Abstract
Target discovery is one of the essential steps in modern drug development, and the identification of promising targets is fundamental for developing first-in-class drug. A variety of methods have emerged for target assessment based on druggability analysis, which refers to the likelihood of a target being effectively modulated by drug-like agents. In the therapeutic target database (TTD), nine categories of established druggability characteristics were thus collected for 426 successful, 1014 clinical trial, 212 preclinical/patented, and 1479 literature-reported targets via systematic review. These characteristic categories were classified into three distinct perspectives: molecular interaction/regulation, human system profile and cell-based expression variation. With the rapid progression of technology and concerted effort in drug discovery, TTD and other databases were highly expected to facilitate the explorations of druggability characteristics for the discovery and validation of innovative drug target. TTD is now freely accessible at: https://idrblab.org/ttd/.
Graphical Abstract
Graphical Abstract.
Introduction
Target discovery is one of the essential steps in modern drug development, and the identification of promising targets lays the foundation for the successful development of first-in-class drug (1). To ensure the success and efficiency of drug development, the quality of the selected target needs to be assessed during the early stage of drug discovery, which has been frequently conducted by evaluating the druggability of target (2–4). The druggability of a target refers to the likelihood of target being effectively modulated by drug-like agents with various evaluation methods proposed (5–7). For example, the presence of suitable binding pocket is crucial for the target's druggability, which is known as one standard procedure in target selection (8); the human system profiles such as human similarity proteins and affiliated pathways have been explored for characterizing target druggability, with their ability to differentiate the targets of rapid (speedy) and slow (non-speedy) clinical development process (9). Moreover, diverse cell-based differential expressions of targets are found informative for identifying new targets that play a crucial role in disease (10).
Notably, the target assessment using single druggability characteristics is often insufficient, and comprehensive evaluation of multiple druggability characteristics is a more helpful approach (9). Therefore, related databases are needed to provide comprehensive druggability characteristics of targets from multiple perspectives. So far, a variety of established databases have been developed to collectively provide drug & target data. Some describe pharmacological information on drugs, such as DrugBank (11), DrugCentral (12), SuperDRUG2 (13), DrugMap (14) and DRESIS (15); some others focus on presenting therapeutic targets, such as TTD (16) and Open Target (17); the remaining offer general molecule and bioactivity information, such as PubChem (18), ChEMBL (19) and BindingDB (20). Although these databases have already accumulated great popularities and substantial impacts on chemical/biological/pharmaceutical communities, the information of target druggability characteristics have not yet be covered by any of the existing databases.
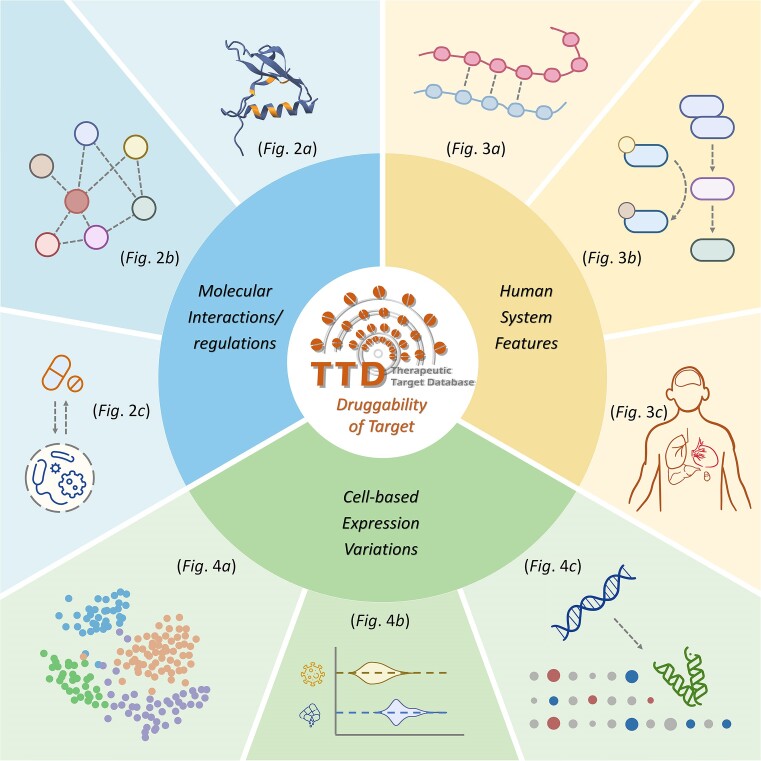
Herein, the therapeutic target database (TTD) was thus significantly updated to its 2024 version, which provided comprehensive information on the druggability characteristics of 426 successful, 1014 clinical trial, 212 preclinical/patented and 1479 literature-reported targets. Particularly, such characteristics were of three perspectives (Figure 1): molecular interactions/regulations, human system profiles and cell-based expression variations. Molecular interactions/regulations offered (1a) ligand-specific spatial structures of target in its drug binding pocket, (1b) network properties of target measured based on protein-protein interactions & (1c) bidirectional regulations between the microbiota and therapeutic agents. Human system profiles provided (2a) similarity profile of target to human proteins outside its families, (2b) involvements of target in well-established life-essential pathways & (2c) distributions of target among a variety of organs in human. Cell-based expression variations described (3a) varied expression of target across cells of different diseases, (3b) differential expressions of target induced by exogenous stimuli & (3c) modified expressions of target altered by human endogenous factors. The statistics of targets & drugs in TTD over the past decade were provided in Table 1, and the detailed data on the major contents integrated into TTD were explicitly described in following sections. With the rapid progression in the techniques of drug discovery, the wealth of druggability data incorporated into TTD are expected to establish a solid foundation for the identification of novel targets and discovery of new therapeutics. TTD is now freely accessible without any login requirement at: https://idrblab.org/ttd/.
Figure 1.
Three major contents integrated into the TTD 2024. A wealth of data was collected to describe target druggability from three distinct perspectives: molecular interactions/regulations, human system features and cell-based expression variations. Each perspective was elaborated in detail through three different sections of data, which were further explicitly described.
Table 1.
Number of drugs and their corresponding therapeutic targets in different versions of TTD over the past decade
| Different versions of TTD published during the past decade | ||||||
|---|---|---|---|---|---|---|
| Statistics of targets and drugs in different versions of TTD | 2024 | 2022 | 2020 | 2018 | 2016 | 2014 |
| All targets | 3730 | 3578 | 3419 | 3101 | 2589 | 2360 |
| Successful targets | 532 | 498 | 461 | 445 | 397 | 388 |
| Clinical trial targets | 1442 | 1342 | 1191 | 1121 | 723 | 461 |
| Preclinical/patented targets | 239 | 185 | 155 | 0 | 0 | 0 |
| Literature-reported targets | 1517 | 1553 | 1612 | 1535 | 1469 | 1467 |
| All drugs | 39 862 | 38 760 | 37 102 | 34 019 | 31 614 | 20 667 |
| Approved drugs | 2895 | 2797 | 2649 | 2544 | 2071 | 2003 |
| Clinical trial drugs | 11 796 | 10 831 | 9465 | 8103 | 7291 | 3147 |
| Preclinical/patented drugs | 5041 | 5009 | 4845 | 0 | 0 | 0 |
| Experimental drugs | 20 130 | 20 123 | 20 143 | 18 923 | 17 803 | 14 856 |
Factual content and data retrieval
Due to the importance of target druggability data (as described above) in modern drug discovery, therapeutic target database (TTD) was mainly updated to its 2024 version by comprehensively providing three types of druggability information for each therapeutic target. As shown in Figure 1, compared with the previous versions, the TTD 2024 updated three major types of druggability: molecular interactions/regulations, human system features and cell-based expression variations. These druggability data were not covered by any of the previous versions of TTD. Each of these three types of druggability was further elaborated using three distinct sub-sections of data, which were explicitly discussed and described as follows.
