Abstract
Traditional Chinese medicine (TCM) is increasingly recognized and utilized worldwide. However, the complex ingredients of TCM and their interactions with the human body make elucidating molecular mechanisms challenging, which greatly hinders the modernization of TCM. In 2016, we developed BATMAN-TCM 1.0, which is an integrated database of TCM ingredient–target protein interaction (TTI) for pharmacology research. Here, to address the growing need for a higher coverage TTI dataset, and using omics data to screen active TCM ingredients or herbs for complex disease treatment, we updated BATMAN-TCM to version 2.0 (http://bionet.ncpsb.org.cn/batman-tcm/). Using the same protocol as version 1.0, we collected 17 068 known TTIs by manual curation (with a 62.3-fold increase), and predicted ∼2.3 million high-confidence TTIs. In addition, we incorporated three new features into the updated version: (i) it enables simultaneous exploration of the target of TCM ingredient for pharmacology research and TCM ingredients binding to target proteins for drug discovery; (ii) it has significantly expanded TTI coverage; and (iii) the website was redesigned for better user experience and higher speed. We believe that BATMAN-TCM 2.0, as a discovery repository, will contribute to the study of TCM molecular mechanisms and the development of new drugs for complex diseases.
Graphical Abstract
Graphical Abstract.
Introduction
Traditional Chinese medicine (TCM), with a history of thousands of years of clinical practice, has gained increasing recognition and application worldwide in recent decades (1). TCM has become a crucial natural template library for new drug development. The active ingredients of TCM have been successfully applied in the development of innovative drugs for the treatment of complex diseases, such as ephedrine for asthma (2) and artemisinin for malaria (3). Despite the important therapeutic value of TCMs, great challenges remain in understanding the pharmacology of TCMs at the molecular level and from a systemic perspective, which greatly hinders the modernization of TCM (4). Studying the interactions between TCM ingredients and target proteins is important for elucidating TCM molecular mechanisms, and screening bioactive ingredients with therapeutic potential.
In 2016, we introduced BATMAN-TCM 1.0 (5), a database of TCM ingredient–target protein interaction (TTI) specially designed for the pharmacological research of TCM. To date, BATMAN-TCM 1.0 has received widespread attention from the community (which has been visited 380 000+ times) and has made a great contribution to the TCM community (6). For example, using BATMAN-TCM, Guo et al. hypothesized that Panax quinquefolium saponins can potentially attenuate myocardial dysfunction induced by chronic ischemia by reducing the expression of PRKCD (protein kinase C delta). This hypothesis has been validated through experiments at the molecular level (7). Meanwhile, we have received numerous inquiries regarding updates to the TTI dataset, as well as the addition of the function for searching therapeutic TCM ingredients by genes of interests (identified from omics datasets or a knowledge base). Based on the investigation of ∼10 000 TCM-related articles in PubMed (2016–2023), we find that there has been an explosive increase in studies on the natural ingredients and their targets, enabling the construction of a larger scale TTI database. Furthermore, screening active TCM ingredients or herbs for complex disease treatment based on disease signature genes from omics data has emerged as an important research direction, such as identifying curcumin for acute myocardial infarction aided by a gene expression profile (8). Consequently, it is promising to establish a high coverage TTI dataset and a novel workflow for screening TCMs based on disease omics data. In fact, great efforts have been devoted to collecting TTIs. In the HIT (Herb Ingredients’ Targets) database (9), Yan et al. established an advanced text-mining algorithms of natural language processing (NLP), and compiled 10 031 compound–target activity pairs from 7100 items in the literature. In the HERB database (10), Fang et al. collected 4815 TTIs from 1966 references by manual curation after a hierarchical filtering. To gather more potential TTIs, the Traditional Chinese Medicine Information Database (TCMID; 11) acquired 205 926 TTIs by the STITCH (12) algorithm. Similarly, the traditional Chinese medicine systems pharmacology database (TCMSP; 13) predicted 54 250 TTIs using random forest and support vector machine (SVM) models. All these efforts presented interaction datasets for precious TCM ingredients and target proteins for the TCM field. However, considering the immense interaction space between TCM ingredients and targets (39 000 × 20 000), the current coverage of TTIs in databases remains relatively limited. There is a substantial need to further construct larger scale TTI datasets. Meanwhile, there is still no bioinformatics resource that can help users screen effective herbal medicines for disease treatment based on disease signature genes obtained from omics data.
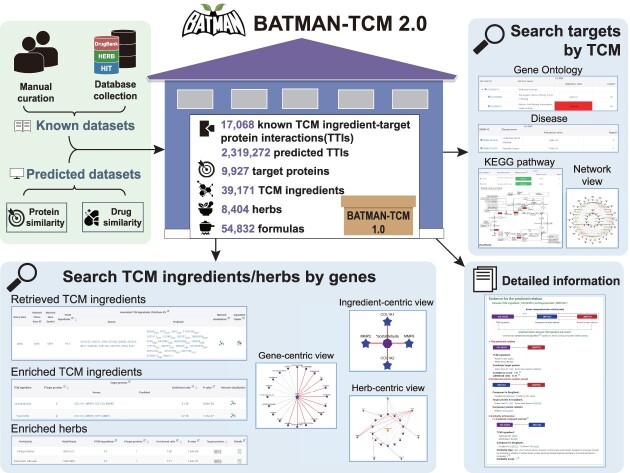
To fill this gap, we upgraded BATMAN-TCM to version 2.0 (Figure 1). Using a similar protocol to the previous version (5), we obtained 17 068 known (62.3-fold increase) and 2 319 272 high-confidence predicted TTIs (3.23-fold increase), together with 54 832 formulae (16.9% increase), 8404 herbs (3% increase) and 39 171 ingredients (215.9% increase). Meanwhile, we added three new features: (i) it allows simultaneous exploration of targets of TCM ingredients for pharmacology research, and TCM ingredients binding to target proteins for drug discovery; (ii) it has significantly increased TTI coverage; and (iii) the website was redesigned for better user experience and high speed, adding functions of ‘Browse’, ‘Download’ and ‘API’ (which enable users to obtain data programmatically).
Figure 1.
