ABSTRACT
Swine H1N1/2009 influenza is a highly infectious respiratory disease in pigs, which poses a great threat to pig production and human health. In this study, we investigated the global expression profiling of swine-encoded genes in response to swine H1N1/2009 influenza A virus (SIV-H1N1/2009) in newborn pig trachea (NPTr) cells. In total, 166 genes were found to be differentially expressed (DE) according to the gene microarray. After analyzing the DE genes which might affect the SIV-H1N1/2009 replication, we focused on polo-like kinase 3 (PLK3). PLK3 is a member of the PLK family, which is a highly conserved serine/threonine kinase in eukaryotes and well known for its role in the regulation of cell cycle and cell division. We validated that the expression of PLK3 was upregulated after SIV-H1N1/2009 infection. Additionally, PLK3 was found to interact with viral nucleoprotein (NP), significantly increased NP phosphorylation and oligomerization, and promoted viral ribonucleoprotein assembly and replication. Furthermore, we identified serine 482 (S482) as the phosphorylated residue on NP by PLK3. The phosphorylation of S482 regulated NP oligomerization, viral polymerase activity and growth. Our findings provide further insights for understanding the replication of influenza A virus.
KEYWORDS: Swine influenza virus, PLK3, nucleoprotein, phosphorylation, virus replication
Introduction
Swine influenza (SI) is an acute and highly contagious respiratory disease of pigs caused by the influenza A virus (IAV) that regularly causes outbreaks of influenza in pigs. SI is often characterized by rapid onset of high fever, lethargy, loss of appetite, laboured abdominal breathing, and coughing, but causes few deaths in pigs [1]. However, swine influenza virus (SIV) infection can readily predispose to secondary bacterial and viral infections, resulting in prolonged respiratory disease and increased mortality. More seriously, SIV poses health challenges to humans on a global scale. Pigs express both human- and avian-type influenza virus receptors on tracheal epithelial cells and can be naturally and experimentally infected with avian influenza viruses and human influenza viruses, supporting the concept of their role as mixing vessel in the generation of novel viruses to which humans are immunologically naïve and highly susceptible [2]. The most recent influenza pandemic emerged from swine in 2009 when a novel reassorted virus, swine origin influenza A (H1N1) virus (S-OIV, now known as pH1N1 2009), spread to more than 200 countries and caused more than 18,000 human deaths worldwide by August 2010 (https://www.who.int/csr/don/2010%2008%2006/en/).
Influenza viruses exploit cell machinery to complete the viral life cycle in the host. Identifying the host molecules that participate in virus replication could provide valuable new targets for antiviral therapy. Genome-wide screens have been used to provide a global view of all the host factors involved in replication of a particular pathogen. For influenza virus, five high-throughput RNA interference or expression profiling screens have been performed in human cells or Drosophila cells, which have defined the cellular network involved in the human influenza virus replication cycle and identified thousands of host factors that may serve as potential antiviral drug targets [3–7]. However, genome-wide interactions between swine influenza virus and swine cells have not been extensively studied.
In this study, we identified PLK3 as a positive regulator for IAV replication. Our studies found that PLK3 significantly promoted virus replication by interacting with and phosphorylating NP protein. Furthermore, we demonstrated that PLK3 phosphorylated NP at S482, which played an important role in polymerase activity and viral ribonucleoprotein assembly. Importantly, we found that phosphorylation of NP S482 was important for influenza virus replication both in vitro and in vivo. Thus, we propose that PLK3 is a positive regulator that promotes virus replication by phosphorylating NP, all these findings suggest that PLK3 is crucial for influenza A virus replication.
Material and methods
Ethics statements
This study was conducted in strict accordance with the recommendations provided in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People’s Republic of China. Animal experiments were approved by the Hubei Administrative Committee for Laboratory Animals (202304250002).
Cells, viruses and reagents
NPTr cells, Porcine kidney cells (PK-15), and Human embryonic kidney HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (HyClone, Logan, UT, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (PAN-Biotech, Germany), and maintained at 37°C in a humidified 5% CO2 incubator (Sanyo, Osaka, Japan). All cells were confirmed to be free of mycoplasma for the entire study. NPTr cells were established following serial culture of primary cells derived from trachea tissues, and various influenza virus subtypes isolated from human, swine and avian species could replicate efficiently with cytopathic effect in NPTr cells [8]. PK-15 and HEK293T were purchased from ATCC (American Type Culture Collection, Manassas, VA, USA). A/swine/Nanchang/F9/2010 (H1N1) (GenBank accession number JF275925-JF275932), a pandemic (H1N1) 2009 influenza virus, was isolated in swine herds by our lab [9]. The virus could replicate efficiently in NPTr cells with 0.25 μg/mL of L-tosylamide-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (Sigma, St. Louis, MO, USA).
siRNA and plasmids construction
The siRNA duplexes against PLK3 (siPLK3) were designed according to previously validated duplexes for human PLK3 [6]. siPLK3 and the negative control duplexes (NC, a nontarget siRNA) were synthesized by GenePharma (Shanghai, China). To construct the PLK3-HA and PLK3-Flag expression plasmids, the coding regions of PLK3 were PCR-amplified from cDNA derived from NPTr cells and cloned into pCAGGS-HA or p3XFLAG-CMV-13 vectors. The cDNA clones of SIV-H1N1/2009 PB1, PB2, PA and NP were separately inserted into pCAGGS-HA vector to generate viral RNA expressing plasmids PB1-HA, PB2-HA, PA-HA and NP-HA. All mutated plasmids of NP (NP S467A, NP S67E, NP S482A and NP S482E) were derived from NP-HA plasmid. The mutation introduced in each construct was confirmed by sequencing.
Transfection and infection
NPTr cells were grown to 80% confluence and subsequently transfected with plasmids and/or siRNA using Lipofectamine 6000 or Lipofectamine 8000 (Invitrogen) according to the manufacturer's protocol. Transfected or untransfected NPTr cells were firstly washed with phosphate-buffered saline (PBS), then infected with SIV-H1N1/2009 at a multiplicity of infection (MOI) of 0.01. After one hour, the cells were re-suspended in DMEM with 0.25 μg/mL of TPCK-treated trypsin, and collected at the indicated time points postinfection. All experiments referring to SIV-H1N1/2009 infection were performed in a biological safety cabinet.
Microarray analysis
To identify the genes expression profile after SIV-H1N1/2009 infection in cells at 36 h, gene microarray was meanwhile performed by Shanghai Biotechnology Corporation (Shanghai, China). Two pools of total RNA were extracted from the infected and uninfected cells at 36 h using TRIzol Reagent (Invitrogen, USA) according to the manufacturer's protocol. After quality control, the total RNA were amplified, labelled and purified by using GeneChip 3’IVT Express Kit (Affymetrix, Santa Clara, CA, US) to obtain the biotin labelled cRNA which was then hybridized to Porcine Gene 1.0 ST Array (Affymetrix, Santa Clara, CA, US) containing a total of 24,192 probe sets which presented 20,201 genes and 220 controls. The hybridization, washing and staining were performed using Affymetrix GeneChip Hybridization, Wash and Stain Kit in the Affymetrix Hybridization Oven 645 and Affymetrix Fluidics Station 450. The slides were scanned by the Affymetrix GeneChip Scanner 3000, and the scanned images were quantified by the Affymetrix Command Console Software 3.1. The raw data were then loaded to the GeneSpring 11.0 (Agilent technologies, Santa Clara, CA, US) and normalized by the Affymetrix Microarray Analysis Suite (MAS) 5.0 algorithm. Expression profiles were analyzed using R programming language of the SBC Analysis System 2.9 software package (Shanghai Biotechnology Corporation, Shanghai, China), and p < 0.05 was considered statistically significant. The fold change (FC) ≥ 2 was identified to be differentially expressed in infected cells compared with that in uninfected cells. The differentially expressed (DE) genes were grouped into functional categories based on the Gene Ontology database (GO: http://www.geneontology.org/), and the KEGG pathway analysis was performed with the online SBC analysis system. The gene microarray data had been deposited in NCBI's Gene Expression Omnibus (GEO) database and were accessible through GEO Series accession number GSE96952.
