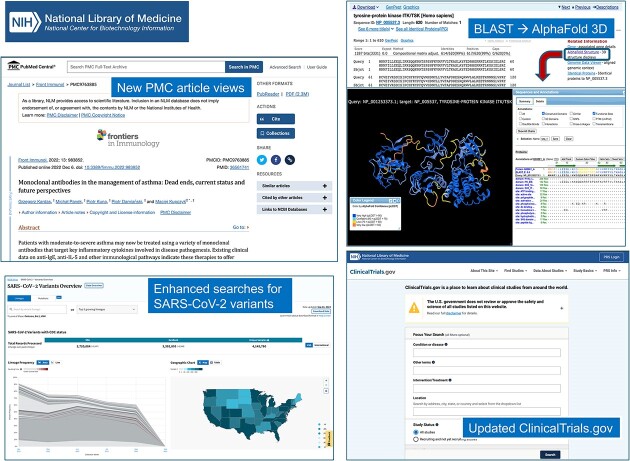
Abstract
The National Center for Biotechnology Information (NCBI) provides online information resources for biology, including the GenBank® nucleic acid sequence database and the PubMed® database of citations and abstracts published in life science journals. NCBI provides search and retrieval operations for most of these data from 35 distinct databases. The E-utilities serve as the programming interface for most of these databases. Resources receiving significant updates in the past year include PubMed, PMC, Bookshelf, SciENcv, the NIH Comparative Genomics Resource (CGR), NCBI Virus, SRA, RefSeq, foreign contamination screening tools, Taxonomy, iCn3D, ClinVar, GTR, MedGen, dbSNP, ALFA, ClinicalTrials.gov, Pathogen Detection, antimicrobial resistance resources, and PubChem. These resources can be accessed through the NCBI home page at https://www.ncbi.nlm.nih.gov.
Graphical Abstract
Graphical Abstract.
Introduction
NCBI overview
The National Center for Biotechnology Information (NCBI), a center within the National Library of Medicine (NLM) at the National Institutes of Health (NIH), was created in 1988 to develop information systems for molecular biology (1). In this article we provide a brief overview of the NCBI collection of databases, followed by a summary of resources that we significantly updated in the past year. We provide more complete discussions of NCBI resources on the home pages of individual databases and in the NCBI Handbook (https://www.ncbi.nlm.nih.gov/books/NBK143764/).
NCBI databases
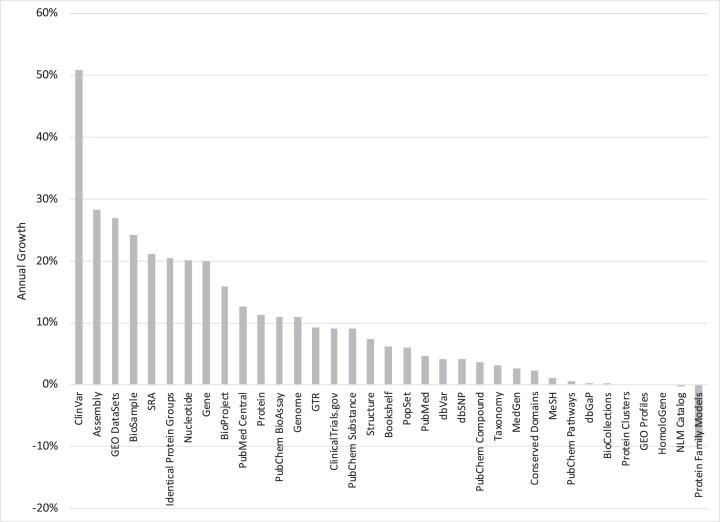
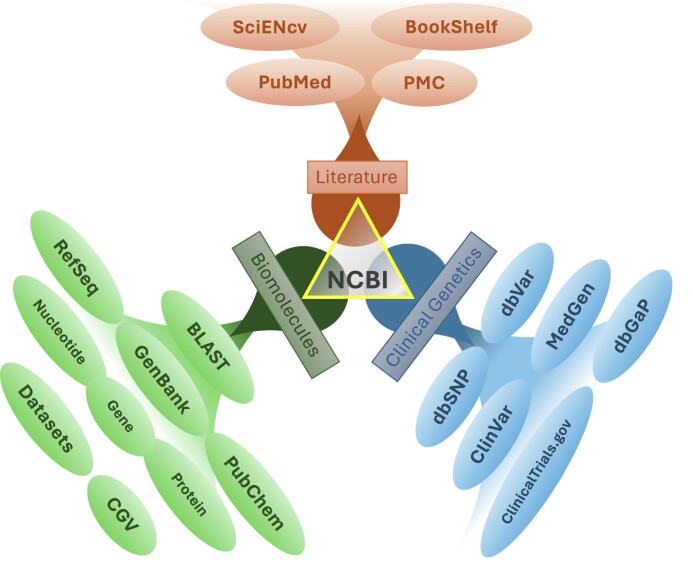
NCBI maintains a diverse set of 35 databases that together contain 4.3 billion records (Table 1 and Figure 1), most of which are available through the Entrez retrieval system (2) at https://www.ncbi.nlm.nih.gov/search/. Figure 2 represents several of these databases graphically, clustering them in three groups: literature, biomolecules, and clinical genetics. These databases will be discussed in that order below. Each database supports text searching using simple Boolean queries, downloading of data in various formats, and linking records between databases based on asserted relationships. Records retrieved in Entrez can be displayed in many formats and downloaded singly or in batches. An Application Programming Interface for Entrez functions (the E-utilities) is available, and detailed documentation is provided at https://eutils.ncbi.nlm.nih.gov/.
Table 1.