Druggability illustrated by molecular interactions or regulations
Ligand-specific spatial structure of a target within drug binding pocket
The drug binding site of therapeutic target was usually considered to be indispensable for modern drug discovery (21–25). The binding pocket structure of established targets was essential for drug design and lead optimization (26), and the binding pocket of promising new targets could further expand the druggable genome and enable development of new strategies for targeted therapeutics (8). Among the >80 FDA-approved kinase inhibitors, many of them were inspired by the binding pocket structure of the catalytic domain of kinases (27). In other words, it is essential to have the valuable drug-specific spatial structures of studied target within its drug binding pocket.
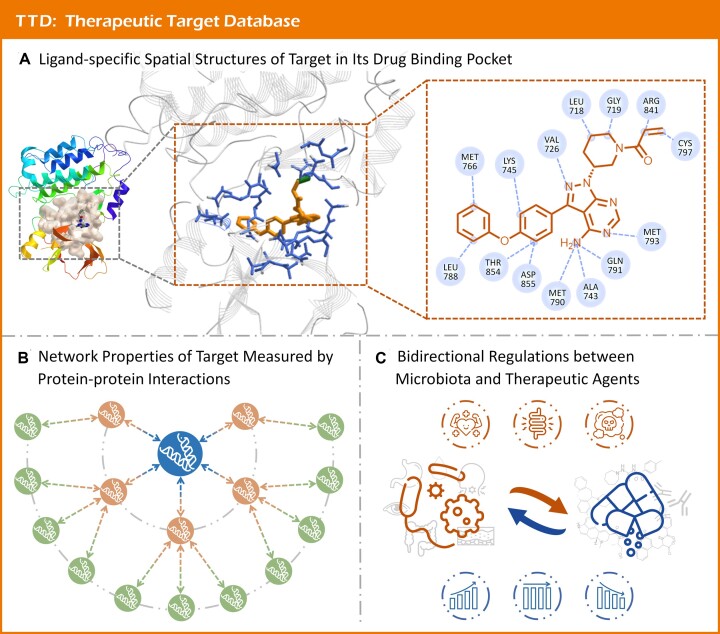
Such structures of drug binding pocket were systematically collected to TTD using the following procedure. First, a comprehensive search of all TTD targets in PDB (28) was realized based on the name and synonyms of the targets. Second, all retrieved structures were carefully checked to remove false matches, which resulted in >25 000 target crystal structures. Third, the availability of drug binding to these structures was investigated, and the corresponding drugs were identified. Forth, the co-crystal structures containing both target and its interacting drug were obtained, and the distance between drug and each residue was calculated based on biopython (29). All residues that interacted with drug at a distance of <5 Å were defined as the ‘drug binding pocket’ (30). As shown in Figure 2A, the binding pocket information was provided in ligand-specific manner for any studied target. For certain complex, the pocket residues together with detailed distances were provided in TTD and highlighted based on their van der Waals surface calculated by iCn3D (31). Additional information (such as structure resolution, sequence, and mutation) was also provided in online TTD. As a result, the ligand-specific binding pockets of 319 successful (targeted by at least one FDA-approved drug), 427 clinical trial (not targeted by any approved drug, but targeted by at least one clinical trial drug), 116 preclinical/patented (not targeted by any approved/clinical trial drug, but targeted by at least one preclinical/patented drug), 375 literature-reported (targeted by experimental drugs only) targets were identified from 22 431 complex structures.
Figure 2.
Druggability of therapeutic target illustrated by molecular interactions and regulations. (A) ligand-specific spatial structure of targets in their drug binding pocket. The crystal structures complexed with ligands were comprehensively collected for a target, and the residues interacting with drugs at a distance of <5 Å were defined as drug binding pockets and highlighted using their van der Waals surface for each complex. (B) network properties of target measured using protein-protein interactions. The human protein-protein interaction network consisting of 9309 proteins and 52 713 interactions was constructed based on the STRING data with confidence score ≥0.95, and diverse network descriptors (degree, connectivity, etc.) were calculated for each target based on PPI network. (C) bidirectional regulations between microbiota and therapeutic agents. On the one hand, microbiota in diverse human tissue or organs (eye, lung, oral cavity, etc.) can alter the bioavailability, bioactivity, and toxicity of therapeutic agent; on the other hand, therapeutic agent can also change the abundance and composition of microbiota.
Network properties of target measured by protein–protein interactions
Target's network properties derived from complex connections among numerous protein–protein interactions (PPI) have been extensively employed for evaluating the target druggability (32–36). Proteins demonstrating high node degrees are posited to exert considerable influence on network function due to the huge amount of interactions (37), while proteins exhibiting high betweenness centrality are considered pivotal in network communication and signaling information flow (38). A multitude of network descriptors have been reported as potential indicators to differentiate the targets of rapid (speedy) and slow (non-speedy) clinical development process (9).
The collection of target's network properties to TTD was accomplished in the following manner. First, PPIs with high confidence score (≥0.95) were collected from STRING database (39), and a human PPI network consisting of 9309 proteins and 52 713 PPIs was then constructed. Second, nine representative network properties (including: betweenness centrality, clustering coefficient, etc.) were calculated for each target (40). As shown in Figure 2B, a two-layer PPI network for a target was illustrated, together with a downloadable file of the complete human PPI network. As an outcome of this process, a variety of network properties for 426 successful, 727 clinical trial, 143 preclinical/patented, and 867 literature-reported targets were provided in TTD 2024.
Bidirectional regulations between microbiota and targeted agents
The regulation between microbiota and targeted agents is complex and bidirectional (41). On the one hand, microbiota can modulate bioavailability, bioactivity and toxicity of drugs; on the other hand, drugs can impact growth, composition, and function of microbiota (42). Taking irinotecan (one medication for treating colon cancer) as an example, it is metabolized to SN-38 glucuronide by beta-glucuronidase of gut microbiota, which resulted in the great elevation of gastrointestinal toxicity (43), and the selective inhibition of bacterial beta-glucuronidase showed the potential to alleviate drug-induced toxicities (44). In other words, unraveling such regulations is anticipated to shed light on the identification of novel therapeutic targets, the discovery of new therapies and the potential modification of existing drug prescription methodologies (45,46).
Bidirectional regulation data between microbiota and targeted drugs were collected to TTD using the following procedure. First, systematic literature review was conducted in PubMed (47) using such keyword combinations as ‘drug + microbiota’, ‘drug + microbe’, ‘drug + microbiome’, etc. All retrieved literatures were carefully reviewed, and the interactions between drugs and microbe were manually recorded. As illustrated in Figure 2C, all the interactions were classified into two categories: microbes affecting drug metabolism & drugs regulating microbe abundance. For the former interaction type, the detailed information (such as involved microbial enzymes, metabolic reactions of studied microbiota, resulting metabolites, and metabolic effects) was also explicitly described. For the latter type of interaction, the detailed information (such as abundance variation of microbe, a variety of species origins and specific experimental samples) was further extracted. As a result, a total of 9812 interactions between 663 drugs and 686 microbes were collected to TTD, which came from 20 phyla, 36 classes, 59 orders, 101 families and 135 genera.
Druggability characterized by different human system profiles
Similarity profile of target to human proteins outside Its family
Drug candidates are typically designed to selectively interact with their intended target, and their interactions with other proteins outside target's biochemical family should therefore be carefully evaluated at the early stage of drug development (48). As reported, the target having fewer human similarity proteins outside its biochemical family is commonly regarded to have greater capacity to avoid undesired interaction and thus increase the possibility of discovering drug-like molecule (9). Therefore, it is highly demanded to have the valuable data on the number of human similarity proteins outside target's functional family to assess the off-target collateral interactions (9).
