Abstract
We examined the pattern of FtsZ localization in a Bacillus subtilis minCD mutant. When grown in minimal medium, the majority (∼89%) of the minCD mutant cells with an FtsZ ring had a single, medially positioned FtsZ ring. These results indicate that genes in addition to minCD function to restrict the number and position of FtsZ rings. When grown in rich medium, greater than 50% of the minCD mutant cells had multiple FtsZ rings, indicating significant differences in regulation of FtsZ ring formation based on growth medium.
Many bacteria divide by binary fission, forming medially positioned septa and identical daughter cells. The process of cell division is governed by several gene products, the most conserved of which appears to be the tubulin-like protein FtsZ (23, 24). Prior to septation, FtsZ polymerizes as a ring structure at the nascent division site (5). Formation of the FtsZ ring is essential but not sufficient for invagination of the cell wall (1, 20).
In Escherichia coli, the minC, minD, and minE gene products are involved in regulating the placement of the division site (2, 11). It is postulated that rod-shaped cells have three potential division sites: the “new” site at midcell and the “old” sites near the cell poles (10, 12, 31, 33). Previous work suggests that MinC and MinD function in tandem as division inhibitors blocking formation of the FtsZ ring at all sites. MinE relieves the MinCD cell division block at the midcell site, allowing for binary fission. In the absence of MinC and/or MinD, all three division sites appear to be available, and FtsZ rings can form at polar and medial positions. The use of polar sites results in the formation of anucleate minicells (4, 6, 12). MinE has recently been shown to localize to a medial position, bolstering the model that it functions to relieve the division block at midcell (30).
In Bacillus subtilis, the position of the cell division site switches from medial to polar during the initiation of sporulation (32). Polar septation results in the formation of two distinct cell types: the larger mother cell and the germ line forespore (3, 26). The switch in the position of the division site is preceded by a shift in the localization of FtsZ from a medial pattern to one in which the protein forms two rings, one at each potential polar division site (19). B. subtilis has homologs of MinC and MinD, but not MinE (17, 18, 21, 34). However, the divIVA gene product may serve as a functional analog of MinE (8, 13, 31). It has been suggested that the Min system may play a role in the switch from medial to polar septation during the initiation of sporulation (8, 13, 18, 21, 31, 34).
To investigate the role of the min genes in division site selection in B. subtilis, we determined the effect of a minCD-null mutation on the pattern of FtsZ localization and cell division during exponential growth in both a minimal and a rich growth medium. More specifically, we were interested in answering two questions: (i) what is the frequency of polar FtsZ ring formation and polar division in the absence of MinC and MinD, and (ii) does a minCD-null mutation cause the formation of multiple rings of FtsZ in a single cell?
We found that the majority of cells of a B. subtilis minCD-null mutant grown in minimal medium formed medially positioned FtsZ rings and that ring formation was limited to a single site per cell. Most of these cells divided by binary fission. These results strongly indicate that factors in addition to the min gene products function to ensure the medial localization of FtsZ and the restriction of FtsZ ring formation to a single site per cell. To our surprise, we found dramatically different results in cells grown in rich (Luria-Bertani [LB]) medium. Over 50% of the minCD mutant cells had multiple FtsZ rings, and of the cells with a ring, only 23% had a single medially positioned ring.
FtsZ ring formation in the minCD mutant in minimal medium.
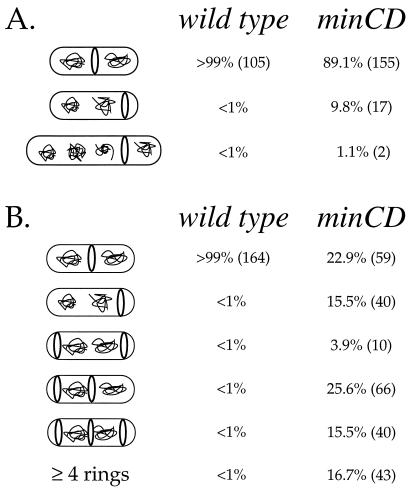
Using immunofluorescence microscopy, we visualized the location and number of FtsZ rings in wild-type and minCD mutant cells during exponential growth in minimal medium (Fig. 1A to F). Of the wild-type cells examined, ∼52% (105 of 201) had FtsZ rings, a number corresponding to that from previous experiments conducted under similar conditions (22). As expected, a medial pattern of FtsZ localization was observed exclusively in these cells, putting the frequency of polar ring formation at less than 1% (Fig. 1A to C and 2A).
