Abstract
Background:
Eosinophilic esophagitis (EoE) involves a chronic immune-mediated response to dietary antigens. Recent work identifies T-cell clonality in children with EoE, however, it is unknown whether this is true in adults or whether there is a restricted food-specific T-cell repertoire. We sought to confirm T-cell receptor (TCR) clonality in EoE and assess for differences with specific food triggers.
Methods:
Bulk TCR sequencing was performed on mRNA isolated from esophageal biopsies obtained from adults and children with EoE (n = 15) who had food triggers confirmed by endoscopic evaluation. Non-EoE adult and pediatric controls (n = 10) were included. Differences in TCR clonality by disease and treatment status were assessed. Shared and similar V-J-CDR3s were assessed based on specific food triggers.
Results:
Active EoE biopsies from children but not adults displayed decreased unique TCRα/β clonotypes and increased relative abundance of TCRs comprising >1% of the total compared to non-EoE controls and paired inactive EoE samples. Among patients in which baseline, post diet elimination, and food trigger reintroduction samples (n = 6) were obtained, we observed ~1% of TCRs were shared only between pre-diet elimination and trigger reintroduction. Patients with a shared EoE trigger (milk) had a greater degree of shared and similar TCRs compared to patients with differing triggers (seafood, wheat, egg, soy).
Conclusion:
We confirmed relative clonality in children but not adults with active EoE and identified potential food-specific TCRs, particularly for milk-triggered EoE. Further studies are needed to better identify the broad TCR repertoire relevant to food triggers.
Keywords: EoE, TCR, biomarker, diet elimination, food trigger
Graphical Abstract
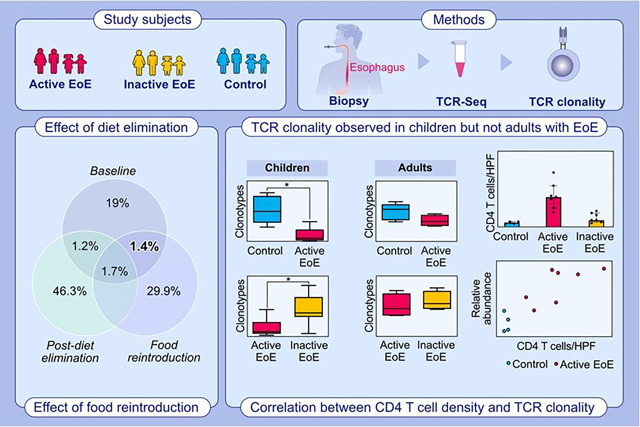
TCR Sequencing was performed on total RNA from esophageal biopsies from children and adults with EoE and non-EoE controls. TCR clonality was identified in children but not adults with EoE. TCRs disappear with food elimination and recur with reintroduction. The relative abundance of TCR clones occupying more than 1% of total correlated with intraepithelial peak CD4 T-cells/hpf in pediatric esophageal biopsies.
Abbreviations: EoE, eosinophilic esophagitis; hpf, high power field; TCR, T cell receptor
1. INTRODUCTION
Eosinophilic esophagitis (EoE) is a chronic food antigen-mediated inflammatory disease.1, 2 Diet elimination of EoE triggers is effective treatment and systematic food reintroduction has facilitated identification of specific triggers.1 Cow’s milk protein is the most common trigger of EoE, and between 30% and 60% of patients demonstrate histologic remission with 1-food elimination diets (1FED).3, 4 Prior studies of food-specific IgG4 and ex vivo T-cell proliferation demonstrate promise as biomarkers for food triggers,5 however, dietary therapy in EoE is currently empiric since these biomarkers have yet to be validated in larger studies.
Numerous studies have established the role of specific dietary proteins in driving T-helper cell type 2 (TH2) mediated recruitment of eosinophils.2, 6–8 Prior studies have found activated T-helper type 2 (TH2) cells with antigen-specific stimulation supporting a role for food-specific T-cells.6, 8 CD4+ T-cells identify immune targets through direct interaction of the T-cell receptor (TCR) with antigen-derived peptide presented on the cell surface by MHC molecules.9 The TCR is a heterodimer of α and β or γ and δ chains, each encoded by V, D (β only), J, and C (constant region) genes with random insertion and deletion in the VDJ junction.10 The complementarity-determining region 3 (CDR3) produced from the rearrangement of the V(D)J segment interacts closely with the epitope during antigen presentation, and reflects diversity, clonality, antigenic recognition, and specificity of the T-cell response.11 Evidence of T-cell clonality in EoE has been mixed at the single cell level. Wen et al. found a trend for increased diversity among TCR clonal repertoires in active EoE and no evidence of clonality. However, Morgan et al. identified evidence of clonality among GATA3-expressing pathogenic effector Type 2 T-cells (peTH2).8 A limitation of these studies was the focus on TCRβ, and analysis of TCRα is lacking. In addition, no study has assessed TCR clonality in adults with EoE and single cell sequencing captures a relatively small number of cells compared to what is present in a biopsy. Thus, we sought to determine whether bulk RNA TCR sequencing could detect T-cell clonality in EoE and ask whether a food-specific TCR repertoire is present among children and adults with endoscopically confirmed food-triggered EoE.
2. METHODS
2.1. Patient samples
Forty-four esophageal biopsies from 11 pediatric and 4 adult patients with EoE based on 2011 consensus criteria,12 and five pediatric and five adult non-EoE controls undergoing endoscopy for symptoms of esophageal dysfunction with histologically normal biopsies were included in this study. Informed consent was obtained from all subjects (≥18 years of age) or their caregiver (<18 years of age) prior to obtaining a research biopsy. All EoE patients included in the study underwent diet elimination with resulting inactive EoE (<15 eos/hpf) along with identification of one or more food triggers. Biopsies from inactive EoE patients had a paired active EoE specimen, and six patients had a trio of baseline, post-diet elimination, and food reintroduction biopsy specimens (in which active EoE recurred). The biopsies were stored in RNAlater (adult samples) or snap frozen in liquid nitrogen (pediatric samples). Pediatric samples were obtained as part of a prior longitudinal study to assess biomarkers,13 and chart review was performed to identify the foods consumed or removed at each endoscopy. Adult EoE samples were collected prior to and following Six Food Elimination Diet and during food reintroduction when a trigger was identified as part of a longitudinal study to assess genetic phenotype. Diet history was collected at the time of sample collection for adult patients. The study was approved by IRB #2010–14,155 (Lurie) and #STU00021171 (Northwestern).
2.2. T-cell immunohistochemistry
Archived unstained slides were obtained from standard-of-care esophageal biopsies of pediatric EoE and control subjects taken at the time of research biopsy collection. Sections were stained for CD4 (PA0427, Leica) and CD8 (PA0183, Leica) on a BOND-MAX machine (Leica, Germany) utilizing Bond Polymer Refine Detection Kit (DS9800, Leica) following manufacturer’s instructions. Peak CD4 and CD8 cells per high power field (400× magnification) were assessed in a blinded fashion in the intraepithelial regions in at least three fields.
