Abstract
The cytoplasmic membrane proteins ExbB and ExbD support TonB-dependent active transport of iron siderophores and vitamin B12 across the essentially unenergized outer membrane of Escherichia coli. In this study, in vivo formaldehyde cross-linking analysis was used to investigate the interactions of T7 epitope-tagged ExbB or ExbD proteins. ExbB and ExbD each formed two unique cross-linked complexes which were not dependent on the presence of TonB, the outer membrane receptor protein FepA, or the other Exb protein. Cross-linking analysis of ExbB- and ExbD-derived size variants demonstrated instead that these ExbB and ExbD complexes were homodimers and homotrimers and suggested that ExbB also interacted with an unidentified protein(s). Cross-linking analysis of epitope-tagged ExbB and ExbD proteins with TonB antisera afforded detection of a previously unrecognized TonB-ExbD cross-linked complex and confirmed the composition of the TonB-ExbB cross-linked complex. The implications of these findings for the mechanism of TonB-dependent energy transduction are discussed.
While the outer membrane of gram-negative bacteria protects the cell from certain harmful environmental components, its presence creates an additional barrier through which nutrients must pass. Unlike most nutrients, iron siderophores and vitamin B12 are too large and scarce to passively diffuse through porin proteins (600-Da size exclusion) which permeate the outer membrane. The TonB-dependent energy transduction complex circumvents this problem by coupling cytoplasmic membrane proton motive force to active transport of iron siderophores and vitamin B12 across the outer membrane, which is essentially unenergized (5, 6).
TonB-dependent energy transduction can be considered an all-purpose system for the delivery of energy to the outer membrane for either import or efflux of important molecules. TonB homologues have been found widely distributed among gram-negative bacteria, where they also energize the transport of iron obtained from mammalian iron-binding proteins (4, 17, 43) and iron chelated to heme (18, 45) and, apparently, the efflux of the toxin aerobactin (16).
The TonB-dependent energy transduction complex consists of, at least, TonB, ExbB, and ExbD. It has previously been postulated that an ion-translocating protein might also be part of the complex and would serve as the means by which the cytoplasmic membrane proton electrochemical potential is harnessed. Resultant conformational changes in TonB could then be transmitted to outer membrane receptors to energize active transport (33, 34). Such a model would have similarities to the mechanism by which light-induced conformational changes in sensory rhodopsin are transduced to drive conformational changes in the closely associated Htr protein (42). The TonB-dependent ion-translocating protein has never been identified in genetic selections. It may be encoded by an essential gene or by a gene encoding a protein with a redundant function, or perhaps known proteins ExbB and ExbD fulfill this function.
Based on available evidence, it appears that energized TonB physically contacts a ligand-bound outer membrane receptor (3, 9, 13, 15, 23, 32, 36, 38, 40, 47) and transduces its energy, driving a conformational change in the outer membrane receptor (19) which results in pumping of ligand into the periplasmic space (36, 48). During this process, TonB appears to undergo significant cyclic changes in affinity for the cytoplasmic and outer membranes. ExbB and ExbD function together to cycle TonB from its high-affinity outer membrane association to its high-affinity cytoplasmic membrane association. Based on those observations, we have hypothesized that following return to a high-affinity cytoplasmic membrane state, TonB can be reenergized by the proton motive force for another round of energy transduction (28).
To understand the mechanism of TonB-dependent energy transduction, it is necessary to understand ExbB and ExbD interactions. ExbB and ExbD are encoded by the exb operon, where mutations in either gene produce the same phenotype: loss of approximately 90% of TonB-dependent activity (2, 5, 10, 40). TolQ and TolR proteins, which can partially substitute for ExbB and ExbD, are responsible for the residual activity, so that when both ExbBD and TolQR are absent, chromosomally encoded TonB has undetectable activity (5, 7). Both ExbB and ExbD also contribute to the stability of TonB (2, 12, 40), and ExbB may serve as a chaperone for the membrane insertion of newly synthesized TonB (22).
ExbB and ExbD are topologically partitioned to opposite sides of the cytoplasmic membrane, with the bulk of ExbB occupying the cytoplasm and the bulk of ExbD occupying the periplasm. ExbB has three transmembrane domains (21, 22), while like TonB (37, 41), ExbD has only a single transmembrane domain consisting of its signal anchor (20). We have speculated that ExbB and ExbD together function as a signal transducer (34), where the prominent soluble domains of each protein could serve as interaction sites with other proteins: perhaps TonB in the case of ExbD and unknown cytoplasmic proteins in the case of ExbB. Identification of these potential interactions is essential for a complete understanding of the mechanism of TonB-dependent energy transduction.
We have previously used in vivo formaldehyde cross-linking to examine TonB interactions with the outer membrane receptor FepA and with ExbB (24, 26, 40). In the present study, this approach is applied to ExbB and ExbD, demonstrating their homodimeric and homotrimeric interactions and providing evidence for the association of ExbB with an additional, as yet unidentified, protein(s). We also demonstrate a physical interaction between TonB and ExbD and confirm that TonB cross-links to ExbB.
MATERIALS AND METHODS
Materials.
Medium components were purchased from Difco Laboratories (Detroit, Mich.). TA cloning kit was purchased from InVitrogen Corp. (Carlsbad, Calif.). Extralong PCR was performed with a GeneAmp XL PCR Kit purchased from Perkin-Elmer (Norwalk, Conn.) or Elongase mix purchased from Gibco BRL (Grand Island, N.Y.). Qiaex II Gel Extraction Kit was purchased from Qiagen Inc. (Santa Clarita, Calif.). The remaining molecular biology enzymes were purchased from New England Biolabs (Beverly, Mass.) or Gibco BRL. Oligonucleotides were purchased from Ransom Hill Bioscience, Inc. (Ramona, Calif.) or the Washington State University Biotechnology Center (Pullman, Wash.). Sequenase kit (version 2.0) was purchased from United States Biochemicals Corp. (Cleveland, Ohio). Trichloroacetic acid (TCA) and standard 37% formaldehyde were purchased from the J. T. Baker Chemical Co. (Phillipsburg, N.J.). Monomeric 16% formaldehyde reconstituted from paraformaldehyde was purchased from Electron Microscopy Sciences (Ft. Washington, Pa.). Acrylamide was purchased from Aldrich Chemical Co., Inc. (Milwaukee, Wis.). Bisacrylamide and ammonium persulfate were purchased from Bio-Rad Laboratories (Richmond, Calif.). Sodium dodecyl sulfate (SDS) was purchased from Gibco BRL. Radioisotopes and the Renaissance chemiluminescence immunoblot kit were purchased from NEN Life Sciences (Boston, Mass.). Immobilon-P polyvinylidene difluoride (PVDF) membrane was purchased from Millipore Corp. (Bedford, Mass.). Anti-T7 monoclonal antibody, pET expression system, and λDE3 lysogenization kit were purchased from Novagen, Inc. (Madison, Wis.). Horseradish peroxidase (HRPO)-conjugated sheep and goat anti-mouse immunoglobulin were purchased from Amersham Corp. (Arlington Heights, Ill.) and Caltag Laboratories (Burlingame, Calif.), respectively. All other supplies and reagents were purchased from Sigma Chemical Co. (St. Louis, Mo.).