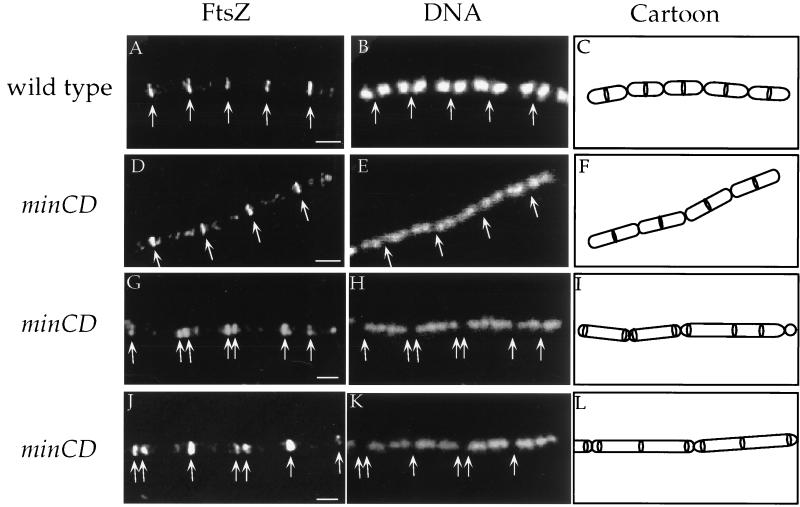
FIG. 1.
Immunolocalization of FtsZ in wild-type and minCD mutant cells. Cells were grown at 37°C in minimal glucose medium with 1.0% glucose, 0.1% glutamine, and required amino acids (16) (A to F) or in LB medium (G to L). Samples were taken during mid-exponential growth, fixed, and stained for immunofluorescence microscopy, essentially as described elsewhere (15, 19, 28). Arrows indicate the positions of FtsZ rings. Bars = 1 μm. Strains used included JH642 (trp phe) and the congenic minCD mutants PL820 (trp phe minCDΔ1::cat) and PL990 (trp phe ΔminCD990::spc). The minCDΔ1::cat allele (18) is a deletion from codon 118 of minC (of 227 codons total) through codon 187 of minD (of 268 codons total), with a chloramphenicol resistance cassette inserted. The ΔminCD990::spc allele deletes all of minC and all but the last two codons of minD with a spectinomycin resistance cassette inserted. Similar results were obtained with each of the two minCD mutants. (A, D, G, and J) Visualization of FtsZ with affinity-purified antibodies (against the B. subtilis FtsZ) and a secondary antibody conjugated to the red fluorophore Cy-3 (Jackson Immunoresearch) (19). (B, E, H, and K) Visualization of DNA with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). (C, F, I, and L) Cartoons of images indicating outline of cells and FtsZ rings. Cell walls were visualized (not shown) with wheat germ agglutinin (lectin) conjugated to the green fluorophore fluorescein isothiocyanate (Molecular Probes) (29). (A to C) Wild-type strain JH642 (trp phe) grown in minimal medium. Note the evenly spaced rings of FtsZ flanked by single nucleoids. (D to F) minCD mutant strain PL820 (trp phe minCDΔ1::cat) grown in minimal medium. A medial pattern of FtsZ localization similar to that of the wild-type strain is observed in these cells. Only one ring of FtsZ is observed per cell length. The minCD mutant cells are ∼30% longer, on average, than the wild-type cells. The significance of this is unclear. (G to L) minCD mutant strain PL820 (trp phe minCDΔ1::cat) grown in LB medium. Most of the cells in these fields have multiple rings of FtsZ.
The frequency of FtsZ ring formation was not altered in the minCD mutant. Approximately 52% (174 of 332) of the minCDΔ1 mutant cells had FtsZ rings. Among the mutant cells with an FtsZ ring, ∼89% (155 of 174) had the FtsZ ring at midcell (Fig. 1D to F and 2). Approximately 10% (17 of 174) had FtsZ rings positioned immediately adjacent to one pole of the cell, and an additional 1.1% (2 of 174) had FtsZ rings that were inset from the poles by approximately one-fourth the length of the cell (Fig. 2). When grown in minimal medium, none of the minCD mutant cells that we examined had more than one FtsZ ring.
FIG. 2.
Analysis of FtsZ ring position in wild-type (JH642) and minCD mutant (PL820) cells. Only cells with visible FtsZ rings were counted. Numbers in parentheses indicate the total numbers of cells observed in each class. Actual cell sizes vary, although for simplicity most cells are drawn similarly. (A) Cells grown in minimal medium. The percentages of cells with FtsZ rings were virtually the same in wild-type and minCDΔ1 mutant cells, 52.2% (105 of 201) and 52.4% (174 of 332), respectively. The small percentage of minCD mutant cells that have rings at the quarter positions may result from previous rounds of cell division in which polar septation occurred at the expense of binary fission, leaving both quarter positions (the medial sites of the previous generation) available in the newly formed daughter cells. (B) Cells grown in LB medium. The percentage of cells with FtsZ rings was 90 to 95% for both wild type and the minCD mutant.