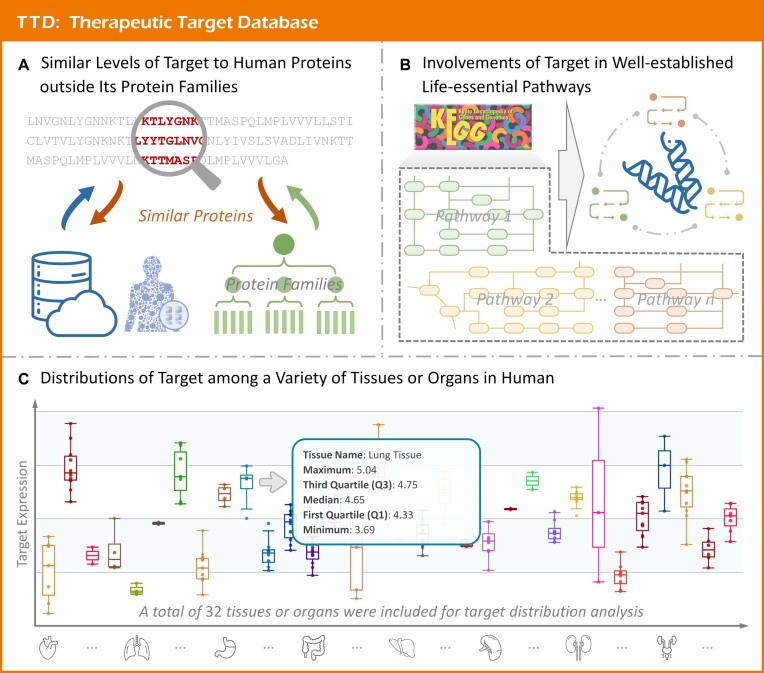
As shown in Figure 3A, such similarity profiles were included into TTD. First, the sequences of TTD targets and all human proteins were extracted from UniProt (49). Then, the protein families to which each protein belonged were obtained from InterPro (50). For a TTD target, its similarity to human proteins was calculated using BLAST (51–53). The similarity proteins of a target were defined as those with E-value <0.005 and outside the protein families of the target. On the target page, the data of protein name, protein family, BLAST identities, and E-values were listed. As a result, the similarity profile information was made available for 389 successful, 933 clinical trial, 204 preclinical/patented and 1479 literature-reported targets in TTD 2024.
Figure 3.
Druggability of therapeutic targets illustrated by human system features. (A) Similarity profile of target to human proteins outside its family. The degree of similarity between target and all human proteins was calculated using BLAST, and the cutoff of E-values was set to 0.005 (the similar proteins of targets were defined as those with E-value <0.005 and outside those functional families of the targets). (B) Involvements of target in the well-established life-essential pathways. All life-essential pathways involved by a target were described on the TTD website with detailed information provided, such as the name, hierarchy & map of, and other targets belonging to these pathways. (C) Distributions of target among a variety of tissues/organs in human. The expressions of studied target across different tissues/organs were provided in the boxplot format, and detailed data (such as tissue name and various statistic values describing the boxplot) were specified.
Involvements of Target in the Well-established Life-essential Pathways
Targets affiliating with fewer life-essential pathways were reported to have greater likelihood of success, while those associated with more signaling pathways were found to increase the chances of undesirable interferences with other human process (48). The target-directed toxicity had been identified as originating from the participation of the targets in potentially harmful pathways (1). Furthermore, in circumstances where the understanding of drug targets’ functions is inadequate, the valuable information of target-affiliating pathways can be very informative (54).
As illustrated in Figure 3B, the life-essential pathways that TTD targets involved were gathered. First, all available pathway information for each target was collected from KEGG (55). Second, the target-affiliated pathways were double-checked based on two criteria: (Ca) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (Cb) the studied targets should occupy an upstream position in pathway, and thus are capable of regulating biological function. Finally, 241 life-essential pathways were included. For a target, all affiliated pathways were integrated to online TTD with the detailed data of pathway hierarchy. Moreover, other targets that belonged to the same pathway were also fully described. All in all, a variety of target-affiliating life-essential pathway data were made available for 373 successful, 897 clinical trial, 196 preclinical/patented, and 1415 literature-reported targets in TTD 2024.
Distributions of target among a variety of tissues or organs in human
The distribution of targets among different tissues/organs needs to be carefully considered, when assessing the target druggability, as it is widely known that the wider the target distributions, the greater the concern over adverse drug reaction (1,56). A previous study on the distribution of 158 successful targets identified that over 50% of these targets were distributed in no more than three tissues, indicating the significance of tissue selectivity in target discovery (48).
Considering the substantial discordance in target's expression at the levels of proteins and RNAs (57), the distributions of TTD targets among various tissues/organs were determined. A landmark study that quantified over 12 000 genes across 32 normal human tissues at both protein and RNA levels was adopted to fulfill our research needs (57). For a target, the relative expressions among tissues/organs were provided, which were displayed in boxplot format together with the detailed abundances (Figure 3C). As a result, the distributions across 32 human tissues of 338 successful, 600 clinical trial, 143 preclinical/patented and 920 literature-reported targets were provided.
Druggability reflected by diverse cell-based expression variations
Varied expressions of target across different cells of diverse diseases
Recent studies had indicated that the heterogeneity among cell types could result in distinct drug responses (58). For instance, FGFR2-amplified gastric cancer cell lines (KatoIII & SNU16) were sensitive to AZD4547 (an FGFR2 inhibitor), while those without FGFR2 amplification (AGS & SNU1) were reported insensitive to AZD4547 (59). In other words, understanding the pattern of target expression among cell types is essential for the selection of representative cell line models and the understanding of the mechanism underlying drug response or resistance (60–64).
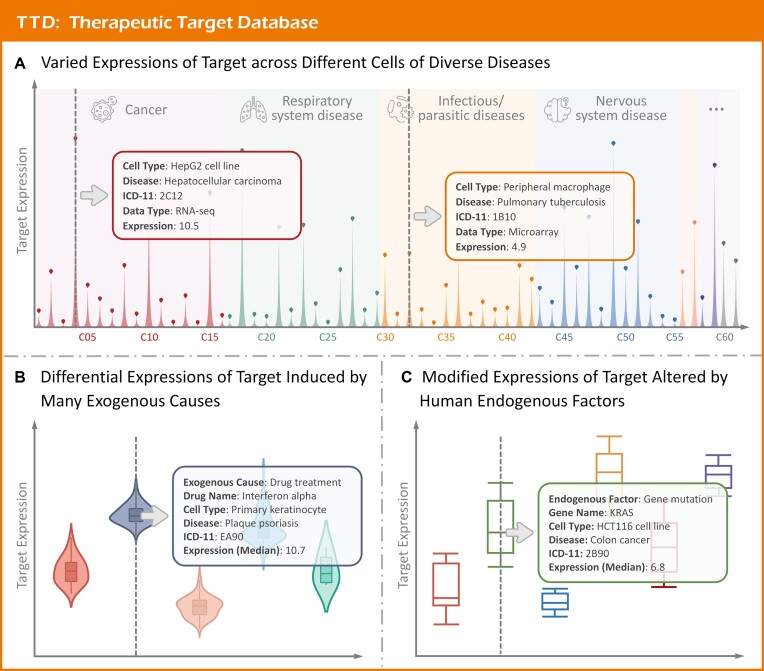
Such expression pattern among different cell lines were collected using the following procedure. First, an exhaustive review was carried out in GEO (65) & Expression Atlas (66) employing the keyword combinations of ‘cell line + expression’, ‘cell type + expression’, ‘cell line + differential expression’, etc. This approach generated a total of 226 datasets containing the expression profile of thousands of proteins across cell lines. Second, detailed information for each dataset, including cell type, disease, etc., was meticulously recorded, which resulted in a total of 1742 types of cell lines from 7289 samples, spanning 121 disease classes as defined by the WHO ICD-11 (such as tuberculosis, skin cancer, allergic rhinitis, and ulcerative colitis). Third, various OMIC data types were processed independently. For microarray data, the original CEL files were downloaded and processed using the RMA function in oligo package (67) to calculate the gene expression matrix; for RNA-seq data, the raw count data were normalized using DESeq2 package (68). For a studied target, its varying expression levels across diverse cell types were visually represented (as shown in Figure 4A). In summary, varying expressions across various cell types for 347 successful, 939 clinical trial, 188 preclinical/patented, and 1371 literature-reported targets were provided.