2.3. RNA isolation
A single frozen biopsy sample for each timepoint was disrupted with a Biomasher (9790A, Takara Bio Inc, Kusatsu, Japan). RNA was isolated using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s protocol. RNA concentration and RNA integrity number (RIN) was measured using the TapeStation Bioanalyzer. All samples had RIN >8. The eluted RNA was stored at −80°C until sequencing was performed.
2.4. TCR library preparation/sequencing
De-identified (coded) RNA samples were sent to iRepertoire, Inc. (Huntsville, AL, USA) for library preparation and sequencing. The CDR3 region was amplified with the iRepertoire multiplex primer sets in the iR-Processor and iR-Cassette. A separate library was generated for each TCR chain. TCR sequencing was performed using an Illumina HiSeq Rapid SBS v2, 500-cycle (250 bp paired-end reads) Reagent Kit (Illumina Inc.), with average read depth of 2.5 million reads per sample.14, 15
2.5. TCR repertoire
The raw sequence files (FASTQ format) were processed with MiXCR v3.0.1216 and aligned to the reference human V, D, J, and C genes of the T- cell receptors, with no adapters. The clone sequences containing the CDR3 region were extracted from the aligned reads and grouped by the assembler to build α, β, γ, and δ full length clonotypes expressed as amino acid sequences. The non-functional sequences (out of frame sequences or those containing stop codons) were filtered out. Duplicated clonotypes with identical CDR3 amino acid sequences along with V and J gene for TCRα and V, D, and J gene for TCRβ were merged and counts/proportions summed to a single clonotype.
2.6. Immune repertoire statistics
The immune repertoire was assessed using Immunarch v0.7.0 (https://cloud.r-project.org/package=immunarch). For each sample, the number of unique clonotypes was determined based on the specific CDR3 amino acid sequence, V and J genes (D gene included for TCRβ), excluding ambiguous gene assignments. TCR diversity was estimated by D50 which represents the minimum number of unique clonotypes than constitutes 50% of the total17 along with Simpson’s diversity index. The relative abundance of clonotypes was assessed at 10-fold intervals. Mean CDR3 sequence length was calculated and compared. The overlap in clonotypes among individuals with tri-paired timepoints was determined by generating a string consisting of the CDR3 amino acid sequence, V/J gene (D gene included for TCRβ). A Venn diagram was plotted for each patient comparing the three timepoints and the data was summarized for the group. To examine the overlap in clonotypes between milk-triggered and non-milk-triggered EoE, we identified the clonotypes in which the CDR3 sequence, V and J-gene combination was shared between at least two of the patients. The Hamming distance was determined for CDR3 amino acid sequences among common clonotypes for patients with milk-triggered EoE vs non-milk-triggered active EoE as previously described, and the relative abundance was compared.8
2.7. Statistics
Descriptive statistics, test for normality, Student’s t-test, unpaired/paired one-way ANOVA with Bonferroni Post Hoc adjustment, Spearman correlation, and proportions test were used to determine statistical significance with GraphPad Prism 9.0. The data is presented as mean ± standard deviation. A p-value <.05 was considered significant.
3. RESULTS
3.1. Patient characteristics/comparisons
Demographic and clinical characteristics of the pediatric and adult cohorts are described in Table 1. Nine patients had cow’s milk-triggered EoE while six had a different food trigger (e.g., soy, wheat). The comparisons of TCR repertoires in this study are depicted in Figure 1: active EoE compared to non-EoE controls (Figure 1A), paired active to inactive EoE (Figure 1B), tri-paired EoE patients with active disease before diet elimination, inactive disease after diet elimination, then active disease with reintroduction of a food (Figure 1C). Finally, we compared milk-triggered and non-milk-triggered EoE (Figure 1D).
TABLE 1.
Patient cohort characteristics.
| Pediatric cohort (n = 16) |
Adult cohort (n = 9) |
||||
|---|---|---|---|---|---|
| EoE (11) | Controls (5) | EoE (4) | Controls (5) | ||
|
| |||||
| Gender (n, % female) | 3 (27) | 2 (40) | 0 (0) | 4 (80) | |
| Age (mean ± SD) | 9 ± 5 | 11 ± 5 | 37 ± 14 | 40 ± 16 | |
| Multiple atopic diseases (n, %) | 3 (27) | 4 (80) | 2 (50) | 0 (0) | |
| IgE-mediated food allergy | 3 (27) | 0 (0) | 2 (50) | 0 (0) | |
| Asthma | 4 (36) | 2 (40) | 0 (0) | 0 (0) | |
| Eczema | 4 (36) | 2 (40) | 0 (0) | 0 (0) | |
| Rhinitis | 6 (55) | 4 (80) | 3 (75) | 0 (0) | |
| PPI use (n, %) | 9 (82) | 3 (60) | 4 (100) | 4 (80) | |
| Endoscopic phenotype (n, %) | |||||
| Normal appearance | 4 (36) | 5 (100) | 0 (0) | 5 (100) | |
| Inflammatory | 5 (45) | 0 (0) | 1 (25) | 0 (0) | |
| Fibrostenotic | 2 (18) | 0 (0) | 3 (75) | 0 (0) | |
| Milk-triggered | 7 | NA | 2 | NA | |
| EoE (n) | |||||
| Non-milk-triggered EoE (n) | 4 | NA | 2 | NA | |
FIGURE 1.