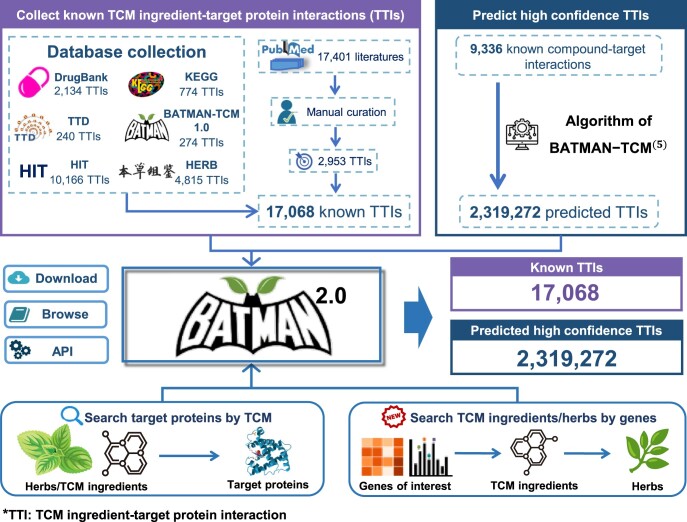
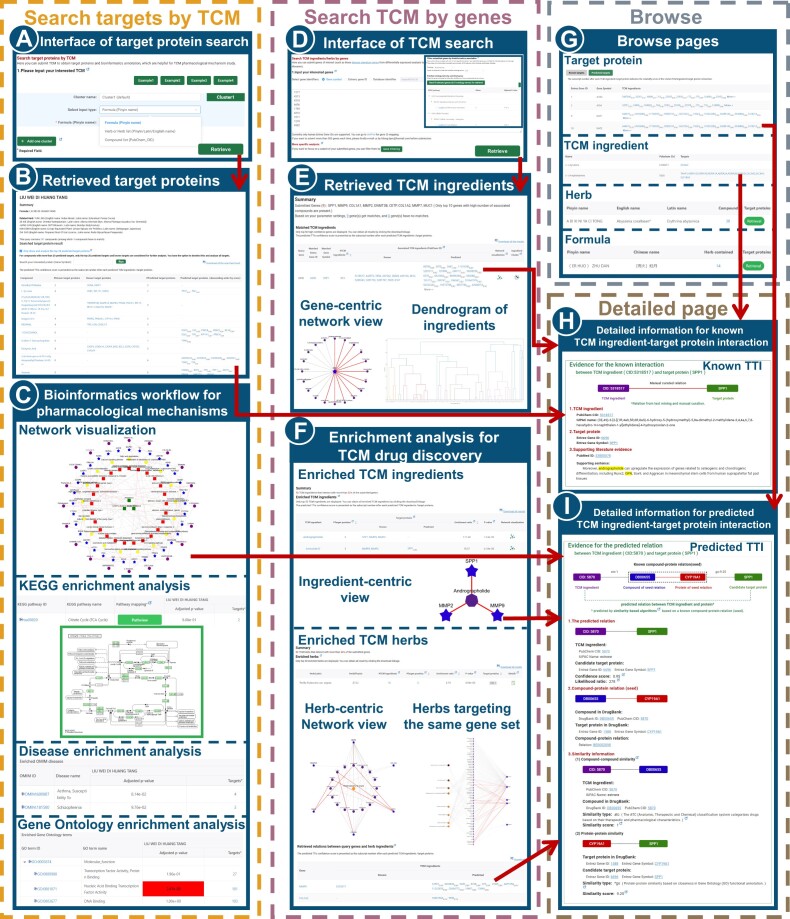
Overview of BATMAN-TCM 2.0, a comprehensive resource for proteome-wide known and predicted TTIs. Known TTI data were manually curated from the literature and other TCM databases including DrugBank, KEGG, TTD, HIT and HERB. High-confidence TTIs were predicted by our published protocol in BATMAN-TCM 1.0 (5). While retaining BATMAN-TCM 1.0’s query function of TCM ingredients/herbs for target proteins to study TCM molecular mechanisms, a new query mode was added to take disease-specific signature genes as input targets and return potential TCM ingredients/herbs that may regulate these signatures, aiming to facilitate drug discovery for treating complex diseases. API, application programming interface.
Improved expansion and new features
Data expansion and statistics
Data expansion
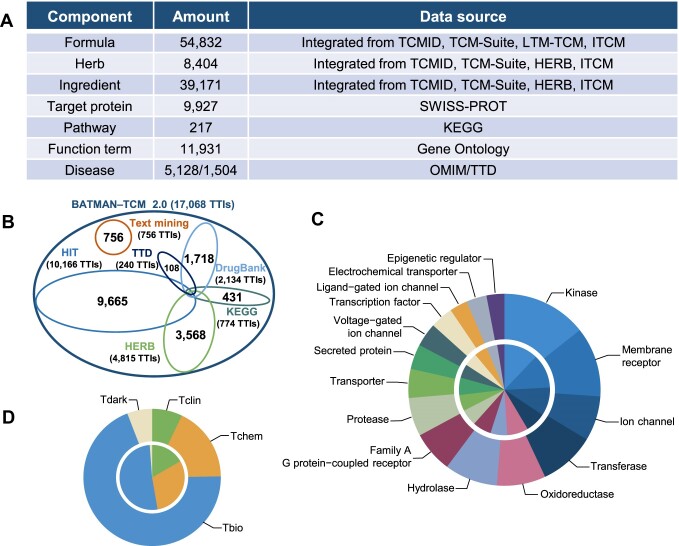
The number of known/predicted TTIs in BATMAN-TCM 2.0 significantly increased from 274/711 828 to 17 068/2 319 272 compared with version 1.0 (Figure 1), together with the addition of 54 832 formulas (16.9% increase), 8404 herbs (3% increase) and 39 171 ingredients (215.9% increase) (Figure 2A).
Figure 2.
Data statistics of BATMAN-TCM 2.0. (A) Data entries of each data category. (B) Number of known TTIs across multiple TCM databases; 756 TTIs were included in the database for the first time from our text mining. (C) The classification of ingredient target proteins by different function categories according to the ChEMBL category scheme. (D) The classification of ingredient target proteins by the target development level. For (C) and (D), from inside to outside, circles correspond to known and predicted target proteins.
