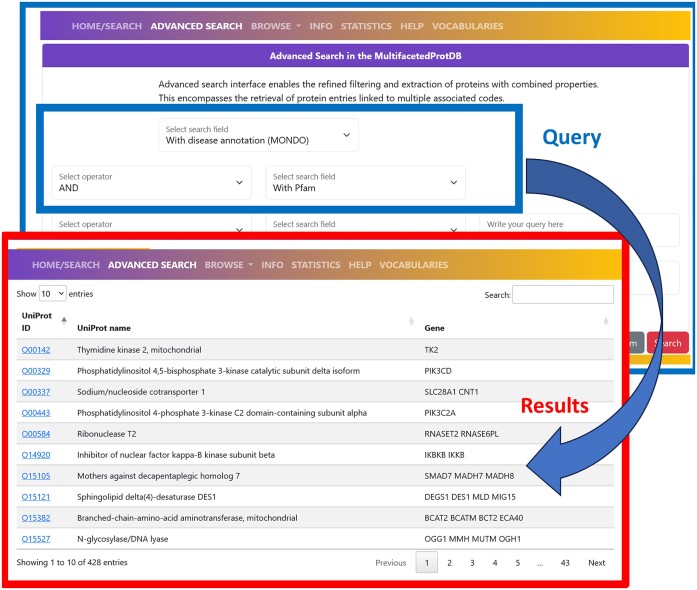
For example, is it possible to search for multifunctional proteins associated to MONDO diseases and endowed with Pfam annotations, obtaining as a result a list of 428 entries that can be explored in detail (Figure 1). Searching for specific Pfam domains highlights those mostly associated with diseases. These are listed in Table 3. Specifically, the Protein kinase domain (PF00069), the cytochrome P450 domain (PF00067), the Connexin domain (PF00029) and the β/γ crystallin domain (PF00030) are most frequently associated with diseases in multifunctional proteins. By inspecting the entries obtained with the advanced search, it is possible to retrieve and integrate information on the diseases, their classification and the structural and functional features of the highlighted proteins including their participation to Reactome pathways.
Figure 1.
Example of advanced search in MultifacetedProtDB. Complex search can be performed by combining different searches with Boolean connectors AND, OR, NOT. In the case illustrated in the figure, proteins endowed with a Pfam annotation and associated with MONDO diseases are retrieved. The search shows a list of 428 entries that can be inspected for obtaining detailed information.
Table 3.
Protein domains mostly related to disease
| Pfam domain | Proteins in DB (#) | Proteins related to diseases (#) | Diseases (#) | ICD10 chapters | Reactome pathways (#)a | Reactome rootsa |
|---|---|---|---|---|---|---|
| PF00069 | 32 | 13 | 24 | II: Neoplasm | 104 | Cell cycle |
| Protein kinase domain | III: Diseases of the blood and blood-forming organs and certain disorders involving the immune | Cellular responses to stimuli | ||||
| mechanism | Circadian clock | |||||
| IV: Endocrine, nutritional and metabolic diseases | Developmental biology | |||||
| V: Mental and behavioural disorders | Gene expression | |||||
| VI: Diseases of the nervous system | Hemostasis | |||||
| IX: Diseases of the circulatory system | Immune system | |||||
| XIII: Diseases of the musculoskeletal system and connective tissue | Metabolism | |||||
| XVII: Congenital malformations, deformations and chromosomal abnormalities | Metabolism of RNA | |||||
| Metabolism of proteins | ||||||
| Neuronal system | ||||||
| Organelle biogenesis and maintenance | ||||||
| Programmed cell death | ||||||
| Signal transduction | ||||||
| PF00067 | 21 | 11 | 19 | IV: Endocrine, nutritional and metabolic diseases | 25 | Metabolism |
| Cytochrome P450 | VI: Diseases of the nervous system | Sensory perception | ||||
| VII: Diseases of the eye and adnexa | ||||||
| IX: Diseases of the circulatory system | ||||||
| XI: Diseases of the digestive system | ||||||
| XVII: Congenital malformations, deformations and chromosomal abnormalities | ||||||
| PF00029 | 21 | 10 | 49 | IV: Endocrine, nutritional and metabolic diseases | 10 | Vesicle-mediated transport |
| Connexin | VI: Diseases of the nervous system | Neural system | ||||
| VIII: Diseases of the ear and mastoid process | Signal transduction | |||||
| IX: Diseases of the circulatory system | ||||||
| XVII: Congenital malformations, deformations and chromosomal abnormalities | ||||||
| PF00030 | 13 | 10 | 21 | VII: Diseases of the eye and adnexa | 0 | - |
| β/γ crystallin | XVII: Congenital malformations, deformations and chromosomal abnormalities |
#: number of
a: Reactome pathways associated to disease related proteins, excluding pathways collected under the roots ‘Disease’ and ‘Drug ADME’.
