Abstract
The mmr gene, cloned from Mycobacterium tuberculosis, was shown to confer to Mycobacterium smegmatis resistance to tetraphenylphosphonium (TPP), erythromycin, ethidium bromide, acriflavine, safranin O, and pyronin Y. The gene appears to code for a protein containing four transmembrane domains. Studies of [3H]TPP intracellular accumulation strongly suggest that the resistance mediated by the Mmr protein involves active extrusion of TPP.
Development of multiple-drug resistance among prokaryotes and eukaryotes is a serious medical problem. Bacteria, for instance, demonstrate the ability to extrude a variety of lipophilic drugs out of the cytoplasm (19). In Bacillus subtilis (17), Escherichia coli (13), and Staphylococcus aureus (10), bmr, emr, and qac genes, respectively, encoding an efflux-mediated multidrug resistance have been identified and sequenced. The Bmr, EmrB, and QacA/B proteins, containing 12 to 14 transmembrane domains (13, 17, 20), belong to the major facilitator superfamily (MFS) (19), which catalyzes transport of sugars, metabolic intermediates, and drugs in organisms ranging from bacteria to humans. There is also a family of small membrane proteins responsible for the efflux-mediated multidrug resistance in bacteria (6, 12). These proteins contain only four transmembrane domains and confer resistance to (i) aromatic dyes, e.g., ethidium bromide; (ii) quaternary amines, e.g., the disinfectant benzalkonium; and (iii) derivatives of tetraphenylphosphonium (TPP) (6). These proteins, belonging to the SMR family (21), include (i) the chromosome-encoded E. coli protein EmrE (previously known as MvrC and Ebr), which is responsible for resistance to ethidium bromide and methyl viologen (11, 15, 22); (ii) the S. aureus Smr protein (also called Ebr and QacC/D) (6, 12); (iii) drug resistance proteins encoded by genes on transmissible plasmids located at the 3′ conserved segment of integrons (18); and (iv) the product of the sugE gene in the E. coli chromosome, which has been shown to suppress the groEL mutation (5). The resurgence of tuberculosis has been characterized by the emergence of a significant number of drug-resistant strains. Furthermore, strains of the Mycobacterium avium complex, opportunistic pathogens common in AIDS patients, are inherently resistant to many traditional antimycobacterial agents (7). Hence, the importance of studies of the antibiotic resistance mechanisms that may aid the development of novel drugs for treatment of atypical infections by M. avium, Mycobacterium intracellulare, and the multiple-drug-resistant strains of Mycobacterium tuberculosis. All known multidrug-resistant strains of M. tuberculosis are explained by sequential accumulation of mutations in several genes involved in resistance to individual antibiotics (16). However, since efflux proteins are known to have important roles in resistance to a variety of unrelated antibacterial compounds in many gram-negative and gram-positive bacteria (19), it has seemed important to identify and characterize efflux proteins in M. tuberculosis. The first efflux pump that has been described for the genus Mycobacterium, the LfrA protein, identified in M. smegmatis, confers resistance to fluoroquinolones, acridine, and some quaternary ammonium compounds (24). We recently identified the energy-dependent efflux pump TetV in M. smegmatis responsible for the resistance to tetracycline (3). These proteins are members of the MFS.
In this work, we describe the characterization of the Mmr multidrug pump of M. tuberculosis, which confers to M. smegmatis, when expressed on a multicopy vector, resistance to TPP, ethidium bromide, erythromycin, acriflavine, safranin O, and pyronin Y. We provide evidence suggesting that Mmr-mediated resistance is correlated with an energy-dependent efflux of the drug. To our knowledge, this is the first protein belonging to the SMR family described in mycobacteria.
Cloning of the M. tuberculosis mmr gene.