NCBI databases (as of 21 August 2023)
| Database | Records | Description |
|---|---|---|
| Literature | ||
| PubMed | 36 100 644 | scientific and medical abstracts/citations |
| PubMed Central | 9 268 952 | full-text journal articles |
| NLM Catalog | 1 634 653 | index of NLM collections |
| Bookshelf | 983 634 | books and reports |
| MeSH | 353 699 | ontology used for PubMed indexing |
| DNA/RNA | ||
| Nucleotide | 605 293 217 | DNA and RNA sequences from GenBank and RefSeq |
| BioSample | 34 796 756 | descriptions of biological source materials |
| SRA | 28 858 671 | high-throughput DNA/RNA sequence read archive |
| Taxonomy | 2 653 432 | taxonomic classification and nomenclature catalog |
| Assembly | 1 782 091 | genome assembly information |
| BioProject | 712 423 | biological projects providing data to NCBI |
| Genome | 79 671 | genome sequencing projects by organism |
| BioCollections | 8497 | museum, herbaria, and biorepository collections |
| Genes | ||
| GEO Profiles | 128 414 055 | gene expression and molecular abundance profiles |
| Gene | 47 059 151 | collected information about gene loci |
| GEO DataSets | 6 874 686 | functional genomics studies |
| PopSet | 404 340 | sequence sets from phylogenetic/population studies |
| HomoloGene | 141 268 | homologous gene sets for selected organisms |
| Proteins | ||
| Protein | 1 194 803 871 | protein sequences from GenBank and RefSeq |
| Identical Protein Groups | 629 076 260 | protein sequences grouped by identity |
| Protein Clusters | 1 137 329 | sequence similarity-based protein clusters |
| Structure | 208 741 | experimentally-determined biomolecular structures |
| Protein Family Models | 166 131 | conserved domain architectures, HMMs, and BlastRules |
| Conserved Domains | 64 234 | conserved protein domains |
| Chemicals | ||
| PubChem Substance | 307 634 967 | deposited substance and chemical information |
| PubChem Compound | 115 669 131 | chemical information with structures, information, and links |
| PubChem BioAssay | 1 626 630 | bioactivity screening studies |
| PubChem Pathways | 240 671 | molecular pathways with links to genes, proteins and chemicals |
| Clinical Genetics | ||
| dbSNP | 1 121 739 543 | short genetic variations |
| dbVar | 7 749 330 | genome structural variation studies |
| ClinVar | 2 339 222 | human variations of clinical significance |
| ClinicalTrials.gov | 463 200 | registry of clinical studies |
| MedGen | 216 373 | medical genetics literature and links |
| GTR | 81 209 | genetic testing registry |
| dbGaP | 1406 | genotype/phenotype interaction studies |
Figure 1.
Annual growth rates of the number of records in each NCBI database as of 21 August 2023.
Figure 2.
Selected NCBI databases and tools clustered into three broad categories as discussed in the text.
Data sources and collaborations
NCBI receives data from three sources: direct submissions from researchers, national and international collaborations or agreements with data providers and research consortia, and internal curation efforts. For example, NCBI manages the GenBank database (3) and participates with the EMBL-EBI European Nucleotide Archive (ENA) (4) and the DNA Data Bank of Japan (DDBJ) (5) as a partner in the International Nucleotide Sequence Database Collaboration (INSDC) (6). Details about direct submission processes are available from the NCBI Submit page (https://www.ncbi.nlm.nih.gov/home/submit.shtml) and from the resource home pages (e.g. the GenBank page, https://www.ncbi.nlm.nih.gov/genbank/). More information about the various collaborations, agreements, and curation efforts are also available through the home pages of the individual resources.
Literature updates
PubMed
PubMed provides free online access to citations and abstracts for biomedical literature and facilitates searching across the MEDLINE, PubMed Central, and Bookshelf literature resources. In the past year, PubMed added over 1.4 million citations, growing the database to >36 million total citations in 2023. PubMed now offers proximity searching in selected search fields (https://www.nlm.nih.gov/pubs/techbull/nd22/nd22_pubmed_proximity_search_available.html). This highly requested feature supports searching for terms appearing within a specified distance of each other, providing a powerful new way to search PubMed for concepts that may be represented in multiple ways, or to capture variations of a phrase (https://pubmed.ncbi.nlm.nih.gov/help/#proximity-searching). For example, a proximity search for ‘rationing healthcare’ can also capture variations such as healthcare rationing, rationing of healthcare, rationing in healthcare, rationing universal healthcare, rationing strategies in healthcare, rationing limited healthcare, and more without needing to search for each of these phrases individually. Additionally, the PubMed E-utilities API was updated to use the same technology stack that supports the PubMed web interface (https://ncbiinsights.ncbi.nlm.nih.gov/2022/11/22/updated-pubmed-eutilities-live/). This update aligns the functions of the PubMed E-utilities API with those of the website to provide consistent behavior and search results.
Our Best Match algorithm (7) reflects both the relevance of the article to the query and user preferences for types of articles. As user preferences change, the model is updated to adapt. In the past these updates required manual review and so were infrequent. Now a robust, automated system can determine whether a new model is reliable, so the updates are more frequent. We have also streamlined the author name disambiguation process (8) such that computed authors in PubMed are continually updated on a weekly basis.
PubMed Central (PMC)
PMC is NLM’s free full-text archive of biomedical and life sciences journal literature. In 2023, the PMC archive surpassed 9 million publicly available full-text journal articles, author manuscripts, and preprints. In 2023, PMC launched the second phase of the NIH Preprint Pilot (https://www.ncbi.nlm.nih.gov/pmc/about/nihpreprints/) expanding the scope of the Pilot to include preprints resulting from NIH-funded research from eligible preprint servers (https://ncbiinsights.ncbi.nlm.nih.gov/2023/01/09/next-phase-preprint-pilot/). A project of NLM, the NIH Preprint Pilot was launched in 2020 to explore new approaches to increase the discoverability of NIH-supported research results, with the first phase focusing on NIH-supported research on COVID-19 and the SARS-CoV-2 virus. As of August 2023, nearly 13 000 preprint records had been added to PMC under the Pilot, accelerating and expanding discovery of NIH research.
Building on a successful March 2022 launch of a modernized PMC website, PMC released several improvements to the article display to support users’ most needed activities, informed by direct feedback and user research from users and stakeholders. These updates include improvements to how citation information is presented on desktop and mobile devices; an easy way to directly add an article to specific My NCBI collections; and an enhanced ‘Resources’ section providing easy access to similar articles, ‘cited by’ articles, and related data records in other NCBI databases (https://ncbiinsights.ncbi.nlm.nih.gov/2023/02/27/enhancements-pmc-website/).
Additionally, PMC made several updates to clarify its role as a digital archive and improve how PMC content is described, displayed, and shared by a large and diverse user base with varying levels of knowledge about NLM, NIH, and the scholarly publishing process (https://ncbiinsights.ncbi.nlm.nih.gov/2023/04/18/pmc-as-an-archive/). These contextual updates include a prominent note on all PMC article pages to clarify the relationship of NLM to the articles it archives in PMC; an update to the default social media display when articles from PMC are shared; and a new infographic on documentation pages showing the different types of content in PMC and how each fit into the scholarly publishing process. With COVID-19 public health emergency declarations expiring in the United States and worldwide, PMC transitioned its COVID-19 Public Health Emergency Initiative to the PMC COVID-19 Collection (https://www.ncbi.nlm.nih.gov/pmc/about/covid-19/), remaining committed to ensuring perpetual access to >350 000 articles deposited by >50 publishers (https://www.nlm.nih.gov/pubs/techbull/ja23/ja23_pmc_covid_collection.html). Finally, in February NLM expanded the eligibility requirements for PMC to consider applications from non-MEDLINE journals published primarily in Spanish, a first step towards aligning the scope of PMC more closely with that of MEDLINE (https://www.nlm.nih.gov/medline/medline_overview.html) and the broader NLM Collection (https://www.ncbi.nlm.nih.gov/books/NBK518693/, https://www.nlm.nih.gov/pubs/techbull/jf23/jf23_pmc_spanish_language_journals.html).
