Abstract
One challenge in the development of novel drugs is their interaction with potential off-targets, which can cause unintended side-effects, that can lead to the subsequent withdrawal of approved drugs. At the same time, these off-targets may also present a chance for the repositioning of withdrawn drugs for new indications, which are potentially rare or more severe than the original indication and where certain adverse reactions may be avoidable or tolerable. To enable further insights into this topic, we updated our database Withdrawn by adding pharmacovigilance data from the FDA Adverse Event Reporting System (FAERS), as well as mechanism of action and human disease pathway prediction features for drugs that are or were temporarily withdrawn or discontinued in at least one country. As withdrawal data are still spread over dozens of national websites, we are continuously updating our lists of discontinued or withdrawn drugs and related (off-)targets. Furthermore, new systematic entry points for browsing the data, such as an ATC tree, were added, increasing the accessibility of the database in a user-friendly way. Withdrawn 2.0 is publicly available without the need for registration or login at https://bioinformatics.charite.de/withdrawn_3/index.php.
Graphical Abstract
Graphical Abstract.
Introduction
The process of drug discovery has consistently been marked by a quest for efficacy, yet the drug discovery pipeline often experiences setbacks due to unanticipated adverse drug reactions (ADRs), and a significant amount of effort goes into their prediction and analysis (1). While some of these reactions can indeed be attributed to the modulation of primary therapeutic targets, unintended off-target interactions pose a significant challenge, often culminating in market withdrawals or the introduction of stringent black-box warnings for prescription drugs. One classical instance is the modulation of the hERG channel, which, when unintentionally inhibited, led to the withdrawal of the antihistaminic drug terfenadine due to severe arrhythmias. Such cases have historically resulted in the financial onus on pharmaceutical industries, with a significant number of new chemical entities (NCEs) facing recalls after regulatory approval, as highlighted in our earlier paper (2).
However, amid these challenges lies a potential silver lining. The very off-target interactions leading to ADRs might also present opportunities for drug repositioning. Drugs withdrawn for certain adverse effects in one context might find renewed applications in more severe indications, where the therapeutic benefits might outweigh the risks, potentially making re-approval feasible. This avenue not only offers hope for drug repurposing but also opens a cost-effective and time-saving alternative compared to the de novo development of novel drugs (3). Especially for rare and neglected conditions that might otherwise not justify the research cost, finding novel therapeutic interactions for known drugs, including withdrawn, and abandoned candidates, may provide a hopeful path (4).
With the evolving drug discovery landscape and the increasing emphasis on repositioning, we present an updated version of the WITHDRAWN database. This novel iteration explores the potential new uses for withdrawn drugs, with a sharp focus on understanding the side effects and the underlying reasons for withdrawal. Extending our earlier focus on withdrawal reasons and potential drug toxicity, the current version now integrates data from the FDA Adverse Event Reporting System (FAERS, https://open.fda.gov/data/faers/), offering a comprehensive perspective on ADRs (5). FAERS provides a vast corpus of post-market drug safety surveillance and captures a broad spectrum of real-world scenarios. However, known challenges of pharmacovigilance systems are the possibility of under-reporting and a varying level of quality, detail and insight (6), as well as issues establishing direct cause-and-effect relationships without additional data. Nonetheless, it is a valuable supporting data source that often remains untapped, and can give significant insight regarding ADRs with proper analysis (6).
In addition to adverse effect data, the updated database is now enriched with detailed mechanisms of action and pathway features, offering further insights into possible drug repositioning avenues. Recognizing the multifaceted reasons for ADRs, from off-target interactions to genetic polymorphisms, this feature enables researchers to unravel potential new therapeutic applications for withdrawn drugs by elucidating how they might interact in the biological milieu.
To ensure a seamless user experience and to facilitate systematic analyses, the updated database now incorporates systematic entry points, such as the Anatomical Therapeutic Chemical (ATC) classification tree. This aids in categorizing drugs and understanding their broader pharmacological roles. Moreover, users can now identify drugs based on their associated targets, making the database an indispensable tool for professionals interested in drug development and repositioning.