A genomic library of M. tuberculosis H37Rv was transformed into E. coli, and transformants were selected on Luria-Bertani agar containing 50 μg of kanamycin per ml. Colonies were pooled, and cosmid DNA was isolated and electroporated into M. smegmatis mc2155 (8), which was then plated on Middlebrook 7H11 agar (Difco) supplemented with 10% Middlebrook OADC enrichment (Difco), 0.2% glycerol, 25 μg of kanamycin per ml, and 5 μg of TPP per ml. Analysis of cosmid DNAs from three of the many colonies, recovered by electroporation into E. coli (2), showed that they had overlapping inserts of 40, 45, and 42 kb. M. smegmatis strains, containing subclones of one of these cosmids on vector pMD31 (4), were tested for TPP resistance. The region required for the expression of this phenotype was present in subclone pMtb15, which contained a 1,738-bp BamHI fragment of the original insert.
Sequence analysis of the mmr gene.
The 1.7-kb fragment was sequenced by the dideoxy chain termination method with a Sequenase kit (U.S. Biochemicals) and universal, reverse, and custom-designed internal primers. A computer search of available sequence databases with the BLAST program (1) revealed that the cloned fragment is identical to nucleotides 15918 to 17655 of the M. tuberculosis cosmid MTCY22D7 (accession no. Z83866), already sequenced within the M. tuberculosis sequencing project. Sequence analysis showed that the 1.7-kb insert of pMtb15 contained two open reading frames (ORFs). Of these ORFs, ORF1, extending from nucleotides 290 to 611, was preceded by a putative ribosome-binding site (AAGGAGG). Potential −10 (TACATT) and −35 (TGGACG) ς70-type promoter sequences, located 17 bp apart, were recognized by comparison with the putative ς70-type Mycobacterium promoters (9). ORF1 is responsible for TPP resistance as shown by subcloning experiments. The 984-bp PstI-BamHI fragment, subcloned into pMD31 (pMtb312), which contains the complete orf1 as well as a truncated orf2, was sufficient for the TPP resistance phenotype. orf1 was designated mmr for mycobacterium multidrug resistance gene (the same gene was termed emrE by S. T. Cole et al. [2a]). It is 321 bp in length and its putative product has 107 amino acid residues with a predicted molecular mass of 11,084 Da. The hydropathy analysis of its sequence suggests the presence of four transmembrane regions (data not shown). Screening of the EMBL and GenBank databases with the BLASTP program revealed that the Mmr protein shows the highest degrees of homology with bacterial proteins belonging to the SMR family such as QacE of Klebsiella pneumoniae (18), EmrE and Ebr of E. coli (15, 22, 23), and Qac and Smr of S. aureus (Qac protein accession no., U81980) (6, 12). Pairwise alignment of amino acid sequences (Fig. 1) showed that Mmr was 49.5, 43, 41, and 41.1% identical (80.3, 75.7, 74.7, and 71% similarities) to QacE, EmrE, Ebr, and Qac, respectively. Homology was also found with the E. coli SugE protein (32.7% identity in a 104-amino-acid overlap) (5). All these proteins are members of the SMR family; this group contains three specific signature sequences (21). In the Mmr protein, motif A (WIXLVIAILLEV) is represented by YLYLLCAIFAEV, motif B (KXSEGFTRLXPS) is represented by KSTEGFTRLWPT, and motif C (PVGTAYAVWTGLG) is represented by QTDVAYALWSAIG.
FIG. 1.
Sequence alignment of M. tuberculosis Mmr, K. pneumoniae QacE, E. coli EmrE and Ebr, and S. aureus Qac proteins. The consensus sequence is shown at the bottom. Identical amino acids are highlighted in black boxes; conserved amino acid substitutions are shown in shaded boxes.
Drug resistance of M. smegmatis transformed with a plasmid carrying the mmr gene.