RNA extraction, reverse transcription (RT) and quantitative real-time PCR (qRT-PCR)
RNA extraction was performed using TRIzol Reagent (Invitrogen, USA) following the manufacturer's protocol. DNase I (Ambion, Austin, TX, USA) digestion was carried out after RNA isolation. Both RNA concentration and purity (A260/280) were evaluated by a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Equal amounts of RNA were reverse transcribed to cDNA using AMV reverse transcriptase (Takara, Kyoto, Japan) in accordance with the manufacturer's instructions. The RT primers for genes were universal primers oligo (dT)18. Otherwise, RNA samples were stored at −80°C until further use. Microarray results were validated by qRT-PCR using FastStart Universal SYBR Green Master (Roche, Mannheim, Germany) on an ABI ViiA7 instrument (Applied Biosystems, Foster City, CA, USA). The 20 μL reactions mixture included 1 μL cDNA products, 0.5 μL forward primers and reverse primers, 10 μL SYBR green mix, and 8 μL DNase/RNase-free deionized water. All primers were designed using Primer Premier 5.0 software based on the sequences of corresponding mRNAs in GeneBank (Table S1). PCR reactions were performed in triplicate to guarantee the reproducibility of amplification of each sample. The relative expression levels were measured in terms of threshold cycle value and normalized to GAPDH expression using 2−△△CT method [10].
Western blotting and coimmunoprecipitation assays
After transfection and/or infection, cells were collected and lysed with Mammalian Protein Extraction Reagent (Cwbio, Beijing, China). Then the concentration of total proteins was determined using Bradford Protein Assay Kit (Beyotime, Shanghai, China). Equal quantity of protein was separated on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to the nitrocellulose membrane (GE Life Science, Piscataway, NJ, USA). Followed by being blocked in 1% bovine serum albumin (BSA), the membranes were then incubated with relevant antibodies. The antibodies against PLK3 and GAPDH were purchased from Abclonal Technology (Wuhan, China). Rabbit polyclonal antibody against SIV-H1N1/2009 NP was purchased from GeneTex, Inc (San Antonio, TX, USA). Horseradish peroxidase-conjugated anti-mouse/rabbit secondary antibodies (Abclonal, Wuhan, China) were used to visualize the proteins, and the protein blots were detected using enhanced chemiluminescence reagent (Advansta, Menlo Park, CA, USA). For coimmunoprecipitation assays, collected cells were washed with PBS and lysed with lysis buffer (Beyotime, Shanghai, China) containing CompleteTM protease inhibitor cocktail (B14001, Biotool, USA) at 24 h post transfection. Then each lysate was incubated with the relevant antibodies at 4°C overnight with gentle shaking. Protein A/G agarose beads (Bimake, Houston, TX, USA) were added, and the samples were incubated for another 3 h. The agarose beads were subsequently collected by centrifugation and washed three times with lysis buffer. The bound proteins were eluted by boiling with SDS-PAGE loading buffer for 5 min and then subjected to western blotting.
Minireplicon assay
Polymerase activity was measured by a minireplicon assay. vRNP recombinant system expression plasmids (pCDNA3.1-PB1, pCDNA3.1-PB2, pCDNA3.1-PA), tagged plasmid (NP-HA or its mutants NP S482A-HA, NP S482E-HA), pPolI-luc and Renilla fluorescence expression plasmids were transfected into HEK293T cells. The luciferase activity was then determined according to the manufacturer's protocol using the Dual-Luciferase Reporter Assay System (Promega, USA) at 24 h posttransfection. Renilla luciferase was used as a standardized internal control. All data were shown as relative firefly luciferase activity normalized to Renilla luciferase activity.
Virus rescue and growth curves
The recombinant virus including wild-type (WT) NP and NP mutants (NP S482A and NP S482E) from SIV-H1N1/2009 or Hunan-H1N1/2015 were generated using an 8-plasimid (PB1, PB2, PA, HA, NA, M, NS from PR8, and NP S482A and NP S482E) reverse genetic system [11]. Briefly, HEK293T cells grown to 70–80% confluence were transfected with the 8 indicated plasmids of the virus rescue system and incubated overnight at 37°C. Then, MDCK cells were added and transferred to a small flask with HEK293T cells. Six hours later, the medium was replaced with serum-free medium containing 2.5 μg/mL TPCK-treated trypsin. The cells were further cultured for 3–4 days at 37°C in 5% CO2, and the supernatant containing the recombinant viruses was harvested and centrifuged at 3000 rpm for 10 min to remove cell debris. Amplification of viruses was performed in embryonated chicken eggs, and the identities of the viruses were confirmed by sequencing the NP gene. To determine the growth kinetics of the rescued viruses, confluent monolayers of PK-15 were infected at a MOI of 0.01 and incubated at 37°C. At 12, 24, and 36 h postinfection, 150 μL of the cellular supernatant was harvested and stored at −80°C until further analysis. TCID50 assays were performed in PK-15 cells to determine virus titres of SIV-H1N1/2009 and the recombinant viruses.
Cell viability assay
In order to ensure the effect of PLK3 silencing on cell proliferation, the cell viability of NPTr cells transfected with siPLK3 or NC were monitored by Cell Counting Kit-8 (CCK-8, Dojindo Molecular Technologies, Kumamoto, Japan) analysis according to the manufacturer's instructions. In brief, NPTr cells in 96-well plates were transfected with siPLK3 or NC, and the cell viability was measured at 24, 36, and 48 h posttransfection. CCK-8 reagent was added to each well of a plate, and the absorbance at 450 nm was detected by a microplate reader after 1 h of incubation.
Pathogenicity study in mice
The virus titres of NP WT, NP S482A and NP S482E were determined by calculating log10TCID50/mL using the Reed and Muench method, then uniformly infected mice with 104 TCID50/mL. Groups of ten 5-week-old female BALB/c mice were lightly anaesthetized with CO2 and inoculated intranasally with NP WT, NP S482A, NP S482E and PBS in a volume of 200 μL. Mice were monitored daily for weight loss and mortality for 14 days. Mice that lost ≥30% of initial weight were humanely euthanized. To determine virus replication, groups of six BALB/c mice were intranasally inoculated with the indicated doses of NP WT, NP S482A and NP S482E diluted in PBS and were euthanized on 3 and 5 days postinfection.
Statistical analysis
All assays were performed in three biological replicates and the results shown were representative of at least three independent experiments. Data were shown as means ± standard deviation. Statistical analysis was performed by Student's t-test using the GraphPad Prism 7 software (GraphPad Software Inc., San Diego, CA, USA). A p value below 0.05 was considered statistically significant and the asterisks in all figures are defined, * p < 0.05, ** p < 0.01, *** p < 0.001.
Results
Screening host genes involved in SIV-H1N1/2009 infection
In order to screen the host genes involved in SIV-HIN1/2009 infection, the genome-wide expression profile was investigated by carrying out a gene microarray. Based on threshold of fold change ≥2 and P-value <0.05, a total of 554 transcripts were identified to be differentially expressed in infected cells compared with that in uninfected cells after quantile normalization and statistical analysis. However, only 166 out of 554 DE transcripts could be functionally annotated, and among these functional host genes, 89 were upregulated and 77 were downregulated (Table S2). Gene ontology (GO) and pathway enrichment analysis were conducted to explore functions of these 166 DE genes in response to SIV-H1N1/2009 infection. In order to confirm the above analysis of overall changes in host genes expression after SIV-H1N1/2009 infection, we selected 10 DE genes randomly to confirm their expression levels by a qRT-PCR method. As shown in Figure S1, VNN2, CSF2, JUN, FOS, MYBL2, E2F1, PLK3 and DUSP5 in infected cells expressed higher than that in uninfected cells, whereas ADAMTS1 and PRSS35 were downregulated compared with the uninfected control (Figure S1A), which were consistent with the gene microarray analysis.
To identify which of these DE genes may play an important role in influenza virus replication, we compared the 166 DE genes selected by the gene microarray with those identified genes in previous screens [3–7,12], and a total of 24 DE genes were found to be potential genes involved in SIV-H1N1/2009 replication (Table S3). Of note, PLK3 and JUN, among these 24 DE genes, were found in two genome-wide RNAi screens [6,7]. Additionally, PLK3 had been validated to promote influenza virus replication [13], but the exact mechanism was still unknown. Therefore, we selected PLK3 for further analysis.