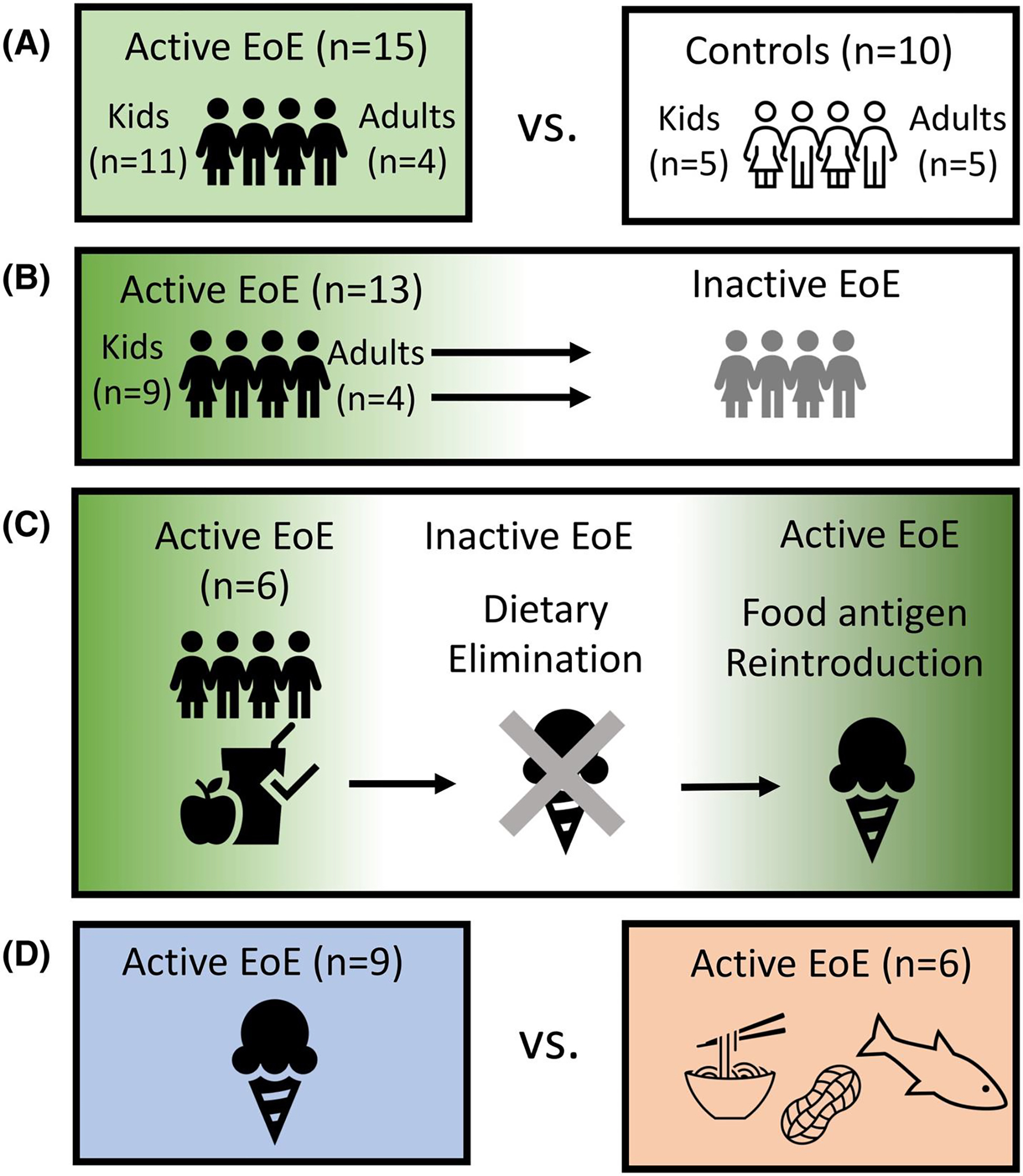
Patient Cohorts. (A) Active EoE (≥15 eos/hpf) patient biopsies were compared to non-EoE controls. (B) Active EoE biopsies were compared to paired inactive EoE (<15 eos/hpf). (C) Tri-paired EoE biopsies consisting of baseline (active EoE), post-diet elimination (inactive EoE), and post-food re-introduction (active EoE) were compared. (D) Milk-triggered active EoE biopsies were compared to non-milk-triggered active EoE.
3.2. Clonality comparison by TCR chain type and age
Initially we examined TCR characteristics between α, β, γ, and δ TCRs. A greater number of unique clonotypes and diversity were observed for TCRα and TCRβ compared to TCRδ and TCRγ (Figure S1A–C). TCRδ and TCRγ had greater relative abundance of clonotypes comprising >1% of the total (Figure S1D–H). We focused our analysis on TCRα/β as clonality was found in pathogenic effector Th2 cells, which express TCRα/β.8 We next examined whether there were clonality differences between children and adults. We found no differences in the number of unique clonotypes, diversity, nor relative abundance by clonal proportion between adult and pediatric controls (Figure S2). Pediatric active EoE had significantly reduced unique clonotypes and increased relative abundance of clonotypes comprising >1% of the total, however no differences were seen for diversity (Figure S3). Given these age-specific differences, further analysis was performed separately for children and adults.
3.3. Clonality in pediatric but not adult EoE
We next investigated differences between active EoE and controls for children and adults (Figure 1A). A summary of data is found in Table S1. Among children with active EoE, we found significantly fewer unique clonotypes than non-EoE controls for TCRα (mean ± SD: 2157 ± 2076 vs. 10,583 ± 4583, p < .0001) and TCRβ (1161 ± 1030 vs. 4134 ± 2438, p = .029) (Figure 2A). Similarly, children with active EoE had less diversity than non-EoE controls (Figure S4A/C). Adults with EoE showed no difference in unique clonotypes nor diversity compared to controls (Figure 2B, Figure S4B/D). The relative abundance of clonotypes comprising >1% of the total was significantly increased in pediatric active EoE compared to controls (Figure 2C). Among clonotypes that were less frequent, pediatric controls had greater relative abundance than active EoE (Figure S5A,B). No differences were seen in relative abundance in adults (Figure 2D, Figure S5C,D).
FIGURE 2.
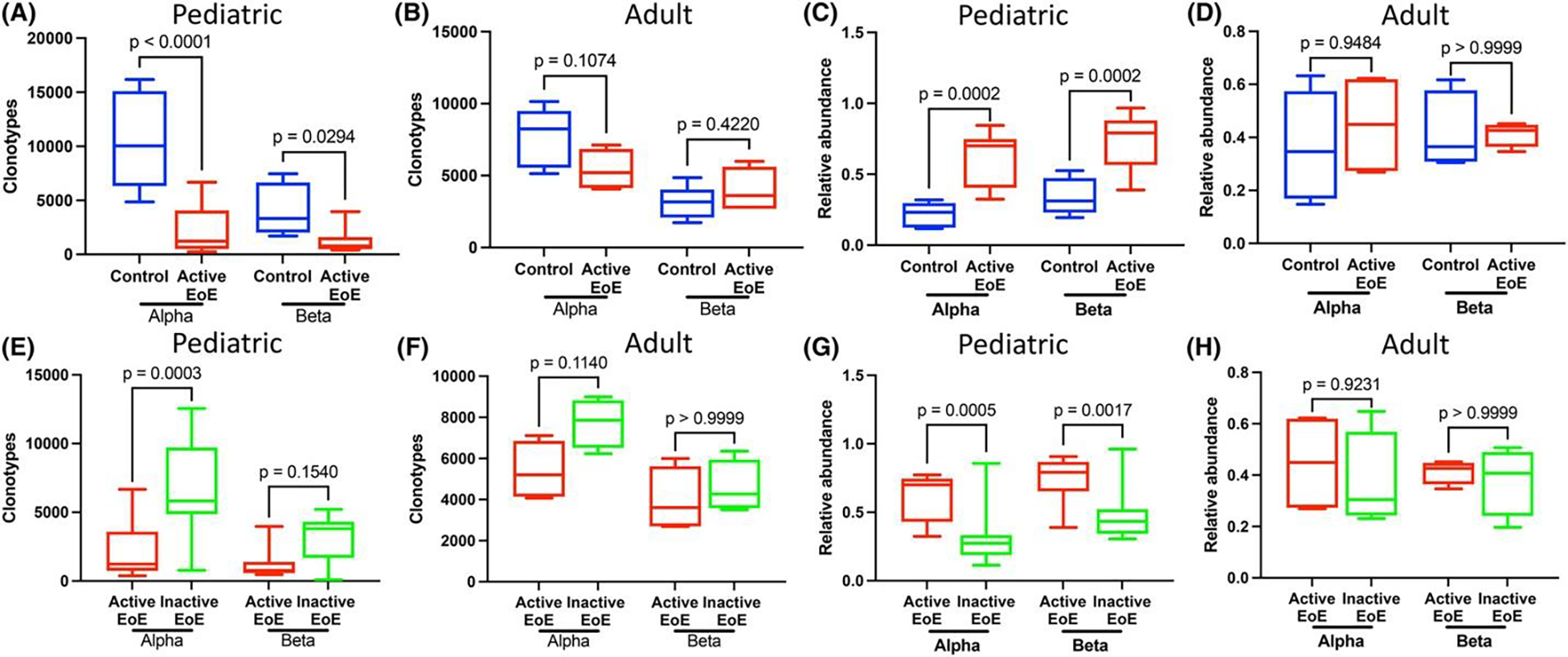
Bulk TCR sequencing confirms relative clonality with pediatric but not adult active EoE in TCRα and TCRβ. Children with active EoE biopsies have reduced unique clonotypes compared to controls (A) and inactive EoE (E). Adults with active EoE have no differences in unique clonotypes compared to controls (B) or inactive EoE (F). Children with active EoE have increased abundance of clones comprising >1% of the total compared to controls (C) or inactive EoE (G). No differences in the abundance of clones comprising >1% of the total in adults with active EoE compared to controls (D) or inactive EoE (H). Comparison of mean total unique clonotypes and relative clonotype abundance by 1-way ANOVA with Bonferroni post hoc.