Bookshelf
The NCBI Bookshelf provides free online access to full-text books and documents in the life sciences, healthcare, and medicine. In the past year, Bookshelf added over 1000 books, growing the repository to over 11 600 total books from over 150 content providers. Significant new peer-reviewed collections added in 2023 were in the subjects of toxicology, health disparities, nursing, and public health. Bookshelf also initiated a pilot to collect Open Educational Resources (OER). The first set of open textbooks added as part of this pilot were those created and updated as part of the Open RN project (https://www.ncbi.nlm.nih.gov/books/NBK590025/) led by Chippewa Valley Technical College, which is funded in part by the Department of Education. To aid users to more easily find all the textbooks and OER resources available in Bookshelf, currently just under 150 books, Bookshelf added a search filter for this collection and added a link to its list of results on all pages of textbooks accessed on the site.
SciENcv
SciENcv (Science Experts Network Curriculum Vitae, https://www.ncbi.nlm.nih.gov/sciencv), is a helpful resource for those seeking federal research grants from federal organizations such as NIH, NSF, and the Institute of Educational Sciences at the Department of Education. By associating an ORCID account to SciENcv, users gain access to a range of benefits, including the inclusion of a persistent identifier on documents, auto-population of fields from the ORCID profile, and the ability to seamlessly incorporate citations from the ORCID profile into biographical sketches.
SciENcv has recently undergone interface enhancements tailored to user needs, encompassing features such as error validation for mandatory fields, user-friendly date entry through calendars, and character counters to ensure adherence to policy-mandated text limits. SciENcv will remain dynamic, updating to accommodate the evolving requirements of federal agencies as they seek more comprehensive applicant data and move towards standardized application forms.
Biomolecule updates
DNA/RNA
NIH comparative genomics resource
The NIH Comparative Genomics Resource (CGR) (https://www.ncbi.nlm.nih.gov/datasets/cgr/) maximizes the impact of eukaryotic research organisms and their genomic data to biomedical research (9). CGR facilitates reliable comparative genomics analyses for all eukaryotic organisms through community collaboration and an NCBI genomics toolkit. Community collaboration is critical to CGR’s success, as it identifies opportunities to connect more genome-related data and metadata with NCBI’s genomics toolkit and provides valuable feedback to drive further development. The toolkit includes high-quality genomics-related data through interconnected databases with access points enabling seamless navigation of NCBI content and tools that can integrate into users’ workflows.
We released or updated several components of this toolkit in the past year. These include a new experimental BLAST (10) database restricted to eukaryotic sequences (nt_euk), along with databases restricted to prokaryotic (nt_prok), viral (nt_viruses) and other sequences (nt_others). These smaller databases require less time to download, reduce search times, and focus searches to sequences of interest. Since September 2022, we added >4500 curated and published domain architectures (specific and superfamily) to SPARCLE (11) and these are available in Conserved Domain Search (CD-Search) results and the Protein Families database. These architectures provide protein product names and protein attributions, such as Gene Ontology (GO) terms (12), Enzyme Commission (EC) numbers (13), PubMed IDs, and IDs from other resources such as the Transporter Classification Database (TCDB) (14), MEROPS (the peptidase database) (15), and CAZy (16), a database of carbohydrate-active enzymes. This supports more accurate and comprehensive comparative protein analyses and renders the data more consistent with FAIR principles by improving the interoperability of classification resources.
We updated the Eukaryotic Genome Annotation Pipeline (EGAP), another toolkit component, in several ways that improve outcomes and make the resulting annotation sets more useful. Additional filtering of alignments generated by the STAR (17) and minimap2 (18) aligners improves annotations in tandemly repeated gene clusters, and implementation refinements improve cross-species alignment rates. Additionally, InterProScan (19) is now used to assign GO terms to annotated genes with data available through FTP and NCBI Gene. Lastly, EGAP now calculates expression per RNA-Seq run and per gene, using Subread featureCounts software (20).
The Comparative Genome Viewer (CGV) (https://ncbi.nlm.nih.gov/genome/cgv/) allows users to visually inspect two genomes based on their alignments to one another, and since the beginning of 2023 has added data from >200 species and 375 alignments, many of which are mammalian cross-species examples. In addition to alignments generated by NCBI, these new data include alignments generated at the UCSC Genomics Institute and minimally processed by NCBI for display. We added a dot plot (2D) display to the tool to facilitate the detection of regions with large genome rearrangements such as translocations or segmental duplications.
Because of our work on CGR, NCBI has also made major advances in providing tools and analyses to improve genome sequence quality. Following last year's beta release, a stable release of a Foreign Contamination Screening (FCS) tool suite (https://github.com/ncbi/fcs) that detects adaptors and cross-species contamination in assembled genomes is now available for download, enabling genome submitters to improve their genome quality before submission (21). The tool supports screening of both eukaryotic and prokaryotic genomes. The genome submission portal has been updated to use the rapid and higher sensitivity FCS-GX screen, accelerating the process and helping to reduce errors in newly submitted genomes. Contamination data for over 1.5 million existing genomes is also available on FTP (https://ftp.ncbi.nlm.nih.gov/genomes/TOOLS/FCS/reports/). We further analyze prokaryote genomes for contamination using Average Nucleotide Identity (ANI) analyses, and we use the combination of FCS and ANI results to flag some eukaryote and prokaryote genomes as ‘contaminated’ in NCBI’s genome resources and exclude them from the NCBI RefSeq collection. Additional summary reports are available on FTP (https://ftp.ncbi.nlm.nih.gov/genomes/ASSEMBLY_REPORTS/), including assembly_summary files with an expanded set of genome and annotation statistics. Ongoing efforts will further expose sequence quality information and help users make informed decisions on which data to use for their studies.