On the basis of sequence analysis, we predicted that this gene encodes a multidrug transporter. In order to verify this hypothesis, we examined the drug resistance of M. smegmatis mc2155 transformed with plasmid pMtb312. The MICs of toxic compounds were determined as follows. Cultures diluted to 105 CFU/ml were grown on supplemented Middlebrook 7H11 agar (Difco) plates containing different concentrations of the compounds. The plates were incubated at 37°C for up to 5 days. As shown in Table 1, the mmr gene confers resistance to TPP, ethidium bromide, erythromycin, acriflavine, safranin O, and pyronin Y. Susceptibility to streptomycin, ciprofloxacin, doxorubicin, rhodamine 123, rifampin, chloramphenicol, tetracycline, proflavine, sulfadiazine, or cetyltrimethylammonium bromide was not affected (data not shown). The finding that CCCP (carbonyl cyanide m-chlorophenylhydrazone), an energy uncoupler that has been shown to inhibit the action of other efflux pumps (11), greatly reduced the MICs of all compounds shown in Table 1 suggested that this resistance was linked to the Mmr pump. The resistance phenotype is almost certainly due to the overexpression of the wild-type M. tuberculosis mmr gene on the multicopy vector pMD31 (20 to 30 copies per cell). This result does not necessarily indicate that this gene contributes to the resistance phenotype of wild-type M. tuberculosis.
TABLE 1.
MICs for M. smegmatis mc2155 carrying the mmr-containing plasmid pMtb312 or the cloning vector pMD31
| Substance | MICs (μg/ml) for:
|
|||
|---|---|---|---|---|
| mc2155/pMD31
|
mc2155/pMtb312
|
|||
| Without CCCP | With CCCPa | Without CCCP | With CCCPa | |
| TPP | 0.5 | 0.3 | 5 | 0.4 |
| Ethidium bromide | 8 | 4 | 40 | 10 |
| Acriflavine | 25 | 10 | 100 | 15 |
| Erythromycin | 15 | 10 | 60 | 15 |
| Safranin O | 5 | 3 | 30 | 3 |
| Pyronin Y | 10 | 5 | 150 | 15 |
CCCP was added at a concentration of 15 μg/ml.
Distribution of the mmr gene among mycobacteria.
The distribution of the mmr gene among other Mycobacterium species was examined by PCR and confirmed by hybridization experiments. DNAs from several mycobacterial species were extracted as previously described (14). The DNA amplification was performed with the oligonucleotide primers RG148 (5′-ATCTTCGCGGAAGTG-3′) and RG149 (5′-CGACCACAGCGCATA-3′) designed against the M. tuberculosis mmr gene. Amplification of DNA from M. simiae, M. gordonae, M. marinum, and M. bovis produced the expected 174-bp fragment that hybridized to the M. tuberculosis mmr probe (data not shown). No mmr fragments could be amplified from DNA of M. chelonae, M. flavescens, M. fortuitum, M. kansasii, M. xenopi, M. terrae, M. avium, or M. smegmatis. In a Southern hybridization experiment with genomic DNA from M. tuberculosis, M. bovis, M. avium, and M. smegmatis, no signal was detected in the case of the last two strains, while strong signals were observed for M. tuberculosis and M. bovis (data not shown). Thus, the mmr gene is present in M. simiae, M. gordonae, M. marinum, M. bovis, and M. tuberculosis.
[3H]TPP accumulation by M. smegmatis cells.
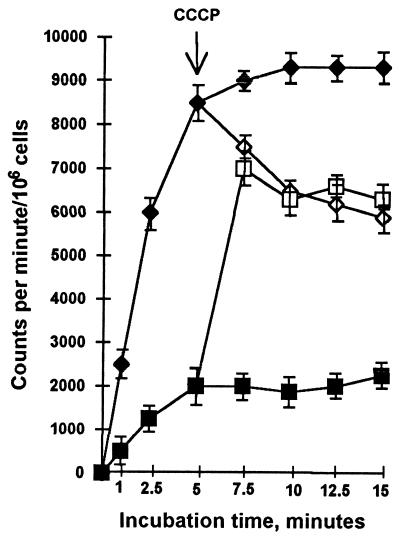