Figure 4.
Targets’ druggability showed by cell-based expression variation. (A) Varied expressions of target across different cells of diverse diseases. Targets’ expression data for different cell types in normal and untreated conditions were collected and illustrated by pictorial bar chart, and 1742 cell types from 7289 experimental samples were reported, which covered several (a total of 121) disease classes (such as skin cancers, tuberculosis, allergic rhinitis, and ulcerative colitis) defined by ICD-11. (B) Differential expressions of a target induced by exogenous stimuli. Cell type-based differential expressions of targets induced by exogenous causes (a total of 625 exogenous causes, such as interferon treatment, influenza infection and hypoxia) among 313 cell types were shown. (C) modified expressions of a target altered by human endogenous factor. Cell type-based target's expression modifications mediated by exogenous causes (a total of 447 endogenous factors, such as KRAS mutation and MYC over-expression) among 198 cell types were explicitly described.
Differential expressions of target induced by many exogenous causes
Different cell types manifest diverse perturbation signals in response to exogenous stimuli, such as drug administration (69). For the same stimulus (such as particular kinase inhibitor), a variety of cell lines were reported to be phenotypically responsive, but the transcriptomic profiles among these cell lines after the stimulation (such as the treatment by kinase inhibitor) were identified to be extremely different (70). Such perturbation signals were valuable for providing novel insights into understanding drug mechanisms of action and identifying potential drug targets (70–73).
The target's expression profiles induced by exogenous stimuli were collected and provided using the following procedure. First, the differential expression data induced by exogenous cause were retrieved from GEO (65) & Expression Atlas (66) using the keyword combination of ‘cell line + drug’, ‘cell line + exogenous causes’, ‘cell line + therapy’, ‘cell line + environment’, etc. Second, all retrieved datasets were carefully examined, and the detailed exogenous stimuli were recorded, which were classified into three groups: treatment with drugs, infection by bacterium/virus and stimulation by environmental factors. Moreover, the explicit description of each dataset was also provided, which included cell line, disease, perturbation factor, etc. Third, different OMIC-based data types were processed independently. For microarray data, the CEL files were processed with the RMA function of oligo package (67) to normalize expression matrix; for RNA-seq data, raw count data were normalized using DESeq2 package (68). Finally, the cell line-specific expression profile was shown in TTD using violin plots for any studied target (shown in Figure 4B). All in all, a total of 625 exogenous stimuli (hypoxia, interferon treatment, influenza infection, etc.) that modified the expression profiles of 357 successful, 926 clinical trial, 197 preclinical/patented & 1382 literature-reported targets among 313 cell lines were made available in TTD 2024.
Modified expressions of target altered by human endogenous factors
Given that a gene can play distinct roles in different contexts, particularly where the cell-specific function is involved, human endogenous gene perturbation (mutation, expression variation, etc.) is considered as a powerful way to explore target functions under different cell contexts (74–77). In other words, cell line-specific gene perturbations are valuable for understanding the molecular mechanism underlying target differential expression among cell lines, which can help to identify new cell-specific functions, protein-protein interactions and regulatory cascades (78–81).
Such target's expressions regulated by endogenous factors were collected based on the following procedure. First, the GEO (65) & Expression Atlas (66) were manually reviewed to retrieve gene expression data altered by diverse human endogenous factors, which included protein mutations, expression variations, etc. Second, detailed information of each dataset (such as cell line, disease, and human endogenous factor) was meticulously extracted, which resulted in over 400 types of human endogenous factors (such as KRAS mutation, and MYC over-expression), and the factor-induced expression variation was also illustrated in Figure 4C. The process and normalization of OMIC-based raw data were conducted by following the same procedure as that was discussed in the above two sections. All in all, the expression profile of 352 successful, 934 clinical trial, 192 preclinical/patented, and 1363 literature-reported targets among 198 cell lines were provided.
Regular update on the drug & target data and diverse functions
The integration of newly emerged drugs and targets to TTD was also routinely conducted in this update. First, the drugs approved during the past two years were collected from two authoritative publications (82,83). Second, new drugs in clinical trials were collected from various established resources, such as ClinicalTrials.gov, PhRMA medicines in development reports, and numerous Drug Pipeline Reports of >200 companies (such as Pfizer, Roche, Sanofi and GlaxoSmithKline). Third, the trial status of drugs available in TTD were continuously updated using the timely data provided in ClinicalTrials.gov, company's official reports, etc. Fourth, the preclinical and patented drugs were collected from diverse data sources, such as company's pipeline reports, large number of patents authorized by the patent offices of many countries, and recent literature reports.
For each of the collected drugs, its corresponding therapeutic target(s) was further validated by following a routine process that showed the functional roles of the target(s) in disease phenotype and the ability of drug-like molecule to modulate the activities of the target to achieve therapeutic efficacies (84). Finally, the status of each therapeutic target was determined based on the highest status of its corresponding drugs, which were then classified to successful target (approved drug), clinical trial target (clinical trial drug), preclinical target (drug in preclinical trial), patented target (drug protected by the authorized patent), and literature-reported target (investigative agent). As a result (provided in Table 1), a total of 3730 targets and 39 862 drugs were finally provided in TTD 2024 and the total numbers of drugs and their corresponding therapeutic targets in different versions of TTD over the past decade was also described.
Moreover, a ‘batch search’ function allowing the upload of a list of TTD drug IDs or Target IDs was implemented to the TTD 2024 (https://db.idrblab.net/ttd/ttd-search/batch-search), and a ‘full download’ function of all search results was also realized by simply clicking the ‘Download the Search Results’ button. Such functions could be very helpful to broad audiences, especially those pharmacologically inclined users. It should be noted that, although it was technically feasible to implement the search function based on multiple types of entries other that drug/target IDs (such as drug/target name/synonyms), that function could have substantial chance to return many false positive search results, which had therefore not been made available in TTD 2024 yet.
Conclusion and perspectives
Identification and validation of therapeutic targets is one of the critical steps in drug development (85). Insufficient analysis of target druggability in the early stage of drug discovery remains one of the key issues of high drug attrition rates, which should therefore be systematically considered and carefully assessed (86). Taking a recent study as an example (9), it identified several essential features of target druggability (such as ‘distribution of target among various tissues or organs in human’, ‘similarity profile of target to human proteins outside its family’, ‘involvements of target in well-established life-essential pathways’ and two ‘network properties of target measured by PPIs’) from 89 successful targets. These features were reported to denote the difference between the targets of rapid and slow clinical progression processes. In the TTD 2024, all those ‘essential features’ of target druggability were collected and significantly extended to 426 successful, 1014 clinical trial, 212 preclinical/patented, and 1479 literature-reported therapeutic targets. All in all, the valuable data on target druggability provided in TTD 2024 (as described in Table 2) together with the future updates of established databases were essential in facilitating the explorations of the druggability characteristics of targets for guiding target and drug discovery.
Table 2.