PLK3 is upregulated after SIV-H1N1/2009 infection and promotes virus replication
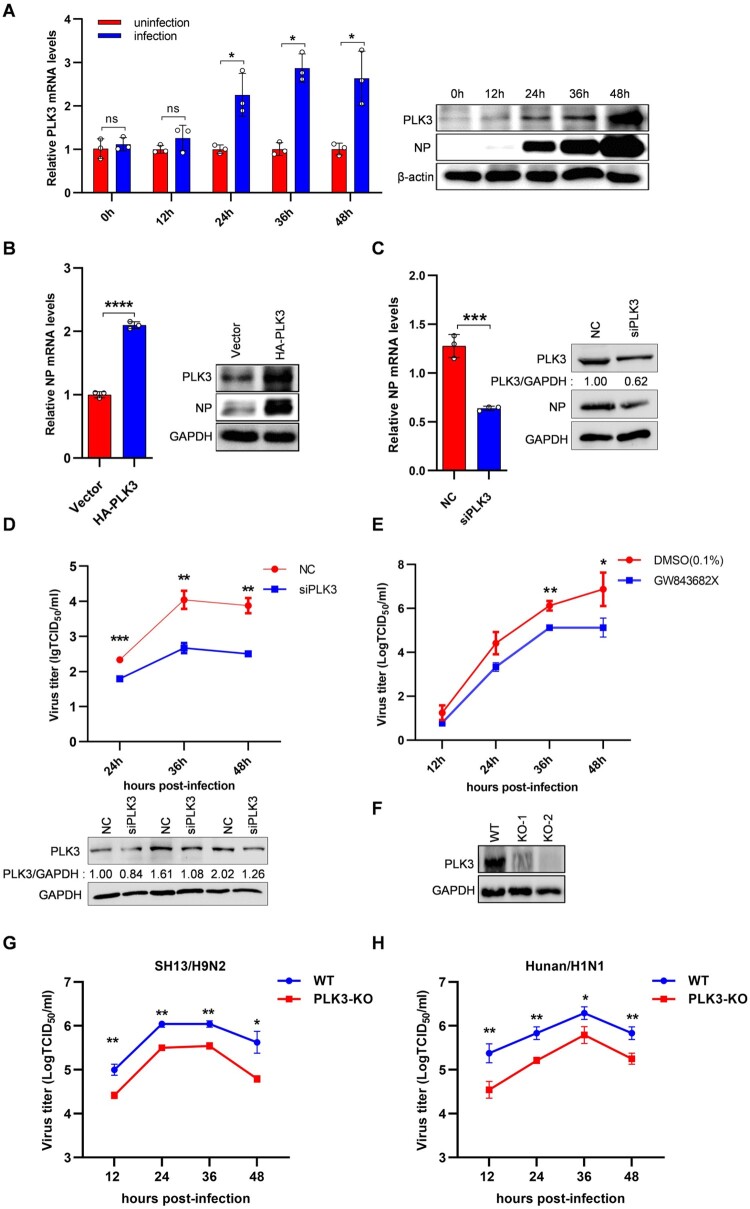
According to the gene microarray, PLK3 was found to be upregulated (FC = 3.985, p = 0.029) (Table S2). To further explore the expression patterns of PLK3 upon influenza virus infection, NPTr cells were infected or uninfected with SIV-H1N1/2009, and the mRNA levels of PLK3 were subsequently measured by qRT-PCR at 0, 12, 24, 36, and 48 h postinfection. Meanwhile, the protein levels of PLK3 were also evaluated by western blotting. These results showed that the mRNA and protein levels of PLK3 were significantly upregulated from 24 to 48 h postinfection compared with that in uninfected cells (Figure 1A), implying that PLK3 might play a role in influenza virus.
Figure 1.
PLK3 promoted SIV-H1N1/2009 replication. (A) PLK3 was upregulated after SIV-H1N1/2009 infection. NPTr cells were uninfected or infected with SIV-H1N1/2009 at a MOI of 0.01 and harvested at 0, 12, 24, 36, and 48 h postinfection, respectively. The mRNA levels of PLK3 were determined by qRT-PCR and normalized to GAPDH. All results were standardized to 1 in uninfected cells for each time point. The protein levels of PLK3 were determined by western blotting and normalized to β-actin. (B) Effects of PLK3 overexpression on NP. NPTr cells were transfected with pCAGGS-HA-PLK3 for 24 h, then infected with SIV-H1N1/2009 at a MOI of 0.01 for 36 h. The mRNA levels and protein levels of viral NP were respectively measured by qRT-PCR and western blotting, and normalized to GAPDH. (C) Effects of siPLK3 on NP. NPTr cells were transfected with siPLK3 for 24 h, then infected with SIV-H1N1/2009 at a MOI of 0.01 for 36 h. The mRNA levels and protein levels of viral NP were respectively measured by qRT-PCR and western blotting, and normalized to GAPDH. (D) The growth curves of SIV-H1N1/2009 in NPTr cells after siPLK3 treated. NPTr cells were transfected with siPLK3 or negative control (NC), then infected with SIV-H1N1/2009 at a MOI of 0.01. Cell supernatants were collected at 24, 36, and 48 h post-infection. The virus titres were assessed by TCID50 assays on PK-15 cells. The protein levels of PLK3 at different time points were measured by western blotting. (E) The growth curves of SIV-H1N1/2009 in PK-15 cells after PLK3 inhibitor (GW843682X) treated. PK-15 cells were treated with GW843682X (10 µM) or DMSO (0.1%) for 2 h, then infected with SIV-H1N1/2009 at a MOI of 0.01. Cell supernatants were collected at 12, 24, 36, and 48 h postinfection. The virus titres were assessed by TCID50 assays on MDCK cells. Data were shown as means ± standard deviations (n = 3). Statistical significance was analyzed by Student's t-test, * p < 0.05, ** p < 0.01, ns represented no significance. (F) PK-15- PLK3-KO- cell lines were obtained by using the CRISPR/Cas9 system. The PLK3-knockout effects were measured by western blotting. (G) The growth curves of SH13/H9N2 in PK-15 cells. PK-15 WT cell line and PLK3-KO cell line were infected with SH13/H9N2 virus at a MOI of 0.01. Cell supernatants were collected at 12, 24, 36, and 48 h post-infection. The virus titres were assessed by TCID50 assays on PK-15 cells. (H) The growth curves of Hunan/H1N1 in PK-15 cells. PK-15 WT cell line and PLK3-KO cell line were infected with Hunan/H1N1 virus at a MOI of 0.01. Cell supernatants were collected at 12, 24, 36, and 48 h post-infection. The virus titres were assessed by TCID50 assays on PK-15 cells.
It was reported that knockdown of PLK3 could reduce the replication of human influenza virus in A549 cells by two genome-wide RNAi screens [6,7], which was validated by Pohl et al. [13]. To confirm the effect of PLK3 on SIV replication in porcine cells, we overexpressed pCAGGS-HA-PLK3 expressing plasmid and silenced PLK3 with PLK3-specific siRNA in NPTr cells, followed by SIV-H1N1/2009 infection. The intracellular mRNA and protein levels of viral nucleoprotein (NP) were respectively determined by qRT-PCR and western blotting at 36 h postinfection. In PLK3-overexpression cells, viral NP mRNA and protein levels were dramatically elevated (Figure 1B), whereas knockdown of PLK3 had the contrary effect on viral NP levels (Figure 1C). In addition, the supernatants were harvested and subjected to TCID50 analysis to evaluate the titres of SIV-H1N1/2009 at different time points (24, 36, and 48 h) postinfection. The results revealed that viral growth was remarkably reduced in the PLK3-silenced cells (Figure 1D). To exclude that the inhibition of viral growth might be due to reduced cell viability, we assessed the impact of PLK3 silencing on cell viability in the absence of virus infection. The data showed that the cell viability of PLK3-silenced NPTr cells were comparable with that of mock-treated cells at 24, 36, and 48 h posttransfection (Figure S2A). In addition, we used a PLK3 inhibitor GW843682X [14], which was a special inhibitor of PLK3 kinase activity, to verify the proliferation of SIV-H1N1/2009 when treated with the inhibitor. In order to select an appropriate concentration, we used 0.1, 1, and 10 µM GW843682X and DMSO as a control to treat PK-15 cells, after treatment of 48 h, CCK-8 reagent was used to detect cell viability. The results showed that 0.1, 1, and 10 µM GW843682X had no obvious effects on cell viability (Figure S2B). Therefore, we chose the concentration of 10 µM for subsequent experiments. PK-15 cells were pretreated with DMSO (0.1%) or GW843682X (10 µM) for 2 h, then infected with SIV-H1N1/2009 (0.01 MOI), the supernatants at 12, 24, 36, and 48 h postinfection were collected for TCID50 determination to evaluate the titres of SIV-H1N1/2009. The results showed that after treated with GW843682X, the virus titres were significantly reduced (Figure 1E). Taken together, these results demonstrate a positive regulatory role of PLK3 in SIV-H1N1/2009 replication.