Materials and methods
Server architecture
The database is hosted as a relational MySQL database on the Charité IT system. The back-end of the database consists of a lab-based LAMP (Linux/Apache/MySQL/PHP) server, and as the back-end language, PHP is used. The connection to the database server is established via a MySQL interface, whereas front-end data is delivered by a mixture of AJAX requests and HTML form submission responses.
For the handling of chemical information on the website, and during the preprocessing of the data and creation of the database, the Python package RDKit (http://www.rdkit.org/) was used. Website functionalities are implemented using JavaScript and its plugin jQuery (https://jquery.com/), as well as the CSS framework Bootstrap 4 (https://getbootstrap.com/). Tables on the website are displayed with the jQuery plugin DataTables (https://datatables.net/), additionally using the absolute sorting extension (https://datatables.net/plug-ins/sorting/absolute). For the chemistry interface of structural search options, the JavaScript library ChemDoodle Web components (7) was used.
As security measures, Fail2Ban (https://www.fail2ban.org/wiki/index.php/Main_Page) and an App-Firewall are activated on the server, which protect the database from possible attacks such as automated mass scans, session hijacking, MySQL injection and more by adding bans for perpetrators. Incorrectly using the website can, in rare cases, lead to the block of users by these security mechanisms. Such a block will prevent the website from loading for a short period for this specific IP address.
The use of a JavaScript-capable browser is essential for the correct functionality, and the website was tested on the most recent version of Google Chrome. No user registration or email address is necessary to access all functionalities of the webserver.
Data collection
Mechanism of action
The mechanism of action prediction for withdrawn drugs and chemical structures is based on the ChEMBL 29 database (8), which was filtered and standardized to retain only highly accurate, direct interactions between human proteins and small molecule compounds. To compare user-entered structures of interest to ChEMBL structures, Morgan fingerprints of length 128 from the Python library RDKit are used.
Pathways
Pathway evaluations are based on different data attributes, which were extracted from the ChEMBL database. Filtering for human targets was done by mapping all human UniProt-IDs (9) to the ChEMBL datasets, as well as mapping relations to different databases such as KEGG (10). Optimal binders were filtered as described in Peon et al. (11) with regard to binding strengths, confidence scores and IC50/EC50 values. The data concerning pathways for human diseases and infections and the selected human genes/pathways was derived from KEGG as well. Information about druggable genes was extracted from IDG (https://druggablegenome.net/) and further enriched by 43 additional genes from the Therapeutic Target database TTD (12), which are targets of approved small molecule drugs. Lastly, the KEGG mapper was used to derive a table containing all information for KEGG pathways related to human genes and diseases.
FAERS side effect data
To include the data from FAERS, all quarterly reports until June 2023 were downloaded and integrated as a MySQL database. Data filtering was done by only retaining reports that were submitted by medical professionals. Data cleaning steps included the deletion of report primary IDs, that were marked as ‘deleted’ in further FAERS reports. Additionally, for primary IDs that are combined with different case IDs, only one was kept, and only the last primary ID for a case ID was used. This ensures that counts of drugs or outcomes of the same case are not counted twice if there were several reports to FAERS at different times for the same adverse reaction.
Reported outcomes were matched to MedDRA (https://www.meddra.org/) lowest-level terms and further mapped to preferred terms. A case is only counted for a preferred term if at least one lowest-level term is present in the FAERS reaction report.
To map withdrawn drugs to FAERS drugs, a collection of drug names, synonyms, products, and brands was created and mapped to its ingredients (in the case of combination drugs). Synonym data was collected from TTD (single drugs and drugs identified as combinations), DrugBank (13) (drugs, synonyms, external identifier, products, mixtures, international brands), NDA (https://www.fda.gov/drugs/types-applications/new-drug-application-nda, tradenames) and a FAERS list of explicit combinations of drug names that often occur in FAERS data.
Database features
Drug search
Searching for a specific withdrawn drug is possible by either using its withdrawn ID or the drug name, which displays a selectable dropdown menu containing all relevant entries of the database.