To determine the mechanism of TPP resistance, TPP accumulation was measured in M. smegmatis cells with and without the mmr-containing plasmid. The intracellular accumulation of TPP was determined by adding 0.4 μM [3H]TPP (24.0 Ci/mmol; Amersham) to cells grown to mid-exponential phase in supplemented Middlebrook 7H9 medium, which were then resuspended in 0.1 M phosphate buffer (pH 7.0) and incubated at 37°C with shaking. At various time intervals, 50 μl of the suspension was removed, diluted in 1 ml of ice-cold 0.1 M LiCl, and immediately filtered through a 0.45-μm-pore-size filter (Millipore). The filter was washed rapidly twice with 2 ml of the same solution and dried, and the radioactivity was determined with a liquid scintillation counter. As shown in Fig. 2, M. smegmatis cells containing multiple copies of the mmr gene accumulated less than 20% of the TPP accumulated by the cells carrying the vector alone. A reduced accumulation of the drug may be caused either by a decreased drug permeability or by active drug extrusion through the cytoplasmic membrane. Since antibiotic efflux pump systems are energy dependent, antibiotic accumulation by bacterial cells increases in the presence of uncouplers (11). To test whether or not this occurs in the presence of the mmr gene, the effect of uncoupler CCCP on [3H]TPP accumulation by M. smegmatis strains carrying the pMD31 or pMtb312 plasmid was tested. As shown in Fig. 2, the addition of CCCP significantly increased TPP accumulation by M. smegmatis cells expressing the mmr gene. The level of accumulation rapidly approached the level found in M. smegmatis cells containing the cloning vector pMD31 only. In the presence of the uncoupler, the uptake of labelled TPP in M. smegmatis, transformed with the vector pMD31, is reduced. This is expected because TPP accumulates passively in response to membrane potential and CCCP abolishes this potential. These results strongly suggest that M. tuberculosis Mmr protein actively pumps out TPP, probably by using the proton motive force as an energy source.
FIG. 2.
Effect of the addition of CCCP on the uptake of [3H]TPP by M. smegmatis cells carrying plasmid pMtb312 (■, no CCCP addition; □, addition of CCCP) or the cloning vector pMD31 (⧫, no CCCP addition; ◊, addition of CCCP). CCCP at a concentration of 0.1 mM was added at the time indicated by the arrow. The results are the averages of three replicates, and error bars indicate standard deviations.
Concluding remarks.
The M. tuberculosis chromosomal gene mmr, cloned on a multicopy plasmid, confers to M. smegmatis resistance to TPP, ethidium bromide, erythromycin, acriflavine, safranin O, and pyronin Y. The protein coded by this gene is a member of the SMR protein family, which transports different antiseptics, drugs, and intercalating dyes. The Mmr protein from M. tuberculosis appears to be a multidrug efflux pump since it confers a multidrug-resistant phenotype to M. smegmatis, it is homologous to other membrane efflux proteins, and it prevents intracellular [3H]TPP accumulation. The resistant phenotype is mediated by an energy-dependent efflux pump and results not from a mutation but from the elevated expression of a wild-type gene, as reported also for other mycobacteria (3, 24).
Gene disruption experiments will allow the physiological role of the M. tuberculosis Mmr protein to be determined.
Acknowledgments
This study was supported by the European Union BIOMED project BMH4-CT96-1241 and by the National Tuberculosis Project, contract no. 96/D/T56. R.C. has a fellowship from the University of Pavia, Italy.
We thank S. T. Cole for providing the M. tuberculosis H37Rv library.