Conclusions and perspectives
The current version of MultifacetedProtDB increases four folds the number of multifunctional proteins with respect to others specialized databases available for the same subject. The database is useful when searching for a multitasking protein in relation to its involvement in cell molecular complexity and possibly in diseases. A list of drugs is provided, when available. The actual content of 1103 proteins, inclusive of 812 enzymes, is a possible underestimation of the total number of multitasking proteins that characterizes the human reference proteome, most likely due to lack of knowledge that future researches will fill, particularly in relation to different isoforms and their involvement in disease (45). Presently, data in our DB indicate that multifunctionality is not exclusively related to multiple subcellular locations, and/or association to diseases. Proteins with specific domains (Figure 1 and Table 3) seems more involved than others in diseases. However, data may change as more experimental knowledge will be acquired. For this reason, we will update the database every year, integrating literature and new UniProt and other main database releases.
Contributor Information
Elisa Bertolini, Biocomputing Group, Dept. of Pharmacy and Biotechnology, University of Bologna, Italy.
Giulia Babbi, Biocomputing Group, Dept. of Pharmacy and Biotechnology, University of Bologna, Italy.
Castrense Savojardo, Biocomputing Group, Dept. of Pharmacy and Biotechnology, University of Bologna, Italy.
Pier Luigi Martelli, Biocomputing Group, Dept. of Pharmacy and Biotechnology, University of Bologna, Italy.
Rita Casadio, Biocomputing Group, Dept. of Pharmacy and Biotechnology, University of Bologna, Italy.
Data availability
The data discussed in this paper are publicly available at https://multifacetedprotdb.biocomp.unibo.it/.
Funding
PRIN2017 [2017483NH8_002 to C.S.] from the Italian Ministry of University and Research; ‘Consolidation of the Italian Infrastructure for Omics Data and Bioinformatics (ElixirxNextGenerationIT)’ [IR0000010, Italian Ministry of University and Research, EU funding within the NextGeneration EU]; ‘National Centre for HPC, Big Data and Quantum Computing’ [CN00000013, Italian Ministry of University and Research, EU funding within the NextGeneration EU]. Funding for open access charge: PRIN2017 [2017483NH8_002 to C.S.] from the Italian Ministry of University and Research; ‘Consolidation of the Italian Infrastructure for Omics Data and Bioinformatics (ElixirxNextGenerationIT)’ [IR0000010, Italian Ministry of University and Research, EU funding within the NextGeneration EU]; ‘National Centre for HPC, Big Data and Quantum Computing’ [CN00000013, Italian Ministry of University and Research, EU funding within the NextGeneration EU].
Conflict of interest statement. None declared.