NCBI virus
The NCBI Virus resource (https://www.ncbi.nlm.nih.gov/labs/virus/) serves as a user-friendly platform for searching and accessing viral genomic sequences and normalized metadata. To ensure consistent and accurate evaluation of genetic variations, NCBI has developed an analytical pipeline (https://www.ncbi.nlm.nih.gov/sra/docs/sars-cov-2-variant-calling/) designed to systematically identify nucleotide and protein alterations within the collection of over 14.8 million SARS-CoV-2 sequence samples archived within the Sequence Read Archive (SRA) and GenBank repositories. The SARS-CoV-2 Variants Overview dashboard (https://www.ncbi.nlm.nih.gov/activ), available as part of the NCBI Virus resource, harnesses insights derived from the NCBI SARS-CoV-2 variation analysis pipeline. Recently improved, this dashboard now offers the ability to interrogate SARS-CoV-2 records utilizing sequence variations or genetic lineages as search parameters. Following a search, the interface provides sequence records and metadata, viewable in the interface or as downloadable files. The SARS-CoV-2 Variants Overview also includes visualizations of the geographical locations and frequencies of lineages for countries and US states, as well as lineage-defining mutations. This interface was developed in collaboration with the NIH Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) Tracking Resistance and Coronavirus Evolution (TRACE) initiative (https://www.nih.gov/research-training/medical-research-initiatives/activ/tracking-resistance-coronavirus-evolution-trace).
Sequence read archive (SRA)
NCBI celebrates three years of SRA data in the cloud (22) comprising over 25 petabytes. Included are more than 27 000 000 public SRA files in both Normalized and Lite format, associated metadata, and STAT analyses (23) available in Amazon Web Services Open Data and Google Cloud Platform Public Data Set programs (https://www.ncbi.nlm.nih.gov/sra/docs/sra-cloud/). Our goal of promoting discovery at petabyte scale by increasing ease of identification and access is materializing. Using SRA cloud data Edgar et al. (24) dramatically expanded our knowledge of virus diversity. Investigators in the public health sphere are actively leveraging STAT results in the cloud to both monitor their own submissions as well as surveil diseases such as polio and measles. Noting that ‘typical homology-based search methods (e.g. BLAST) could not be applied at such a large scale’, Hodgins et al. (25) identified ancient Clostridium tetani-related sequences in metagenomic archeological samples searching ‘the NCBI-stat database on 15 March 2021 for matches to tax_id = 1513 using Google's Big Query API’. NCBI provides open access to the original source files, SRA files, and VCF files generated from them by our participation in the NIH ACTIV TRACE program (26) focusing on the SARS-CoV-2 subset of SRA data. Finally, these public data stimulated exploration of VCFs for population genomics during a recent codeathon (https://ncbiinsights.ncbi.nlm.nih.gov/event/vcf-for-population-genomics-codeathon).
RefSeq
The NCBI RefSeq collection now includes 311 967 prokaryote and 1735 eukaryote genomes as of 11 August 2023, representing yearly growth of 21% for both sets. A separate article describing the improvements in the prokaryotic collections can be found in this issue (27). Within the eukaryotic collection, genomes from 1056 species are now annotated with NCBI’s Eukaryotic Genome Annotation Pipeline (EGAP), incorporating extensive manual curation efforts in human, mouse, rat, and other taxa. We have revised the naming system for new EGAP annotation runs: these names are based on the assembly accession and have a date-based suffix corresponding to the annotation run, e.g. GCF_000001405.40-RS_2023_03 for the March 2023 human GRCh38.p14 annotation. This improves clarity for reporting the assembly and annotation data and makes the data more FAIR (Findable, Accessible, Interoperable, and Reusable). The RefSeq annotation of the human genome prominently incorporates the Matched Annotation from the NCBI and EMBL-EBI (MANE) dataset (28). The latest MANE release (v1.2) includes transcripts for 99.4% of protein-coding genes to serve as universal standards for clinical variant reporting. We encourage adoption of MANE transcripts to increase the consistency of clinical reporting, streamline clinical interpretation, and facilitate the comparison and exchange of data between resources. We have also added alignment and annotation data for historical human RefSeq transcripts to aid clinical groups with migrating legacy datasets to the GRCh38.p14 reference genome (https://ftp.ncbi.nlm.nih.gov/refseq/H_sapiens/historical/). In addition to the comprehensive annotations provided for the human GRCh38.p14 and T2T-CHM13v2.0 genomes, we now provide annotation of curated genes on additional genomes such as those from the human HPRC consortium as a pilot project for pan-genome resources (https://ftp.ncbi.nlm.nih.gov/genomes/all/pilot/annotation_releases/).
The human and mouse genome annotations included tremendous increases in the number of RefSeq Functional Elements (RefSeqFEs) that catalog diverse and functionally important non-genic elements, such as gene regulatory elements and other genomic regions that have been experimentally validated in the literature (29). The GCF_000001405.40-RS_2023_03 and GCF_009914755.1-RS_2023_03 annotations for human GRCh38.p14 and T2T-CHM13v2.0 added over 78 000 and 66 000 new RefSeqFE features, respectively, since our 2022 annotations on those assemblies. The mouse GCF_000001635.27-RS_2023_04 annotation on GRCm39 added over 3900 new RefSeqFE features since our last mouse annotation in 2020. Other RefSeqFE improvements in 2023 included the provision of extractable cell type activity data for annotated features, additional fields for data mining in download files, additional target gene linkages for gene regulatory elements, and updates to the RefSeqFE track hub on the GRCh38.p14, T2T-CHM13v2.0 and GRCm39 assemblies. Further details, including data access options, are available on the RefSeqFE webpage (https://www.ncbi.nlm.nih.gov/refseq/functionalelements/). We encourage the use of this rapidly growing dataset as a reference resource for experimentally validated non-genic regions.
Taxonomy
NCBI continues to curate prokaryotic type strains and their genomes (30) to support ANI analyses (31). We have introduced new files to the Taxonomy FTP site (https://ftp.ncbi.nlm.nih.gov/pub/taxonomy/new_taxdump) for excluded types previously listed incorrectly in the literature and public resources (excludedfromtype.dmp). We have also demonstrated the usefulness of ANI as a tool to assess the validity of taxonomic merges. When two independently described taxa are identified as belonging to the same species, they are merged, and the subsequently described taxon becomes a heterotypic synonym of the initially described taxon. We therefore expect assemblies derived from heterotypic synonyms to show high ANI values. If genomes of heterotypic synonyms exhibit low identity or low ANI values, this may imply that the species in question are distinct and should not have been merged. We have collected such potentially problematic taxonomic merges and their related ANI values (ftp.ncbi.nlm.nih.gov/genomes/ASSEMBLY_REPORTS/prokaryote_ANI_suspect_heterotypic_synonyms.txt). Since ANI processes rely on high quality genome sequences of type strains, their potential is limited when such data are unavailable for type strains. Unfortunately, there are many species that still do not have any genomes from type material, and so we strongly encourage sequencing and submitting genomes for these species (ftp.ncbi.nlm.nih.gov/genomes/ASSEMBLY_REPORTS/prokaryote_without_type_assembly.txt).