Three major contents and their corresponding statistics integrated to this version of TTD, which included: target druggability illustrated by molecular interactions or regulations, characterized by different human system features and reflected by diverse cell-based expression variations
| Target druggability illustrated by molecular interactions or regulations | |||||
| ☆ Ligand-specific spatial structures of target in its drug binding pocket | |||||
| No. of targets with drug binding sites information | No. of ligands | No. of structures | |||
| Successful | Clinical trial | Preclinical/patent | Literature-reported | ||
| 319 | 427 | 116 | 375 | 16373 | 22431 |
| ☆ Network properties of target measured by protein–protein interactions | |||||
| No. of targets with protein–protein interaction information | No. of Interacting Protein | No. of protein–protein interactions | |||
| Successful | Clinical trial | Preclinical/patent | Literature-reported | ||
| 426 | 727 | 143 | 867 | 9309 | 52713 |
| ☆ Bidirectional regulations between microbiota and therapeutic agents | |||||
| No. of microbe(s) affecting the metabolism of drugs | No. of drugs | No. of microbe and drug interactions | |||
| Order | Family | Genus | Species | ||
| 59 | 101 | 135 | 194 | 663 | 9812 |
| Target druggability characterized by different human system features | |||||
| ☆ Similarity profile of target to human proteins outside its amily | |||||
| No. of targets with human similarity proteins outside the target families | No. of similarity proteins | No. of protein families | |||
| Successful | Clinical trial | Preclinical/patent | Literature-reported | ||
| 389 | 933 | 204 | 1479 | 3128 | 1004 |
| ☆ Involvements of target in the well-established life-essential pathways | |||||
| No. of targets with affiliated life-essential pathways information | No. of life-essential pathways | No. of targets with only one pathway | |||
| Successful | Clinical trial | Preclinical/patent | Literature-reported | ||
| 373 | 897 | 196 | 1415 | 241 | 679 |
| ☆ Distributions of target among a variety of tissues or organs in human | |||||
| No. of targets with human tissues or organs distribution information | No. of tissues/organs | No. of experimental samples | |||
| Successful | Clinical trial | Preclinical/patent | Literature-reported | ||
| 338 | 600 | 143 | 920 | 32 | 201 |
| Target druggability reflected by diverse cell-based expression variations | |||||
| ☆ Varied expressions of target across different cells of diverse diseases | |||||
| No. of targets with varied expressions across different cell types | No. of cell types | No. of disease classes | |||
| Successful | Clinical trial | Preclinical/patent | Literature-reported | ||
| 347 | 939 | 188 | 1371 | 1742 | 121 |
| ☆ Differential expressions of target induced by many exogenous causes | |||||
| No. of targets with differential expressions induced by exogenous causes | No. of cell types | No. of exogenous causes | |||
| Successful | Clinical trial | Preclinical/patent | Literature-reported | ||
| 357 | 926 | 197 | 1382 | 313 | 625 |
| ☆ Modified expressions of target altered by human endogenous factors | |||||
| No. of targets with modified expressions altered by endogenous factors | No. of cell types | No. of endogenous factors | |||
| Successful | Clinical trial | Preclinical/patent | Literature-reported | ||
| 352 | 934 | 192 | 1363 | 198 | 447 |
TTD has been committing to provide comprehensive data on therapeutic targets to facilitate new drug and target discovery. Since the beginning of the 21st century, it has undergone many updates (16,87–89) and accumulated worldwide impacts during the past twenty years. Particularly, there were many online tools that adopted TTD data for server development. Some of them used TTD data to establish servers facilitating drug repurposing, such as LigAdvisor (90) & DrugRep (91); discovery of drug/target, such as CoVex (92) & DeepCancerMap (93); prediction of adverse drug reaction/synergistic combination, such as MEDICASCY (94) & H-RACS (95); compound-based functional enrichments, such as MBROLE3 (96) & MMEASE (97). Moreover, TTD information has also been adopted by recent studies to promote various scientific discoveries. Some identified the association between genetic variant and disease (98–103); some others revealed the molecular characteristics crucial in virus infection (104), target variability key in determining drug response (62), and target promising in discovering antifungal therapy (105). With the rapid progression of modern technology and concerted effort in current drug discovery, the wealth of data amassed in TTD and other databases (11–20) over the past decades collectively established solid foundations for the identification of novel targets and the discovery of new therapeutics (98–100).
Contributor Information
Ying Zhou, College of Pharmaceutical Sciences, The Second Affiliated Hospital, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou 310058, China; National Key Laboratory of Diagnosis and Treatment of Severe Infectious Disease, National Clinical Research Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Zhejiang Provincial Key Laboratory for Drug Clinical Research and Evaluation, The First Affiliated Hospital, Zhejiang University, Hangzhou, Zhejiang 310000, China; Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University, Alibaba-Zhejiang University Joint Research Center of Future Digital Healthcare, Hangzhou 330110, China.
Yintao Zhang, College of Pharmaceutical Sciences, The Second Affiliated Hospital, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou 310058, China.
Donghai Zhao, College of Pharmaceutical Sciences, The Second Affiliated Hospital, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou 310058, China.
Xinyuan Yu, College of Pharmaceutical Sciences, The Second Affiliated Hospital, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou 310058, China.
Xinyi Shen, Department of Environmental Health Sciences, Yale School of Public Health, Yale University, New Haven 06510, USA.
Yuan Zhou, College of Pharmaceutical Sciences, The Second Affiliated Hospital, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou 310058, China.
Shanshan Wang, Qian Xuesen Collaborative Research Center of Astrochemistry and Space Life Sciences, Institute of Drug Discovery Technology, Ningbo University, Ningbo 315211, China.
Yunqing Qiu, College of Pharmaceutical Sciences, The Second Affiliated Hospital, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou 310058, China; National Key Laboratory of Diagnosis and Treatment of Severe Infectious Disease, National Clinical Research Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Zhejiang Provincial Key Laboratory for Drug Clinical Research and Evaluation, The First Affiliated Hospital, Zhejiang University, Hangzhou, Zhejiang 310000, China.
Yuzong Chen, State Key Laboratory of Chemical Oncogenomics, Key Laboratory of Chemical Biology, The Graduate School at Shenzhen, Tsinghua University, Shenzhen 518055, China; Institute of Biomedical Health Technology and Engineering, Shenzhen Bay Laboratory, Shenzhen 518000, China.
Feng Zhu, College of Pharmaceutical Sciences, The Second Affiliated Hospital, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou 310058, China; Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University, Alibaba-Zhejiang University Joint Research Center of Future Digital Healthcare, Hangzhou 330110, China.
Data availability
TTD is freely accessible to all users without any login requirement at: https://idrblab.org/ttd/.
Funding
Natural Science Foundation of Zhejiang Province [LR21H300001]; National Key R&D Program of China Synthetic Biology Research [2019YFA0905900]; National Natural Science Foundation of China [82373790, 22220102001, U1909208, 81872798, 81971982]; Scientific Research Grant of Ningbo University [215-432000282]; Ningbo Top Talent Project [215-432094250]; National Key R&D Program of China [2022YFC3400501]; Leading Talent of ‘Ten Thousand Plan’ National High-Level Talents Special Support Plans of China; Westlake Laboratory (Westlake Laboratory of Life Sciences and Biomedicine); Fundamental Research Funds for Central Universities [2018QNA7023]; ‘Double Top-Class’ University Projects [181201*194232101]; Key R&D Programs of Zhejiang Province [2020C03010]; Alibaba-Zhejiang University Joint Research Center of Future Digital Healthcare; Alibaba Cloud; The information technology center of Zhejiang University. Funding for open access charge: Natural Science Foundation of Zhejiang Province [LR21H300001].
Conflict of interest statement. None declared.