References
- 1. Gupta M.N., Uversky V.N.. Moonlighting enzymes: when cellular context defines specificity. Cell. Mol. Life Sci. 2023; 24:130–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haage A., Dhasarathy A. Working a second job: cell adhesion proteins that moonlight in the nucleus. Front. Cell Dev. Biol. 2023; 11:1163553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodríguez-Saavedra C., Morgado-Martínez L.E., Burgos-Palacios A., King-Díaz B., López-Coria M., Sánchez-Nieto S.. Moonlighting proteins: the case of the hexokinases. Front. Mol. Biosci. 2021; 8:701975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeffery C.J. Enzymes, pseudoenzymes, and moonlighting proteins: diversity of function in protein superfamilies. FEBS J. 2020; 287:4141–4149. [DOI] [PubMed] [Google Scholar]
- 5. Gurevich V.V. Protein multi-functionality: introduction. Cell. Mol. Life Sci. 2019; 76:4405–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Espinosa-Cantú A., Cruz-Bonilla E., Noda-Garcia L., DeLuna A.. Multiple forms of multifunctional proteins in health and disease. Front. Cell Dev. Biol. 2020; 8:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huerta M., Franco-Serrano L., Amela I., Perez-Pons J.A., Piñol J., Mozo-Villarías A., Querol E., Cedano J.. Role of moonlighting proteins in disease: analyzing the contribution of canonical and moonlighting functions in disease progression. Cells. 2023; 12:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kang J., Brajanovski N., Chan K.T., Xuan J., Pearson R.B., Sanij E.. Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy. Signal Transduct. Target Ther. 2021; 6:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wygrecka M., Kosanovic D., Kwapiszewska G., Preissner K.T.. Editorial: multitasking biomolecules in human pathologies: known players on their unexpected journeys. Front. Med. (Lausanne). 2020; 7:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Molavi G., Samadi N., Hosseingholi E.Z.. The roles of moonlight ribosomal proteins in the development of human cancers. J. Cell. Physiol. 2019; 234:8327–8341. [DOI] [PubMed] [Google Scholar]
- 11. Jühlen R., Fahrenkrog B.. Moonlighting nuclear pore proteins: tissue-specific nucleoporin function in health and disease. Histochem. Cell Biol. 2018; 150:593–605. [DOI] [PubMed] [Google Scholar]
- 12. Sissler M., González-Serrano L.E., Westhof E.. Recent advances in mitochondrial aminoacyl-tRNA synthetases and disease. Trends Mol. Med. 2017; 23:693–708. [DOI] [PubMed] [Google Scholar]
- 13. Piatigorsky J., Wistow G.J.. Enzyme/crystallins: gene sharing as an evolutionary strategy. Cell. 1989; 57:197–199. [DOI] [PubMed] [Google Scholar]
- 14. Wistow G., Piatigorsky J.. Recruitment of enzymes as lens structural proteins. Science. 1987; 236:1554–1556. [DOI] [PubMed] [Google Scholar]
- 15. Jeffery C.J. Moonlighting proteins. Trends Biochem. Sci. 1999; 24:8–11. [DOI] [PubMed] [Google Scholar]
- 16. Piatigorsky J. Gene Sharing and Evolution. 2007; Cambridge, MA: Harvard University Press. [Google Scholar]
- 17. Jeffery C.J. Multifunctional proteins: examples of gene sharing. Ann. Med. 2003; 35:28–35. [DOI] [PubMed] [Google Scholar]
- 18. Tawfik D.S. Accuracy-rate tradeoffs: how do enzymes meet demands of selectivity and catalytic efficiency?. Curr. Opin. Chem. Biol. 2014; 21:73–80. [DOI] [PubMed] [Google Scholar]
- 19. Copley S.D. An evolutionary biochemist's perspective on promiscuity. Trends Biochem. Sci. 2015; 40:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeffery C.J. Protein moonlighting: what is it, and why is it important?. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018; 373:20160523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Das S., Khan I., Kihara D., Orengo C.. Henderson B. Exploring structure–function relationships in moonlighting proteins. Moonlighting Proteins: Novel Virulence Factors in Bacterial Infections. 2017; Wiley; 21–43. [Google Scholar]
- 22. Peracchi A. The limits of enzyme specificity and the evolution of metabolism. Trends Biochem. Sci. 2018; 43:984–996. [DOI] [PubMed] [Google Scholar]
- 23. Uzdensky A.B. Multifunctional proteins. Biophysics. 2020; 65:390–403. [Google Scholar]