Besides, we would like to investigate the impact of PLK3 on the replication of influenza viruses from various sources. Initially, we conducted a comparison of the homology between swine PLK3 and human PLK3 sequences. According to the analysis performed on the Uniprot website, the amino acid sequence homology between swine PLK3 and human PLK3 was determined to be 92.26%. This high level of homology suggests that PLK3 potentially plays a significant role in the replication process of influenza viruses from diverse origins. Next, we investigated the effects of PLK3 on influenza virus replication in different hosts, including avian influenza (SH13/H9N2) and human influenza (Hunan/H1N1) viruses. We used CRISPR-Cas9 gene editing technology to knock out PLK3 of the PK-15 cells, as shown in Figure 1F, the monoclonal cell line PLK3-KO-2 with better knockout effect was selected for the follow-up experiments (Figure 1F). We infected the two different influenza viruses respectively with PK-15-WT or PK-15-PLK3-KO cell lines, then collected the supernatants and performed TCID50 analysis to evaluate titres at different time points (12, 24, 36, and 48 h) after infection. The results showed that both viral growth of avian influenza (SH13/H9N2) and human influenza (Hunan/H1N1) were significantly reduced in PLK3-KO cell lines compared to WT cell lines (Figure 1G,H), suggesting that PLK3 plays an important regulatory role in the replication of influenza viruses in different hosts.
PLK3 interacts with PB1, PB2 and NP proteins, and promotes NP phosphorylation level
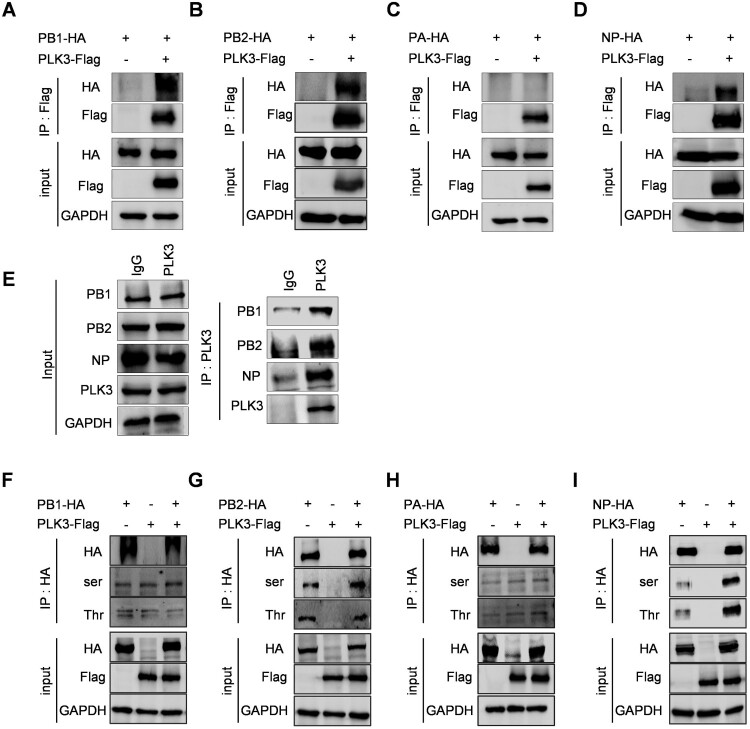
As known, influenza viral ribonucleoprotein plays an important role in virus replication. In order to explore whether PLK3 positively regulating SIV-H1N1/2009 replication is related with viral ribonucleoprotein, we first determined whether PLK3 could interact with the components of ribonucleoprotein. HEK293T cells were cotransfected with PLK3-Flag and PB1-HA, PB2-HA, PA-HA, or NP-HA, respectively, followed by performing co-immunoprecipitation (Co-IP) assays using anti-Flag monoclonal antibody. Western blot analysis revealed that PB1, PB2, and NP were coprecipitated by PLK3, whereas PA was not coprecipitated by PLK3 (Figure 2A–D), demonstrating that PLK3 interacted with PB1, PB2, and NP. In addition, we also performed the interaction between endogenous PLK3 and viral proteins in the context of viral infections. Consistent with previous results, endogenous PLK3 can interact with PB1, PB2 and NP in the context of viral infections (Figure 2E), while it has no obvious interaction with other viral proteins such as PA, NA, HA, M2, and NS1 (data not shown).
Figure 2.
PLK3 interacted with NP and promoted NP phosphorylation. (A, B, C, and D) Interactions between PLK3 and viral RNP components. The expression plasmid of PLK3-Flag was respectively cotransfected with PB1-HA (A), PB2-HA (B), PA-HA (C), or NP-HA (D) in HEK293T cells. The cell lysates were collected at 24 h post-transfection. Co-IP assays were performed using an anti-Flag antibody to precipitate viral RNP components which interacted with PLK3, followed by western blotting to detect PLK3 using an anti-Flag antibody and PB1, PB2, PA, and NP using an anti-HA antibody. GAPDH served as a loading control in all of these experiments. (E) Interactions between endogenous PLK3 and viral RNP components in the context of viral infections. NPTr cells were infected with SIV-H1N1/2009 at a MOI of 0.01. The cell lysates were collected at 24 h post-infection. Co-IP assays were performed using anti-IgG and anti-PLK3 antibodies to precipitate viral RNP components which interacted with PLK3, followed by western blotting to detect PLK3 using an anti-PLK3 antibody and PB1, PB2, PA, and NP using endogenous antibodies. GAPDH served as a loading control in all of these experiments. (F, G, H, and I) Effects of PLK3 on the phosphorylation of viral RNP components. The expression plasmid of PLK3-Flag was respectively cotransfected with PB1-HA (F), PB2-HA (G), PA-HA (H), or NP-HA (I) in HEK293T cells. The cell lysates were collected at 24 h posttransfection, then, PB1-HA, PB2-HA, PA-HA, and NP-HA were immunoprecipitated with an anti-HA antibody, and the associated phosphorylated serine/threonine of PB1, PB2, PA, and NP were detected using anti-p-serine/threonine antibodies. GAPDH served as a loading control in all of these experiments.
Considering that PLK3 is a serine/threonine kinase and phosphorylation of viral proteins regulates the life cycle of influenza virus, we then wondered whether PLK3 had an impact on the phosphorylation of viral ribonucleoprotein subunits. Firstly, PLK3-Flag were cotransfected with PB1-HA, PB2-HA, PA-HA, or NP-HA in HEK293T cells, respectively. Then, all cell lysates were immunoprecipitated with anti-HA agarose, and the phosphorylation of PB1, PB2, PA, and NP were detected by immunoblotting with the anti-serine/threonine pan-phosphorylated antibodies. As the results shown, PLK3 had no significant effect on the phosphorylation levels of PB1 and PA, had only a little effect on the phosphorylation levels of PB2, but significantly increased the serine/threonine phosphorylation levels of NP (Figure 2F–I). In conclusion, PLK3 interacts with NP and stimulates NP phosphorylation. Hence, we hypothesize that PLK3 might execute its proviral role through the regulation of influenza viral NP functions.