We next analyzed clonality differences between paired active and inactive EoE samples for children and adults (Figure 1B). Among children with active EoE, we found significantly reduced unique clonotypes compared to inactive EoE in TCRα (2155 ± 2113 vs. 6754 ± 3536, p = .0003) but not TCRβ (Figure 2E). Active EoE had reduced D50 diversity in TCRα but not TCRβ compared to inactive EoE (Figure S4E/G). No differences were seen in unique clonotypes nor diversity in adults with EoE compared to inactive EoE (Figure 2F, Figure S4F/H). The relative abundance of clonotypes comprising >1% of the total was significantly greater in active EoE compared to inactive EoE in both TCRs (Figure 2G). For clonotypes comprising <1% of the total, active EoE had lower relative abundance than inactive EoE in children (Figure S5E,F). No differences in relative abundance were seen between adults with active and inactive EoE (Figure 2H, Figure S5G,H). There were no significant differences between non-EoE controls and inactive EoE (data not shown).
We next examined CDR3 amino acid length and found no differences between active EoE compared to controls or inactive EoE (Figure S6A–D). TCRβ had increased CDR3 length in control children compared to control adults but no differences were seen for TCRα (Figure S6E,F). Taken together, this supports disease- and age-specific TCR clonality. In children, clonality was reversed with elimination of dietary antigens.
3.4. CD4 T-cells and TCR clonality
We next sought to understand the relationship between intraepithelial T-cells and TCR clonality. We assessed the changes in CD4 and CD8 T-cells between pediatric active EoE compared to controls and inactive EoE by immunohistochemistry. We found CD4 and CD8 T-cells were increased in active EoE (mean ± standard deviation, CD4: 29 ± 13, CD8: 45 ± 22) compared to inactive EoE (CD4: 8 ± 5, CD8: 13 ± 3) and controls (CD4: 4 ± 1, CD8: 11 ± 2) (Figure 3A,B). We next examined the correlation of unique TCRα/β clonotype count with CD4 and CD8 T-cell counts in the esophageal epithelium. We found a strong inverse correlation between CD4 T-cell counts and the number of unique clonotypes for both TCR chains (Figure 3C,D). No correlation was seen with CD8 T-cell counts (data not shown). Given the increased relative abundance of clonotypes comprising >1% of the total in active EoE, we examined the correlation of relative abundance with CD4 T-cell counts. We found a strong correlation between the relative abundance of clonotypes comprising >1% of the total and CD4 cell counts for both TCRs (Figure 3E,F). Thus, CD4 T-cell density correlates with TCR clonality.
FIGURE 3.
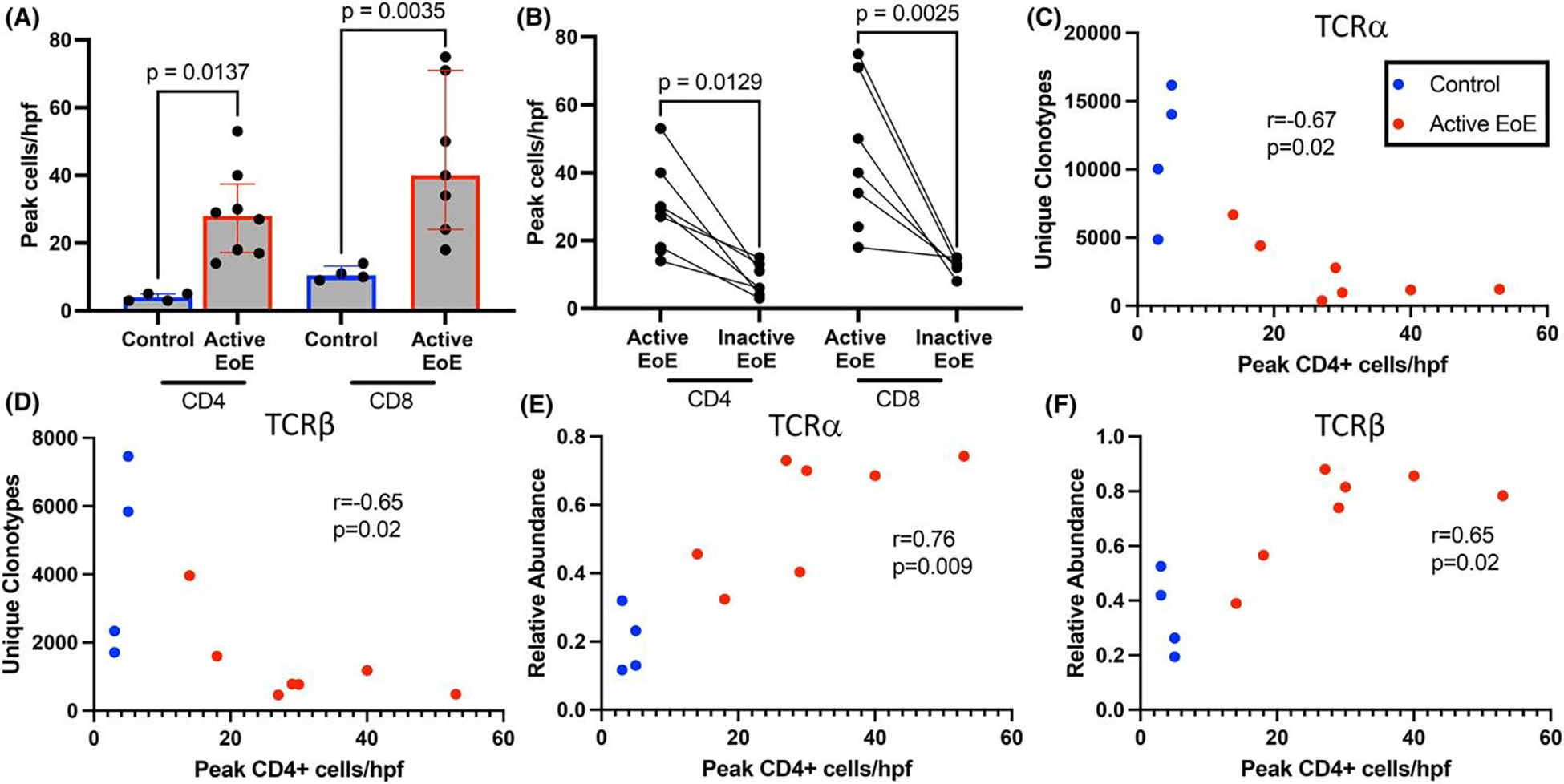
CD4 T-cells are increased in active EoE and correlate with measures of TCR clonality. Increased peak intraepithelial CD4 and CD8 T-cells per (400x) high power field (hpf) in active EoE compared to control (A) and inactive EoE (B). At least 3 hpfs were assessed per biopsy. Comparison by one-way ANOVA with Bonferroni post hoc. Spearman correlation of CD4+ cells per hpf with unique clonotypes for TCRα (C) and TCRβ (D) and relative abundance of TCRs comprising >1% of the total for TCRα (E) and TCRβ (F).