- 24. The UniProt Consortium UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023; 51:D523–D531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chao H., Chen L., Craig P.A., Crichlow G.V., Dalenberg K., Duarte J.M.et al.. RCSB Protein Data Bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023; 51:D488–D508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chapple C.E., Robisson B., Spinelli L., Guien C., Becker E., Brun C.. Extreme multifunctional proteins identified from a human protein interaction network. Nat. Commun. 2015; 6:7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ribeiro D.M., Briere G., Bely B., Spinelli L., Brun C.. MoonDB 2.0: an updated database of extreme multifunctional and moonlighting proteins. Nucleic Acids Res. 2019; 47:D398–D402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen C., Liu H., Zabad S., Rivera N., Rowin E., Hassan M., Gomez De Jesus S.M., Llinás Santos P.S., Kravchenko K., Mikhova M.et al.. MoonProt 3.0: an update of the moonlighting proteins database. Nucleic Acids Res. 2021; 49:D368–D372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Franco-Serrano L., Hernández S., Calvo A., Severi M.A., Ferragut G., Pérez-Pons J., Piñol J., Pich Ò., Mozo-Villarias Á., Amela I.et al.. MultitaskProtDB-II: an update of a database of multitasking/moonlighting proteins. Nucleic Acids Res. 2018; 46:D645–D648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y.et al.. The GeneCards Suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics. 2016; 54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 31. Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A.et al.. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022; 50:D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paysan-Lafosse T., Blum M., Chuguransky S., Grego T., Pinto B.L., Salazar G.A., Bileschi M.L., Bork P., Bridge A., Colwell L.et al.. InterPro in 2022. Nucleic Acids Res. 2023; 51:D418–D427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ontology Consortium G., Aleksander S.A., Balhoff J., Carbon S., Cherry J.M., Drabkin H.J., Ebert D., Feuermann M., Gaudet P., Harris N.L.et al.. The Gene Ontology knowledgebase in 2023. Genetics. 2023; 224:iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gillespie M., Jassal B., Stephan R., Milacic M., Rothfels K., Senff-Ribeiro A., Griss J., Sevilla C., Matthews L., Gong C.et al.. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022; 50:D687–D692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shefchek K.A., Harris N.L., Gargano M., Matentzoglu N., Unni D., Brush M., Keith D., Conlin T., Vasilevsky N., Zhang X.A.et al.. The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 2020; 48:D704–D715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W.et al.. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018; 46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vasilevsky N.A., Matentzoglu N.A., Toro S., Flack IV J.E., Hegde H., Unni D.R., Alyea G.F., Amberger J.S., Babb L., Balhoff J.P.et al.. Mondo: unifying diseases for the world, by the world. 2022; medRxiv doi:03 May 2022, preprint: not peer reviewed 10.1101/2022.04.13.22273750. [DOI] [Google Scholar]
- 38. Amberger. J.S., Bocchini C.A., Scott A.F., Hamosh A. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019; 47:D1038–D1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Köhler S., Gargano M., Matentzoglu N., Carmody L.C., Lewis-Smith D., Vasilevsky N.A., Danis D., Balagura G., Baynam G., Brower A.M.et al.. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2021; 49:D1207–D1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Steinegger M., Söding J.. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017; 35:1026–1028. [DOI] [PubMed] [Google Scholar]
- 41. Baldazzi D., Savojardo C., Martelli P.L., Casadio R.. BENZ WS: the Bologna ENZyme Web Server for four-level EC number annotation. Nucleic Acids Res. 2021; 49:W60–W66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Savojardo C., Baldazzi D., Babbi G., Martelli P.L., Casadio R.. Mapping human disease-associated enzymes into Reactome allows characterization of disease groups and their interactions. Sci. Rep. 2022; 12:17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao L.N., Kaldis P.. Pairing structural reconstruction with catalytic competence to evaluate the mechanisms of key enzymes in the folate-mediated one-carbon pathway. FEBS J. 2023; 290:2279–2291. [DOI] [PubMed] [Google Scholar]
- 44. Leandro J., Khamrui S., Suebsuwong C., Chen P.J., Secor C., Dodatko T., Yu C., Sanchez R., DeVita R.J., Houten S.M.et al.. Characterization and structure of the human lysine-2-oxoglutarate reductase domain, a novel therapeutic target for treatment of glutaric aciduria type 1. Open Biol. 2022; 12:220179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Babbi G., Savojardo C., Baldazzi D., Martelli P.L., Casadio R.. Pathogenic variation types in human genes relate to diseases through Pfam and InterPro mapping. Front. Mol. Biosci. 2022; 9:966927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data discussed in this paper are publicly available at https://multifacetedprotdb.biocomp.unibo.it/.