While inheriting all known TTIs of BATMAN-TCM 1.0, we integrated known TTI datasets from multiple published databases (Figure 1), including the Kyoto Encyclopedia of Genes and Genomes (KEGG; 14), DrugBank (15), the Therapeutic Target Database (TTD; 16), HIT (9) and HERB (10). The overlap of TTI in these databases is notably limited (Figure 2B), with distinct databases demonstrating complementarity; therefore, it is imperative to consolidate these disperse TTI datasets. Considering that all the latest TTI identifications after 2020 have not been included in any of these databases, we performed text mining and manual curation following a keyword co-occurrence protocol proposed by Yan et al. (9). First, we downloaded 17 401 PubMed abstracts (published from 2020 to 2023) with a TCM ingredient name. Subsequently, an in-house python script was used to filter those sentences which may contain TTI information based on the rules (‘TCM ingredient name’ AND ‘keywords used to describe interactions’ AND ‘target protein’). Keywords used here can be found in the Supplementary Methods. Next, 3078 sentences with 6806 candidate TTIs were manually checked to extract known TTIs by 10 experienced researchers, and then these selected TTIs and their supporting sentences were manually reviewed by three senior experts. Finally, we obtained 2953 manually curated TTIs supported by 925 peer-reviewed articles, among which 756 TTIs were included in the database for the first time (Supplementary Methods).
To further expand the potential interaction space between TCM ingredients and target proteins, benefiting from more public TCM ingredient information and more known compound–protein target interaction data (seeds), BATMAN-TCM 2.0 predicted 2 319 272 putative TTIs with high confidence, based on the similarity algorithm we developed in BATMAN-TCM 1.0 (5). Specifically, the similarity between potential TTIs and seed interaction was calculated as the product of the compound similarity score and the protein similarity score. We selected eight similarity features, i.e. ATC-GO, FP2-closeness, STITCH-sequence, expression-closeness, ATC-sequence, functional_group-sequence, functional_group-GO and side_effect-sequence, and used the likelihood ratio (LR) to measure the efficacy of each feature. The maximum LR was used to measure the confidence of potential TTIs for prediction (Supplementary Methods). Overall, BATMAN-TCM 2.0 comprises 17 068 known TTIs and 2 319 272 predicted TTIs (Figure 1). Compared with similar TCM-related databases, BATMAN-TCM 2.0 has the most comprehensive known and predicted TTI dataset, providing a valuable complement (Table 1).
Table 1.
Comparison with other TCM databases
| TCM ingredient–target protein interactions | Database entities | ||||
|---|---|---|---|---|---|
| Database | Published year | Literature-described | Computationally predicted | TCM ingredients | Target proteins |
| BATMAN-TCM 2.0 | 2023 | 17 068 | 2 31927 2a | 39 171 | 9927 |
| BATMAN-TCM | 2016 | 274 | 711 828a | 12 398 | 7080 |
| HIT 2.0 | 2021 | 10 166 | —— | 1237 | 2208 |
| HERB | 2020 | 4815 | —— | 49 258 | 12 933 |
| TCMID 2.0 | 2018 | —— | 205 926 | 43 413 | 17 602 |
| TCMSP | 2014 | 3970 | 84 260 | 13 729 | 3339 |
aLikelihood ratio > 10.
Data statistics
BATMAN-TCM 2.0 includes 3279 known and 9493 predicted TCM target proteins, providing more potential targets for drug discovery. In order to comprehensively understand these target proteins, we classified them by the ChEMBL functional category scheme, which is a manually curated family hierarchy according to nomenclature commonly used by drug discovery scientists (17). Compared with known TCM ingredient target proteins, the predicted targets share a similar functional distribution, covering major target protein categories (kinase, membrane receptor, ion channel and transferase), suggesting no distinct function bias in the predicted TTI dataset (Figure 2C). Additionally, following the Target Development Level (TDL) classification scheme developed by Oprea et al. (18), we classified both known and predicted target proteins into four categories, namely Tclin (clinic), Tchem (chemistry), Tbio (biology) and Tdark (dark genome) (18). As shown in Figure 2D, proteins from both Tbio and Tdark categories constitute larger proportions in the predicted target proteins compared with those in known target proteins (Fisher's exact test, P < 2.2e-16). This indicates that BATMAN-TCM 2.0 could provide more information on potential target proteins for drug development.
Newly developed retrieval pipeline for screening active TCM ingredients based on disease-specific signatures
Drug discovery is a time-consuming, expensive and high-risk process (19). Emerging omics technologies such as proteomics can streamline and expedite this process at multiple stages including drug target discovery, drug screening, pharmacological analysis and efficacy evaluation (19,20). Remarkably, based on disease-specific signatures (genes specifically expressed in certain disease samples identified by omics analysis or known disease genes from the literature/databases), researchers have successfully discovered some drugs by screening a western pharmaceutical library for treating malignant peripheral nerve sheath tumor (21). In fact, TCM has been widely employed for the treatment of various human diseases, presenting a valuable resource for modern drug discovery (22). However, there is still no bioinformatics resource available for the discovery of TCMs to treat complex disease based on the disease-specific signature genes. Laborious and time-intensive manual reviewing of the massive volume of TCM publications has to be undertaken to find the most promising TCM ingredients and herbs that are intended to target disease-specific gene signatures.
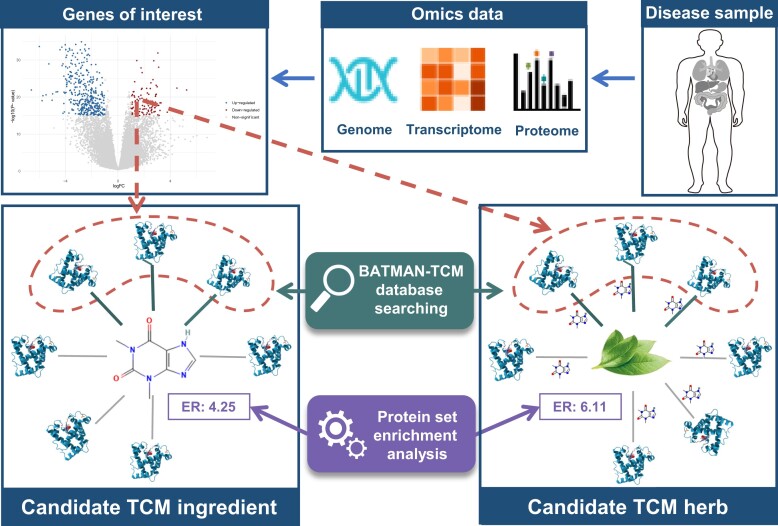
To meet this need, we developed a new retrieval pipeline (Figure 3). Disease-specific signature genes were first mapped to their associated TCM ingredients in BATMAN-TCM 2.0. Then, for each matched TCM ingredient, the numbers of its binding proteins in the submitted lists and those in the background TTI dataset were calculated. Finally, a hypergeometric distribution test was performed to identify those TCM ingredients which are statistically over-represented in interactions for the submitted list. For example, if 33.33% (3/9) of the target proteins in the submitted obesity-related gene signatures are acted upon by the compound epigallocatechin gallate (EGCG), compared with 0.71% (135/19 016) of all target proteins in the database (the population background), the calculated P-value of 2.98e-07 from the hypergeometric distribution test indicates that protein targets of EGCG are significantly enriched in the submitted disease signatures [enrichment ratio = (3/9) ÷ (135/19 016) = 46.95], and therefore EGCG is promising to act as a candidate drug for obesity. Following the same protocol, we can also identify the enriched herbs for the query disease signatures based on our TTI datasets (the correlations between the candidate herb and the submitted protein are mediated by TCM ingredients) (Figure 3).