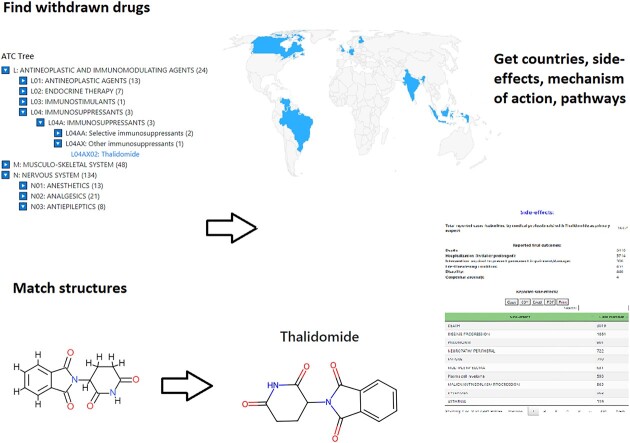
Additionally, it is possible to search for withdrawn drugs that have a similar molecular structure as a user-submitted structure of interest. These can be entered by using its PubChem (14) name or a SMILES (Simplified Molecular-Input Line-Entry System) string of the compound, or by uploading a standard molecule file. Once a valid structure is entered, it will be displayed in the ChemDoodle structure viewer (7), where it can be further modified, such as deleting or adding single atoms or substructures. Alternatively, it is also possible to draw a molecular structure completely from scratch using the provided drawing tools. The finished structure is translated to a Morgan fingerprint using RDKit and compared to all structures in the database. As a result, the ten most similar withdrawn drugs are displayed, sorted by Tanimoto similarity (15) to the query structure, with a Tanimoto similarity of 1 indicating identical structures.
Searching for withdrawn drugs that contain a specific substructure works completely analogous, with the difference that only a small substructure of interest is entered, and the result page displays all withdrawn drugs that contain the specific substructure.
Browse by name/ATC code
In addition to searching for specific drugs or molecular structures, it is also possible to gain a general overview of the Withdrawn database by browsing the contained drugs, either according to their name or their assigned code from the Anatomical Therapeutic Chemical (ATC) classification system (16). Since its first publication in 1976, it has been continuously updated to account for new findings and is still prevalently used to classify approved drugs according to their anatomical, therapeutic, pharmacological, and chemical properties. The five hierarchical levels contain groups of drugs with similar properties, and the ATC tree of all withdrawn drugs in the database can be browsed to gain insights into broader pharmacological categories.
Target search
As an alternative to starting from a drug/molecular compound, the option to search for targets of withdrawn drugs was newly implemented. By entering (part of) a target name, a substring search is performed over all targets of withdrawn drugs in the database, and all human protein targets that fit the required query are reported back, along with their assigned withdrawn drugs.
Mechanism of action
A mechanism of action prediction can be done for both withdrawn drugs contained in the database, and user-entered molecular structures, which can be submitted as described for the drug search option. For the submitted structures, Morgan fingerprints are calculated and compared to the fingerprints of compounds from the ChEMBL database. Subsequently, the five most similar small-molecule compounds (according to the Tanimoto coefficient) are displayed, along with their filtered interactions with human protein targets and UniProt information.
Pathways
To give insights into the involvement of withdrawn drugs in human disease pathways, a pathway enrichment analysis was performed, evaluating the number of distinct druggable targets on KEGG disease pathways. Additionally, the number of distinct druggable KEGG-IDs for each molecule, the number of distinct druggable KEGG-IDs for each pathway, and the number of distinct druggable KEGG-IDs appearing in any disease-related pathway were assessed and used to evaluate the probability of a specific compound to act on the evaluated pathways.
Entering a query structure finds all structurally similar ChEMBL compounds (ranked by the Tanimoto coefficient) that act on human disease pathways, along with the P- and e-values, which were calculated using the cumulative hypergeometric distribution. The P-value represents the probability that one has at least as many interactions within the given pathways only by chance. This way, a binding within a pathway containing only two druggable genes is ranked up, compared to a few bindings within pathways with a lot of druggable genes, and a single binding in one pathway of a compound with a lot of interactions is ranked down. The e-value gives the probability that an interaction will be found within a pathway by chance if you search within several pathways, for which the total number of pathways with druggable targets is taken into account.