Strains and plasmids.
The principal bacteria and plasmids used are listed in Table 1. All bacteria are derivatives of Escherichia coli K-12. KP1345 was constructed by transduction of GUC41 with λDE3, which encodes T7 RNA polymerase, and associated phage lysates, as described in the λDE3 lysogenization kit. To allow assays of bacteriophage φ80 sensitivity, a wild-type fhuA gene was restored to KP1345 by P1vir cotransduction (31) with Tn10 from CAG12025, selected by tetracycline resistance, and screened for sensitivity to φ80, creating KP1269. KP1346 was constructed by P1vir transduction of ΔtonB::blaM from KP1344, where the tonB sequence has been precisely replaced by a blaM gene (25).
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristic | Reference or source |
|---|---|---|
| Strains | ||
| GM1 | ara Δ(pro-lac) thi F′ pro lac | 44 |
| GUC41 | thr leu fhuA lacY supE44 Δ(exbBD) met thi | 14 |
| KP1345 | GUC41(λDE3) | This study |
| KP1269 | GUC41(λDE3) fhuA+ | This study |
| KP1346 | KP1269 Δ(tonB)::blaM | 25 |
| CAG12025 | MG1655 Tn10 at 03.5 min | 39 |
| Plasmids | ||
| pCRII | kan amp lacZα | InVitrogen |
| pKP298 | pACYC184 expressing ExbB/D | 2 |
| pKP360 | pKP298 expressing only ExbD | This study |
| pKP361 | pKP298 expressing only ExbB | This study |
| pKP323 | pET-24(a)+ expressing T7-ExbD | This study |
| pKP339 | pET-24(a)+ expressing T7-ExbB | This study |
| pKP353 | pET-24(a)+ expressing T7-ExbB-C19 | This study |
| pKP376 | pET-24(a)+ expressing T7-ExbB-C41 | This study |
| pKP375 | pET-24(a)+ expressing T7-ExbD-C19 | This study |
| pKP377 | pET-24(a)+ expressing T7-ExbD-C41 | This study |
| Phages | ||
| φ80vir | 29 | |
| λDE3 | T7 RNA polymerase under lacUV5 control | Novagen |
| P1vir | 31 |
Plasmids pKP339 (T7-ExbB) and pK323 (T7-ExbD) were constructed as follows: exbB or exbD PCR products (base pairs 582 to 1316 or 1326 to 1748 of the published sequence, respectively) were amplified from plasmid pKP298 containing the exbBD operon. Purified amplimers were cloned into pCRII plasmids, transformed into INV αF′ E. coli, and selected on Luria-Bertani (LB) medium supplemented with 50 μg of kanamycin ml−1 and 50 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) ml−1 (for blue and white screening). Inserts were sequenced by chain-termination reactions using the Sequenase version 2.0 kit and a set of exbBD- and plasmid-specific primers. Verified inserts were then subcloned into pET-24a(+) by using EcoRI restriction sites located within the primers and the vector such that the ExbB or ExbD protein was expressed from the lac-controlled T7 promoter and fused at its amino terminus with a T7 epitope tag of the following sequence: MASMTGGQQMGRGSEF.
Compatible plasmids pKP361 (ExbB) and pKP360 (ExbD) were constructed by extralong inverse PCR of the entire pKP298 vector (contains exbBD operon), excluding either exbD or exbB, respectively. The PCR products were then recircularized, resulting in excision of either exb gene from the operon. To excise exbD (pKP361), we used primers corresponding to base pairs 1316 to 1292 (oKP162) of the exbBD operon antisense strand and 1749 to 1771 (oKP163) of the sense strand (10). To excise exbB (pKP360), primers corresponding to base pairs 581 to 560 (oKP164) of the antisense strand and 1323 to 1346 (oKP165) of the sense strand were used. Purified amplimers were treated with Klenow fragment, T4 polynucleotide kinase, and T4 DNA ligase and were transformed into DH5α-competent cells (pKP360) or GM1-competent cells (pKP361).
Plasmids pKP360 and pKP361 (pACYC184 ori: p15A) were compatible with the pET-24a(+)-derived plasmids pKP339 and pKP323 (ColE1 ori: pMB1). The plasmid target gene (exbB or exbD) was constructed to be regulated by the wild-type exb promoter-operator region and the Fur DNA-binding protein in response to iron. The ATG start codon for the second gene in the operon (exbD) was designed to be 6 bp downstream from the ribosome binding site of the first gene in the operon (exbB). This distance mimicked the location of the putative GTG start codon (base pairs 582 to 584) for the exbB gene (10). Sequence analysis of pKP361 determined that part of the exb 3′ untranslated region was missing; however, based on the complementation results, the absence of this region did not appear to unduly influence expression.
Plasmids pKP353 (T7-ExbB-C19) and pKP375 (T7-ExbD-C19) were constructed by the method outlined for pKP339 and pKP323, except the following sequences were used: base pairs 585 to 1312 (exbB) or 1326 to 1745 (exbD) of the published exbBD sequence such that the stop codon of each gene was deleted, allowing readthrough of pET-24(a)+ until the ensuing stop codon at position 139 of the vector. The added protein sequence was NSSSVDKLAAALEHHHHHH.
Plasmids pKP376 (T7-ExbB-C41) and pKP377 (T7-ExbD-C41) were constructed from pKP353 and pKP375, respectively, by deletion of the final base pair of the stop codon [base pair 137 of pET-24(a)+], allowing readthrough of the pET-24(a)+ plasmid until the ensuing stop codon at base pair 76. pET-24(a)+ base pair 137 was deleted by extralong inverse PCR of pKP353 or pKP375 with 5′ phosphorylated primers oKP236 (5′ T TGT TAG CAG CCG GAT CCA GTG G 3) and oKP237 (5′ AG CCC GAA AGG AAG CTG AGT TGG C). Purified amplimers were recircularized with T4 DNA ligase and transformed by electroporation into DH5α. Colonies were pooled, and plasmid DNA was isolated and then retransformed into competent KP1269. Resulting colonies were picked, induced with 0.01 mM isopropyl β-d-thiogalactopyranoside (IPTG), and screened for expected protein size by anti-T7 monoclonal antibody immunoblotting.
Culture conditions.
Strains were maintained on LB agar. Liquid cultures were grown with aeration at 37°C in LB broth or in M9 minimal salts medium supplemented with 0.4% glucose, 40 μg of tryptophan ml−1, 0.4 μg of thiamine ml−1, 1 mM MgSO4, 0.5 mM CaCl2, 0.2% Casamino Acids, and 1.85 μM FeCl3 · 6H2O. Antibiotics were used where appropriate in LB agar plates, LB broth, or supplemented M9 minimal broth at the following concentrations (except where otherwise indicated): ampicillin, 100 μg ml−1; chloramphenicol, 34 μg ml−1; kanamycin, 50 μg ml−1.
Analysis of T7 epitope-tagged-ExbB and -ExbD.