PLK3 facilitates NP oligomerization and viral ribonucleoprotein assembly
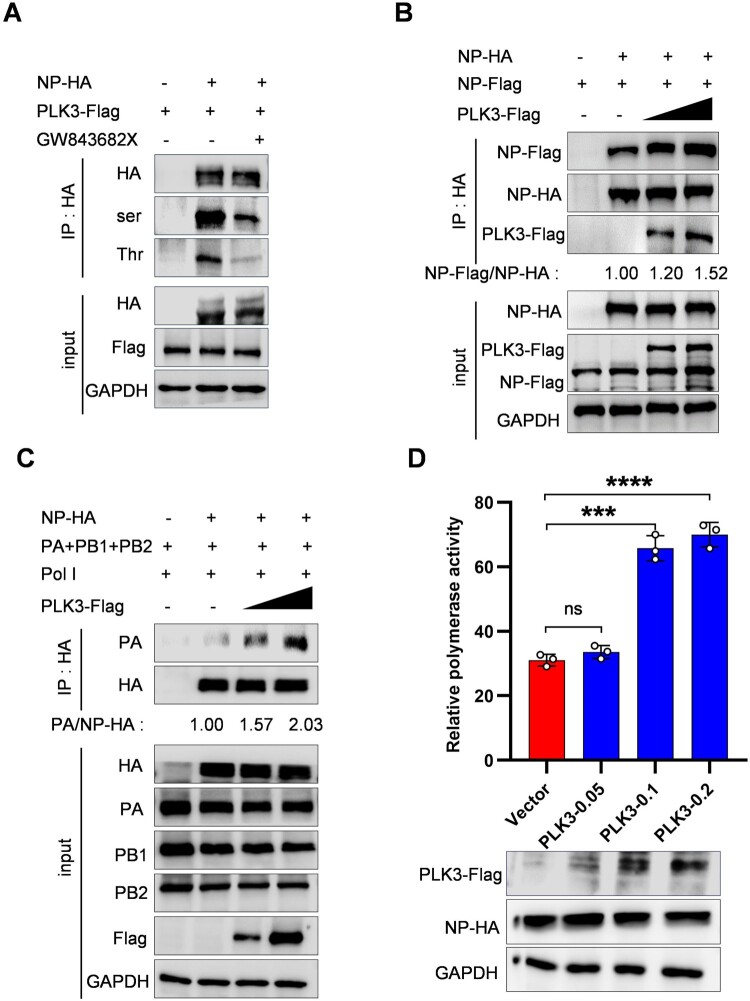
To further explore the phosphorylation between PLK3-targeted NP, we assessed the impact of PLK3 on NP phosphorylation levels while using the inhibitor GW843682X [14]. The data showed that the level of NP phosphorylation was significantly reduced with the inhibitor GW843682X treatment compared with the control group (Figure 3A). The results indicate that PLK3 can regulate the phosphorylation level of NP.
Figure 3.
PLK3 promoted NP oligomerization, vRNP assembly and viral polymerase activity. (A) PLK3 inhibitor (GW843682X) blocked NP phosphorylation. The expression plasmid of NP-HA was co-transfected with PLK3-Flag in HEK293T cells with or without GW843682X treatment for 2 h posttransfection. Co-IP assays were performed using an anti-HA antibody. The phosphorylated serine/threonine of NP were detected by western blotting with an anti-p-serine/threonine antibodies, and the total NP was detected with an anti-HA antibody. (B) The effect of PLK3 on NP oligomerization. Expression plasmids of PLK3-Flag (0, 0.5, and 1 µg), NP-Flag and NP-HA were cotransfected in HEK293T cells which were then lysed at 24 h posttransfection. Co-IP was performed using an anti-HA antibody followed by western blotting. The band intensities were quantified by ImageJ, and the relative precipitated NP-Flag/NP-HA ratios were shown below. (C) The effect of PLK3 on vRNP assembly. HEK293T cells were transfected with the vRNP reconstitution plasmids (pCDNA-PA, pCDNA-PB1, pCDNA-PB2, pCAGGS-HA-NP), and pPol I plasmid together with PLK3-Flag (0, 0.5, and 1 µg). Cells were then lysed at 24 h post-transfection. Co-IP was performed using an anti-HA antibody followed by western blotting. The band intensities were quantified by ImageJ, and the relative precipitated PA/NP-HA ratios were shown below. GAPDH was used as a loading control in all of these experiments. (D) The effect of PLK3 on viral polymerase activity. HEK293T cells were transfected with plasmids (pCDNA-PA, pCDNA-PB1, pCDNA-PB2, pCAGGS-HA-NP), pPolI-firefly and Renilla luciferase expression plasmids along with Flag-PLK3 (0.05, 0.1 and 0.2 µg or vector). The luciferase activity was measured at 24 h after the transfection. The proteins expression of NP and PLK3 were detected by western blotting. GAPDH was used as a control. Data were shown as means ± standard deviations (n = 3). Statistical significance was analyzed by Student's t-test, * p < 0.05, ** p < 0.01, ns represented no significance.
It was reported that NP phosphorylation regulated its oligomerization, this led us to question whether PLK3 influences NP oligomerization [15]. To validate this idea, Co-IP of NP-HA and NP-Flag with or without PLK3 was performed. HA-tagged beads were used to coprecipitate NP-Flag which was associated with NP-HA. The results showed that more NP-Flag was detected in a dose-dependent manner in the presence of PLK3 compared with that in the absence of PLK3 (Figure 3B), suggesting that PLK3 enhanced NP oligomerization.
NP assembles with viral RNA and three subunits of the polymerase into a vRNP complex, and NP polymerization is crucial for maintaining the structure of the vRNP complex [16]. Therefore, we hypothesized that PLK3 might affect the assembly of vRNP. A vRNP reconstitution system in which NP was substituted with an expression plasmid NP-HA was used to imitate the functional vRNA complex, then a Co-IP assay was carried out using HA-tagged beads to precipitate PA and the amount of precipitated PA represented the efficiency of vRNP assembly (the role of the Pol I plasmid is to provide vRNAs, which together with the viral polymerase to forms vRNP). As expected, the amount of PA precipitated by NP-HA was clearly increased with an increasing dose of PLK3 (Figure 3C), indicating that PLK3 can facilitate vRNP assembly. Furthermore, we verified the effect of PLK3 on viral polymerase activity, a minireplicon assay was carried out to determine the effect of PLK3 on the viral polymerase activity. The data showed that PLK3 promoted viral polymerase activity in a dose-dependent manner (Figure 3D). Taken together, these findings suggest that PLK3 is capable of enhancing the vRNP assembly and the polymerase activity of SIV.
PLK3 phosphorylated NP protein at serine 482
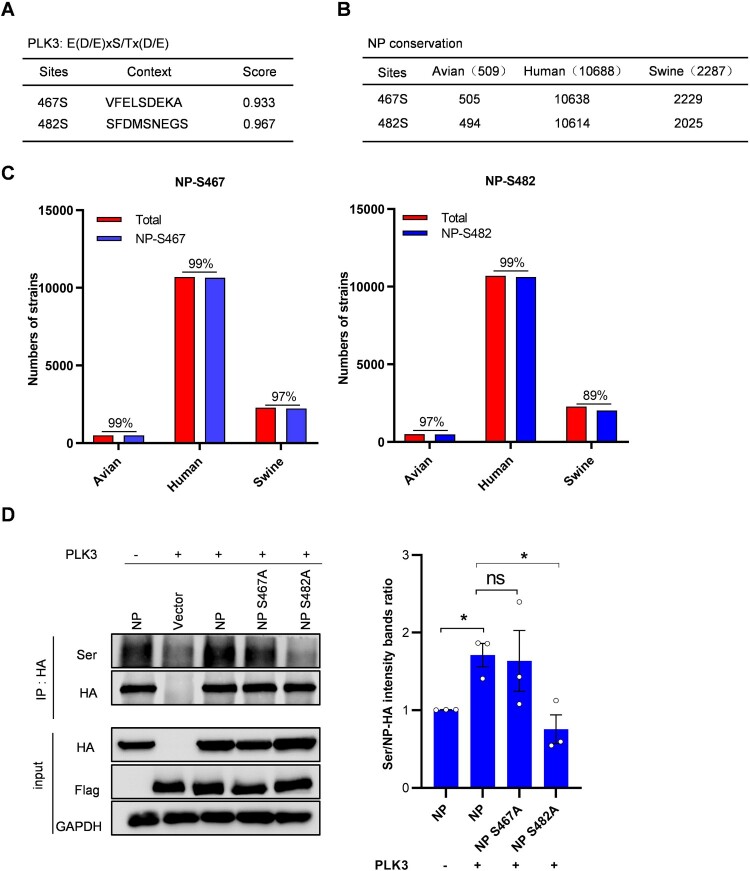
It has been confirmed in the previous section of this study that PLK3 can phosphorylate NP. To further investigate this, we predicted the potential phosphorylation sites on NP. We used the NetPhos 3.1 server (http://www.cbs.dtu.dk/services/NetPhos/), which employs ensembles of neural networks to perform this prediction. Higher scores indicate a greater potential for residues to be phosphorylated, with scores above 0.5 being considered as the threshold. As a result, a total of 64 potential serine/threonine phosphorylated sites in SIV-H1N1/2009 viral NP were predicted (data not shown), of which 18 sites with a score of >0.5 were considered to get phosphorylated with high potential (Table S4). In addition, the consensus sequence for PLK3 phosphorylation was reported as E(D/E)xS/Tx(D/E) [17]. Among the 18 residue sites, only serine 467 and serine 482 were correspondent with the consensus sequence, indicating that PLK3 might phosphorylate NP at serine 467 or/and serine 482 (Figure 4A).