References
- 1. Emmerich C.H., Gamboa L.M., Hofmann M.C.J., Bonin-Andresen M., Arbach O., Schendel P., Gerlach B., Hempel K., Bespalov A., Dirnagl U.et al.. Improving target assessment in biomedical research: the GOT-IT recommendations. Nat. Rev. Drug Discov. 2021; 20:64–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Solier S., Muller S., Caneque T., Versini A., Mansart A., Sindikubwabo F., Baron L., Emam L., Gestraud P., Pantos G.D.et al.. A druggable copper-signalling pathway that drives inflammation. Nature. 2023; 617:386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stanford S.M., Bottini N.. Targeting protein phosphatases in cancer immunotherapy and autoimmune disorders. Nat. Rev. Drug Discov. 2023; 22:273–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hagemann S., Misiak D., Bell J.L., Fuchs T., Lederer M.I., Bley N., Hammerle M., Ghazy E., Sippl W., Schulte J.H.et al.. IGF2BP1 induces neuroblastoma via a druggable feedforward loop with MYCN promoting 17q oncogene expression. Mol. Cancer. 2023; 22:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Padroni G., Bikaki M., Novakovic M., Wolter A.C., Rudisser S.H., Gossert A.D., Leitner A., Allain F.H.. A hybrid structure determination approach to investigate the druggability of the nucleocapsid protein of SARS-CoV-2. Nucleic Acids Res. 2023; 51:4555–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jiang J., Yu Y.. Pharmacologically targeting transient receptor potential channels for seizures and epilepsy: emerging preclinical evidence of druggability. Pharmacol. Ther. 2023; 244:108384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sutkeviciute I., Lee J.Y., White A.D., Maria C.S., Pena K.A., Savransky S., Doruker P., Li H., Lei S., Kaynak B.et al.. Precise druggability of the PTH type 1 receptor. Nat. Chem. Biol. 2022; 18:272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kozlovskii I., Popov P.. Spatiotemporal identification of druggable binding sites using deep learning. Commun. Biol. 2020; 3:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y.H., Li X.X., Hong J.J., Wang Y.X., Fu J.B., Yang H., Yu C.Y., Li F.C., Hu J., Xue W.W.et al.. Clinical trials, progression-speed differentiating features and swiftness rule of the innovative targets of first-in-class drugs. Brief. Bioinform. 2020; 21:649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fa B., Wei T., Zhou Y., Johnston L., Yuan X., Ma Y., Zhang Y., Yu Z.. GapClust is a light-weight approach distinguishing rare cells from voluminous single cell expression profiles. Nat. Commun. 2021; 12:4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z.et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Avram S., Bologa C.G., Holmes J., Bocci G., Wilson T.B., Nguyen D.T., Curpan R., Halip L., Bora A., Yang J.J.et al.. DrugCentral 2021 supports drug discovery and repositioning. Nucleic Acids Res. 2021; 49:D1160–D1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Siramshetty V.B., Eckert O.A., Gohlke B.O., Goede A., Chen Q., Devarakonda P., Preissner S., Preissner R.. SuperDRUG2: a one stop resource for approved/marketed drugs. Nucleic Acids Res. 2018; 46:D1137–D1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li F., Yin J., Lu M., Mou M., Li Z., Zeng Z., Tan Y., Wang S., Chu X., Dai H.et al.. DrugMAP: molecular atlas and pharma-information of all drugs. Nucleic Acids Res. 2023; 51:D1288–D1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun X., Zhang Y., Li H., Zhou Y., Shi S., Chen Z., He X., Zhang H., Li F., Yin J.et al.. DRESIS: the first comprehensive landscape of drug resistance information. Nucleic Acids Res. 2023; 51:D1263–D1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou Y., Zhang Y., Lian X., Li F., Wang C., Zhu F., Qiu Y., Chen Y.. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022; 50:D1398–D1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ochoa D., Hercules A., Carmona M., Suveges D., Baker J., Malangone C., Lopez I., Miranda A., Cruz-Castillo C., Fumis L.et al.. The next-generation open targets platform: reimagined, redesigned, rebuilt. Nucleic Acids Res. 2023; 51:D1353–D1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem 2023 update. Nucleic Acids Res. 2023; 51:D1373–D1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Felix E., Magarinos M.P., Mosquera J.F., Mutowo P., Nowotka M.et al.. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019; 47:D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J.. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016; 44:D1045–D1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tinivella A., Nwachukwu J.C., Angeli A., Foschi F., Benatti A.L., Pinzi L., Izard T., Ferraroni M., Erumbi R., Christodoulou M.S.et al.. Design, synthesis, biological evaluation and crystal structure determination of dual modulators of carbonic anhydrases and estrogen receptors. Eur. J. Med. Chem. 2023; 246:115011. [DOI] [PubMed] [Google Scholar]
- 22. Pinzi L., Tinivella A., Rastelli G.. Chemoinformatics analyses of tau ligands reveal key molecular requirements for the identification of potential drug candidates against tauopathies. Molecules. 2021; 26:5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pinzi L., Rastelli G.. Identification of target associations for polypharmacology from analysis of crystallographic ligands of the protein data bank. J. Chem. Inf. Model. 2020; 60:372–390. [DOI] [PubMed] [Google Scholar]
- 24. Yin J., Li F., Zhou Y., Mou M., Lu Y., Chen K., Xue J., Luo Y., Fu J., He X.et al.. INTEDE: interactome of drug-metabolizing enzymes. Nucleic Acids Res. 2021; 49:D1233–D1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fu T., Li F., Zhang Y., Yin J., Qiu W., Li X., Liu X., Xin W., Wang C., Yu L.et al.. VARIDT 2.0: structural variability of drug transporter. Nucleic Acids Res. 2022; 50:D1417–D1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jakubec D., Skoda P., Krivak R., Novotny M., Hoksza D. PrankWeb 3: accelerated ligand-binding site predictions for experimental and modelled protein structures. Nucleic Acids Res. 2022; 50:W593–W597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Attwood M.M., Fabbro D., Sokolov A.V., Knapp S., Schioth H.B.. Trends in kinase drug discovery: targets, indications and inhibitor design. Nat. Rev. Drug Discov. 2021; 20:839–861. [DOI] [PubMed] [Google Scholar]
- 28. Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chao H., Chen L., Craig P.A., Crichlow G.V., Dalenberg K., Duarte J.M.et al.. RCSB protein data bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023; 51:D488–D508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cock P.J., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B.et al.. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009; 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krapp L.F., Abriata L.A., Cortes Rodriguez F., Dal Peraro M.. PeSTo: parameter-free geometric deep learning for accurate prediction of protein binding interfaces. Nat. Commun. 2023; 14:2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang J., Youkharibache P., Zhang D., Lanczycki C.J., Geer R.C., Madej T., Phan L., Ward M., Lu S., Marchler G.H.et al.. iCn3D, a web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures. Bioinformatics. 2020; 36:131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. You Y., Lai X., Pan Y., Zheng H., Vera J., Liu S., Deng S., Zhang L.. Artificial intelligence in cancer target identification and drug discovery. Signal Transduct. Target Ther. 2022; 7:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li X.X., Yin J., Tang J., Li Y., Yang Q., Xiao Z., Zhang R., Wang Y., Hong J., Tao L.et al.. Determining the balance between drug efficacy and safety by the network and biological system profile of its therapeutic target. Front. Pharmacol. 2018; 9:1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Muslu O., Hoyt C.T., Lacerda M., Hofmann-Apitius M., Frohlich H.. GuiltyTargets: prioritization of novel therapeutic targets with network representation learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022; 19:491–500. [DOI] [PubMed] [Google Scholar]