Expression of T7-ExbB or T7-ExbD was induced as follows: 0.01 mM IPTG was added to cultures at an A550 of 0.3 (path length, 1.5 cm; measured with a Spectronic 20 spectrophotometer), and the cultures were then further incubated to an A550 of 0.5. Proteins were precipitated from whole cells by addition of TCA to 10%, washed with 100 mM Tris-HCl (pH 8.0), and solubilized in 2× Laemmli sample buffer (LSB). Proteins were then resolved by SDS-polyacrylamide gel electrophoresis on 11% polyacrylamide gels, transferred to PVDF membrane for 200 V · h (46) and probed with an anti-T7 monoclonal antibody at 1:10,000 and then goat anti-mouse immunoglobulin G (IgG)-HRPO secondary antibody at 1:10,000. Immunoreactive proteins were detected by chemiluminescence.
Complementation assays.
Strains to be tested were subcultured 1:100 from saturated cultures into LB broth plus appropriate antibiotics and grown to an A550 of 0.3. To induce expression of the T7-promoted genes, IPTG was added to a concentration of 0.01 mM and cultures were further grown to an A550 of 0.5. Bacteriophage φ80, colicin B, and colicin D spot titer analyses were performed as previously described (27), except the bottom and top agar contained 0.01 mM IPTG and appropriate antibiotics and were made from LB agar. φ80 stock (approximately 9 × 1010 PFU ml−1) and colicins were prepared as previously described (27). Five-microliter volumes of φ80 (10-fold serial dilutions) or colicin B or colicin D (5-fold serial dilutions) were spotted onto top agar overlays. Plates were incubated at 37°C for approximately 18 h, and activity was scored as the greatest dilution resulting in either plaques generated in (φ80) or cleared from (colicins) the bacterial lawn. Assays were performed in triplicate.
In vivo cross-linking analysis.
Various strains expressing T7 epitope-tagged ExbB or ExbD, with or without respective untagged partner proteins, or carrying vector control plasmids were grown to saturation at 37°C with aeration in LB broth containing appropriate antibiotics, subcultured 1:100 into M9 minimal broth, and grown to an A550 of 0.3. IPTG was added to 0.01 mM, and cultures were incubated a further 40 min. Samples of 0.5 A550-ml equivalents were harvested, and cross-linked as previously described (26). Briefly, cells were pelleted at room temperature, resuspended in 1 ml of sodium phosphate buffer (pH 6.8), and cross-linked for 15 min at room temperature with either 1% formaldehyde or 1% monomeric formaldehyde generated from solubilized paraformaldehyde. The monomeric formaldehyde was stored at 4°C and used within 1 month of opening the glass ampule. Results obtained with monomeric formaldehyde (see Fig. 3) were identical to those obtained with the standard formaldehyde solutions and were also identical to past results (data not shown and references 26 and 40). Cross-linked cells were then pelleted and solubilized in 2× LSB for 5 min at 60°C. Samples of 0.375 A550-ml equivalents were analyzed by immunoblotting as described above except that monoclonal antibody specific for the T7 tag was used at a dilution of 1:5,000, anti-TonB monoclonal antibody was used at a dilution of 1:2,500, and anti-mouse IgG-HRPO secondary antibody was used at a dilution of 1:5,000.
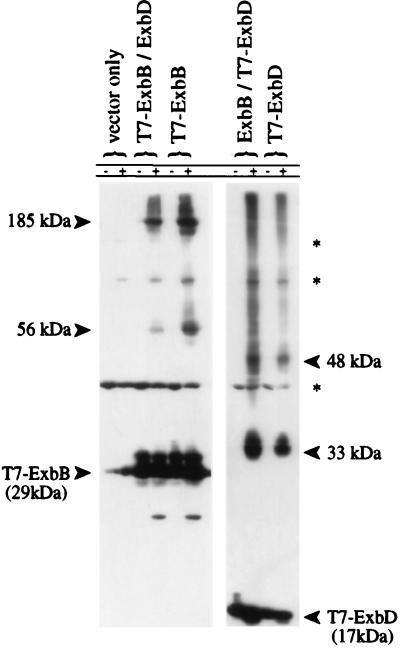
FIG. 3.
In vivo cross-linking of cells expressing T7-ExbB or T7-ExbD. ΔexbBD cells (KP1269) expressing plasmids pACYC184 and pET-24(a)+ (vector controls), pKP360 (ExbD), pKP339 (T7-ExbB), pKP361 (ExbB), or pKP323 (T7-ExbD) were untreated (−) or cross-linked in vivo with monomeric formaldehyde (+) as described in Materials and Methods. Samples representing equal cell numbers were electrophoresed on SDS–11% polyacrylamide gels, and T7 epitope-tagged proteins were detected by immunoblot analysis with an anti-T7 epitope tag monoclonal antibody. The small amount of T7-ExbB monomer signal detected in vector-only controls is due to bleed-through from the adjacent lane and is visible only after extended exposures. The bands denoted by an asterisk at 42, 85, and 120 kDa appear to arise from unidentified cross-reactive proteins since they appear in lanes carrying vector controls.
RESULTS
ExbB and ExbD expressed as T7 epitope tag fusion proteins are functional.
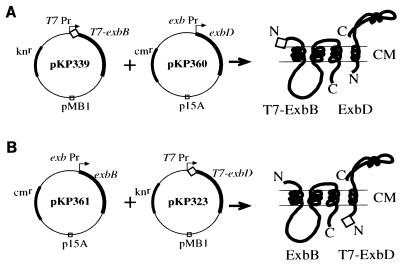
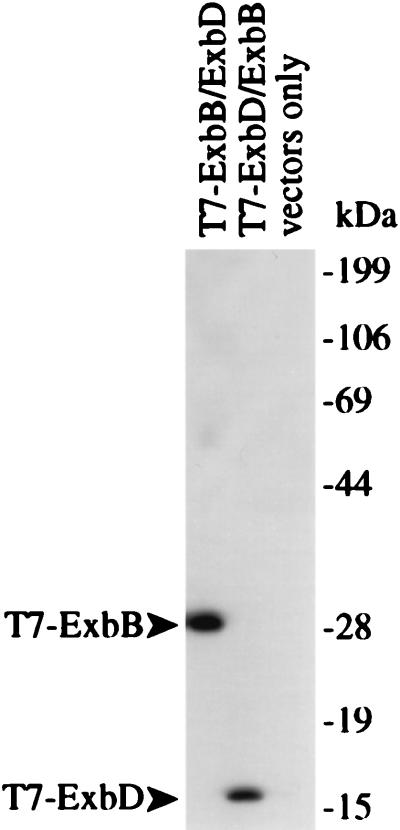
In order to detect ExbB and ExbD, plasmids were constructed to allow IPTG-inducible expression of either ExbB (pKP339) or ExbD (pKP323) fused with an N-terminal T7 epitope tag to which monoclonal antibodies are commercially available. Since both proteins are required for efficient TonB-dependent energy transduction (2), compatible plasmids were also constructed to allow concurrent expression of the untagged partner protein from the exb promoter. Plasmid sets pKP339 and pKP361 were paired to encode T7-ExbB and ExbD, respectively, while pKP323 and pKP360 were paired to encode T7-ExbD and ExbB, respectively (Fig. 1). Immunoblot analysis using an anti-T7 monoclonal antibody of a ΔexbBD strain expressing either T7-ExbB or T7-ExbD detected proteins at 29 and 17 kDa (Fig. 2), respectively, consistent with their predicted molecular masses (11, 20).
FIG. 1.