Finally, we completed two high profile name changes: new phylum names for Bacteria and Archaea and binomial species names for influenza. These changes were prompted by rule changes made by the International Code of Nomenclature for Prokaryotes (ICNP) and the International Code of Virus Classification and Nomenclature (ICVCN). We described these changes in more detail in NCBI Insights blog posts and on the FTP site (https://ftp.ncbi.nih.gov/pub/taxonomy/Major_taxonomic_updates_2023.txt).
Proteins
iCn3D
A large fraction of protein sequences tracked by NCBI can be mapped to experimentally derived or computationally predicted 3D structures. We are continuing to develop the 3D structure viewer iCn3D as a powerful tool for comparative analysis of sequence and structure and for exploring sequence-structure-function relationships. iCn3D can visualize experimentally determined 3D structures as well as predicted structures, and it retrieves 3D coordinate sets from various online resources. We now provide direct visualization links from the results of protein BLAST searches. In the ‘Alignments’ pane of the BLAST results, such links appear under the right-hand ‘Related Information’ list for each sequence in the BLAST result that we can link to experimental or predicted structures. Clicking on the link will open a browser tab with iCn3D displaying the alignment between the user query sequence and the structure-linked sequence, together with the 3D structure or model. iCn3D will retrieve annotations on the template structure, such as conserved domain footprints, functional sites, and sequence variations, and allows the user to compare query-subject conservation patterns vis-à-vis this annotation and the detailed 3D conformation. We continue to update iCn3D frequently, often in response to direct user requests. Updates are documented at https://github.com/ncbi/icn3d/blob/master/CHANGELOG.md, and iCn3D is accessible at https://www.ncbi.nlm.nih.gov/Structure/icn3d.
Chemicals
Over the past year, PubChem (32,33), a public chemical database at NCBI, expanded the scope of its data content and now provides information for more than 115 million compounds collected from >930 data sources. Notably, data from the FDA Global Substance Registration System (GSRS) were integrated to annotate compounds in PubChem, making it easier to access information on chemicals regulated by the FDA. It is also noteworthy that NLM discontinued its chemical information resources, ChemIDPlus and Drug Information Portal, in December 2022 and that data from these resources are available in PubChem (https://www.nlm.nih.gov/pubs/techbull/ja22/ja22_pubchem.html).
In the last year, we made major changes to PubChem web interfaces as summarized in the PubChem Help site (https://pubchem.ncbi.nlm.nih.gov/docs/user-interface-updates-2023). One notable change is the introduction of the consolidated literature table, which shows a list of all papers about a given compound. We generate this list by combining information from various literature data sources, including journals, publishers, and databases. The consolidated literature table allows users to search, sort, and download the data in a single location. We also updated PubChemRDF (34), the machine-readable PubChem data formatted using the Resource Description Framework (RDF; https://www.w3.org/RDF/). The co-occurrence subdomain (35) was added to encode relationships among chemicals, genes/proteins, and diseases based on their occurrences in biomedical literature. This update enables users to identify chemicals, genes/proteins, and diseases mentioned together with a given named entity through SPARQL queries.
Clinical genetics updates
ClinVar
ClinVar is NCBI’s archive of human genetic variants classified for diseases and drug responses. Over the past year, ClinVar added 780 000 new variants to the database, processed from 1 million new submitted records. We added several features to the ClinVar Submission Portal to make it easier for submitters to maintain information related to their organization. Buttons for ‘Edit Submitter Group’ and ‘Edit Personnel’ allow the submitter to easily edit information about their organization's submitters (those who are authorized to submit for the organization) and personnel (those who are listed publicly on the organization's page in ClinVar), respectively. A button for ‘View/add assertion criteria files’ lets the submitter add new files to use in ClinVar submissions as assertion criteria. Files for assertion criteria are now submitted independently from variant submissions, which means submitters only must provide each file for assertion criteria once, and then it is always available for use in future submissions.
The ClinVar team also developed a prototype for somatic variant classifications in ClinVar. New fields for the clinical impact (therapeutic, diagnostic, or prognostic) and the oncogenicity of a somatic variant were designed, distinct from a germline classification for the variant. We also developed new variant pages and submission spreadsheet templates modified for somatic classifications and tested these with users in video interviews. We used feedback from those interviews to modify the variant pages and the spreadsheet template and to inform the design for aggregation of somatic variants. We posted a preview of the expected changes to the ClinVar XML and submission spreadsheet template to GitHub (https://github.com/ncbi/clinvar) to help users and submitters prepare for this change. We will post additional previews as they are available, e.g. expected updates to ClinVar VCF files.
Genetic testing registry (GTR)
The Genetic Testing Registry (GTR, https://www.ncbi.nlm.nih.gov/gtr/) is NCBI’s database of orderable clinical and research genetic tests and molecular and serologic tests for infectious diseases. GTR aims to support healthcare providers by providing access to genetic testing information and bringing transparency to the genetic testing landscape. As of July 2023, GTR contained 77 486 clinical tests and 233 research tests provided by 492 labs from 48 countries, including 279 US labs. Of the clinical tests, labs have assigned CPT® (Current Procedural Terminology) codes to 2237 tests and LOINC® codes to 519 tests. GTR contains 74 973 molecular tests of which nearly 90% are single gene tests and the remaining are multi-gene panels, exome, and genome tests. Next Generation Sequencing (NGS) is the most used technology (72% of tests). GTR also includes cytogenetic tests (2563 tests interrogate 1 314 unique chromosomal regions or mitochondria) and biochemical genetic tests (111 proteins are measured by 137 tests; 69 enzymes are measured by 209 tests; and 2346 analytes are measured by 596 tests).
In the past year, GTR focused on improving the submission experience for data submitters. New features include a redesigned homepage for a more intuitive submission experience. The new page provides quick access to the groups feature where submitters can manage permissions for laboratory staff who can submit data for their lab and a one-click download of all clinical test data. A new test submission page allows submitters to add new tests, download Excel templates, upload spreadsheets, and track API submissions. We also improved the navigation within the GTR submission site, making it easier to move between the home page, lab record, test management tool, and test submission pages. A test management tool provides several benefits: it improves the search and selection of tests the submitter needs to update or delete, it allows submitters to track and update test data more easily, and it provides a way for submitters to download data for a selected number of tests. A new feature allows submitters to update a subset of data fields simultaneously for multiple clinical tests. Finally, submitters can employ a Submission API that supports fully automated test record submissions.
MedGen
MedGen is NCBI’s portal to clinical information about diseases with a genetic component. Its goal is to support integration of clinical genetics in the practice of medicine. Towards this goal, MedGen provides an online portal to information about genetic phenotypes and harmonizes clinical genetics information from authoritative sources in the community. MedGen also serves as the phenotype backbone for ClinVar and GTR. MedGen aims to provide access to the growing knowledge-base of genetics applicable to clinical care so that these data are useful at the point-of-care while also fostering computational interoperability.