3.5. Effect of diet elimination and food reintroduction on esophageal TCR repertoire
Assuming food antigen promotes the presence of food-specific T-cells in active EoE, we sought to characterize the extent to which specific TCRs are dependent on food trigger consumption (Figure 1C) to identify food-specific TCRs. Among patients with tri-paired samples, we examined the overlap of TCR clonotypes (defined as unique CDR3 (VJ) for TCRα and unique CDR3 (VDJ) for TCRβ) between the pre-diet elimination, post-diet elimination, and food reintroduction timepoints for each individual patient (Figure S7). We found ~1% of clonotypes were present in the pre-diet elimination and food reintroduction timepoints but absent in the post-diet elimination timepoint in both TCRα and TCRβ (Figure 4A,B). We examined the change in unique clonotypes between timepoints and observed four of six patients with increased unique clonotypes post-diet elimination compared to the pre-diet elimination or food reintroduction timepoints (Figure 4C). We assessed the change in abundance for clonotypes shared among all three time points that were reduced post-diet elimination but increased pre-diet elimination and with food reintroduction (Figure 4D). For TCRα, all samples had shared TCRs with this pattern, although this was only seen in three of six patients in TCRβ. Thus, we found that during avoidance of EoE food triggers, some TCRs disappear while others have reduced abundance, which supports the occurrence of food-specific TCRs in EoE.
FIGURE 4.
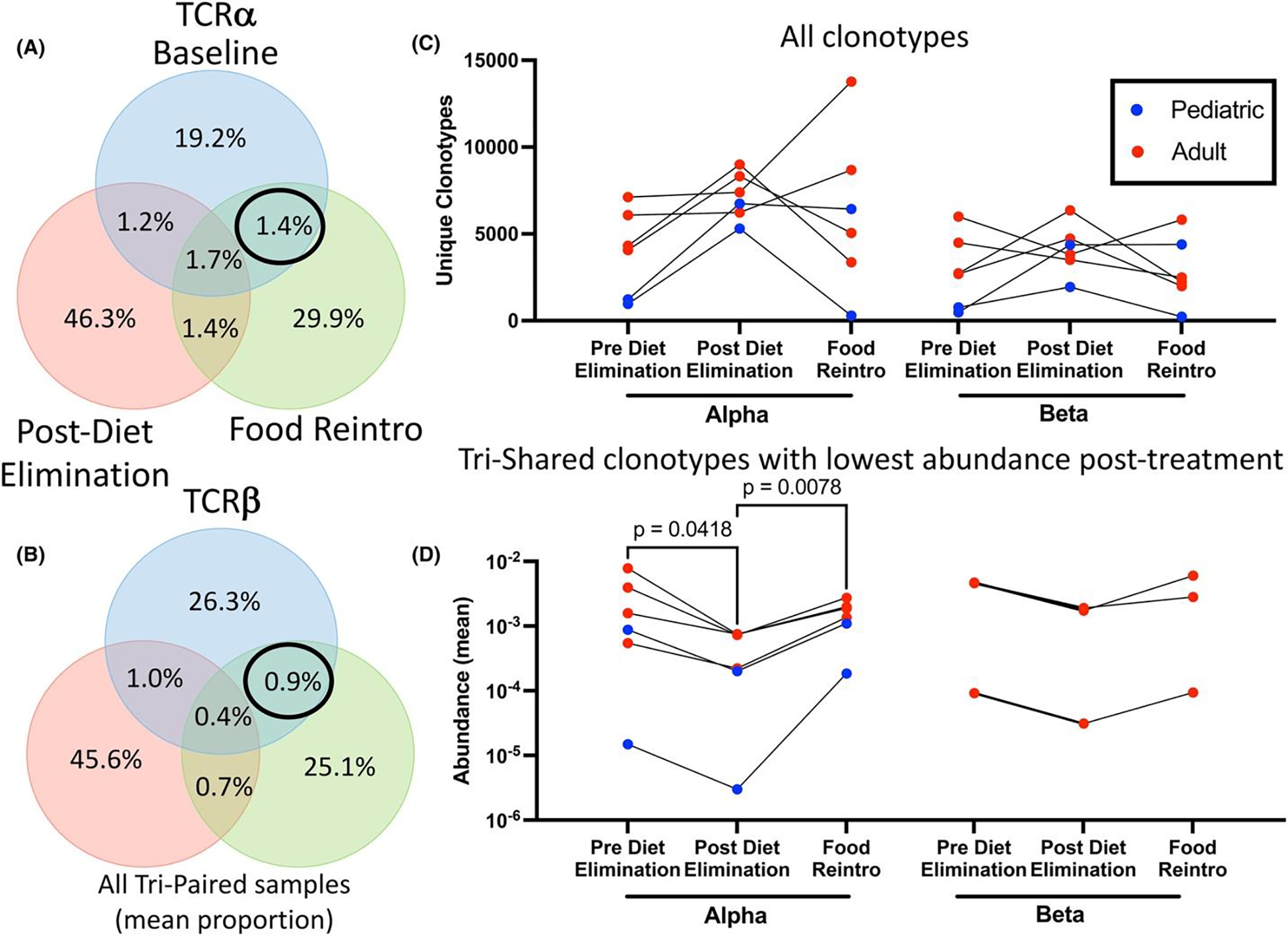
Pathogenic clonotypes recur with food reintroduction and show evidence of food specificity. Venn diagrams showing mean proportion of unique clonotypes for tri-paired patients (n = 6) for TCRα (A) and TCRβ (B). Change in total number of unique clonotypes over time for tri-paired patients for TCRα/β (C). Change in abundance for unique clonotypes shared between all timepoints that were lowest post-diet elimination for TCRα/β (D). Comparison by one-way ANOVA with Bonferroni post hoc.