Plasmids expressing T7 epitope-tagged ExbB or ExbD proteins and the untagged partner proteins. In each case, the T7-tagged protein is expressed from the T7 promoter. T7 RNA polymerase is induced by 0.01 mM IPTG from λDE3 inserted into the host cell chromosome. The partner protein is expressed from the exb promoter. (A) pKP339 (T7-ExbB) and the compatible plasmid pKP360 (ExbD). (B) pKP323 (T7-ExbD) and the compatible plasmid pKP361 (ExbB). Expected protein products are illustrated to the right of each plasmid set.
FIG. 2.
Expression of T7-ExbB and T7-ExbD. T7 epitope-tagged proteins produced from ΔexbBD cells (KP1269) expressing T7-ExbB-ExbD (pKP339-pKP360), T7-ExbD-ExbB (pKP323-pKP361), or vector-only controls [pACYC184-pET-24(a)+] are shown. Equivalent numbers of cells were electrophoresed on SDS–11% polyacrylamide gels and detected by immunoblotting with a T7 epitope tag-specific monoclonal antibody.
We wanted to express either T7-ExbB or T7-ExbD at physiologically relevant levels. However, since we had no means of determining levels of the chromosomally encoded proteins, we decided to induce the T7 plasmids with a concentration of IPTG that did not affect the growth rate of cells carrying each plasmid set and that also produced detectable levels of protein (data not shown). To determine whether addition of an N-terminal epitope tag disrupted activity of either protein under these conditions, these plasmid sets were expressed in an exbBD deletion strain (KP1269) and assayed for sensitivity to bacteriophage φ80 and to colicins B and D, all lethal agents which require TonB-dependent energy transduction to gain entry into the cell. Under the conditions assayed, both plasmid sets conferred significantly more sensitivity to colicins B and D and to φ80 than the vector control, demonstrating that the T7-tagged proteins were functional (Table 2).
TABLE 2.
ExbB and ExbD complementation assaysa
| Strain (plasmid[s]) | Relevant genotype | Dilution resulting in clearance by:
|
||
|---|---|---|---|---|
| Colicin B | Colicin D | Phage φ80 | ||
| GM1 | Wild type | ND | ND | 109 |
| KP1269(pKP298) | exbBD+ | 57 | 56 | 109 |
| KP1269(pKP339/pKP360) | T7exbB+exbD+ | 56 | 55 | 106 |
| KP1269(pKP323/pKP361) | T7exbD+exbB+ | 56 | 55 | 107 |
| KP1269[pET-24(a)+/ pACYC184] | ΔexbBD | 54 | 53 | 104 |
ExbBD activity was measured by sensitivity to colicins B and D and phage φ80. The results for colicins B and D indicate the final dilution that resulted in complete clearing of the indicator strain. The results for phage φ80 indicate the first dilution that resulted in no plaques being formed on the indicator strain. In both cases, lower numbers indicate higher levels of TonB (and thus ExbBD) function. All assays were performed in triplicate and gave identical results. ND, not done.
Cross-linking analysis of ExbB and ExbD proteins.
ExbB and ExbD topologically partition to opposite sides of the cytoplasmic membrane (20–22). ExbB and ExbD have significant extramembrane domains (approximately 88 and 98 residues, respectively), suggesting that each may interact with additional proteins. We chose to analyze potential interactions of proteins with either ExbB or ExbD by in vivo cross-linking analysis of epitope-tagged Exb proteins.
The ΔexbBD strain expressing T7-ExbB and untagged ExbD was cross-linked with 1% formaldehyde in vivo, and ExbB-specific complexes were detected by immunoblotting with anti-T7 monoclonal antibody (Fig. 3). In addition to ExbB monomer detected at approximately 29 kDa, two additional ExbB-dependent complexes were detected at 56 and 185 kDa. ExbD was not required to form either ExbB-dependent cross-linked complex, but its absence appeared to enhance the formation of the 56- and 185-kDa complexes.
Similar analyses were performed on the ΔexbBD strain expressing T7-ExbD and untagged ExbB. ExbD monomer was detected at approximately 17 kDa with ExbD-dependent cross-linked complexes occurring at 33 and 48 kDa (Fig. 3). The absence of ExbB had no effect on formation of the complexes. Surprisingly, the 33-kDa complex was as abundant as the monomer.
Neither TonB nor FepA is involved in formation of the ExbB- or ExbD-dependent complexes.
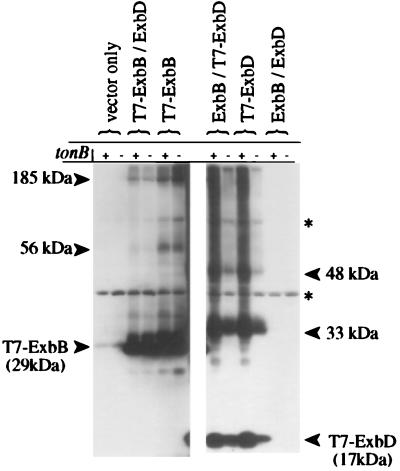
Previously we have identified proteins which cross-link to TonB by using strains with the genes for the candidate proteins ExbB and FepA deleted (40). The identical approach was taken here by using strains bearing mutations in proteins known to be involved in TonB-dependent energy transduction. T7-ExbB and ExbD or T7-ExbD and ExbB were expressed in a ΔtonB ΔexbBD strain and cross-linked as described above. The ExbB- and ExbD-dependent complexes observed in Fig. 3 were still present, suggesting TonB was not a component of any of the complexes (Fig. 4).
FIG. 4.
Effect of TonB on T7-ExbB and T7-ExbD complex formation. Strains KP1269 (ΔexbBD) and KP1346 (ΔexbBD ΔtonB) carrying plasmids pKP360 (ExbD) and/or pKP339 (T7-ExbB), pKP361 (ExbB) and/or pKP323 (T7-ExbD), pACYC184 and pET-24(a)+ (vector controls), or pKP298 and pET-24(a)+ (ExbBD) were cross-linked in vivo by using standard formaldehyde as described in Materials and Methods. Samples representing equal cell numbers were electrophoresed on SDS–11% polyacrylamide gels, and complexes containing T7 epitope-tagged proteins were detected by immunoblot analysis with anti-T7 epitope tag monoclonal antibody. +, strain containing tonB; −, strain in which tonB has been deleted; ∗, cross-reactive band.
The 185-kDa ExbB-dependent complex was sufficiently large to contain the outer membrane receptor FepA, although based on the localization of ExbB and ExbD to the cytoplasmic membrane in sucrose density gradients (10, 11, 28), this possibility was regarded as unlikely. Nonetheless, to determine whether FepA participated in the formation of any novel Exb complexes, the T7-tagged proteins were analyzed as before in a ΔfepA ΔexbBD strain. No ExbB- or ExbD-dependent complexes were altered in the absence of FepA (data not shown). Taken together, these results suggested that ExbB and ExbD each formed interactions with either two novel proteins or with additional ExbB and ExbD monomers.