Figure 3.
Flowchart of the newly added retrieval mode for TCM ingredients/herbs based on genes of interest from disease omics studies. Disease high-throughput experimental studies such as gene expression microarrays or quantitative proteomics can generate disease-specific signature genes (such as differentially expressed genes, which can be illustrated by a volcano plot). These signature genes are mapped to their associated TCM ingredients/herbs in BATMAN-TCM 2.0. The TCM ingredients/herbs over-represented within the signature genes will be identified based on their enrichment ratios (ERs). For each candidate TCM ingredient or herb, the ER was calculated as: ER = (nquery/Nquery) ÷ (nall/Nall), where Nquery and Nallare numbers of all genes in query signatures and those in BATMAN-TCM, while nquery and nall are numbers of genes interacting with this candidate TCM ingredient or herb in query signatures and those in the BATMAN-TCM 2.0 database.
Three results will be returned by this new retrieval pipeline: (i) matched TCM ingredients for query genes, which provide a landscape of the relationship between disease-specific signature genes and TCM ingredients; (ii) enriched TCM ingredients, which have the strongest association with the disease-specific signature gene list and can act on multiple genes; and (iii) enriched herbs, which have the strongest association with the disease-specific signature gene list and may become candidate medicines for treating corresponding disease. For the results of enrichment TCM ingredients and herbs, a network view is provided to help users examine the relationship between enriched TCM ingredients/herbs and the disease-specific signature genes, facilitating the screening of candidates for disease treatment (Figure 3). The following case study section will provide an application example for this new pipeline. In fact, many drug discoveries are based on known disease genes rather than omics data. For instance, numerous drugs targeting the epidermal growth factor receptor (EGFR) have been discovered (23). Users can simply submit such a list of target proteins to this workflow for TCM-related drug discovery.
Enhanced user interface and query speed
To facilitate the usage of the expanded TTI datasets and the newly added search mode, we redesigned user interfaces and optimized the website's technical framework, which have greatly enhanced the user experience and search speed. We also added new features such as ‘Browse’, ‘Download’ and ‘API’.
Search
BATMAN-TCM 2.0 provides simultaneous two-way retrieval and analysis between TCM ingredients and target proteins: (i) to search target proteins by TCM ingredient/herb/formula for pharmacological mechanism study; and (ii) to search TCM ingredient/herb by signature genes for disease omics-aided drug discovery. Users can select either of them according to their requirements. In both retrieval modes, when users set a higher cut-off, the retrieved predicted TTIs will have higher reliability (a lower false-positive rate). Considering that some ingredients are ubiquitous in multiple herbs and act on various protein targets (‘herb homogeneity’), which may confuse pharmacology research and TCM drug discovery, the upgraded website provides an option for users to decide whether to include those ingredients (Supplementary Table S1) and their associated TTIs in their analysis.
Search target proteins by TCM
Three search options are formula, herb and compound (Figure 4A). A table view of the matched TCM target proteins will be presented (Figure 4B). Further, for these target proteins, BATMAN-TCM 2.0 provides a systematic bioinformatics analysis workflow, including KEGG biological pathway, Gene Ontology (GO) functional annotation and OMIM/TTD disease. A network view is provided to present the ‘ingredient–target–pathway/disease’ association, capturing TCM’s complexity in a simple way (Figure 4C).
Figure 4.
Screenshots of the redesigned BATMAN-TCM 2.0 website. BATMAN-TCM 2.0 enabled simultaneous two-way retrieval and analysis between TCM ingredients and target proteins. (A) Interface for querying TCM target proteins. Three options are formula, herb and compound. (B) Table view of the retrieved TCM target proteins. (C) Bioinformatics annotation of retrieved TCM target proteins, including network visualization, KEGG/GO/disease enrichment analysis. (D) Interface of an added gene-based query for TCM, where users can submit disease-specific signature genes (such as differentially expressed genes from a disease omics study). The inset illustrates the gene set filter functionality, through which users can conduct GO, KEGG and disease category enrichment analyses to identify the most promising target proteins for drug discovery. (E) Retrieved TCM ingredients matching the query genes. For each query gene, the matched TCM ingredients in the table view can be illustrated by a popped network view, with the query gene as the central node and the TCM ingredients as surrounding nodes. The dendrogram presents hierarchical clustering of TCM ingredients associated with the specific gene. (F) Enriched TCM ingredients/herbs from enrichment analysis. The ingredient-centric network view presents the retrieved known and predicted TTIs. A herb-centric network connects herb, TCM ingredients and target proteins. For all predicted interactions in network view, the corresponding width of the edges is positively proportional to confidence of the interactions . A further network view was designed for herbs targeting the same gene set, where the enrichment level of herbs is represented by the corresponding node color depth. (G) Interfaces for browsing known and predicted TTIs based on TCM compounds, herbs and formulas, and their target proteins. (H) Supporting literature/database information page for the known TTIs. For our manual curation interactions, the entries of TCM ingredients and target proteins in abstract texts are highlighted in color. (I) Supporting evidence page for the predicted TTIs, including seed compound–protein interaction information and similarity information. Clicking each TCM ingredient/target protein in the table view (B, E, F and G), or clicking on edges of known/predicted interactions in network view (C, E and F), will lead to a detailed page of the known/predicted TTI (H and I), respectively.
In certain TCM queries, some TCMs obtain a large number of predicted target proteins and, in such cases, we provide an option for users to select the top 20 high-confidence predicted targets for presentation and subsequent bioinformatics analysis. In addition, gene druggability (24), which reflects the likelihood of gene products binding to drugs, has been integrated into BATMAN-TCM 2.0 as an additional criterion for filtering predicted targets. Users can set higher druggability score thresholds to select the most likely drug targets.