MedGen is a key player in the community driving standardization of genetic phenotype data. It aggregates and harmonizes human disease names and attributes from authoritative sources including UMLS within NLM, OMIM (36), Mondo (37), HPO (38), Orphanet (https://orpha.net), testing labs that submit test descriptions to GTR, and organizations that submit variant interpretation information to ClinVar. Terminologies are available as flat files (OMIM) or ontologies (Mondo, HPO), and MedGen processes each of them differently to present them in an easily usable format on its website for GTR and ClinVar users, and on reports on the FTP site for use by outside organizations. When a record is needed to support GTR and ClinVar submissions and this record is not available from authoritative resources, MedGen creates a new record and sends monthly reports to UMLS for review. For example, MedGen creates records to represent how individuals may respond to a drug based on their genotype by using the generic drug name and the word response, i.e. ‘drug response’. When processing data from multiple sources, discrepancies are sometimes found and MedGen curators review the problem and either find a solution or report them to the source so the community can benefit from data standardization. MedGen may need to split a record, merge multiple records, or create new records. Some reviews require input from the data source and other community stakeholders. A common data conflicts that prompts curation review is the need for differing concept granularity between the testing labs and the data sources. Additional examples include representing clinical presentations versus specific genetic subtypes, conflicting synonyms (cancer and carcinoma), using terms such as ‘Gene-related disorders’ to refer to multiple distinct phenotypes, and using broad concepts to describe multiple distinct phenotypes. MedGen staff works with the community and provides reports of data discrepancies to harmonize disease concept mappings from multiple sources onto one unified, specific record that can be used by clinicians, clinical labs, researchers and data sources.
dbSNP and ALFA
To commemorate the 25th anniversary of dbSNP in 2023, dbSNP released Build 156 and ALFA Release 3, a significant milestone of over 1 billion RefSNP (rs) records with allele frequencies. dbSNP build 156 merges data from thousands of sources, including large-scale population studies such as 1000Genomes, TOPMed, gnomAD, and NCBI ALFA release 3. dbSNP Build 156 offers population frequencies, molecular insights, ClinVar clinical interpretations, publications, and genomic mappings that focus on human single nucleotide variations, insertions, and deletions. In addition, the release of NCBI ALFA Release 3 (Version 20230706150541) represents a significant achievement, as it incorporates data from a global population exceeding 200 000 subjects. ALFA Release 3 also improved variant analysis over earlier releases by adding genotype frequency data and Hardy-Weinberg equilibrium probabilities. This release aggregated an astounding 5.8 trillion total genotypes, giving rise to 904.7 million unique variations, including 554 000 novel variants unknown in dbSNP Build 156. It is one of the most complete aggregated variation collections with allele and genotype frequencies available for 12 major populations. Information about the projects and data access are available on the dbSNP (https://www.ncbi.nlm.nih.gov/snp/) and ALFA (https://www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/) websites. dbSNP in conjunction with ALFA enhances understanding of genetic diversity for genetics research, driving improvements in personalized medicine and disease genetics for both common variants and clinical mutations.
ClinicalTrials.gov
Launched in 2000, ClinicalTrials.gov (https://clinicaltrials.gov/) is a website and online database of information provided by sponsors or investigators for approximately 460 000 clinical research studies conducted around the world, including summary results for nearly 60 000 of those studies. Since October 2019, NLM has been engaging stakeholders and using feedback to modernize ClinicalTrials.gov to deliver an improved user experience on an updated platform that will accommodate growth and enhance efficiency. In June 2023 NLM launched the modernized ClinicalTrials.gov website. This new design includes simple web components, such as left-side menus and expandable accordions, that improve navigation and make information readily findable. In addition, the modernized website is optimized for use on mobile devices. The modernized website replaces the classic ClinicalTrials.gov, which will remain available until 2024.
In 2022, NLM released the initial version of the beta Protocol Registration and Results System (PRS), the data entry and management system for ClinicalTrials.gov. In 2023, releases to this beta site included all the Protocol Section modules to the PRS. Each of these modules includes a new design, improved navigation, and updated onscreen and slide-out drawer help content. Data entered in PRS Beta will be saved on both the classic and Beta sites, and later in 2023 users will be able to submit their study protocol and obtain their NCT (national clinical trial) number in the modernized version of PRS.
Pathogen detection
The NCBI Pathogen Detection Project (https://www.ncbi.nlm.nih.gov/pathogens/) helps public health scientists investigate disease outbreaks by integrating pathogen genomic sequences obtained from cultured bacterial isolates and quickly clustering and identifying related sequences (1). Investigators have successfully used it to help uncover an international outbreak due to contaminated mushrooms (39) and have demonstrated its significant contributions to reducing illness and the burden of disease in the US for foodborne pathogens (40). As of 10 August 2023, over 1 585 000 pathogen isolates covering 80 bacterial taxa and one emerging fungal pathogen, Candida auris, are actively being analyzed. The analysis results are available in the Isolates Browser daily (https://www.ncbi.nlm.nih.gov/pathogens/isolates).
This near real-time update of comprehensive public data is now central to many bacterial outbreak detection and analysis efforts in the US and internationally. The FDA through the GenomeTrakr project has used NCBI Pathogen Detection to initiate 1056 actions intended to protect consumers from foodborne illness (https://www.fda.gov/food/whole-genome-sequencing-wgs-program/genometrakr-network). It is also used to investigate hospital outbreaks; in one example scientists at Harvard Medical School and multiple public health agencies used Pathogen Detection clustering information to identify cryptic methicillin resistant Staphylococcus aureus (MRSA) outbreaks in NICU patients and AMRFinderPlus results to characterize the isolates’ AMR and virulence genes (41). For more examples showing how NCBI Pathogen Detection resources contribute to public health and research see https://www.ncbi.nlm.nih.gov/pathogens/success_stories.
Antimicrobial resistance
The Pathogen Detection team has continued to improve and release updated resources for antimicrobial resistance (AMR) (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/) (42). The team has curated 7827 total proteins (6757 AMR proteins, 252 stress response proteins and 818 virulence proteins) as well as 1217-point mutations and 3818 publication references for proteins and point mutations in the August 2023 release. AMRFinderPlus software updates (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/AMRFinder/) include automatic parsing of the output from nine common annotation tools and databases as well as an increase in processing speed by over 60% on average. AMRFinderPlus is also part of other scientists' workflows; one example is its inclusion in an ISO-certified pipeline for the detection of AMR determinants from whole genome sequencing data, with outputs adapted for clinical antibiotic susceptibility prediction and public health microbiology reporting (43).