Frequency of minicell formation in the minCD-null mutant.
We determined the frequency of minicell formation in wild-type and minCDΔ1 mutant cells grown in minimal medium. Two types of minicells were observed: those that were floating free in the medium and those that were still attached to the parent cell. As expected, no minicells were visible in the culture of wild-type cells; the frequency of minicells was less than 0.5% (0 of 201).
Assuming that each polar FtsZ ring results in polar cell division and that the percentage of anucleate minicells in a population remains static, then after four generations the number of minicells should be equivalent to 1.875 times the number of polar FtsZ rings observed at a given time. Given that the FtsZ ring was polar in ∼9.8% of minCDΔ1 cells with rings (Fig. 2A), after four generations we expect that ∼18% of the cells should be anucleate minicells.
We visualized living cells from a growing culture of the minCD mutant. Cells were grown in the presence of the membrane dye FM4-64 (200 ng/ml; Molecular Probes) and sampled directly onto a microscope slide (27). By this method, we observed that ∼17% (31 of 178) of the cells in the minCDΔ1 culture were minicells. Similar results were obtained by using a minCD allele (ΔminCD::spc) that removes the entire minC coding region. These findings indicate that most of the polar FtsZ rings that form in minCD mutant cells in minimal medium are used for division.
The frequency of minicell formation that we observed is somewhat lower than that originally described for the B. subtilis min mutants (9). This difference could be due to use of different strain backgrounds, use of different alleles, and different growth conditions.
We also used wheat germ agglutinin (lectin) conjugated to the green fluorophore fluorescein isothiocyanate (Molecular Probes) to visualize the bacterial cell walls (29) and found that minicells comprised ∼12% (41 of 332) of the cells in a culture of the minCD mutant. We suspect that the multiple washes of samples during preparation for lectin staining and immunofluorescence microscopy cause some loss of minicells compared to the minimal preparation with FM4-64.
FtsZ ring formation and minicell production in the minCD mutant in rich medium.
Growth conditions are known to affect the fraction of cells that contain an FtsZ ring. Approximately 90 to 95% of wild-type cells have an FtsZ ring when grown in LB medium, compared to ∼50% when grown in minimal medium (22). As in minimal medium, the minCD mutation did not alter the fraction of cells containing an FtsZ ring. In LB medium, ∼92% (258 of 280) of minCD mutant cells had FtsZ rings.
However, the minCD mutation had a dramatic effect on the number of FtsZ rings per cell in LB medium (Fig. 1G to L and 2B). Of the mutant cells with an FtsZ ring, only 23% (59 of 258) had a single medial ring, 16% (40 of 258) had single polar rings, ∼45% (116 of 258) had two or three FtsZ rings located at polar and medial positions (Fig. 2B), and the remaining 15% (43 of 258) had more than three FtsZ rings. Cells with more than three FtsZ rings tended to be significantly longer than the other cells, and the extra FtsZ rings were usually positioned at the cell quarters, positions that would have served as midcell had the previous round of cell division been completed. Despite the fact that ∼60% (156 of 258) of the cells had at least one polar FtsZ ring, minicells were only ∼17% (193 of 1171) of the total population (counted by using the membrane stain FM4-64), indicating that not all polar rings are used for division.
Conservation of the min genes among the bacteria.
To date, the E. coli min system has provided the primary model for division site selection. However, in contrast to FtsZ, the MinC, -D, and -E proteins are not conserved throughout the eubacteria and archaea. As discussed above, B. subtilis does not possess a gene that has any significant similarity to E. coli minE, although it has been suggested that the divIVA gene product could be a MinE analog (8, 13). Many of the bacterial species whose genome sequences have recently been published, including Haemophilus influenzae and the archaeon Methanococcus jannaschii, do not possess homologs of either E. coli minC or minE (7, 14). The apparent lack of conservation of the min genes at the primary sequence level suggests that the problem of division site selection may have been solved in different ways and that the min system represents only the tip of the proverbial iceberg.
MinD is the most conserved and widespread of the MinCDE trio. MinD is a member of a large family of proteins, some of whose members are involved in plasmid partitioning and transcriptional regulation (25). It will be interesting to determine if MinD shares any functions with other family members, and if so, whether these functions are important for its role in cell division.
Additional factors involved in the frequency and position of FtsZ ring formation.
It is clear that in B. subtilis MinC and MinD are involved in division site selection. Our results indicate that other factors also function to ensure that FtsZ localizes to midcell and that cell division is medial. The function of these factors is most obvious in minimal medium, where, even in the absence of minCD, FtsZ rings are limited to one per cell and the position is usually medial. In rich medium, there are many more rings per cell, but division is still predominantly medial. If all three potential division sites were equally available in cells lacking minCD, then the expected frequency of medial division (and medial FtsZ ring formation) would be only 33%. Therefore, even in the absence of MinC and MinD, something is limiting the number and position of FtsZ ring formation.