3.6. Food-specific repertoire in EoE cohorts
We next wondered whether a restricted food-specific TCR repertoire exists among EoE patients with a shared food trigger. We examined differences in TCR repertoires between patients with a shared food trigger (milk) and compared them to patients with a variety of food triggers (non-milk). We found no differences in the number of unique clonotypes, diversity, nor relative abundance of clonotypes comprising >1% of the total between patients with milk-triggered and non-milk-triggered EoE (Figure S8A–F). To assess for food specificity among TCRs, we identified clonotypes present in at least two patients with identical V-J-CDR3 combinations among clonotypes comprising ≥.001% of the total. We found multiple V-J-CDR3 combinations that were unique to milk-triggered or non-milk-triggered EoE in TCRα and TCRβ (Figure 5A–D, columns with circles or triangles only) along with combinations shared by milk-triggered and non-milk-triggered EoE (Figure 5A–D, columns with circles and triangles). We examined the amino acid pattern for milk-specific and non-milk-specific CDR3s using kmer analysis. We found a trend toward increased acidic residues in milk-triggered EoE (Odds Ratio: 1.97, 95% CI: 0.97–4.02, p = .062, Figure S9).
FIGURE 5.
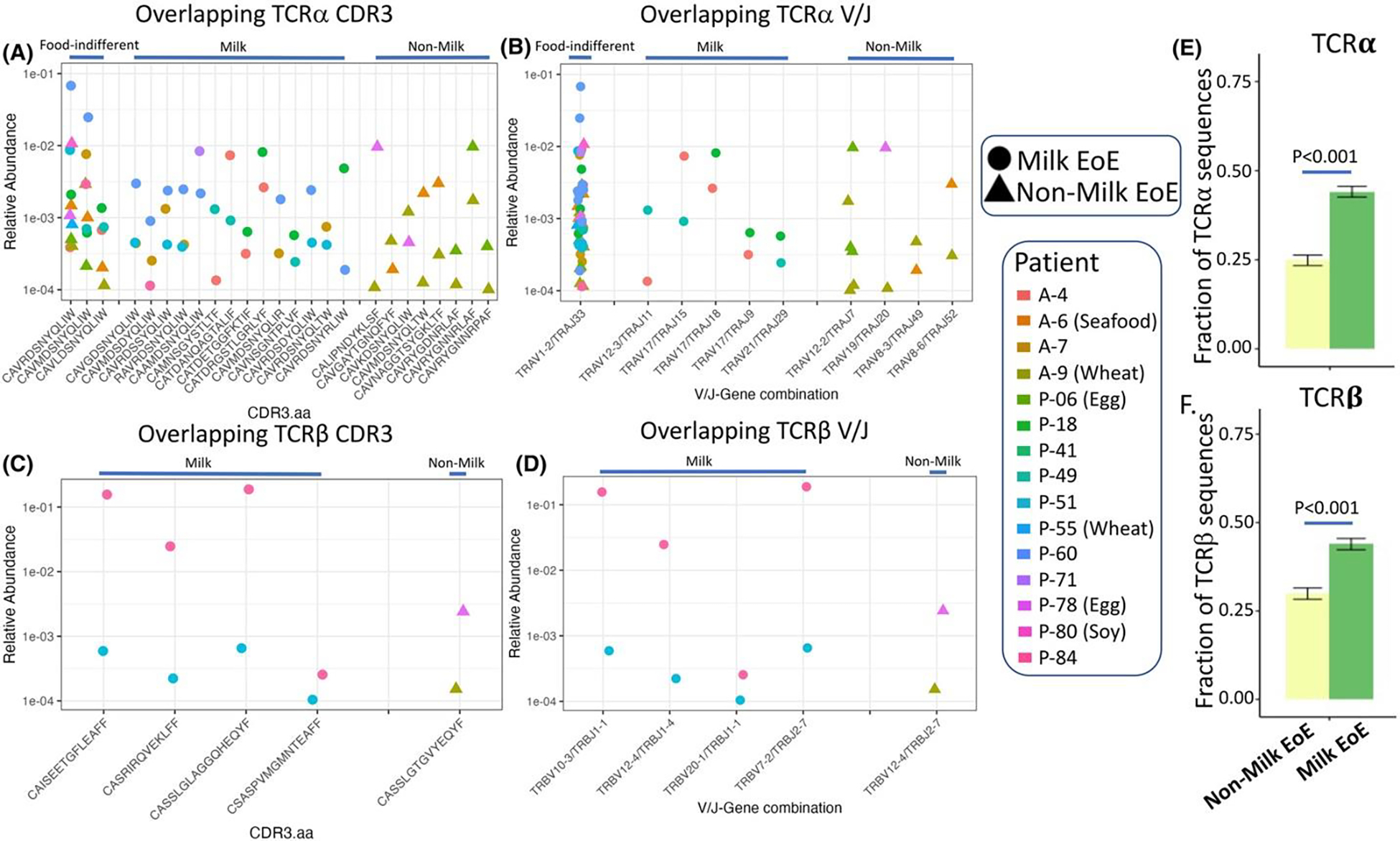
Food-specific and food-indifferent TCRs exist between patients with milk-triggered and non-milk-triggered EoE. Food-specific and food-indifferent CDR3 amino acid sequences were identified for TCRα (A) and TCRβ (C) between patient with milk-triggered and non-milk-triggered EoE. Corresponding V/J family pairs for TCRα (B) and TCRβ (D). Increased unique clonotypes with a Hamming distance of 1 for TCRα (E) and TCRβ (F) among TCRs with greater than .01% abundance from milk-triggered EoE compared to non-milk-triggered EoE. Comparison by proportions test.