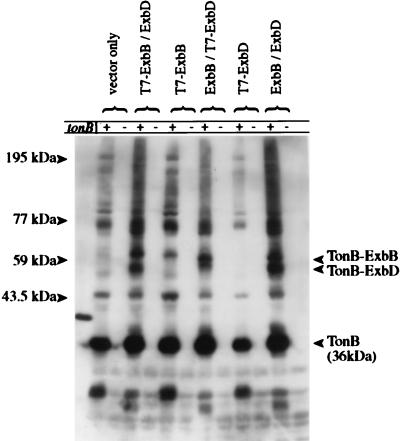
Overexpression of T7-ExbB and T7-ExbD altered the pattern of TonB-specific interactions detected by in vivo cross-linking (Fig. 5). In the strains expressing T7-ExbB, the migration of the 59-kDa complex proposed to consist of TonB-ExbB (26, 40) was retarded, reflecting addition of the T7 epitope tag to ExbB. The difference in migration of this complex provided a final confirmation that ExbB was the other component of the 59-kDa TonB-dependent complex. Based on the apparent molecular mass, this complex must consist of one TonB and one ExbB. In the samples containing overexpressed ExbD, a previously undetected 54-kDa TonB-dependent complex was identified. This 54-kDa complex also showed retarded migration in the samples containing T7 epitope-tagged ExbD, suggesting this complex contained both TonB and ExbD.
FIG. 5.
Effects of T7-ExbB and T7-ExbD on the TonB cross-linking profile. Strains KP1269 (ΔexbBD) and KP1346 (ΔexbBD ΔtonB) carrying plasmids pKP360 (ExbD) and/or pKP339 (T7-ExbB), pKP361 (ExbB) and/or pKP323 (T7-ExbD), pACYC184 and pET-24(a)+ (vector controls), or pKP298 and pET-24(a)+ (ExbBD) were cross-linked in vivo by using standard formaldehyde as described in Materials and Methods. Samples representing equal cell numbers were electrophoresed on SDS–11% polyacrylamide gels, and complexes containing TonB were detected by immunoblot analysis with an anti-TonB monoclonal antibody. The TonB-T7-ExbD complex was not detected in the absence of ExbB presumably because the chemical stabilities of both TonB and ExbD depend upon ExbB. +, strain containing tonB; −, strain in which tonB has been deleted.
The 56-kDa ExbB complex is a homodimer and the 185-kDa complex contains an ExbB homotrimer.
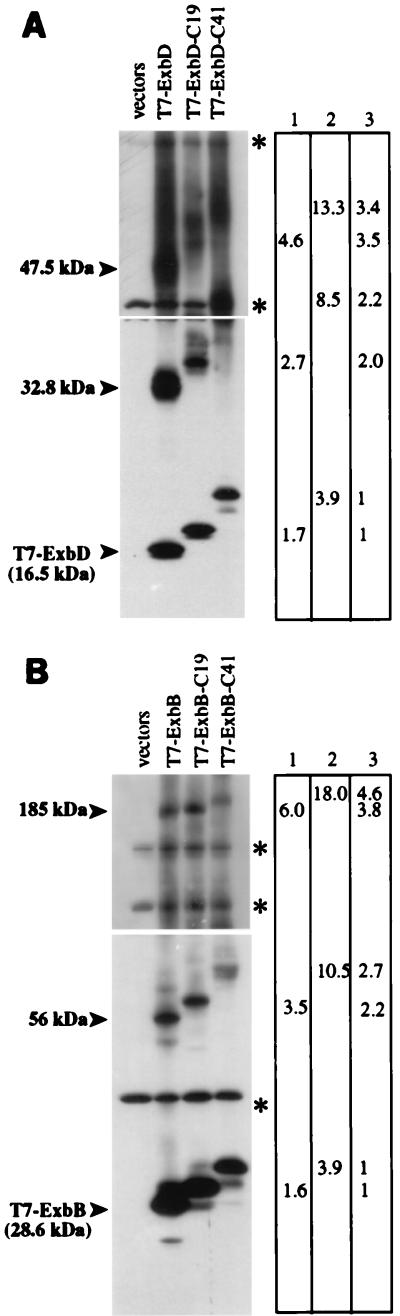
The apparent molecular mass of the 56-kDa ExbB-dependent complex was twice that of the T7-ExbB monomer, which raised the possibility that this complex might represent an ExbB dimer. This possibility was addressed by analyzing T7-ExbB-dependent cross-linked complexes from strains expressing T7-ExbB with an additional either 19 or 41 residues at the C terminus (T7-ExbB-C19 or T7-ExbB-C41, respectively) (Fig. 6). If the 56-kDa complex was a dimer, the difference in migration between the 56-kDa T7-ExbB complex and the equivalent T7-ExbB size variant complex should be twice the difference in migration measured between the ExbB monomer proteins. The molecular masses of T7-ExbB-C19 and T7-ExbB-C41 were measured to be larger than T7-ExbB by 1.6 and 3.9 kDa, respectively. The differences in migration between the 56-kDa T7-ExbB cross-linked complex and the corresponding T7-ExbB-C19 and T7-ExbB-C41 cross-linked complexes were measured to be, respectively, 2.2 and 2.7 times the differences in migration between the monomer proteins. These data suggested that the 56-kDa ExbB-dependent cross-linked complex consisted of an ExbB homodimer. Correspondingly, the difference in migration between the 185-kDa T7-ExbB-dependent complex and the equivalent T7-ExbB-C19 and T7-ExbB-C41 cross-linked complexes was at least threefold (3.8 and 4.6 times, respectively) the difference in migration between the T7-ExbB and T7-ExbB-C19 monomers, suggesting this complex most likely contained a homotrimer (but possibly a homotetramer) of ExbB.
FIG. 6.
In vivo cross-linking of T7-ExbB or T7-ExbD size variants. (A) Strain KP1269 (ΔexbBD) carrying plasmids pKP361 (ExbB) and pKP323 (T7-ExbD), pKP375 (T7-ExbD-C19), or pKP377 (T7-ExbD-C41). (B) Strain KP1269 (ΔexbBD) carrying plasmids pKP360 (ExbD) and pKP339 (T7-ExbB), pKP353 (T7-ExbB-C19), or pKP376 (T7-ExbB-C41). Cells were cross-linked in vivo by using standard formaldehyde as described in Materials and Methods. Samples representing equal cell numbers were electrophoresed on SDS–7 to 20% polyacrylamide gradient gels, and complexes containing T7 epitope-tagged proteins were detected by immunoblot analysis with an anti-T7 epitope tag monoclonal antibody. The molecular masses of complexes were measured relative to those of unstained protein standards (BenchMark; Gibco BRL). Columns: 1, measured differences in molecular mass between T7-Exb-C19 complexes and T7-Exb complexes (in kilodaltons); 2, measured differences in molecular mass between T7-Exb-C41 complexes and T7-Exb complexes (in kilodaltons); 3, factor increase in molecular mass. Longer exposures (top) were necessary for detection of the higher-molecular-mass complexes; shorter exposures (bottom) were necessary for accurate relative mobility measurement of lower-molecular-mass complexes. ∗, cross-reactive band.
The 33- and 48-kDa ExbD complexes are homodimers and homotrimers, respectively.