Acknowledgments
We thank C. Price for the gift of a strain bearing the minCDΔ1::cat mutation and K. Pogliano for suggesting the membrane stain FM4-64. We are grateful to members of our lab, especially K. P. Lemon, for useful discussions and comments on the manuscript, and to N. King and R. Losick for comments on the manuscript.
P.A.L. was supported, in part, by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship (DRG-1397). This work was also supported in part by Public Health Service grant GM41934 from the NIH to A.D.G.
REFERENCES
- 1.Addinall S G, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler H I, Fisher W D, Cohen A, Hardigree A H. Miniature Escherichia coli cells deficient in DNA. Proc Natl Acad Sci USA. 1967;57:321–326. doi: 10.1073/pnas.57.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arigoni F, Duncan L, Alper S, Losick R, Stragier P. SpoIIE governs the phosphorylation state of a protein regulating transcription factor sigma-F during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:3238–3242. doi: 10.1073/pnas.93.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi E, Lutkenhaus J. Interaction between the min locus and ftsZ. J Bacteriol. 1990;172:5610–5616. doi: 10.1128/jb.172.10.5610-5616.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 6.Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 8.Cha J-H, Stewart G C. The divIVA minicell locus of Bacillus subtilis. J Bacteriol. 1997;179:1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coyne S I, Mendelson N H. Clonal analysis of cell division in the Bacillus subtilis divIV-B1 minicell-producing mutant. J Bacteriol. 1974;118:15–20. doi: 10.1128/jb.118.1.15-20.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Boer P A J, Cook W R, Rothfield L I. Bacterial cell division. Annu Rev Genet. 1990;24:249–274. doi: 10.1146/annurev.ge.24.120190.001341. [DOI] [PubMed] [Google Scholar]
- 11.de Boer P A J, Crossley R E, Rothfield L I. Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J Bacteriol. 1988;170:2106–2112. doi: 10.1128/jb.170.5.2106-2112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Boer P A J, Crossley R E, Rothfield L I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 13.Edwards D H, Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- 14.Fleishmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L, Glodek A, Kelly J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 15.Harry E J, Pogliano K, Losick R. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:3386–3393. doi: 10.1128/jb.177.12.3386-3393.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaacks K J, Healy J, Losick R, Grossman A D. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 18.Lee S, Price C W. The minCD locus of Bacillus subtilis lacks the minE determinant that provides topological specificity to cell division. Mol Microbiol. 1993;7:601–610. doi: 10.1111/j.1365-2958.1993.tb01151.x. [DOI] [PubMed] [Google Scholar]
- 19.Levin P A, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 20.Levin P A, Losick R, Stragier P, Arigoni F. Localization of the sporulation protein SpoIIE in Bacillus subtilis is dependent upon the cell division protein FtsZ. Mol Microbiol. 1997;25:839–846. doi: 10.1111/j.1365-2958.1997.mmi505.x. [DOI] [PubMed] [Google Scholar]
- 21.Levin P A, Margolis P, Setlow P, Losick R, Sun D. Identification of Bacillus subtilis genes for septum placement and shape determination. J Bacteriol. 1992;174:6717–6728. doi: 10.1128/jb.174.21.6717-6728.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin D C H, Levin P A, Grossman A D. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc Natl Acad Sci USA. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Löwe J, Amos L A. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 24.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 25.Motallebi-Veshareh M, Rouch D A, Thomas C M. A family of ATPases involved in the active partitioning of diverse bacterial plasmids. Mol Microbiol. 1990;4:1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- 26.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pogliano, K. 1998. Personal communication.
- 28.Pogliano K, Harry E, Losick R. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- 29.Pogliano K, Hofmeister A E M, Losick R. Disappearance of the ςE transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to the establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raskin D M, de Boer P A. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 31.Rothfield L I, Zhao C R. How do bacteria decide where to divide? Cell. 1996;84:183–186. doi: 10.1016/s0092-8674(00)80971-x. [DOI] [PubMed] [Google Scholar]
- 32.Ryter A. Étude morphologique de la sporulation de Bacillus subtilis. Ann Inst Pasteur. 1964;108:40–60. [PubMed] [Google Scholar]
- 33.Teather R M, Collins J F, Donachie W D. Quantal behavior of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli. J Bacteriol. 1974;118:407–413. doi: 10.1128/jb.118.2.407-413.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varley A W, Stewart G C. The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (MinCD) and cell shape (MreBCD) determinants. J Bacteriol. 1992;174:6729–6742. doi: 10.1128/jb.174.21.6729-6742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]