Search TCM ingredients/herbs by genes
Users can submit a signature gene list from a disease omics study (such as differentially expressed genes). We support eight types of gene identifiers, namely Gene symbol, Entrez Gene ID, UniProtKB AC/ID, Ensembl ID, RefSeq accession, HGNC ID, MIM ID and IMGT/GENE-DB ID. An integrated plugin can be used to filter the gene list into more specific functional pathways or diseases for the most promising target proteins for drug discovery (Figure 4D).
All the matched TCM ingredients will be displayed in table form (Figure 4E). TCM ingredients binding the same target protein often have some structural similarity (25). We clustered these TCM ingredients based on their structural similarity, and presented their relationship using dendrograms. This will help users gain comprehensive views for those TCM ingredients with the same target.
Often multiple ingredients/herbs could be associated with the query disease signature genes. To prioritize them, our website employs two widely recognized principles: functionality and specificity (26). Firstly, we prioritize ingredients/herbs binding the most disease signature genes. Secondly, for those binding the same number of genes, we use the ER to measure their specificity and prioritize those with higher ratios. The enriched TCM ingredients and herbs for these results will be presented in separate table views. Network views are also designed to illustrate the relationship between enriched TCM ingredients/herbs and signature genes. In the TCM ingredient-centric view, the central node is the enriched TCM ingredient, the surrounding nodes are the retrieved target proteins and the width of edges for predicted TTIs is positively proportional to TTI confidence. In the herb-centric view, the central node is the herb, the outermost nodes are the signature genes and the middle nodes are the mediated TCM ingredients (Figure 4F). Meanwhile, multiple herbs may target the same set of proteins through different ingredients. To present the complex relationships among herbs, ingredients and target proteins, we developed an intuitive network view (Figure 4F). The depth of node colors of herbs indicates their enrichment levels. Those enriched TCM ingredients and herbs are promising to treat the complex diseases by acting on multiple targets.
Browse
The ‘Browse’ function was added in BATMAN-TCM 2.0 (Figure 4G). On the ‘Browse’ page, there is an overview table of all collected interactions between TCM ingredients and target proteins, as well as the corresponding herbs and formulas. Users can browse based on multiple items including target proteins, TCM ingredients, herbs and formulas. User can click on the hyperlink on the browse page to view the detailed information of each TTI.
Detailed information for TTIs
For each TTI entry presented on the website, a hyperlink can lead to the detailed information page, which illustrates its source databases, or the details of our expandable algorithm (5). This makes all TTIs traceable and verifiable (Figure 4H, I).
Supporting information for the known TTIs
All known TTIs were manually curated from the literature or from other TCM databases (DrugBank, KEGG, TTD, HIT and HERB). For each TTI curated from the literature, the supported sentence is shown with the entries of TCM ingredient and target proteins colored in yellow. For the TTI from other databases, a database evidence hyperlink was provided for tracing back to the source (Figure 4H).
Supporting evidence for the predicted TTIs
BATMAN-TCM 2.0 used a similarity-based algorithm that we developed in version 1.0 to predict potential TTIs (5). The rationale of this algorithm is to rank each potential TTI based on its similarity to the seed compound–protein interaction (5). To help users trace the details of this prediction process, we designed a supporting evidence page for each predicted TTI (Figure 4I). This page presents the confidence score, the original LR and related similarity features, together with the seed interaction and similarity information (compound similarity and protein similarity).
Download
A download function was added to enhance BATMAN-TCM 2.0’s data accessibility. This function enables users to download the dataset for data mining rather than querying it via the web interface. All known and predicted TTIs are available for bulk download from the ‘Download’ page. The files are formatted as tab-delimited text.
API
To enable programmatic access and cross-references, we also developed an application programming interface (API). The API can be called by building a URL containing the selected parameters (including the request type, desired output format and input item), and returns the query result either in a computer-readable JSON format for programming or as a visible hypertext page for external database cross-references (please refer to documentation on our website for details).
Website performance optimization
During this update, we optimized the overall technical framework of the website. We adopted in-memory databases for database access, R: doParallel parallelization technologies for bioinformatics analysis, and deployed the database on a high-performance server (configured with 16 CPU cores, 64 GB of memory). These strategies have significantly reduced the data analysis time. For example, the time spent on querying a herb decreased from 80 s (in version 1.0) to 3 s, greatly improving user experience.
Case studies
BATMAN-TCM 1.0 (5) has been regarded as a discovery resource for understanding TCM therapeutic mechanisms. Some of the TCM targets predicted by BATMAN-TCM and their downstream bioinformatics analysis have contributed to multiple TCM studies. Examples include the finding of the PD-1/IL17A pathway as the target of Xuanfei Baidu Decoction for treating acute lung injury (27), and the finding of SOD/NOX2 as the target of Si-Miao-Yong-An Decoction for treating heart failure (28).
During this update, aided by the more comprehensive TTI dataset and the new gene-based retrieval mode, we can identify potential therapeutic TCM ingredients/herbs for treating complex diseases with the help of omics data. Here, we will demonstrate using BATMAN-TCM 2.0 to explore obesity treatment herbs. Obesity is a complex and chronic non-communicable disease affecting over a third of the global population (29), associated with multiple diseases including heart failure, coronary artery disease and stroke (30). Currently, the Food and Drug Administration (FDA) has approved seven drugs for treating obesity (31). However, all these drugs are frequently associated with various side effects, particularly gastrointestinal adverse effects (32–37). While TCM is a valuable resource for modern drug discovery and development (10), it is promising to identify effective TCMs for obesity treatment by BATMAN-TCM 2.0.
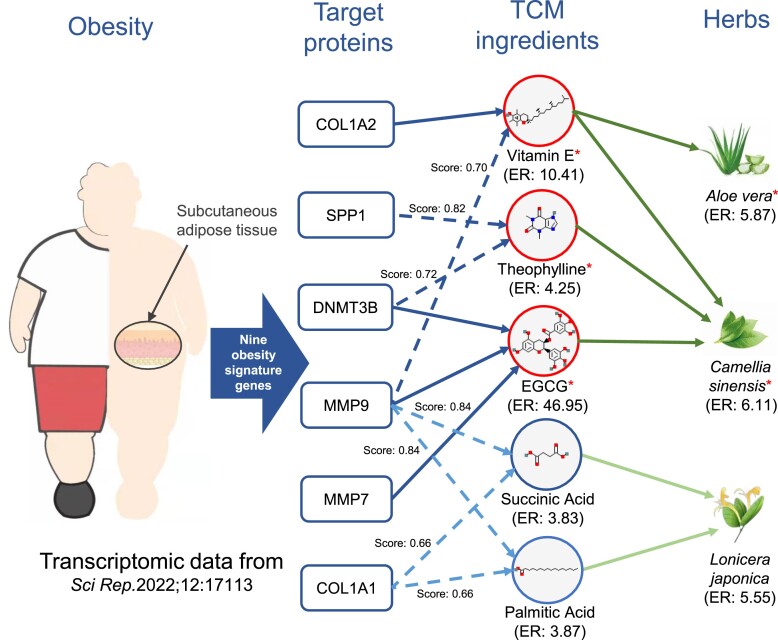
First, we obtained nine obesity-related signature genes from the literature (38). These signatures were identified by differential expression and protein network analysis based on the transcriptomic datasets of subcutaneous adipose tissues from obese patients. Then, we utilized BATMAN-TCM 2.0 to identify TCM ingredients/herbs that showed enriched interactions with these nine signature genes. These enriched TCM ingredients/herbs were considered as potential drug candidates for treating obesity. Supplementary Table S2 lists the top 50 enriched TCM herbs. After manual curation, we found that 40 of these 50 enriched TCM herbs have been reported to be associated with obesity, including two prevalent herbs Aloe vera (LUHUI, ER: 5.87) and Camellia sinensis (CHAYE, ER: 6.11).