One V-J gene family was shared between both milk-triggered and non-milk-triggered EoE (Figure 5B, 1st column, circles, and triangles) for TCRα, although none were shared for TCRβ. The three CDR3 amino acid sequences in the TCRα chain shared by both milk-triggered and non-milk-triggered EoE were CAVRDSNYQLIW, CAVMDSNYQLIW and CAVLDSNYQLIW representing the TRAV1–2/TRAJ33 gene combination. We searched the VDJDB public TCR database using these shared CDR3 peptides and found they all match with MHC class I molecules comprising HLA-A and B2M that represent the epitope LLLGIGILV and the gene BST2, which encodes for Tetherin (Figure S10A). None of the other overlapping CDR3s for either TCR had a match in VDJDB, and non-overlapping TCRs for BST2 were present in TCRβ. We further examined a potential role for BST2 in EoE using EGIDexpress, an online repository of bulk and single cell RNA-Sequencing datasets.18 BST2 expression was increased among EoE patients compared to controls and its expression correlated strongly with CD4 and IL-13 (Figure S10B–D) using previously published RNA-SEQuencing data.19 Among a single cell RNA-SEQ dataset from esophageal biopsies, BST2 was expressed by all cell types (Figure S10E).20 We further examined BST2 expression in epithelial cell subsets and found predominant expression in quiescent, transitioning and proliferating epithelial cells, but lowest in differentiated cells (Figure S10F).21 Analysis of BST2 expression from experiments on esophageal epithelial cells identified increased BST2 with loss of SPINK7 or exposure to IL-13 (Figure S10G,H).22, 23
Finally, we assessed for CDR3s existed that bore significant similarly to each other by only 1 amino acid among patients with a shared trigger (milk) compared to patients with differing triggers (non-milk). We compared the TCR clonotypes with a Hamming distance (HD) of 1 for TCRα and β. We report increased abundance of clonotypes with a HD of 1 (p < .001) among milk-triggered EoE compared to non-milk-triggered EoE patients (Figure 5E,F) suggesting the presence of recently described TCR meta-clusters.11
4. DISCUSSION
Here we report bulk TCR sequencing of esophageal biopsies from children and adults with EoE treated with diet elimination and identified to have milk-triggered and non-milk-triggered disease. Among children, but not adults with active EoE, we found reduced unique clonotypes and increased relative abundance of clonotypes comprising >1% of the total compared to non-EoE controls and inactive EoE. These clonality measures correlated with intraepithelial CD4 T-cell counts. Taken together, this validates the finding of increased T-cell clonality in EoE although this appears restricted to children.8 Among patients consuming food triggers of inflammation, we found specific TCRs that were absent or had decreased abundance when the food was removed. In addition, we found overlapping TCRs between patients with milk-triggered EoE that were absent in patients with non-milk-triggered EoE. Together this raises the prospect of food specificity of the TCR in EoE. We further found evidence of a common antigen regardless of food trigger, BST2, which was expressed by differentiating epithelial cells, and induced by IL-13 or loss of SPINK7. Our study provides important insight into T-cell clonality, food specificity and a potential T-cell target in EoE that warrants further research.
A key finding of our work are differences in clonality between adults and children. We uncovered no differences in clonality measures between adults with EoE and controls. This was starkly different in children and surprising given that all patients in our study were diet responsive. This was unlikely due to sample processing differences as we found no difference between pediatric and adult controls. This may be due to the evolving nature of adult EoE into a more fibrostenotic disease or longer duration of disease giving rise to new clones over time. No prior assessment of TCR in adults with EoE has been performed, and prior studies in children have shown mixed results regarding clonality.8, 24 Morgan et al. identified clonality of peTH2 cells while Wen et al. found no evidence of clonality from isolated CD3 cells.8, 24 Our study uncovers potential age-specific differences in the T-cell response to food antigen between children and adults.
Our study raises questions regarding TCR food specificity in EoE. We found TCRs present in active EoE before diet elimination and with food reintroduction that were absent or had reduced abundance post-diet elimination. We also observed shared clonotypes among patients with milk-triggered EoE that were absent in patients with non-milk-triggered EoE. None of these clonotypes were identifiable in a public database, suggesting they may be food-specific TCRs, although it is possible these TCRs could target something other than food (e.g., auto-antigen, microbe). Further investigation is needed to assess this, particularly whether specific foods drive expansion or activation of these TCRs ex vivo. As food-associated TCRs were uncommonly shared between patients, we assessed the Hamming Distance to understand whether similar TCRs exist among patients with a shared food trigger. We observed an increased number of clonotypes that differed by only one amino acid among EoE patients with a shared trigger (milk) compared to those with heterogenous triggers (non-milk). This finding was similarly noted by Morgan et al8 and raises questions regarding the existence of TCR meta-clusters that may define food specificity, and whether there may be utility for whole biopsy TCR analysis in identifying food triggers.11
A novel finding of our study is the identification of a potential common antigen, BST2, which may be a target of CD4 T-cells in EoE. This was identified by the presence of three clonotypes shared by patients with milk-triggered and non-milk-triggered EoE patients. BST2 has described to have a role in epithelial polarization, endocytosis of antigens, and interferon responses in plasmacytoid dendritic cells, and priming of CD8 T-cells, but has an unknown role in EoE.25–27 We examined publicly available datasets and found BST2 was increased in EoE and correlated with T-cell genes: CD4 and IL-13. We found expression of BST2 in multiple cell types including non-differentiated epithelial cells. More importantly, we found increased BST2 expression in epithelial cells with IL-13 exposure or loss of SPINK7. This supports a potential mechanism whereby loss of SPINK7 leads to increased BST2 which attracts IL-13-producing T-cells that further drives BST2 expression and attracts more pathogenic T-cells.
Our approach utilized bulk RNA Sequencing which allowed for examination of a large quantity of unique TCRs. Single cell sequencing requires digestion and dissociation for which there can be cell loss and is significantly more costly. While limited in the ability to identify α/β chain pairings, bulk TCR sequencing nonetheless can inform single cell analysis of suspected food-specific clonotypes. It was interesting that differences in unique clonotypes were seen only in TCRα. This may be due to the development of a TCRα/β pair as TCRβ rearranges first followed by TCRα. Thus, one TCRβ has opportunity to be paired with multiple TCRα. Furthermore, T-cells only express a single allele for TCRβ (allelic exclusion), two alleles can be expressed by TCRα in 20%–30% of T-cells.28 In addition, we examined TCRγ and TCRδ in a subset of patients. We identified reduced unique clonotypes and diversity compared to α/β, and further investigation is warranted into γ/δ TCRs, which are involved in “innate like” responses.
A key limitation of our study is the lack of patients with three time points, which affects our ability to identify statistical differences. We were unable to match MHC with specific TCRs, a key piece of information that would be beneficial. The sample size of adult patients was small and further analysis of larger cohorts is necessary to validate the age-specific differences seen in our study. The storage of adult and pediatric samples was different (RNAlater vs snap freezing in liquid nitrogen). While it is possible this could affect RNA quality, no differences were seen with RIN nor were there significant differences in unique clonotypes between pediatric and adult controls.