We analyze all bacterial isolates in the Pathogen Detection Isolates Browser with AMRFinderPlus (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/AMRFinder/), and the three categories of genes (AMR, stress, and virulence) are available in the Isolates Browser. Currently over 1 520 000 isolates have at least one identified AMR gene, over 1 280 000 have at least one identified stress response gene, and over 920 000 have at least one identified virulence gene. For the subset of isolates with assemblies in GenBank, detailed information and sequences for over 22 000 000 genes and point mutations identified by AMRFinderPlus in over 1 100 000 assemblies are available in the Microbial Browser for Identification of Genetic and Genomic Elements (MicroBIGG-E; https://www.ncbi.nlm.nih.gov/pathogens/microbigge). An antibiogram template for capturing antibiotic susceptibility data is available and tied to BioSample submission (https://www.ncbi.nlm.nih.gov/pathogens/submit-data/#ast) with the user-submitted S/I/R calls displayed in the Isolates Browser for over 23 000 isolates. Isolate Browser and MicroBIGG-E data are also available on Google Cloud Platform including the contig and protein sequences for all 22 million genes and point mutations in MicroBIGG-E (https://www.ncbi.nlm.nih.gov/pathogens/docs/gcp). A recent NCBI webinar demonstrates the use of these cloud resources (https://www.ncbi.nlm.nih.gov/pathogens/docs/ncbi_minute_230329).
For further information
The resources described here include documentation, other explanatory materials, and references to collaborators and data sources on their respective web sites. The NCBI Help Manual and the NCBI Handbook (www.ncbi.nlm.nih.gov/books/NBK143764/) describe the principal NCBI resources in detail. An Outreach Events page (https://ncbiinsights.ncbi.nlm.nih.gov/ncbi-outreach-events/) provides links to webinars, courses, and upcoming conference exhibits. A variety of video tutorials are available on the NLM YouTube channel that can be accessed through links in the standard NCBI page footer. User-support staff are available to answer questions at info@ncbi.nlm.nih.gov, and users can view support articles at https://support.nlm.nih.gov. Updates on NCBI resources and database enhancements are described on the NCBI Insights blog (https://ncbiinsights.ncbi.nlm.nih.gov/), NCBI social media sites (FaceBook, X and LinkedIn), and the several mailing lists and RSS feeds that provide updates on services and databases. Links to these resources are in the NCBI page footer and on NCBI Insights.
Acknowledgements
The authors would like to thank all the NCBI staff who through their dedicated efforts continue to allow NCBI to provide our full collection of services to the community. This work was supported by the National Center for Biotechnology Information of the National Library of Medicine (NLM), National Institutes of Health.
Contributor Information
Eric W Sayers, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Jeff Beck, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Evan E Bolton, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
J Rodney Brister, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Jessica Chan, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Donald C Comeau, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Ryan Connor, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Michael DiCuccio, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Catherine M Farrell, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Michael Feldgarden, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Anna M Fine, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Kathryn Funk, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Eneida Hatcher, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Marilu Hoeppner, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Megan Kane, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Sivakumar Kannan, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Kenneth S Katz, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Christopher Kelly, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
William Klimke, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Sunghwan Kim, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Avi Kimchi, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Melissa Landrum, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Stacy Lathrop, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Zhiyong Lu, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Adriana Malheiro, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Aron Marchler-Bauer, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Terence D Murphy, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Lon Phan, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Arjun B Prasad, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Shashikant Pujar, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Amanda Sawyer, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Erin Schmieder, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Valerie A Schneider, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Conrad L Schoch, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Shobha Sharma, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Françoise Thibaud-Nissen, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Barton W Trawick, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Thilakam Venkatapathi, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Jiyao Wang, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Kim D Pruitt, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Stephen T Sherry, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Data availability
The resources can be accessed through the NCBI home page at https://www.ncbi.nlm.nih.gov.
Funding
Funding for open access charge: National Center for Biotechnology Information of the National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
References
- 1. Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Farrell C.M., Feldgarden M., Fine A.M., Funk K.et al.. Database resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Res. 2023; 51:D29–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schuler G.D., Epstein J.A., Ohkawa H., Kans J.A.. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996; 266:141–162. [DOI] [PubMed] [Google Scholar]
- 3. Sayers E.W., Cavanaugh M., Clark K., Pruitt K.D., Sherry S.T., Yankie L., Karsch-Mizrachi I.. GenBank 2023 update. Nucleic Acids Res. 2023; 51:D141–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burgin J., Ahamed A., Cummins C., Devraj R., Gueye K., Gupta D., Gupta V., Haseeb M., Ihsan M., Ivanov E.et al.. The European Nucleotide Archive in 2022. Nucleic Acids Res. 2023; 51:D121–D125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tanizawa Y., Fujisawa T., Kodama Y., Kosuge T., Mashima J., Tanjo T., Nakamura Y.. DNA Data Bank of Japan (DDBJ) update report 2022. Nucleic Acids Res. 2023; 51:D101–D105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arita M., Karsch-Mizrachi I., Cochrane G.. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2021; 49:D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fiorini N., Canese K., Starchenko G., Kireev E., Kim W., Miller V., Osipov M., Kholodov M., Ismagilov R., Mohan S.et al.. Best match: new relevance search for PubMed. PLoS Biol. 2018; 16:e2005343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu W., Islamaj Dogan R., Kim S., Comeau D.C., Kim W., Yeganova L., Lu Z., Wilbur W.J.. Author name disambiguation for PubMed. J. Assoc. Inf. Sci. Technol. 2014; 65:765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bornstein K., Gryan G., Chang E.S., Marchler-Bauer A., Schneider V.A.. The NIH Comparative Genomics Resource: addressing the promises and challenges of comparative genomics on human health. BMC Genomics. 