Aloe vera (LUHUI) has been reported to reduce fat accumulation via its protective role against obesity-related metabolic alterations and antioxidant effects (39). In BATMAN-TCM 2.0, we found that A. vera may exert anti-obesity effects through its ingredient vitamin E (ER: 10.41) by targeting COL1A2 and MMP9 (Figure 5). In fact, vitamin E has been utilized as a treatment for obesity (40), and COL1A2 is a known target for vitamin E (41). BATMAN-TCM’s similarity algorithm also predicts the interaction between vitamin E and MMP9 with a confidence score of 0.70. A discovery by Sozen et al. (42) that vitamin E can effectively reduce the expression of MMP9 partially validated this prediction.
Figure 5.
Use cases for BATMAN-TCM 2.0. Discovery of potential TCM ingredients or herbs for the treatment of obesity based on transcriptomic data. From transcriptomic analysis for subcutaneous adipose tissue, nine differentially expressed genes (signature genes) were identified. Through BATMAN-TCM 2.0 analysis, three enriched TCM ingredients (marked with asterisks) and two herbs (marked with asterisks) were identified, all of which have been substantiated in the literature as being associated with obesity. Dashed lines represent predicted TTIs based on BATMAN-TCM similarity algorithms, while solid lines denote known TTIs retrieved from the BATMAN-TCM 2.0 dataset. Here only six of nine differentially expressed genes are presented. ER: enrichment ratio.
It is well known that C. sinensis (CHAYE) is rich in bioactive compounds, endowed with functions including antioxidative, anti-inflammatory and hypoglycemic activities (43,44). Through BATMAN-TCM analysis (Figure 5), we identified three ingredients in C. sinensis, namely theophylline (ER: 4.25), vitamin E (ER: 10.41) and epigallocatechin gallate (EGCG, ER: 46.95), that may be associated with obesity. The literature has also verified that these all three ingredients can reduce fat storage and body weight by promoting lipolysis and lipid metabolism (45–47). Furthermore, based on the known TTI dataset, BATMAN-TCM analysis revealed potential anti-obesity target proteins (MMP9, MMP7 and DNMT3B) for EGCG. BATMAN-TCM also predicted SPP1 (score: 0.82) and DNMT3B (score: 0.72) as potential targets for theophylline. Experimental research has demonstrated that both SPP1 and DNMT3B show a negative correlation with obesity severity (48,49). Further research is still needed to elucidate the specific mechanism of both SPP1 and DNMT3B interacting with theophylline.
Conclusion and future
Using the same protocol as version 1.0 (5), we collected 17 068 known TTIs, and we predicted ∼2.3 million high-confidence TTIs. BATMAN-TCM 2.0 has been greatly improved compared with version 1.0. It contains the most comprehensive TTI dataset, which is derived from other TTI-related databases, as well as through manual curation of PubMed abstracts. A new retrieval mode has been added to screen active herbs based on disease-specific signature genes or treating complex diseases. In addition, we have implemented several new features in the updated version: (i) it enables simultaneous exploration of target proteins by TCM and TCM ingredients/herbs by genes; (ii) it has significantly increased TTI coverage; (iii) it presents a uniform confidence scoring system to rank predicted TTIs, providing guidance to balance different levels of coverage and accuracy; and (iv) the web interfaces have been redesigned for exploring the relationship between TCM ingredients and target proteins.
In the future, we will continue to update and maintain our database by using a more automatic text mining strategy and large language models (50). The TTI dataset will be expanded to cover other experimental animals commonly used for drug discovery. Cross-references to other public databases, such as omics data databases (51), will be added to satisfy different requirements. We believe that the updates (with larger TTI space and new retrieval modes) will enable BATMAN-TCM to be a more comprehensive and useful resource for the TCM community.
Supplementary Material
Acknowledgements
We gratefully acknowledge HIT, HERB, DrugBank, TTD and KEGG for kindly sharing the TTI datasets. We thank the bioinformatics platform at Phoenix Center for the strong and stable IT support.
Contributor Information
Xiangren Kong, State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China.
Chao Liu, State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China.
Zuzhen Zhang, School of Basic Medical Sciences, Anhui Medical University, Hefei 230032, China.
Meiqi Cheng, College of Life Sciences, Hebei University, Baoding 071002, China.
Zhijun Mei, College of Life Sciences, Hebei University, Baoding 071002, China.
Xiangdong Li, College of Life Sciences, Hebei University, Baoding 071002, China.
Peng Liu, College of Life Sciences, Hebei University, Baoding 071002, China.
Lihong Diao, School of Traditional Chinese Medicine, Beijing University of Chinese Medicine, Beijing 100029, China.
Yajie Ma, College of Life Sciences, Hebei University, Baoding 071002, China.
Peng Jiang, Beijing Geneworks Technology Co., Ltd, Beijing 100101, China.
Xiangya Kong, Beijing Geneworks Technology Co., Ltd, Beijing 100101, China.
Shiyan Nie, State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China.
Yingzi Guo, State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China.
Ze Wang, State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China.
Xinlei Zhang, Beijing Geneworks Technology Co., Ltd, Beijing 100101, China.
Yan Wang, State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China.
Liujun Tang, State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China.
Shuzhen Guo, School of Traditional Chinese Medicine, Beijing University of Chinese Medicine, Beijing 100029, China.
Zhongyang Liu, State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China; School of Basic Medical Sciences, Anhui Medical University, Hefei 230032, China; College of Life Sciences, Hebei University, Baoding 071002, China.