In conclusion, we validated the finding of clonality in pediatric active EoE and identified potential TCR markers of milk-triggered EoE. We found evidence that specific TCRs disappear with food elimination and reappear with food trigger reintroduction along with limited overlap of TCRs for specific foods. Further studies are needed to establish the full breadth and restricted nature of the TCR repertoire with regards to EoE triggers. Studies are warranted of paired whole biopsy bulk versus and single cell TCR sequencing.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank all patients who participated in the study. The authors are also grateful to their colleagues and clinical support staff for procuring biopsies and clinical data.
FUNDING INFORMATION
This study was supported by Campaign Urging Research for Eosinophilic Disease (CURED), Digestive Health Foundation (DHF), and the Brickner Foundation.
Abbreviations:
- CURED
Campaign Urging Research for Eosinophilic Diseases
- EoE
eosinophilic esophagitis
- GI
gastrointestinal
- HPF
high powered field
- PPI
proton pump inhibitor
- TCR
T-cell receptor
Footnotes
CONFLICT OF INTEREST STATEMENT
JBW is a consultant for Allakos, Regeneron, Sanofi/Genzyme, AstraZeneca, and Invea Therapeutics. JBW receives clinical trial/research funding from Allakos and Sanofi-Regeneron. FLK receives research funding from AstraZeneca and is a consultant for GlaxoSmithKline. NG is a consultant for Allakos, Sanofi-Regeneron, AstraZeneca, Abbvie, Knopp, Nutritia. NG receives royalties from Up-to-date. IH is a consultant for Adare/Ellodi, Arena Pharmaceuticals, AstraZeneca, Celgene/Receptos/BMS, Regeneron/Sanofi, Esocap, Gossamer Bio, Lilly, Shire/Takeda, Allakos. IH receives research funding from Allakos, Celgene/Bristol Meyers Squibb, Regeneron/Sanofi, Shire/Takeda, AstraZeneca, Arena, Adare/Ellodi.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Kagalwalla AF, Wechsler JB, Amsden K, et al. Efficacy of a 4-food elimination diet for children with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2017;15(11):1698–1707.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonsalves N, Yang GY, Doerfler B, et al. Elimination diet effectively treats eosinophilic esophagitis in adults; food reintroduction identifies causative factors. Gastroenterology. 2012;142:1451–9 e1. [DOI] [PubMed] [Google Scholar]
- 3.Kagalwalla AF, Shah A, Li BU, et al. Identification of specific foods responsible for inflammation in children with eosinophilic esophagitis successfully treated with empiric elimination diet. J Pediatr Gastroenterol Nutr. 2011;53:145–149. [DOI] [PubMed] [Google Scholar]
- 4.Bashaw H, Schwartz S, Kagalwalla AF, Wechsler JB. Tutorial: nutrition therapy in eosinophilic esophagitis-outcomes and deficiencies. JPEN J Parenter Enteral Nutr. 2020;44:600–609. [DOI] [PubMed] [Google Scholar]
- 5.Dellon ES, Guo R, McGee SJ, et al. A novel allergen-specific immune signature-directed approach to dietary elimination in eosinophilic esophagitis. Clin Transl Gastroenterol. 2019;10:e00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cianferoni A, Ruffner MA, Guzek R, et al. Elevated expression of activated TH2 cells and milk-specific TH2 cells in milk-induced eosinophilic esophagitis. Ann Allergy Asthma Immunol. 2018;120(2):177–183.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bullock JZ, Villanueva JM, Blanchard C, et al. Interplay of adaptive th2 immunity with eotaxin-3/c-C chemokine receptor 3 in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2007;45:22–31. [DOI] [PubMed] [Google Scholar]
- 8.Morgan DM, Ruiter B, Smith NP, et al. Clonally expanded, GPR15-expressing pathogenic effector TH2 cells are associated with eosinophilic esophagitis. Sci Immunol. 2021;6:eabi5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Natarajan K, Jiang J, May NA, et al. The role of molecular flexibility in antigen presentation and T cell receptor-mediated signaling. Front Immunol. 2018;9:1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morath A, Schamel WW. Alphabeta and gammadelta T cell receptors: similar but different. J Leukoc Biol. 2020;107:1045–1055. [DOI] [PubMed] [Google Scholar]
- 11.Mayer-Blackwell K, Schattgen S, Cohen-Lavi L, et al. TCR metaclonotypes for biomarker discovery with tcrdist3 enabled identification of public, HLA-restricted clusters of SARS-CoV-2 TCRs. Elife. 2021;10:e68605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liacouras CA, Furuta GT, Hirano I, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20 e6. [DOI] [PubMed] [Google Scholar]
- 13.Wechsler JB, Ackerman SJ, Chehade M, et al. Noninvasive biomarkers identify eosinophilic esophagitis: a prospective longitudinal study in children. Allergy. 2021;76:3755–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C, Sanders CM, Yang Q, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci U S A. 2010;107:1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu SG, Pan W, Liu H, et al. High throughput sequencing of T-cell receptor repertoire using dry blood spots. J Transl Med. 2019;17: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolotin DA, DA, Poslavsky S, Mitrophanov I, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. [DOI] [PubMed] [Google Scholar]
- 17.Lee YN, Frugoni F, Dobbs K, et al. Characterization of T and B cell repertoire diversity in patients with RAG deficiency. Sci Immunol. 2016;1:eaah6109. EGID Express. Accessed on 12/09/22. https://egidexpress.research.cchmc.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherrill JD, Kiran KC, Blanchard C, et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014;15:361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Baruch Morgenstern N, Ballaban AY, Wen T, et al. Single-cell RNA sequencing of mast cells in eosinophilic esophagitis reveals heterogeneity, local proliferation, and activation that persists in remission. J Allergy Clin Immunol. 2022;149:2062–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rochman M, Wen T, Kotliar M, et al. Single-cell RNA-seq of human esophageal epithelium in homeostasis and allergic inflammation. JCI. Insight. 2022;7:e159093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kc K, Rothenberg ME, Sherrill JD. In vitro model for studying esophageal epithelial differentiation and allergic inflammatory responses identifies keratin involvement in eosinophilic esophagitis. PLoS One. 2015;10:e0127755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azouz NP, Ynga-Durand MA, Caldwell JM, et al. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci Transl Med. 2018;10:eaap9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen T, Aronow BJ, Rochman Y, et al. Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest. 2019;129:2014–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epeldegui M, Blom B, Uittenbogaart CH. BST2/Tetherin is constitutively expressed on human thymocytes with the phenotype and function of Treg cells. Eur J Immunol. 2015;45:728–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swiecki M, Omattage NS, Brett TJ. BST-2/tetherin: structural biology, viral antagonism, and immunobiology of a potent host antiviral factor. Mol Immunol. 2013;54:132–139. [DOI] [PubMed] [Google Scholar]
- 26.Rollason R, Korolchuk V, Hamilton C, Jepson M, Banting G. A CD317/tetherin-RICH2 complex plays a critical role in the organization of the subapical Actin cytoskeleton in polarized epithelial cells. J Cell Biol. 2009;184:721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21:133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