Figure 4.
Identification of phosphorylation sites of PLK3 in NP. (A) The consensus sequence of PLK3 phosphorylation. The NP S467 and S482 sites were correspondent with the consensus sequence. (B and C) Residues S467 and S482 are highly conserved among different species of H1N1 influenza A viruses. The sequence alignments of avian, human, and swine H1N1 influenza A viruses were performed and analyzed by MegAlign. (D) Effects of PLK3 on S467 and S482 phosphorylation of NP. The expression plasmid of PLK3-Flag was co-transfected with NP-HA or NP mutants (NP S467A and NP S482A) in HEK293T cells. Co-IP assays were performed using an anti-HA antibody. The phosphorylated serine of NP was detected by western blotting with anti-p-Ser antibody, and the total NP was detected with an anti-HA antibody. GAPDH served as a loading control (left). The band intensities were quantified by ImageJ, and the grayscale values of each experiment was statistically analyzed by Graphpad (right, n = 3).
Then, we performed sequence alignments of avian, human, and swine influenza A viruses using MegAlign. We found that the residue serine 467 was conserved in at least 97% of the analyzed avian, human, and swine isolates. Similarly, the residue serine 482 was also conserved in at least 97% of the analyzed avian and human isolates, and in 89% of the swine isolates (Figure 4B,C). The results demonstrated that residues serine 467 and 482 were both highly conserved among different species of influenza A viruses. To further verify whether residues S467 and S482 were actual phosphorylation sites of PLK3 on NP, an alanine (A) substitution at S467 or S482 was introduced to mimic the dephosphorylated residue. We observed that the impact of PLK3 on NP serine phosphorylation was less significant when the S467 residues were dephosphorylated (S467A) compared to wild-type NP. However, PLK3 significantly suppressed NP serine phosphorylation levels when the S482 residue was dephosphorylated (S482A) (Figure 4D). To quantify these findings, we measured the grayscale values of each experiment. The results indicated that NP S482 was the phosphorylation site targeted by PLK3, while NP S467 was not (Figure 4D). Based on these results, we selected site S482 for further investigation.
Effect of NP S482 phosphorylation on SIV-H1N1/2009 replication
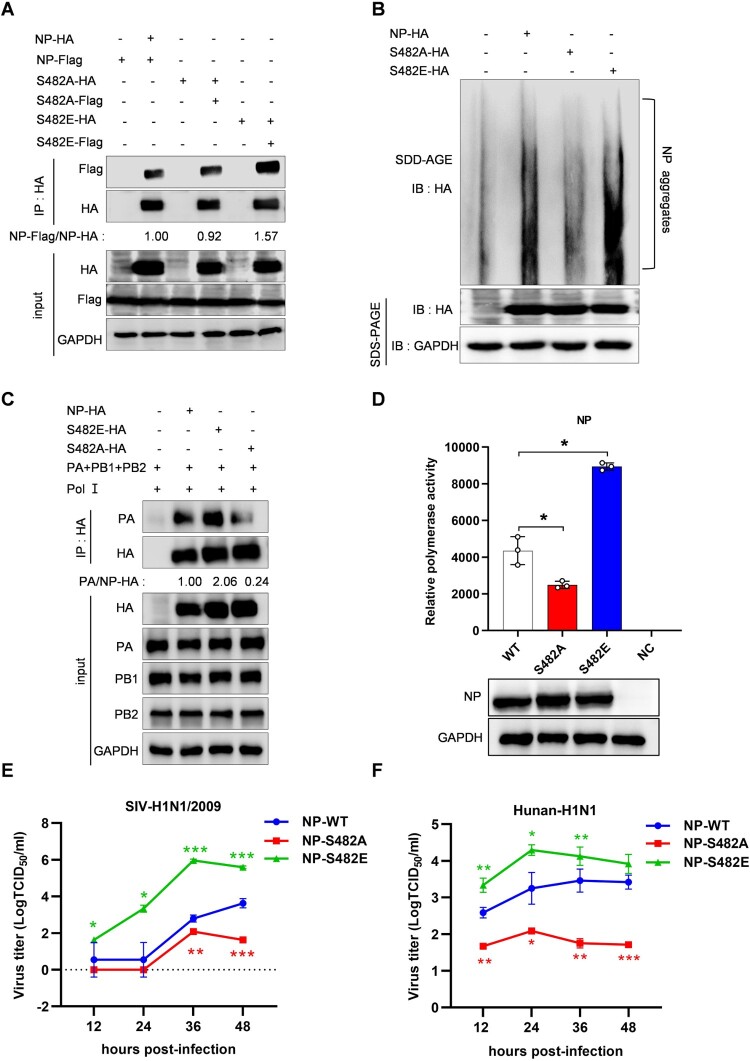
Existing studies suggests that phosphorylation sites (S165, S407 and S486) on opposite sides of the NP: NP interface played important roles in the regulation of NP oligomerization [18], and NP S482 is close to the key region. Therefore, we assessed the effect of S482 phosphorylation on NP oligomerization. We constructed different mutants at the NP S482 site, S482A and S482E, to simulate dephosphorylation and sustained phosphorylation states, respectively. The HA-tagged and Flag-tagged NP expression plasmids or its mutants were cotransfected, then Co-IP assays were performed using HA-tagged beads. As expected, the amount of NP S482A-Flag coprecipitated by NP S482A-HA was slightly less than that of NP-Flag coprecipitated by NP-HA, while the amount of NP S482E-Flag coprecipitated by NP S482E-HA was significantly more than that of NP-Flag coprecipitated by NP-HA (Figure 5A). To further determine whether S482 phosphorylation modulated NP aggregation, we employed the semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) analysis. The results showed that NP S482E could promote NP aggregation while NP S482A significantly attenuated NP aggregation (Figure 5B). Together, these results demonstrate that phosphorylation of NP at S482 stimulates NP oligomerization.
Figure 5.