Use case
An interesting case of a withdrawn drug that has gotten re-approved for other indications is Thalidomide. It was originally approved as a sedative drug and thought to be virtually non-toxic (17). Only a few years after its launch in Germany under the brand name Contergan, it was connected with a series of birth deformities in children of women who had taken the drug during their pregnancy, and subsequently withdrawn from the market in the 1960s (18). Three decades later, it was approved again for the much more serious indications of Erythema nodosum leprosum (ENL), and in 2006 for the treatment of patients suffering from multiple myeloma, a form of blood cancer (19).
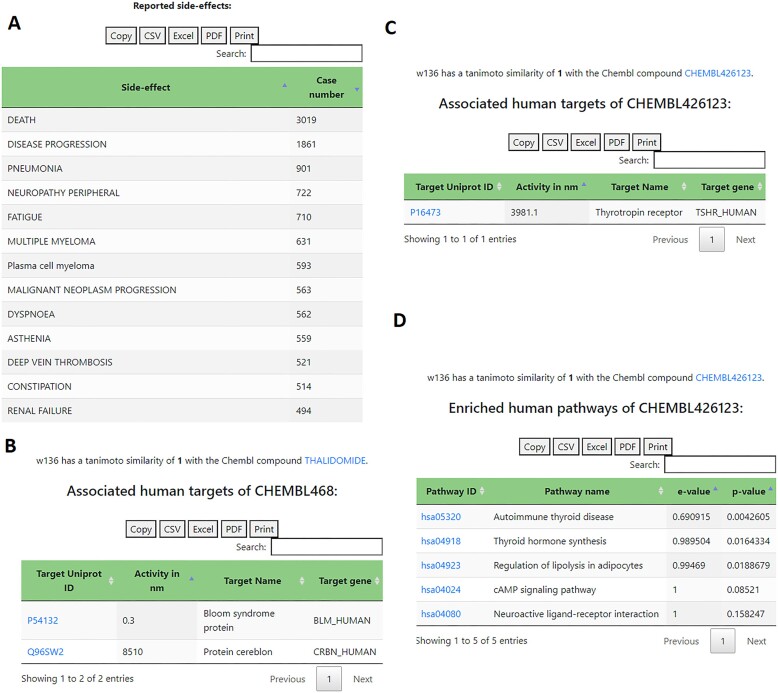
Due to its comparatively recent re-approval, an extensive amount of FAERS reports is available, showing a high number of adverse reactions that were unknown at the time of its original approval but became apparent with ongoing treatments, such as peripheral neuropathy (20–22) and thrombosis (23) (Figure 1A).
Figure 1.
Example outputs for Thalidomide (Withdrawn ID w136). (A) Adverse effects extracted from the FAERS database; (B) extracted and filtered human ChEBML targets of Thalidomide (B) and its S-enantiomer (C) and reported human disease pathways (D) of Thalidomide S-enantiomer, obtained by enriched pathway analysis using KEGG pathways and druggable targets.
A mechanism of action analysis shows Bloom syndrome protein as the most active human protein target of Thalidomide from the filtered ChEMBL data (Figure 1B), which was recently discovered to potentially play a role in the treatment of multiple myeloma (24).
At the same time though, it shows Thalidomide S-enantiomer with a high affinity to the Thyrotropin receptor, which might play a role in cases of severe hypothyroidism after the treatment with Thalidomide (25) (Figure 1C). The enriched human disease pathway analysis further underlines this, by reporting a potential involvement in the autoimmune thyroid disease pathway and thyroid hormone synthesis pathway (Figure 1D).
This shows the crucial role of off-targets in both toxic adverse reactions and potential drug repositioning opportunities, and further stresses the necessity to keep an eye on potential risks when searching for new areas of indication.