The apparent molecular masses of the 33- and 48-kDa T7-ExbD-dependent complexes were approximately twice and three times the molecular mass of T7-ExbD monomer, suggesting that these complexes could consist of ExbD homodimers and homotrimers, respectively. To determine if this was the case, strains expressing size variants of T7-ExbD were constructed, cross-linked, and analyzed as described above (Fig. 6). The differences in migration between the 33-kDa T7-ExbD dependent complex and the equivalent T7-ExbD-19 or T7-ExbD-C41 complex were 2.0 and 2.2 times, respectively, the difference between the monomer proteins, suggesting that the 33-kDa complex contained an ExbD dimer. Similarly, the differences in migration between the 48-kDa T7-ExbD-dependent complex and the equivalent T7-ExbD-C19 or T7-ExbD-C41 complex were 3.5 and 3.4 times, respectively, the difference between the monomer proteins. These data suggested that the 48-kDa ExbD-dependent complex consisted of an ExbD trimer.
DISCUSSION
We have previously used in vivo formaldehyde cross-linking analysis of TonB to define interactions within the TonB-dependent energy transduction complex (24, 26, 40). In this study, we performed a similar analysis to investigate the interactions of ExbB and ExbD with other proteins.
Previous studies from another lab had suggested that formaldehyde as it is usually purchased exists as a solution of variable-length polymers (35). In order to ensure that the formaldehyde used in this study inserted only a methylene bridge between reactive residues and was thus a zero-length cross-linker (30), we also used monomeric formaldehyde reconstituted from paraformaldehyde. To detect ExbB and ExbD, we designed plasmids to allow inducible expression of either ExbB or ExbD fused at the N terminus with a T7 epitope tag to which monoclonal antibodies are commercially available. Since ExbB and ExbD appear to function as a unit (2), we also designed compatible plasmids to allow concurrent expression of the untagged partner protein from the exb promoter region.
Immunoblot analysis using an anti-T7 monoclonal antibody of cells expressing T7-ExbB or T7-ExbD detected proteins at approximately 29 and 17 kDa, respectively, consistent with the predicted molecular masses of these proteins. A level of expression of T7-ExbB or T7-ExbD was chosen which would not affect cell growth rate but would produce sufficient quantities of protein to be detectable by immunoblotting using a chemiluminescence assay. The low level of IPTG used (0.01 mM compared to 1 mM used for maximal induction of the lactose operon [31]) together with the short time of induction (40 min in most cases) suggested that the T7-ExbB and T7-ExbD proteins were not strongly overproduced, yet their presence was sufficient to confer function. The small reduction in TonB-dependent activity in strains expressing the plasmid sets compared to plasmid-encoded wild-type ExbBD may be the result of (i) slight interference by the T7 epitope tag, (ii) a dominant negative effect arising from unbalanced stoichiometry between ExbB and ExbD, or (iii) expression of subchromosomal levels of T7-ExbB or T7-ExbD. Since the pET induction system is significantly leaky and T7 RNA polymerase is highly efficient, we regard the latter explanation as unlikely. Fractionation studies have demonstrated that T7-ExbB and T7-ExbD expressed under these conditions localized to the cytoplasmic membrane fractions as expected (reference 28 and data not shown).
Using monomeric formaldehyde, T7-ExbB could be cross-linked into complexes of 56 and approximately 185 kDa, while T7-ExbD could be cross-linked into complexes of 33 and 48 kDa. Identical results were obtained with the same reagent-grade formaldehyde used in our previous studies (data not shown). We conclude, therefore, that the reagent-grade formaldehyde was also monomeric when diluted to 1%. The absence of TonB, FepA, and the untagged ExbD or ExbB partner had no effect on the presence of the cross-linked complexes. In addition, we realized that, based on the molecular masses of the complexes, the possibility that they contained homomultimers existed.
To test that possibility, plasmids expressing two size variants of both T7-tagged proteins were constructed, and the resultant proteins were cross-linked. The differences in sizes of putative dimers and trimers from all three T7-ExbB and T7-ExbD proteins allowed us to conclude that the 33-kDa ExbD- and 56-kDa ExbB-dependent complexes arose from homodimers, that the 48-kDa ExbD-dependent complex was a trimer, and that the 185-kDa ExbB-dependent complex was most likely a trimer, although an ExbB tetramer could not be excluded. Coexpression of ExbBD from a compatible plasmid decreased the abundance of T7-ExbB dimer and trimer; however, the effect on T7-ExbD complex formation was not determined (data not shown). Unlike all the other complexes we observed in this study, the migration of the 185-kDa complex varied, with migration increasingly retarded at higher percentages of acrylamide, suggesting an unusual composition or shape. The size of the 185-kDa complexes ranged from a minimum of 135 kDa (in an 8% polyacrylamide gel [data not shown]) to a maximum of 185 kDa in an 11% polyacrylamide gel (Fig. 3 and 4). Assuming ExbB to be a trimer, the result left approximately 50 (at least) to 100 kDa (at most) worth of unidentified protein in the complex. It should be noted that, although the apparent molecular mass of the 185-kDa complex varied with the percentage of acrylamide in the gel, the differences in molecular mass between the 185-kDa T7-ExbB and equivalent T7-ExbB-C19 and T7-ExbB-C41 complexes remained relatively constant in gels with different percentages of acrylamide.
Some individual differences in cross-linking efficiency were noted during the course of these experiments. First, in the case of T7-ExbB, the abundance of the 56- and 185-kDa complexes was significantly increased in the absence of ExbD but not of TonB. It seemed likely, therefore, that the increase in cross-linking efficiency was due to the loss of a protein capable of competing with ExbB for its ExbB homomultimeric interactions, i.e., ExbD, which is likely to be as abundant as ExbB. While the ExbD-dependent cross-linked complexes did not appear to be similarly increased in the absence of ExbB, the interpretation of these data becomes complicated because the chemical stability of ExbD is known to be reduced in the absence of ExbB (12). One possibility is that the expected increase in cross-linking efficiency of ExbD-dependent complexes in the absence of ExbB is masked by the reduced levels of T7-ExbD in this strain.
Second, in the case of T7-ExbD, the cross-linked dimer complexes were surprisingly abundant. Whereas with TonB and ExbB the non-cross-linked monomer is by far the most abundant species in a cross-linking profile, very brief exposures of cross-linked ExbD immunoblots reveal that approximately 50% of the detectable ExbD was present as monomer and 50% was present as dimer (data not shown). The amount of trimer under these circumstances is significantly less. These estimated percentages were based on determining that no more than 2% of the ExbD monomer or dimer passed through the PVDF membrane during transfer and that none of the monomer or dimer remained in the gel following electroblotting (data not shown). This efficiency may reflect stability of the complexes or an abundance of correctly positioned cross-linkable residues in the periplasmic domain of ExbD. In addition, we were able to detect a TonB-ExbD complex for the first time with anti-TonB antisera. As above, this interaction is likely to be occurring within the periplasmic domains of the two proteins.
Third, while we can detect TonB-ExbB and TonB-ExbD cross-linked complexes with an anti-TonB monoclonal antibody, we do not correspondingly detect TonB-ExbB or TonB-ExbD complexes with an anti-T7 epitope antibody. This could be due to a combination of factors: (i) ExbB and ExbD may be normally present in greater abundance than TonB (12); (ii) ExbB and ExbD are somewhat overexpressed in our system; and (iii) ExbB and ExbD interactions with TonB constitute a small proportion of possible ExbB and ExbD interactions, whereas TonB interactions with ExbB and ExbD represent approximately 25% of the detectable TonB-specific interactions based on our cross-linking results (40). This seems to be the situation for TonB interactions with outer membrane receptor BtuB (1), where the interaction is more easily detected with an anti-TonB antibody than with antibodies directed against BtuB.