Effects of NP S482 phosphorylation on SIV-H1N1/2009 replication. (A) The effect of NP S482 phosphorylation on NP oligomerization. HEK293T cells were co-transfected with HA-tagged and Flag-tagged expression plasmids of NP or its mutants (NP S482A and NP S482E), then cell lysates were collected at 24 h post-transfection. Co-IP assay was performed using an anti-HA antibody followed by western blotting. The band intensities were quantified by ImageJ, and the relative precipitated NP-Flag (NP-Flag/NP-HA) were shown below. (B) SDD-AGE analysis of NP oligomerization. HEK293T cells were transfected with indicated plasmids for 24 h, cell lysates were analyzed by SDD-AGE and SDS-PAGE assays. (C) The effect of NP S482 on vRNP assembly. HEK293T cells were co-transfected with the vRNP reconstitution plasmids (pCDNA-PA, pCDNA-PB1, pCDNA-PB2) and tagged plasmid (NP-HA or its mutants NP S482E-HA, NP S482A-HA) and p-Pol I plasmid that to provide vRNAs. Cells were then lysed at 24 h posttransfection. Co-IP was performed using an anti-HA antibody followed by western blotting. The band intensities were quantified by ImageJ, and the relative precipitated PA/NP-HA or PA/NP S482 A/E-HA ratios were shown below. GAPDH was used as a loading control in all of these experiments. (D) The effect of NP S482 phosphorylation on viral polymerase activity. HEK293T cells were cotransfected with the vRNP expression plasmids (pCDNA-PA, pCDNA-PB1, pCDNA-PB2), tagged plasmid (NP-HA or its mutants NP S482A-HA, NP S482E-HA), pPolI-firefly and Renilla luciferase expression plasmids. The luciferase activity was measured at 24 h after the transfection. The luciferase activities of vRNPs containing NP mutants were compared with that generated by the vRNP containing WT NP. (E and F) The effects of NP S482 phosphorylation on virus growth. The recombinant viruses including wild-type (WT) NP and NP mutants (NP S482A and NP S482E) from SIV-H1N1/2009 or Hunan-H1N1/2015 were generated, then infected PK-15 cells at a MOI of 0.01. Cell supernatants were collected at 12, 24, 36, and 48 h postinfection. Virus titres were determined by TCID50 on PK-15 cells. Statistical significance was analyzed by Student's t-test, * p < 0.05, ** p < 0.01, *** p < 0.001, ns represented no significance.
Similarly, as phosphorylation of NP at S482 can facilitate NP oligomerization, we proposed that the NP S482 phosphorylation could potentially enhance vRNP assembly. Then a Co-IP experiment was performed in HEK293T cells to verify the effect of NP S482 on vRNP assembly. As expected, the amount of PA precipitated by NP S482E was significantly increased compared with the amount PA precipitated by wild type NP (Figure 5C), indicating that phosphorylation of NP at S482 can promote vRNP assembly. Then, we verified the effect of NP S482 phosphorylation on viral polymerase activity, a minireplicon assay was carried out to determine the effect of NP or its mutants on the viral polymerase activity. The data showed that NP S482A led to a significant reduction of polymerase activity, while NP S482E resulted in remarkable rise of polymerase activity (Figure 5D). These results collectively indicate that phosphorylation of NP at S482 can promote the polymerase activity of SIV.
To investigate the impact of NP S482 phosphorylation on viral growth, the recombinant viruses including WT NP and NP mutants (NP S482A and NP S482E) from SIV-H1N1/2009 or Hunan-H1N1/2015 were generated using an 8-plasimid reverse genetic system. Then, PK-15 cells were infected with WT NP or mutant NP viruses at a MOI of 0.01 to determine the replicative properties of viruses. At 12, 24, 36 and 48 h postinfection, the cellular supernatant was harvested and subjected to TCID50 assays in PK-15 cells for virus titration. These data showed that the growth of mutant virus with NP S482A was significantly attenuated, whereas the growth of mutant virus with NP S482E was clearly accelerated at different time points (Figure 5E,F). To further confirm that the NP S482 is the phosphorylation site targeted by PLK3, we verified the proliferation ability of the 3 recombinant viruses on PK-15 cells after PLK3 knockout. The PLK3-KO cell line was infected with WT NP or mutant NP viruses (0.01 MOI) and the cell supernatant was collected at 12, 24, and 36 h post-infection for TCID50 analysis. The results showed that the NP S482E mutant virus exhibited a replication capacity similar to that of the WT NP virus at different time points. However, the NP S482A virus showed a significant increase in replication capacity (Figure S3A), indicating that NP S482 is an important phosphorylation site targeted by PLK3. In general, these data provides evidence that NP S482 phosphorylation may play an important role in influenza virus replication.
NP S482 phosphorylation on SIV-H1N1/2009 alters virus pathogenicity in mice
Given that NP S482 phosphorylation affects the viral replication in PK-15 cells, next we determined the pathogenicity of the mutated viruses in BALB/c mice. Results showed that mice infected with NP S482E virus led to a rapid loss of weight and 100% mortality, while those infected with the same dose of the NP WT virus only caused 50% mortality (Figure 6A,B). In addition, the body weight of mice infected with NP S482A virus was almost the same as that of PBS group, and did not induce lethal infection (Figure 6A,B).
Figure 6.
NP S482 phosphorylation on SIV-H1N1/2009 alters virus pathogenicity in mice. (A and B) Weight loss and mortality of mice infected with each indicated virus. Body weights of the NP-WT and mutant groups were compared and statistically analyzed. Error bars represent means ± SEM (n = 10). Statistical analysis was used by two-tailed analysis of variance with Bonferroni post test. (C) Virus titres in the lungs of infected mice (n = 3) at 3 and 5 days post-infection. Error bars represent means ± SD. Statistical analysis was performed by using one-tailed method (ND, Not detected; ***, p < 0.001). (D) Pathological lesions in the lungs of mice infected with the indicated virus at 3 and 5 days postinfection with haematoxylin and eosin (H&E) staining. Scale bars, 100 μm. (E) Immunofluorescent staining of lung sections of mice infected with the indicated PBS or virus at 3 and 5 dpi. The viral NP antigen was stained red and the nucleus was stained blue. Scale bars, 100 μm.
Virus titres in the lungs of mice infected with NP S482E were about 10-fold higher at 3 and 5 days postinfection (dpi) than detected in NP WT infected mice (Figure 6C). Furthermore, no virus titre was detected in the lungs of mice infected with NP S482A virus at 3 and 5 dpi. The lungs of mice infected with NP WT and NP S482E had moderate to severe bronchiolar necrosis, pulmonary oedema, and inflammatory cell infiltrates, while the lung of mice infected with NP S482A showed almost no lesions at 3 and 5 dpi (Figure 6D). In addition, the stronger NP antigen signals were detected in the lungs of mice infected with NP S482E virus when compared with those infected with NP WT virus at 3 and 5 dpi, while the NP antigen signals almost not detected in the lungs of mice infected with NP S482A (Figure 6E). These results suggest that mutations in NP phosphorylation sites are critical for the pathogenicity of influenza virus infection in mammals.
Discussion
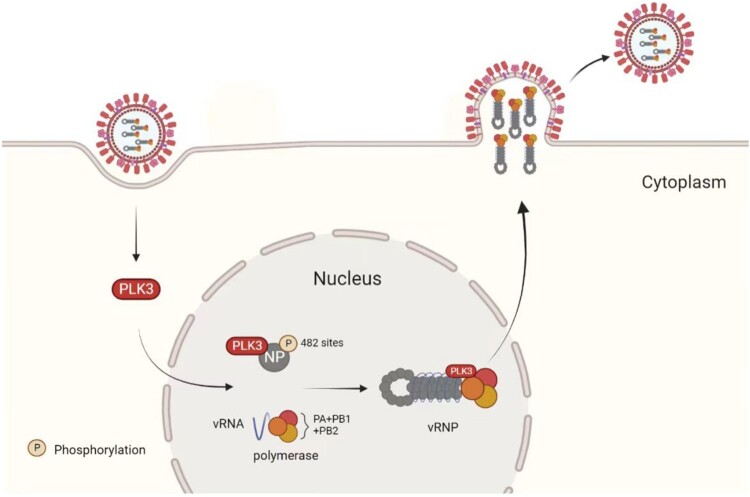
Host factors play a vital role in the life cycle of influenza viruses and many studies have screened and identified host factors involved in the replication and transcription of influenza virus genomes [19–21]. Karlas's Group and Konig's Group also showed that PLK3 is involved in the replication of influenza virus [6,7]. In this study, we also identified an influenza virus-related host factor PLK3 through gene expression profiling. Our study found that PLK3 could promote influenza A virus replication by regulating NP phosphorylation, and we identified a new functional phosphorylation site serine 482 (S482) on NP targeted by PLK3. The results indicated that phosphorylation of NP S482 could enhance viral polymerase activity and vRNP assembly, which plays a crucial role in virus replication (Figure 7). In addition, in this study, we used a PLK3 inhibitor GW843682X, which can inhibit the phosphorylation level of NP and the proliferation of influenza A virus. Thus, our data reveal that PLK3 serves as a positive regulator for influenza A virus to link phosphorylation and virus replication, which provide new ideas for the treatment of influenza virus and the development of antiviral drugs.