Conclusions and future prospects
To facilitate the analysis of withdrawn drugs regarding their adverse effects and withdrawal reasons, but also potential new areas of indication, the Withdrawn database has been reworked to give a higher focus to side effects and human target analysis, while at the same time enhancing the accessibility of the database systematically.
The new ATC tree of withdrawn drugs enables users to quickly gain an overview of the drug categories that withdrawn drugs fall into, and the ability to not only start from a compound but also from a human target further facilitates looking into (off-)targets of withdrawn drugs, that were potentially responsible for adverse reactions, leading to the withdrawal of the compound.
The inclusion of FAERS data and the collection of more detailed drug withdrawal reasons offers further insights into potential harmful side effects. The connection with pharmacovigilance data and mechanisms of action enables surveillance approaches using the prediction of withdrawals (26). Additionally, the mechanism of action prediction and pathway enrichment analysis can be used to discover new areas of indication for withdrawn drugs, where the therapeutic benefits might outweigh the potential treatment risks, while still retaining a cautious look at potentially harmful effects.
To ensure the relevance of these features, the database will be regularly updated and the analyses, such as FAERS mapping, pathway enrichment analysis and mechanism of action prediction, will be re-done for the most recent version of the underlying databases every year.
Contributor Information
Kathleen Gallo, Charité - Universitätsmedizin Berlin, Institute of Physiology and GB IT, Science IT, Corporate Member of Freie Universität Berlin, Berlin Institute of Health, Humboldt-Universität zu Berlin, 10117 Berlin, Germany.
Andrean Goede, Charité - Universitätsmedizin Berlin, Institute of Physiology and GB IT, Science IT, Corporate Member of Freie Universität Berlin, Berlin Institute of Health, Humboldt-Universität zu Berlin, 10117 Berlin, Germany.
Oliver-Andreas Eckert, Charité - Universitätsmedizin Berlin, Institute of Physiology and GB IT, Science IT, Corporate Member of Freie Universität Berlin, Berlin Institute of Health, Humboldt-Universität zu Berlin, 10117 Berlin, Germany.
Bjoern-Oliver Gohlke, Charité - Universitätsmedizin Berlin, Institute of Physiology and GB IT, Science IT, Corporate Member of Freie Universität Berlin, Berlin Institute of Health, Humboldt-Universität zu Berlin, 10117 Berlin, Germany.
Robert Preissner, Charité - Universitätsmedizin Berlin, Institute of Physiology and GB IT, Science IT, Corporate Member of Freie Universität Berlin, Berlin Institute of Health, Humboldt-Universität zu Berlin, 10117 Berlin, Germany.
Data availability
The Withdrawn 2.0 database is publicly available via: https://bioinformatics.charite.de/withdrawn_3/index.php, and completely accessible to all users without the need for a login or registration. Furthermore, results are displayed immediately on the website without the need to provide an e-mail address. Additionally, the general information on all drugs contained in the database is available as a bulk download in the website FAQs as a CSV file.
Funding
Deutsche Forschungsgemeinschaft [TRR295]. Funding for open access charge: University funds.
Conflict of interest statement. None declared.