2023; 24:575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., Madden T.L., Matten W.T., McGinnis S.D., Merezhuk Y.et al.. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013; 41:W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S.et al.. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020; 48:D265–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gene Ontology Consortium The Gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021; 49:D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000; 28:304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saier M.H., Reddy V.S., Moreno-Hagelsieb G., Hendargo K.J., Zhang Y., Iddamsetty V., Lam K.J.K., Tian N., Russum S., Wang J.et al.. The Transporter Classification Database (TCDB): 2021 update. Nucleic Acids Res. 2021; 49:D461–D467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rawlings N.D., Barrett A.J., Bateman A.. MEROPS: the peptidase database. Nucleic Acids Res. 2010; 38:D227–D233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B.. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014; 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G.et al.. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014; 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 21. Astashyn A., Tvedte E.S., Sweeney D., Sapojnikov V., Bouk N., Joukov V., Mozes E., Strope P.K., Sylla P.M., Wagner L.et al.. Rapid and sensitive detection of genome contamination at scale with FCS-GX. 2023; bioRxiv doi:06 June 2023, preprint: not peer reviewed 10.1101/2023.06.02.543519. [DOI] [PMC free article] [PubMed]
- 22. Sayers E.W., Beck J., Bolton E.E., Bourexis D., Brister J.R., Canese K., Comeau D.C., Funk K., Kim S., Klimke W.et al.. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021; 49:D10–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katz K.S., Shutov O., Lapoint R., Kimelman M., Brister J.R., O'Sullivan C.. STAT: a fast, scalable, MinHash-based k-mer tool to assess Sequence Read Archive next-generation sequence submissions. Genome Biol. 2021; 22:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edgar R.C., Taylor J., Lin V., Altman T., Barbera P., Meleshko D., Lohr D., Novakovsky G., Buchfink B., Al-Shayeb B.et al.. Petabase-scale sequence alignment catalyses viral discovery. Nature. 2022; 602:142–147. [DOI] [PubMed] [Google Scholar]
- 25. Hodgins H.P., Chen P., Lobb B., Wei X., Tremblay B.J.M., Mansfield M.J., Lee V.C.Y., Lee P.G., Coffin J., Duggan A.T.et al.. Ancient Clostridium DNA and variants of tetanus neurotoxins associated with human archaeological remains. Nat. Commun. 2023; 14:5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Connor R., Yarmosh D.A., Maier W., Shakya M., Martin R., Bradford R., Brister J.R., Chain P.S., Copeland C.A., Iulio J.d.et al.. Towards increased accuracy and reproducibility in SARS-CoV-2 next generation sequence analysis for public health surveillance. 2022; bioRxiv doi:03 November 2022, preprint: not peer reviewed 10.1101/2022.11.03.515010. [DOI]
- 27. Haft D.H., Badretdin A., Coulouris G., Dicuccio M., Durkin A.S., Jovenitti E., Li W., Mersha M., O’Neill K.R., Virothaisakun J.et al.. RefSeq and the prokaryotic genome annotation pipeline in the age of metagenomes. Nucleic Acids Res. 2023; 10.1093/nar/gkad988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morales J., Pujar S., Loveland J.E., Astashyn A., Bennett R., Berry A., Cox E., Davidson C., Ermolaeva O., Farrell C.M.et al.. A joint NCBI and EMBL-EBI transcript set for clinical genomics and research. Nature. 2022; 604:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farrell C.M., Goldfarb T., Rangwala S.H., Astashyn A., Ermolaeva O.D., Hem V., Katz K.S., Kodali V.K., Ludwig F., Wallin C.L.et al.. RefSeq Functional Elements as experimentally assayed nongenic reference standards and functional interactions in human and mouse. Genome Res. 2022; 32:175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kannan S., Sharma S., Ciufo S., Clark K., Turner S., Kitts P.A., Schoch C.L., DiCuccio M., Kimchi A.. Collection and curation of prokaryotic genome assemblies from type strains at NCBI. Int. J. Syst. Evol. Microbiol. 2023; 73:005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ciufo S., Kannan S., Sharma S., Badretdin A., Clark K., Turner S., Brover S., Schoch C.L., Kimchi A., DiCuccio M.. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018; 68:2386–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem 2023 update. Nucleic Acids Res. 2023; 51:D1373–D1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim S. Exploring chemical information in PubChem. Curr Protoc. 2021; 1:e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fu G., Batchelor C., Dumontier M., Hastings J., Willighagen E., Bolton E.. PubChemRDF: towards the semantic annotation of PubChem compound and substance databases. J Cheminform. 2015; 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Q., Kim S., Zaslavsky L., Cheng T., Yu B., Bolton E. Yamaguchi A., Splendiani A., Marshall M.S., Baker C., Bolleman J., Burger A., Castro L., Eigenbrod O., Österle S., Romacker M.et al.. Resource description framework (RDF) modeling of named entity co-occurrences derived from biomedical literature in the PubChemRDF. 14th International Conference on Semantic Web Applications and Tools for Health Care and Life Sciences (SWAT4HCLS 2023). 2023; Basel, Switzerland: 32–41.CEUR-WS.org. [Google Scholar]
- 36. Amberger J.S., Hamosh A.. Searching online mendelian inheritance in man (OMIM): a knowledgebase of Human genes and genetic phenotypes. Curr. Protoc. Bioinformatics. 2017; 58:1.2.1–1.2.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vasilevsky N.A., Matentzoglu N.A., Toro S., Flack J.E. IV, Hegde H., Unni D.R., Alyea G.F., Amberger J.S., Babb L., Balhoff J.P.et al.. Mondo: unifying diseases for the world, by the world. 2022; medRxiv doi:03 May 2022, preprint: not peer reviewed 10.1101/2022.04.13.22273750. [DOI]
- 38. Kohler S., Gargano M., Matentzoglu N., Carmody L.C., Lewis-Smith D., Vasilevsky N.A., Danis D., Balagura G., Baynam G., Brower A.M.et al.. The Human phenotype ontology in 2021. Nucleic Acids Res. 2021; 49:D1207–D1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pereira E., Conrad A., Tesfai A., Palacios A., Kandar R., Kearney A., Locas A., Jamieson F., Elliot E., Otto M.et al.. Multinational outbreak of Listeria monocytogenes infections linked to Enoki mushrooms imported from the Republic of Korea 2016-2020. J. Food Prot. 2023; 86:100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brown B., Allard M., Bazaco M.C., Blankenship J., Minor T.. An economic evaluation of the Whole Genome Sequencing source tracking program in the U.S. PLoS One. 2021; 16:e0258262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Worley J.N., Crothers J.W., Wolfgang W.J., Venkata S.L.G., Hoffmann M., Jayeola V., Klompas M., Allard M., Bry L.. Prospective genomic surveillance reveals cryptic MRSA outbreaks with local to international origins among NICU patients. J. Clin. Microbiol. 2023; 61:e0001423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Feldgarden M., Brover V., Fedorov B., Haft D.H., Prasad A.B., Klimke W.. Curation of the AMRFinderPlus databases: applications, functionality and impact. Microb Genom. 2022; 8:mgen000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sherry N.L., Horan K.A., Ballard S.A., Gonçalves da Silva A., Gorrie C.L., Schultz M.B., Stevens K., Valcanis M., Sait M.L., Stinear T.P.et al.. An ISO-certified genomics workflow for identification and surveillance of antimicrobial resistance. Nat. Commun. 2023; 14:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The resources can be accessed through the NCBI home page at https://www.ncbi.nlm.nih.gov.