- 35. Conte F., Paci P.. Alzheimer's disease: insights from a network medicine perspective. Sci. Rep. 2022; 12:16846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pati S.K., Gupta M.K., Banerjee A., Mallik S., Zhao Z.. PPIGCF: a protein-protein interaction-based gene correlation filter for optimal gene selection. Genes. 2023; 14:1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paci P., Fiscon G.. SWIMmeR: an R-based software to unveiling crucial nodes in complex biological networks. Bioinformatics. 2022; 38:586–588. [DOI] [PubMed] [Google Scholar]
- 38. Murakami Y., Tripathi L.P., Prathipati P., Mizuguchi K.. Network analysis and in silico prediction of protein-protein interactions with applications in drug discovery. Curr. Opin. Struct. Biol. 2017; 44:134–142. [DOI] [PubMed] [Google Scholar]
- 39. Szklarczyk D., Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S.et al.. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023; 51:D638–D646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang P., Tao L., Zeng X., Qin C., Chen S.Y., Zhu F., Yang S.Y., Li Z.R., Chen W.P., Chen Y.Z.. PROFEAT update: a protein features web server with added facility to compute network descriptors for studying omics-derived networks. J. Mol. Biol. 2017; 429:416–425. [DOI] [PubMed] [Google Scholar]
- 41. de Vos W.M., Tilg H., Van Hul M., Cani P.D.. Gut microbiome and health: mechanistic insights. Gut. 2022; 71:1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lindell A.E., Zimmermann-Kogadeeva M., Patil K.R.. Multimodal interactions of drugs, natural compounds and pollutants with the gut microbiota. Nat. Rev. Micro. 2022; 20:431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chrysostomou D., Roberts L.A., Marchesi J.R., Kinross J.M.. Gut microbiota modulation of efficacy and toxicity of cancer chemotherapy and immunotherapy. Gastroenterology. 2023; 164:198–213. [DOI] [PubMed] [Google Scholar]
- 44. Zhang J., Zhang J., Wang R.. Gut microbiota modulates drug pharmacokinetics. Drug Metab. Rev. 2018; 50:357–368. [DOI] [PubMed] [Google Scholar]
- 45. Savage N. The complex relationship between drugs and the microbiome. Nature. 2020; 577:10–11. [DOI] [PubMed] [Google Scholar]
- 46. Tomofuji Y., Maeda Y., Oguro-Igashira E., Kishikawa T., Yamamoto K., Sonehara K., Motooka D., Matsumoto Y., Matsuoka H., Yoshimura M.et al.. Metagenome-wide association study revealed disease-specific landscape of the gut microbiome of systemic lupus erythematosus in Japanese. Ann. Rheum. Dis. 2021; 80:1575–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Connor R., Funk K., Kelly C., Kim S.et al.. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022; 50:D20–D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zheng C.J., Han L.Y., Yap C.W., Ji Z.L., Cao Z.W., Chen Y.Z.. Therapeutic targets: progress of their exploration and investigation of their characteristics. Pharmacol. Rev. 2006; 58:259–279. [DOI] [PubMed] [Google Scholar]
- 49. UniProt C. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res. 2023; 51:D523–D531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paysan-Lafosse T., Blum M., Chuguransky S., Grego T., Pinto B.L., Salazar G.A., Bileschi M.L., Bork P., Bridge A., Colwell L.et al.. InterPro in 2022. Nucleic Acids Res. 2023; 51:D418–D427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Camacho C., Boratyn G.M., Joukov V., Vera Alvarez R., Madden T.L.. ElasticBLAST: accelerating sequence search via cloud computing. BMC Bioinf. 2023; 24:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., Madden T.L., Matten W.T., McGinnis S.D., Merezhuk Y.et al.. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013; 41:W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 54. Wright S.C., Lauschke V.M.. Rewiring of catecholamine-induced calcium signalling is an early event in non-alcoholic fatty liver disease. J. Physiol. 2023; 601:1317–1318. [DOI] [PubMed] [Google Scholar]
- 55. Kanehisa M., Furumichi M., Sato Y., Kawashima M., Ishiguro-Watanabe M.. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023; 51:D587–D592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou Y., Nevosadova L., Eliasson E., Lauschke V.M.. Global distribution of functionally important CYP2C9 alleles and their inferred metabolic consequences. Hum. Genomics. 2023; 17:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jiang L., Wang M., Lin S., Jian R., Li X., Chan J., Dong G., Fang H., Robinson A.E., Snyder M.P.. A quantitative proteome map of the human body. Cell. 2020; 183:269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ben-David U., Siranosian B., Ha G., Tang H., Oren Y., Hinohara K., Strathdee C.A., Dempster J., Lyons N.J., Burns R.et al.. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018; 560:325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen J., Bell J., Lau B.T., Whittaker T., Stapleton D., Ji H.P.. A functional CRISPR/Cas9 screen identifies kinases that modulate FGFR inhibitor response in gastric cancer. Oncogenesis. 2019; 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yu K., Chen B., Aran D., Charalel J., Yau C., Wolf D.M., van ’t Veer L.J., Butte A.J., Goldstein T., Sirota M.. Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types. Nat. Commun. 2019; 10:3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jia P., Hu R., Zhao Z.. Benchmark of embedding-based methods for accurate and transferable prediction of drug response. Brief. Bioinform. 2023; 24:bbad098. [DOI] [PubMed] [Google Scholar]
- 62. Zhou Y., Arribas G.H., Turku A., Jurgenson T., Mkrtchian S., Krebs K., Wang Y., Svobodova B., Milani L., Schulte G.et al.. Rare genetic variability in human drug target genes modulates drug response and can guide precision medicine. Sci. Adv. 2021; 7:eabi6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fu J., Zhang Y., Wang Y., Zhang H., Liu J., Tang J., Yang Q., Sun H., Qiu W., Ma Y.et al.. Optimization of metabolomic data processing using NOREVA. Nat. Protoc. 2022; 17:129–151. [DOI] [PubMed] [Google Scholar]
- 64. Zhang Y., Sun H., Lian X., Tang J., Zhu F.. ANPELA: significantly enhanced quantification tool for cytometry-based single-cell proteomics. Adv. Sci. (Weinh). 2023; 10:e2207061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Papatheodorou I., Moreno P., Manning J., Fuentes A.M., George N., Fexova S., Fonseca N.A., Fullgrabe A., Green M., Huang N.et al.. Expression atlas update: from tissues to single cells. Nucleic Acids Res. 2020; 48:D77–D83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Carvalho B.S., Irizarry R.A.. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010; 26:2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K.et al.. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017; 171:1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhu J., Wang J., Wang X., Gao M., Guo B., Gao M., Liu J., Yu Y., Wang L., Kong W.et al.. Prediction of drug efficacy from transcriptional profiles with deep learning. Nat. Biotechnol. 2021; 39:1444–1452. [DOI] [PubMed] [Google Scholar]
- 71. Raschka T., Sood M., Schultz B., Altay A., Ebeling C., Frohlich H.. AI reveals insights into link between CD33 and cognitive impairment in Alzheimer's Disease. PLoS Comput. Biol. 2023; 19:e1009894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li F., Zhou Y., Zhang Y., Yin J., Qiu Y., Gao J., Zhu F.. POSREG: proteomic signature discovered by simultaneously optimizing its reproducibility and generalizability. Brief Bioinform. 2022; 23:bbac040. [DOI] [PubMed] [Google Scholar]