Dong Li, State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences (Beijing), Beijing Institute of Lifeomics, Beijing 102206, China; School of Basic Medical Sciences, Anhui Medical University, Hefei 230032, China; College of Life Sciences, Hebei University, Baoding 071002, China.
Data availability
BATMAN-TCM 2.0 can be accessed at http://bionet.ncpsb.org.cn/batman-tcm/.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
The National Natural Science Foundation of China [32088101 and 32271518]; and the National key Research and Development Program of China [2021YFA1301603 and 2020YFE0202200].
Conflict of interest statement. None declared.
References
- 1. Qiu J. China plans to modernize traditional medicine. Nature. 2007; 446:590–591. [DOI] [PubMed] [Google Scholar]
- 2. Chen K.K. A pharmacognostic and chemical study of ma huang (Ephedra vulgaris var. helvetica). 1925. J. Am. Pharm. Assoc. (2003). 2012; 52:406–412. [DOI] [PubMed] [Google Scholar]
- 3. Tu Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 2011; 17:1217–1220. [DOI] [PubMed] [Google Scholar]
- 4. Lv C., Wu X., Wang X., Su J., Zeng H., Zhao J., Lin S., Liu R., Li H., Li X.et al.. The gene expression profiles in response to 102 traditional Chinese medicine (TCM) components: a general template for research on TCMs. Sci. Rep. 2017; 7:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Z., Guo F., Wang Y., Li C., Zhang X., Li H., Diao L., Gu J., Wang W., Li D.et al.. BATMAN-TCM: a bioinformatics analysis tool for molecular mechanism of traditional Chinese medicine. Sci. Rep. 2016; 6:21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li T., Zhong Y., Tang T., Luo J., Cui H., Fan R., Wang Y., Wang D.. Formononetin induces vasorelaxation in rat thoracic aorta via regulation of the PI3K/PTEN/Akt signaling pathway. Drug Des. Dev. Ther. 2018; 12:3675–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo M., Liu J., Guo F., Shi J., Wang C., Bible P.W., Yang M., Tian Y., Wei L., Wang P.et al.. Panax quinquefolium saponins attenuate myocardial dysfunction induced by chronic ischemia. Cell. Physiol. Biochem. 2018; 49:1277–1288. [DOI] [PubMed] [Google Scholar]
- 8. Jiang F., Zhang W., Lu H., Tan M., Zeng Z., Song Y., Ke X., Lin F.. Prediction of herbal medicines based on immune cell infiltration and immune- and ferroptosis-related gene expression levels to treat valvular atrial fibrillation. Front. Genet. 2022; 13:886860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yan D., Zheng G., Wang C., Chen Z., Mao T., Gao J., Yan Y., Chen X., Ji X., Yu J.et al.. HIT 2.0: an enhanced platform for Herbal Ingredients' Targets. Nucleic Acids Res. 2022; 50:D1238–D1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fang S., Dong L., Liu L., Guo J., Zhao L., Zhang J., Bu D., Liu X., Huo P., Cao W.et al.. HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 2021; 49:D1197–D1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang L., Xie D., Yu Y., Liu H., Shi Y., Shi T., Wen C.. TCMID 2.0: a comprehensive resource for TCM. Nucleic Acids Res. 2018; 46:D1117–D1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Szklarczyk D., Santos A., von Mering C., Jensen L.J., Bork P., Kuhn M.. STITCH 5: augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016; 44:D380–D384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ru J., Li P., Wang J., Zhou W., Li B., Huang C., Li P., Guo Z., Tao W., Yang Y.et al.. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014; 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kanehisa M., Furumichi M., Sato Y., Kawashima M., Ishiguro-Watanabe M.. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023; 51:D587–D592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z.et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou Y., Zhang Y., Lian X., Li F., Wang C., Zhu F., Qiu Y., Chen Y.. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022; 50:D1398–D1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños M.P., Mosquera J.F., Mutowo P., Nowotka M.et al.. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019; 47:D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oprea T.I., Bologa C.G., Brunak S., Campbell A., Gan G.N., Gaulton A., Gomez S.M., Guha R., Hersey A., Holmes J.et al.. Unexplored therapeutic opportunities in the human genome. Nat. Rev. Drug Discov. 2018; 17:317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amiri-Dashatan N., Koushki M., Abbaszadeh H.-A., Rostami-Nejad M., Rezaei-Tavirani M.. Proteomics applications in health: biomarker and drug discovery and food industry. Iran. J. Pharm. Res. 2018; 17:1523. [PMC free article] [PubMed] [Google Scholar]
- 20. Paananen J., Fortino V.. An omics perspective on drug target discovery platforms. Brief. Bioinform. 2020; 21:1937–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsuchiya R., Yoshimatsu Y., Noguchi R., Sin Y., Ono T., Akiyama T., Kosako H., Yoshida A., Ohtori S., Kawai A.et al.. Integrating analysis of proteome profile and drug screening identifies therapeutic potential of MET pathway for the treatment of malignant peripheral nerve sheath tumor. Expert Rev. Proteomics. 2023; 20:109–119. [DOI] [PubMed] [Google Scholar]
- 22. Zhang Y., Li X., Shi Y., Chen T., Xu Z., Wang P., Yu M., Chen W., Li B., Jing Z.et al.. ETCM v2.0: an update with comprehensive resource and rich annotations for traditional Chinese medicine. Acta Pharm. Sin. B. 2023; 13:2559–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uribe M.L., Marrocco I., Yarden Y.. EGFR in cancer: signaling mechanisms, drugs, and acquired resistance. Cancers (Basel). 2021; 13:2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cunningham M., Pins D., Dezső Z., Torrent M., Vasanthakumar A., Pandey A.. PINNED: identifying characteristics of druggable human proteins using an interpretable neural network. J. Cheminform. 2023; 15:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Periwal V., Bassler S., Andrejev S., Gabrielli N., Patil K.R., Typas A., Patil K.R.. Bioactivity assessment of natural compounds using machine learning models trained on target similarity between drugs. PLoS Comput. Biol. 2022; 18:e1010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dai W., Chen J., Lu P., Gao Y., Chen L., Liu X., Song J., Xu H., Chen D., Yang Y.et al.. Pathway pattern-based prediction of active drug components and gene targets from H1N1 influenza's treatment with maxingshigan-yinqiaosan formula. Mol. Biosyst. 2013; 9:375–385. [DOI] [PubMed] [Google Scholar]