References
- 1. Nguyen D.A., Nguyen C.H., Mamitsuka H.. A survey on adverse drug reaction studies: data, tasks and machine learning methods. Brief. Bioinform. 2021; 22:164–177. [DOI] [PubMed] [Google Scholar]
- 2. Siramshetty V.B., Nickel J., Omieczynski C., Gohlke B.-O., Drwal M.N., Preissner R.. WITHDRAWN—a resource for withdrawn and discontinued drugs. Nucleic Acids Res. 2016; 44:D1080–D1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jourdan J.P., Bureau R., Rochais C., Dallemagne P.. Drug repositioning: a brief overview. J. Pharm. Pharmacol. 2020; 72:1145–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bellera C.L., Alberca L.N., Sbaraglini M.L., Talevi A.. In Silico Drug Repositioning for Chagas Disease. Curr. Med. Chem. 2020; 27:662–675. [DOI] [PubMed] [Google Scholar]
- 5. Montané E., Santesmases J.. Adverse drug reactions. Med. Clin. (Barc). 2020; 154:178–184. [DOI] [PubMed] [Google Scholar]
- 6. Zhan C., Roughead E., Liu L., Pratt N., Li J.. Detecting high-quality signals of adverse drug-drug interactions from spontaneous reporting data. J. Biomed. Inform. 2020; 112:103603. [DOI] [PubMed] [Google Scholar]
- 7. Burger M.C. ChemDoodle Web Components: HTML5 toolkit for chemical graphics, interfaces, and informatics. J. Cheminform. 2015; 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gaulton A., Hersey A., Nowotka M., Bento A.P., Chambers J., Mendez D., Mutowo P., Atkinson F., Bellis L.J., Cibrián-Uhalte E.et al.. The ChEMBL database in 2017. Nucleic Acids Res. 2017; 45:D945–D954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. The UniProt Consortium UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023; 51:D523–D531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M.. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021; 49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peón A., Naulaerts S., Ballester P.J.. Predicting the reliability of drug-target interaction predictions with maximum coverage of. Target Space. Sci Rep. 2017; 7:3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qin C., Zhang C., Zhu F., Xu F., Chen S.Y., Zhang P., Li Y.H., Yang S.Y., Wei Y.Q., Tao L.et al.. Therapeutic target database update 2014: a resource for targeted therapeutics. Nucleic Acids Res. 2014; 42:D1118–D1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z.et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019; 47:D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bajusz D., Rácz A., Héberger K.. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations?. J. Cheminform. 2015; 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. World Health Organization The selection and use of essential medicines. 2006; 933:1–119.Report of the WHO expert committee, 2005 (including the 14th model list of essential medicines). [PubMed] [Google Scholar]
- 17. Botting J. The History of Thalidomide. Drug News Perspect. 2002; 15:604–611. [DOI] [PubMed] [Google Scholar]
- 18. Kim J.H., Scialli A.R.. Thalidomide: the tragedy of birth defects and the effective treatment of disease. Toxicol. Sci. 2011; 122:1–6. [DOI] [PubMed] [Google Scholar]
- 19. Rehman W., Arfons L.M., Lazarus H.M.. The rise, fall and subsequent triumph of thalidomide: lessons learned in drug development. Ther. Adv. Hematol. 2011; 2:291–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xue H.X., Fu W.Y., Cui H.D., Yang L.L., Zhang N., Zhao L.J.. High-dose thalidomide increases the risk of peripheral neuropathy in the treatment of ankylosing spondylitis. Neural Regen. Res. 2015; 10:814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chaudhry V., Cornblath D.R., Corse A., Freimer M., Simmons-O’Brien E., Vogelsang G.. Thalidomide-induced neuropathy. Neurology. 2002; 59:1872–1875. [DOI] [PubMed] [Google Scholar]
- 22. Ghobrial I.M., Rajkumar S.V.. Management of thalidomide toxicity. J. Support Oncol. 2003; 1:194–205. [PMC free article] [PubMed] [Google Scholar]
- 23. Palumbo A., Palladino C.. Venous and arterial thrombotic risks with thalidomide: evidence and practical guidance. Ther. Adv. Drug Saf. 2012; 3:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ovejero S., Viziteu E., Dutrieux L., Devin J., Lin Y.L., Alaterre E., Jourdan M., Basbous J., Requirand G., Robert N.et al.. The BLM helicase is a new therapeutic target in multiple myeloma involved in replication stress survival and drug resistance. Front. Immunol. 2022; 13:983181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Savary N., Lee R., Vaidya B.. Severe hypothyroidism after thalidomide treatment. J. R. Soc. Med. 2004; 97:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mazuz E., Shtar G., Shapira B.. Pretrained transformer models for predicting the withdrawal of drugs from the market. Bioinformatics. 2023; 39:btad519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Withdrawn 2.0 database is publicly available via: https://bioinformatics.charite.de/withdrawn_3/index.php, and completely accessible to all users without the need for a login or registration. Furthermore, results are displayed immediately on the website without the need to provide an e-mail address. Additionally, the general information on all drugs contained in the database is available as a bulk download in the website FAQs as a CSV file.