We did not detect ExbB and ExbD interactions by our cross-linking assays. This could be due to the fact that ExbD contains no formaldehyde-reactive residues in its putative transmembrane signal anchor, the probable domain for interaction with ExbB, or to the fact that there are no cross-linkable residues positioned closely enough between the two proteins to allow cross-linking. However, there is good evidence that the two proteins do interact: ExbB stabilizes ExbD to exposure to endogenous proteases in vivo and to exogenous protease in spheroplasts (12) and both TonB and ExbD (independently) copurify with His6-ExbB on a nickel affinity column (8).
Taken together, our results here and those demonstrating interactions between ExbB and ExbD from Braun’s lab (8, 12) suggest the possibility that a complex might exist which consists of three ExbB proteins and three ExbD proteins. Two alternative, and overlapping, predictions of ExbB membrane-spanning segments have been made with the important difference that in one instance the transmembrane segments have no charged residues (22), while in the other model they contained charged residues (21). In addition, an essential aspartate (D25, conferring a dominant negative phenotype) is found in the ExbD transmembrane segment (8). It will be important to determine the transmembrane segment boundaries more precisely since the presence or absence of charged residues in the transmembrane regions of either protein would have important implications for function.
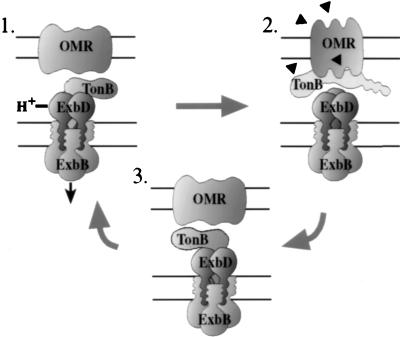
While direct biochemical evidence for a heterohexamer complex is lacking, the proposed complex would contain 12 membrane-spanning segments and would thus have interesting implications for the TonB-dependent energy transduction mechanism. The proposed heterohexamer complex could serve either or both of two possible functions. (i) It could act as a protein channel to allow docking of the TonB N-terminal transmembrane segment in the cytoplasmic membrane. This arrangement of the proteins might offer a means by which, consistent with our previous data, TonB could be released from the cytoplasmic membrane to shuttle to the outer membrane (28). (ii) Our recent results suggest that ExbB and ExbD are necessary for TonB to achieve an energy-dependent conformation (25). The proposed ExbB and ExbD heterohexamer could potentially form an ion-translocating channel which could couple proton motive force to TonB conformational changes and subsequently to active transport of siderophores and vitamin B12 across the outer membrane. A model incorporating both these possibilities is presented in Fig. 7.
FIG. 7.
Model of TonB-dependent energy transduction. ExbB and ExbD may exist as a heterohexamer complex (three ExbB and three ExbD) in the cytoplasmic membrane. ExbB and ExbD probably interact through transmembrane segments and together may provide a protein channel for the transmembrane segment of TonB. Step 1. TonB is energized by passage of a proton through the proposed ExbBD heterohexamer (25). Step 2. TonB either shuttles to or becomes more strongly associated with the outer membrane (28) and physically contacts a TonB-dependent outer membrane receptor (OMR) (40), triggering a conformational change in the OMR (19) such that ligand is pumped into the periplasmic space (36, 48). Step 3. ExbB and ExbD cycle TonB that is in a tight association with the outer membrane back to a high-affinity interaction with the cytoplasmic membrane energy transduction complex, allowing the energy transduction process to be repeated (28).
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institute of General Medical Sciences (GM42416) awarded to K.P.
We gratefully acknowledge Ray Larsen for construction of pKP353 and Lisa Helms for construction of pKP375. We thank Ray Larsen, Micky Thomas, and Tracy Letain for helpful discussions and Ray Larsen for critical reading of the manuscript.
REFERENCES
- 1.Ahmer, B., and K. Postle. Unpublished data.
- 2.Ahmer B M M, Thomas M G, Larsen R A, Postle K. Characterization of the exbBD operon of Escherichia coli and the role of ExbB and ExbD in TonB function and stability. J Bacteriol. 1995;177:4742–4747. doi: 10.1128/jb.177.16.4742-4747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell P E, Nau C D, Brown J T, Konisky J, Kadner R J. Genetic suppression demonstrates direct interaction of TonB protein with outer membrane transport proteins in Escherichia coli. J Bacteriol. 1990;172:3826–3829. doi: 10.1128/jb.172.7.3826-3829.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas G D, Anderson J E, Sparling P F. Cloning and functional characterization of Neisseria gonorrhoeae tonB, exbB and exbD genes. Mol Microbiol. 1997;24:169–179. doi: 10.1046/j.1365-2958.1997.3421692.x. [DOI] [PubMed] [Google Scholar]
- 5.Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J Bacteriol. 1993;175:3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun V. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 7.Braun V. The structurally related exbB and tolQ genes are interchangeable in conferring tonB-dependent colicin, bacteriophage, and albomycin sensitivity. J Bacteriol. 1989;171:6387–6390. doi: 10.1128/jb.171.11.6387-6390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun V, Gaisser S, Herrman C, Kampfenkel K, Killman H, Traub I. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J Bacteriol. 1996;178:2836–2845. doi: 10.1128/jb.178.10.2836-2845.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun V, Günter K, Hantke K. Transport of iron across the outer membrane. Biol Metals. 1991;4:14–22. doi: 10.1007/BF01135552. [DOI] [PubMed] [Google Scholar]
- 10.Eick-Helmerich K, Braun V. Import of biopolymers into Escherichia coli: nucleotide sequences of the exbB and exbD genes are homologous to those of the tolQ and tolR genes, respectively. J Bacteriol. 1989;171:5117–5126. doi: 10.1128/jb.171.9.5117-5126.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eick-Helmerich K, Hantke K, Braun V. Cloning and expression of the exbB gene of Escherichia coli K-12. Mol Gen Genet. 1987;206:246–251. doi: 10.1007/BF00333580. [DOI] [PubMed] [Google Scholar]
- 12.Fischer E, Günter K, Braun V. Involvement of ExbB and TonB in transport across the outer membrane of Escherichia coli: phenotypic complementation of exb mutants by overexpressed tonB and physical stabilization of TonB by ExbB. J Bacteriol. 1989;171:5127–5134. doi: 10.1128/jb.171.9.5127-5134.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Günter K, Braun V. In vivo evidence for FhuA outer membrane receptor interaction with the TonB inner membrane protein from Escherichia coli. FEBS Lett. 1990;274:85–88. doi: 10.1016/0014-5793(90)81335-l. [DOI] [PubMed] [Google Scholar]
- 14.Gutermann S K, Dann L. Excretion of enterochelin by exbA and exbB mutants of Escherichia coli. J Bacteriol. 1973;114:1225–1230. doi: 10.1128/jb.114.3.1225-1230.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heller K J, Kadner R J, Günter K. Suppression of the btuB451 mutation by mutations in the tonB gene suggests a direct interaction between TonB and TonB-dependent receptor proteins in the outer membrane of Escherichia coli. Gene. 1988;64:147–153. doi: 10.1016/0378-1119(88)90488-x. [DOI] [PubMed] [Google Scholar]