- 73. Yang Q., Wang Y., Zhang Y., Li F., Xia W., Zhou Y., Qiu Y., Li H., Zhu F.. NOREVA: enhanced normalization and evaluation of time-course and multi-class metabolomic data. Nucleic Acids Res. 2020; 48:W436–W448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Niepel M., Hafner M., Duan Q., Wang Z., Paull E.O., Chung M., Lu X., Stuart J.M., Golub T.R., Subramanian A.et al.. Common and cell-type specific responses to anti-cancer drugs revealed by high throughput transcript profiling. Nat. Commun. 2017; 8:1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pan S., Liu X., Liu T., Zhao Z., Dai Y., Wang Y.Y., Jia P., Liu F.. Causal inference of genetic variants and genes in amyotrophic lateral sclerosis. Front. Genet. 2022; 13:917142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yang Q., Li B., Tang J., Cui X., Wang Y., Li X., Hu J., Chen Y., Xue W., Lou Y.et al.. Consistent gene signature of schizophrenia identified by a novel feature selection strategy from comprehensive sets of transcriptomic data. Brief. Bioinform. 2020; 21:1058–1068. [DOI] [PubMed] [Google Scholar]
- 77. Yang Q., Gong Y., Zhu F.. Critical assessment of the biomarker discovery and classification methods for multiclass metabolomics. Anal. Chem. 2023; 95:5542–5552. [DOI] [PubMed] [Google Scholar]
- 78. Xiao Y., Gong Y., Lv Y., Lan Y., Hu J., Li F., Xu J., Bai J., Deng Y., Liu L.et al.. Gene perturbation atlas (GPA): a single-gene perturbation repository for characterizing functional mechanisms of coding and non-coding genes. Sci. Rep. 2015; 5:10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Paci P., Fiscon G., Conte F., Wang R.S., Farina L., Loscalzo J.. Gene co-expression in the interactome: moving from correlation toward causation via an integrated approach to disease module discovery. NPJ Syst. Biol. Appl. 2021; 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Konuma T., Ogawa K., Okada Y.. Integration of genetically regulated gene expression and pharmacological library provides therapeutic drug candidates. Hum. Mol. Genet. 2021; 30:294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tang J., Fu J., Wang Y., Li B., Li Y., Yang Q., Cui X., Hong J., Li X., Chen Y.et al.. ANPELA: analysis and performance assessment of the label-free quantification workflow for metaproteomic studies. Brief Bioinform. 2020; 21:621–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mullard A. 2022 FDA approvals. Nat. Rev. Drug Discov. 2023; 22:83–88. [DOI] [PubMed] [Google Scholar]
- 83. Mullard A. 2021 FDA approvals. Nat. Rev. Drug Discov. 2022; 21:83–88. [DOI] [PubMed] [Google Scholar]
- 84. Zhu F., Shi Z., Qin C., Tao L., Liu X., Xu F., Zhang L., Song Y., Liu X., Zhang J.et al.. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012; 40:D1128–D1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Meissner F., Geddes-McAlister J., Mann M., Bantscheff M.. The emerging role of mass spectrometry-based proteomics in drug discovery. Nat. Rev. Drug Discov. 2022; 21:637–654. [DOI] [PubMed] [Google Scholar]
- 86. Jones L.H., Bunnage M.E.. Applications of chemogenomic library screening in drug discovery. Nat. Rev. Drug Discov. 2017; 16:285–296. [DOI] [PubMed] [Google Scholar]
- 87. Wang Y., Zhang S., Li F., Zhou Y., Zhang Y., Wang Z., Zhang R., Zhu J., Ren Y., Tan Y.et al.. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020; 48:D1031–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li Y.H., Yu C.Y., Li X.X., Zhang P., Tang J., Yang Q., Fu T., Zhang X., Cui X., Tu G.et al.. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018; 46:D1121–D1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Yang H., Qin C., Li Y.H., Tao L., Zhou J., Yu C.Y., Xu F., Chen Z., Zhu F., Chen Y.Z.. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016; 44:D1069–D1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pinzi L., Tinivella A., Gagliardelli L., Beneventano D., Rastelli G.. LigAdvisor: a versatile and user-friendly web-platform for drug design. Nucleic Acids Res. 2021; 49:W326–W335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gan J.H., Liu J.X., Liu Y., Chen S.W., Dai W.T., Xiao Z.X., Cao Y.. DrugRep: an automatic virtual screening server for drug repurposing. Acta Pharmacol. Sin. 2023; 44:888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sadegh S., Matschinske J., Blumenthal D.B., Galindez G., Kacprowski T., List M., Nasirigerdeh R., Oubounyt M., Pichlmair A., Rose T.D.et al.. Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nat. Commun. 2020; 11:3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wu J., Xiao Y., Lin M., Cai H., Zhao D., Li Y., Luo H., Tang C., Wang L.. DeepCancerMap: a versatile deep learning platform for target- and cell-based anticancer drug discovery. Eur. J. Med. Chem. 2023; 255:115401. [DOI] [PubMed] [Google Scholar]
- 94. Zhou H., Cao H., Matyunina L., Shelby M., Cassels L., McDonald J.F., Skolnick J.. MEDICASCY: a machine learning approach for predicting small-molecule drug side effects, indications, efficacy, and modes of action. Mol. Pharm. 2020; 17:1558–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yan X., Yang Y., Chen Z., Yin Z., Deng Z., Qiu T., Tang K., Cao Z.. H-RACS: a handy tool to rank anti-cancer synergistic drugs. Aging. 2020; 12:21504–21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Lopez-Ibanez J., Pazos F., Chagoyen M.. MBROLE3: improved functional enrichment of chemical compounds for metabolomics data analysis. Nucleic Acids Res. 2023; 51:W305–W309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yang Q., Li B., Chen S., Tang J., Li Y., Li Y., Zhang S., Shi C., Zhang Y., Mou M.et al.. MMEASE: online meta-analysis of metabolomic data by enhanced metabolite annotation, marker selection and enrichment analysis. J. Proteomics. 2021; 232:104023. [DOI] [PubMed] [Google Scholar]
- 98. Sun B.B., Kurki M.I., Foley C.N., Mechakra A., Chen C.Y., Marshall E., Wilk J.B., Biogen Biobank T., Chahine M., Chevalier P.et al.. Genetic associations of protein-coding variants in human disease. Nature. 2022; 603:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ferkingstad E., Sulem P., Atlason B.A., Sveinbjornsson G., Magnusson M.I., Styrmisdottir E.L., Gunnarsdottir K., Helgason A., Oddsson A., Halldorsson B.V.et al.. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021; 53:1712–1721. [DOI] [PubMed] [Google Scholar]
- 100. Shirai Y., Nakanishi Y., Suzuki A., Konaka H., Nishikawa R., Sonehara K., Namba S., Tanaka H., Masuda T., Yaga M.et al.. Multi-trait and cross-population genome-wide association studies across autoimmune and allergic diseases identify shared and distinct genetic component. Ann. Rheum. Dis. 2022; 81:1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kanoni S., Graham S.E., Wang Y., Surakka I., Ramdas S., Zhu X., Clarke S.L., Bhatti K.F., Vedantam S., Winkler T.W.et al.. Implicating genes, pleiotropy, and sexual dimorphism at blood lipid loci through multi-ancestry meta-analysis. Genome Biol. 2022; 23:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Surapaneni A., Schlosser P., Zhou L., Liu C., Chatterjee N., Arking D.E., Dutta D., Coresh J., Rhee E.P., Grams M.E.. Identification of 969 protein quantitative trait loci in an African American population with kidney disease attributed to hypertension. Kidney Int. 2022; 102:1167–1177. [DOI] [PubMed] [Google Scholar]
- 103. Wang X., Li F., Qiu W., Xu B., Li Y., Lian X., Yu H., Zhang Z., Wang J., Li Z.et al.. SYNBIP: synthetic binding proteins for research, diagnosis and therapy. Nucleic Acids Res. 2022; 50:D560–D570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Grodzki M., Bluhm A.P., Schaefer M., Tagmount A., Russo M., Sobh A., Rafiee R., Vulpe C.D., Karst S.M., Norris M.H.. Genome-scale CRISPR screens identify host factors that promote human coronavirus infection. Genome Med. 2022; 14:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fu C., Zhang X., Veri A.O., Iyer K.R., Lash E., Xue A., Yan H., Revie N.M., Wong C., Lin Z.Y.et al.. Leveraging machine learning essentiality predictions and chemogenomic interactions to identify antifungal targets. Nat. Commun. 2021; 12:6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
TTD is freely accessible to all users without any login requirement at: https://idrblab.org/ttd/.