- 27. Wang Y., Wang X., Li Y., Xue Z., Shao R., Li L., Zhu Y., Zhang H., Yang J.. Xuanfei Baidu decoction reduces acute lung injury by regulating infiltration of neutrophils and macrophages via PD-1/IL17A pathway. Pharmacol. Res. 2022; 176:106083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ren Y., Chen X., Li P., Zhang H., Su C., Zeng Z., Wu Y., Xie X., Wang Q., Han J.et al.. Si-Miao-Yong-An decoction ameliorates cardiac function through restoring the equilibrium of SOD and NOX2 in heart failure mice. Pharmacol. Res. 2019; 146:104318. [DOI] [PubMed] [Google Scholar]
- 29. Uranga R.M., Keller J.N.. The complex interactions between obesity, metabolism and the brain. Front. Neurosci. 2019; 13:513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Derosa G., Maffioli P.. Anti-obesity drugs: a review about their effects and their safety. Expert Opin. Drug Saf. 2012; 11:459–471. [DOI] [PubMed] [Google Scholar]
- 31. Chakhtoura M., Haber R., Ghezzawi M., Rhayem C., Tcheroyan R., Mantzoros C.S.. Pharmacotherapy of obesity: an update on the available medications and drugs under investigation. EClinicalMedicine. 2023; 58:101882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kakkar A.K., Dahiya N.. Drug treatment of obesity: current status and future prospects. Eur. J. Intern. Med. 2015; 26:89–94. [DOI] [PubMed] [Google Scholar]
- 33. Srivastava G., Apovian C.. Future pharmacotherapy for obesity: new anti-obesity drugs on the horizon. Curr. Obes. Rep. 2018; 7:147–161. [DOI] [PubMed] [Google Scholar]
- 34. Clément K., Mosbah H., Poitou C.. Rare genetic forms of obesity: from gene to therapy. Physiol. Behav. 2020; 227:113134. [DOI] [PubMed] [Google Scholar]
- 35. Calderon G., Gonzalez-Izundegui D., Shan K.L., Garcia-Valencia O.A., Cifuentes L., Campos A., Collazo-Clavell M.L., Shah M., Hurley D.L., Abu Lebdeh H.S.et al.. Effectiveness of anti-obesity medications approved for long-term use in a multidisciplinary weight management program: a multi-center clinical experience. Int. J. Obes. 2021; 46:555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trivedi M.H., Walker R., Ling W., dela Cruz A., Sharma G., Carmody T., Ghitza U.E., Wahle A., Kim M., Shores-Wilson K.et al.. Bupropion and naltrexone in methamphetamine use disorder. N. Engl. J. Med. 2021; 384:140–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Idrees Z., Cancarevic I., Huang L.. FDA-approved pharmacotherapy for weight loss over the last decade. Cureus. 2022; 14:e29262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tai Y., Tian H., Yang X., Feng S., Chen S., Zhong C., Gao T., Gang X., Liu M.. Identification of hub genes and candidate herbal treatment in obesity through integrated bioinformatic analysis and reverse network pharmacology. Sci. Rep. 2022; 12:17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shakib Z., Shahraki N., Razavi B.M., Hosseinzadeh H.. Aloe vera as an herbal medicine in the treatment of metabolic syndrome: a review. Phytother. Res. 2019; 33:2649–2660. [DOI] [PubMed] [Google Scholar]
- 40. Alcalá M., Sánchez-Vera I., Sevillano J., Herrero L., Serra D., Ramos M.P., Viana M.. Vitamin E reduces adipose tissue fibrosis, inflammation, and oxidative stress and improves metabolic profile in obesity. Obesity (Silver Spring). 2015; 23:1598–1606. [DOI] [PubMed] [Google Scholar]
- 41. Pickett-Blakely O., Young K., Carr R.M.. Micronutrients in nonalcoholic fatty liver disease pathogenesis. Cell. Mol. Gastroenterol. Hepatol. 2018; 6:451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sozen E., Karademir B., Yazgan B., Bozaykut P., Ozer N.K.. Potential role of proteasome on c-jun related signaling in hypercholesterolemia induced atherosclerosis. Redox Biol. 2014; 2:732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shang A., Li J., Zhou D.-D., Gan R.-Y., Li H.-B.. Molecular mechanisms underlying health benefits of tea compounds. Free Radic. Biol. Med. 2021; 172:181–200. [DOI] [PubMed] [Google Scholar]
- 44. Bag S., Mondal A., Majumder A., Banik A.. Tea and its phytochemicals: hidden health benefits & modulation of signaling cascade by phytochemicals. Food Chem. 2022; 371:131098. [DOI] [PubMed] [Google Scholar]
- 45. Wong S.K., Chin K.-Y., Suhaimi F.H., Ahmad F., Ima-Nirwana S.. Vitamin E as a potential interventional treatment for metabolic syndrome: evidence from animal and human studies. Front. Pharmacol. 2017; 8:444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Javaid M.S., Latief N., Ijaz B., Ashfaq U.A.. Epigallocatechin gallate as an anti-obesity therapeutic compound: an in silico approach for structure-based drug designing. Nat. Prod. Res. 2018; 32:2121–2125. [DOI] [PubMed] [Google Scholar]
- 47. Liu T.-T., Liu X.-T., Huang G.-L., Liu L., Chen Q.-X., Wang Q.. Theophylline extracted from Fu brick tea affects the metabolism of preadipocytes and body fat in mice as a pancreatic lipase inhibitor. Int. J. Mol. Sci. 2022; 23:2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang S., Cao Q., Cui X., Jing J., Li F., Shi H., Xue B., Shi H.. Dnmt3b deficiency in Myf5+-brown fat precursor cells promotes obesity in female mice. Biomolecules. 2021; 11:1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Imbert A., Vialaneix N., Marquis J., Vion J., Charpagne A., Metairon S., Laurens C., Moro C., Boulet N., Walter O.et al.. Network analyses reveal negative link between changes in adipose tissue GDF15 and BMI during dietary-induced weight loss. J. Clin. Endocrinol. Metab. 2022; 107:e130–e142. [DOI] [PubMed] [Google Scholar]
- 50. Trieu H.-L., Miwa M., Ananiadou S.. BioVAE: a pre-trained latent variable language model for biomedical text mining. Bioinformatics. 2022; 38:872–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Clough E., Barrett T.. The Gene Expression Omnibus database. Methods Mol. Biol. 2016; 1418:93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BATMAN-TCM 2.0 can be accessed at http://bionet.ncpsb.org.cn/batman-tcm/.