REFERENCES
- 1.Altschul S F, Boguski M S, Gish W, Wooton J C. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 2.Baulard A, Jourdan C, Mercenier A, Locht C. Rapid mycobacterial plasmid analysis by electroduction between Mycobacterium spp. and Escherichia coli. Nucleic Acids Res. 1992;20:4105. doi: 10.1093/nar/20.15.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaya F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborche J, Quail M A, Rajanolream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 3.De Rossi E, Blokpoel M C J, Cantoni R, Branzoni M, Riccardi G, Young D B, De Smet K A L, Ciferri O. Molecular cloning and functional analysis of a novel tetracycline resistance determinant, tet(V), from Mycobacterium smegmatis. Antimicrob Agents Chemother. 1998;42:1931–1937. doi: 10.1128/aac.42.8.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnelly-Wu M K, Jacobs W R, Jr, Hatfull G F. Superinfection immunity of mycobacteriophage L5: applications for genetic transformation of mycobacteria. Mol Microbiol. 1993;7:407–417. doi: 10.1111/j.1365-2958.1993.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 5.Greener T, Govezensky D, Zamir A. A novel multicopy suppressor of groEL mutation includes two nested open reading frames transcribed from different promoters. EMBO J. 1993;12:889–896. doi: 10.1002/j.1460-2075.1993.tb05729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grinius L L, Dreguniene G, Goldberg E B, Liao C H, Projan S J. A staphylococcal multidrug resistance gene product is a member of a new protein family. Plasmid. 1992;27:119–129. doi: 10.1016/0147-619x(92)90012-y. [DOI] [PubMed] [Google Scholar]
- 7.Inderlied C B, Kemper C A, Bermudez C E M. The Mycobacterium avium complex. Clin Microbiol Rev. 1993;6:266–310. doi: 10.1128/cmr.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs W R, Jr, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B. Genetic system for mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 9.Kremer L, Baulard A, Estaquier J, Content J, Capron A, Locht C. Analysis of the Mycobacterium tuberculosis 85A antigen promoter region. J Bacteriol. 1995;177:642–653. doi: 10.1128/jb.177.3.642-653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leelaporn A, Paulsen I T, Tennent J M, Littlejohn T G, Skurray R A. Multidrug resistance to antiseptics and disinfectants in coagulase-negative staphylococci. J Med Microbiol. 1994;40:214–220. doi: 10.1099/00222615-40-3-214. [DOI] [PubMed] [Google Scholar]
- 11.Lewis K. Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem Sci. 1994;19:119–123. doi: 10.1016/0968-0004(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 12.Littlejohn T G, DiBerardino D, Messerotti L J, Spiers S J, Skurray R A. Structure and evolution of a family of genes encoding antiseptic and disinfectant resistance in Staphylococcus aureus. Gene. 1991;101:59–66. doi: 10.1016/0378-1119(91)90224-y. [DOI] [PubMed] [Google Scholar]
- 13.Lomovskaya O, Lewis K. Emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci USA. 1992;89:8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milano A, De Rossi E, Gusberti L, Heym B, Marone P, Riccardi G. The katE gene, which encodes the catalase HPII of Mycobacterium avium. Mol Microbiol. 1996;19:113–123. doi: 10.1046/j.1365-2958.1996.352876.x. [DOI] [PubMed] [Google Scholar]
- 15.Morimyo M, Hongo E, Hama-Inaba H, Machida I. Cloning and characterization of the mvrC gene of Escherichia coli K-12 which confers resistance against methyl viologen toxicity. Nucleic Acids Res. 1992;20:3159–3165. doi: 10.1093/nar/20.12.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musser J M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neyfakh A A, Bidnenko V E, Chen L B. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc Natl Acad Sci USA. 1991;88:4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulsen I T, Littlejohn T G, Radstroem P, Sundstroem L, Skoeld O, Swedberg G, Skurray R A. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulsen I T, Brown M H, Littlejohn T G, Mitchell B A, Skurray R A. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci USA. 1996;93:3630–3635. doi: 10.1073/pnas.93.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulsen I T, Skurray R A, Tam R, Saier M H, Jr, Turner R J, Weiner J H, Goldberg E B, Grinius L L. The SMR family: a novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol Microbiol. 1996;19:1167–1175. doi: 10.1111/j.1365-2958.1996.tb02462.x. [DOI] [PubMed] [Google Scholar]
- 22.Purewal A S. Nucleotide sequence of the ethidium efflux gene from Escherichia coli. FEMS Microbiol Lett. 1991;82:229–232. doi: 10.1016/0378-1097(91)90338-b. [DOI] [PubMed] [Google Scholar]
- 23.Sundstroem L, Radstroem P, Swedberg G, Skoeld O. Site-specific recombination promotes linkage between trimethoprim and sulfonamide resistance genes. Sequence characterization of dhfrV and sull and a recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 24.Takiff H, Cimino M, Musso M C, Weisbrod T, Martinez R, Delgado M B, Salazar L, Bloom B R, Jacobs W R., Jr Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacterium smegmatis. Proc Natl Acad Sci USA. 1996;93:362–366. doi: 10.1073/pnas.93.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]