- 16.Howard S P, Meiklejohn H G, Shivak D, Jahagirdar R. A TonB-like protein and a novel membrane protein containing an ATP-binding cassette function together in exotoxin secretion. Mol Microbiol. 1996;22:595–604. doi: 10.1046/j.1365-2958.1996.d01-1713.x. [DOI] [PubMed] [Google Scholar]
- 17.Jarosik G P, Maciver I, Hansen E J. Utilization of transferrin-bound iron by Haemophilus influenzae requires an intact tonB gene. Infect Immun. 1995;63:710–713. doi: 10.1128/iai.63.2.710-713.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarosik G P, Sanders J D, Cope L D, Muller-Eberhard U, Hansen E J. A functional tonB gene is required for both utilization of heme and virulence expression by Haemophilus influenzae type b. Infect Immun. 1994;62:2470–2477. doi: 10.1128/iai.62.6.2470-2477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, Payne M A, Cao Z, Foster S B, Feix J B, Newton S C, Klebba P E. Ligand-specific opening of a gated porin channel in the outer membrane of living bacteria. Science. 1997;276:1261–1264. doi: 10.1126/science.276.5316.1261. [DOI] [PubMed] [Google Scholar]
- 20.Kampfenkel K, Braun V. Membrane topology of the Escherichia coli ExbD protein. J Bacteriol. 1992;174:5485–5487. doi: 10.1128/jb.174.16.5485-5487.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kampfenkel K, Braun V. Topology of the ExbB protein in the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1993;268:6050–6057. [PubMed] [Google Scholar]
- 22.Karlsson M, Hannavy K, Higgins C F. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol Microbiol. 1993;8:389–396. doi: 10.1111/j.1365-2958.1993.tb01582.x. [DOI] [PubMed] [Google Scholar]
- 23.Larsen R A, Foster-Hartnett D, McIntosh M A, Postle K. Regions of Escherichia coli TonB and FepA proteins essential for in vivo physical interactions. J Bacteriol. 1997;179:3213–3221. doi: 10.1128/jb.179.10.3213-3221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen R A, Myers P S, Skare J T, Seachord C L, Darveau R P, Postle K. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J Bacteriol. 1996;178:1363–1373. doi: 10.1128/jb.178.5.1363-1373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen, R. A., M. G. Thomas, and K. Postle. Unpublished data.
- 26.Larsen R A, Thomas M T, Wood G E, Postle K. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (ΔV17) by a missense mutation in ExbB. Mol Microbiol. 1994;13:627–640. doi: 10.1111/j.1365-2958.1994.tb00457.x. [DOI] [PubMed] [Google Scholar]
- 27.Larsen R A, Wood G E, Postle K. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol Microbiol. 1993;10:943–953. doi: 10.1111/j.1365-2958.1993.tb00966.x. [DOI] [PubMed] [Google Scholar]
- 28.Letain T E, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Gram-negative bacteria. Mol Microbiol. 1997;24:271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- 29.Matsushiro A. Specialized transduction of tryptophan markers in Escherichia coli K-12 by bacteriophage φ80. Virology. 1963;19:475–482. doi: 10.1016/0042-6822(63)90041-2. [DOI] [PubMed] [Google Scholar]
- 30.Means G E, Feeney R E. Chemical modification of proteins. San Francisco, Calif: Holden-Day, Inc.; 1971. [Google Scholar]
- 31.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 32.Moeck G S, Coulton J W, Postle K. Cell envelope signaling in Escherichia coli. Ligand binding to the ferrichrome-iron receptor FhuA promotes interaction with the energy-transducing protein TonB. J Biol Chem. 1997;272:28391–28397. doi: 10.1074/jbc.272.45.28391. [DOI] [PubMed] [Google Scholar]
- 33.Postle K. TonB and the gram-negative dilemma. Mol Microbiol. 1990;4:2019–2025. doi: 10.1111/j.1365-2958.1990.tb00561.x. [DOI] [PubMed] [Google Scholar]
- 34.Postle K. TonB protein and energy transduction between membranes. J Bioenerg Biomembr. 1993;25:591–601. doi: 10.1007/BF00770246. [DOI] [PubMed] [Google Scholar]
- 35.Prossnitz E, Nikaido K, Ulbrich S J, Ames G F-L. Formaldehyde and photoactivatable cross-linking of the periplasmic binding protein to a membrane component of the histidine transport system of Salmonella typhimurium. J Biol Chem. 1988;263:17917–17920. [PubMed] [Google Scholar]
- 36.Reynolds P R, Mottur G P, Bradbeer C. Transport of vitamin B12 in Escherichia coli. Some observations on the roles of the gene products of btuC and tonB. J Biol Chem. 1980;255:4313–4319. [PubMed] [Google Scholar]
- 37.Roof S K, Allard J D, Bertrand K P, Postle K. Analysis of Escherichia coli TonB membrane topology by use of PhoA fusions. J Bacteriol. 1991;173:5554–5557. doi: 10.1128/jb.173.17.5554-5557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schöffler H, Braun V. Transport across the outer membrane of Escherichia coli K12 via the FhuA receptor is regulated by the TonB protein of the cytoplasmic membrane. Mol Gen Genet. 1989;217:378–383. doi: 10.1007/BF02464907. [DOI] [PubMed] [Google Scholar]
- 39.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skare J T, Ahmer B M M, Seachord C L, Darveau R P, Postle K. Energy transduction between membranes—TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J Biol Chem. 1993;268:16302–16308. [PubMed] [Google Scholar]
- 41.Skare J T, Roof S K, Postle K. A mutation in the amino terminus of a hybrid TrpC-TonB protein relieves overproduction lethality and results in cytoplasmic accumulation. J Bacteriol. 1989;171:4442–4447. doi: 10.1128/jb.171.8.4442-4447.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spudich J L. Variations on a molecular switch: transport and sensory signalling by archaeal rhodopsins. Mol Microbiol. 1998;28:1051–1058. doi: 10.1046/j.1365-2958.1998.00859.x. [DOI] [PubMed] [Google Scholar]
- 43.Stojiljkovic I, Srinivasan N. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein-bound iron in neisseriae. J Bacteriol. 1997;179:805–812. doi: 10.1128/jb.179.3.805-812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun T P, Webster R E. fii, a bacterial locus required for filamentous phage infection and its relation to colicin-tolerant tolA and tolB. J Bacteriol. 1986;165:107–115. doi: 10.1128/jb.165.1.107-115.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torres A G, Payne S M. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1997;23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 46.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuckman M, Osburne M S. In vivo inhibition of TonB-dependent processes by a TonB box consensus pentapeptide. J Bacteriol. 1992;174:320–323. doi: 10.1128/jb.174.1.320-323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wooldridge K G, Morrisey J A, Williams P H. Transport of ferric aerobactin into the periplasm and cytoplasm of Escherichia coli K12: role of envelope-associated proteins and effect of endogenous siderophores. J Gen Microbiol. 1992;138:597–603. doi: 10.1099/00221287-138-3-597. [DOI] [PubMed] [Google Scholar]