Figure 7.
Summary. PLK3 as a novel positive regulator of influenza A virus and can promote virus replication. Further mechanism revealed that PLK3 can interact with NP and promote NP phosphorylation, thereby promoting NP oligomerization and vRNP assembly. More importantly, we identified a PLK3-targeted NP-specific phosphorylation site S482, the phosphorylation of S482 is vital of importance for influenza virus replication both in vitro and in vivo.
Polo-like kinases (PLKs) belong to serine and threonine kinases which are important for cell cycle regulation and they are characterized by the presence of a common 28 amino acid sequence called a polo-box domain [22]. In mammalian cells, the PLKs are composed of five members: PLK1, PLK2 [5], PLK3 [6,7], PLK4 [6], and the last identified member PLK5 [23]. Among these members, PLK1 has been the most thoroughly investigated [24–29]. PLK1 played a vital role in the regulation of the cell cycle and the occurrence and development of tumours [25,27]. However, the other members of PLK family are less studied and their functional roles are still poorly understood. PLK2 localized to the centrosome and might participate in centrosome biology and S-phase checkpoints [30]. PLK4 was a vital regulator required for centriole duplication [31,32]. PLK5 executed a specific role in neurone differentiation and glioblastoma suppression [23]. Besides the multiple well-studied roles in cellular function, PLKs had been also involved in virus infection. For example, PLK1 is a proviral factor in the replication of HBV and HCV, and HBV X protein can regulate PLK1 expression [24,26,29]. Moreover, PLK1 can down-regulate the expression of parainfluenza virus 5 [28]. In addition, some other viruses such as chikungunya virus [33], mumps virus [34], and human Cytomegalovirus [35] are also related to the PLKs family. Of note, studies have shown that PLK1, PLK2, PLK3, and PLK4 became novel targets for anti-influenza drugs, and found that knockdown of PLK1, PLK3, and PLK4 can inhibit the replication of influenza virus [13]. Most studies on PLK3 focused on cell cycle regulation [36–39], but the interaction between PLK3 and influenza virus was still unknown. Our study discovered that PLK3 interacts with the influenza virus nucleoprotein NP and stimulates NP phosphorylation, which in turn promotes NP oligomerization and vRNP assembly. In summary, the host kinase PLK3 can promote influenza growth by modulating the phosphorylation of NP. Similarly, the study reported that the host factor CDC25B also had a similar function, affecting the growth of influenza virus by regulating the phosphorylation of NP [40,41], besides, S191 and S198 of the same family member CDC25C can be phosphorylated by PLK3 causing nuclear translocation [42]. Currently, there are not many reports on host factors that regulate viral replication by affecting NP phosphorylation, but host factors such as Moloney leukaemia virus 10 (MOV10) and Cellular nuclear transport factor 2 (NTF2), all targeted NP to regulate influenza virus replication [43,44]. Host factors such as PLSCR1, HMGB1, and BinCARD1 can regulate influenza virus replication by interacting with NP. PLSCR1 could interact with influenza A virus NP to impair its nuclear import and inhibit virus replication [45]. HMGB1 interacted with NP to enhance viral polymerase activity, thus promoting influenza virus replication [46]. BinCARD1 interacted with NP to promote NP binding to importin α7, and IAV leverages BinCARD1 as an important host factor that promotes viral replication [47]. In addition, recent studies have shown that PKCδ, a member of the human protein kinase C (PKC) family, can interact with the polymerase subunit PB2 after activation, and it also can regulate the oligomerization of NP and vRNP assembly through phosphorylation to further affect influenza virus growth [48]. Our study found PLK3 can promote NP phosphorylation thereby facilitating influenza growth. All these findings reinforce the positive role of PLK3 during influenza virus replication. However, the process by which PLK3 activates NP phosphorylation post-virus infection remains unclear, and needs further investigation.
The nuclear protein NP of influenza virus is a multifunctional protein that plays an important role in viral transcription and replication [49,50]. It has been shown that nucleoprotein NP can interact with influenza virus polymerases PB1 and PB2, thereby regulating virus growth [51,52]. NP is the main structural protein of influenza virus which determines vRNP structure [53]. In addition, NP is important for vRNP assembly which is an essential process in the life cycle of influenza virus. Research has shown that the host protein LYAR can promote the replication of influenza virus by facilitating the assembly of vRNP [54]. Similarly, our research showed that PLK3 could interact with influenza virus RNPs components including PB1, PB2 and NP but not interacted with PA. Furthermore, PLK3 also can promote vRNP assembly by elevating NP phosphorylation and oligomerization. However, the reasons why PLK3 interacts with other RNP components but not PA deserve further investigation, additionally, the method by which PLK3 regulates NP oligomerization by influencing NP phosphorylation is also unknown, and the underlying mechanism requires further exploration.
Phosphorylation of proteins, a significant form of post-translational protein modification, also plays a crucial role in influenza virus infection. Research has demonstrated that phosphorylation can influence NP oligomerization and govern the nuclear-cytoplasmic shuttling of NP [18]. Moreover, phosphorylation of viral proteins can affect the life cycle of the virus, and current research indicated that phosphorylation sites were almost identified in all viral proteins of influenza virus [55], it has been reported that there were many phosphorylation sites on NP and some of them had important effects on virus replication. Phosphorylation at NP S165, S407, and S486 affected the oligomerization of NP [15]. Phosphorylation of NP S9, Y10, Y296, and Y78 can regulate the nuclear shuttle of NP [56,57]. In this study, we determined a functional phosphorylation site S482 on NP targeted by PLK3. These results showed that the phosphorylation of S482 can affect the viral polymerase activity and vRNP assembly, ultimately regulated the replication of influenza virus both in vitro and in vivo. In addition to the NP S482 site, we also identified a PLK3-targeted NP S467 site, and found that PLK3 had little effect on the phosphorylation levels of S467A. Therefore, we selected site S482 for follow-up studies. Additionally, our findings also revealed that PLK3 impacts the threonine phosphorylation of NP, suggesting the potential existence of other undiscovered consensus sequences of PLK3. The threonine phosphorylation sites may also have significant roles in the replication process of influenza viruses. These aspects are all worth further investigation. Interestingly, we found that the impacts of NP phosphorylation sites on influenza virus were not completely consistent. Studies have reported that NP S9E, Y10E, Y296E, and T188E significantly reduced the activity of viral polymerase, while the effects of NP S9A, Y10F, Y296F, and T188A on viral polymerase activity were not obvious [56,58]. However, our results showed that NP S482E enhanced viral polymerase activity and vRNP assembly, whereas NP S482A exerted a negative effect on influenza virus. So we supposed that the effect of phosphorylation sites on the virus was not absolute, but the specific mechanism influencing viral growth remains unclear. Moreover, we have confirmed that PLK3 can interact with NP and enhance NP phosphorylation. Regardless, it remains uncertain whether the effect of PLK3 on NP phosphorylation is direct or indirect, making it a subject worthy of further research.
Interestingly, our study found that PLK3 plays a regulatory role in the replication of influenza viruses in different hosts. In PLK3-KO cell lines, the proliferation of both avian influenza (SH13/H9N2) and human influenza (Hunan/H1N1) viruses were inhibited. So, does PLK3 affect the phosphorylation of NP proteins from other sources of influenza viruses? This is worthy of our further study and exploration. Given that we found that NP of influenza virus is generally very conserved at 482, and we detected the effect of PLK3 on NP phosphorylation through the co-transfection of PLK3 and NP protein, independent of virus infection, so we hypothesized that PLK3 might also influence the phosphorylation of NP protein in other host influenza viruses. However, we have no relevant evidence so far, which will be the focus of our follow-up studies.
In summary, our studies found the host kinase PLK3 plays a crucial role in the life cycle of influenza A virus, which can promote replication of influenza A virus by regulating NP phosphorylation. In addition, we identified the serine phosphorylation site S482 on NP targeted by PLK3, and found that the phosphorylation of S482 regulated polymerase activity and assembly of vRNP, which in turn affected the replication of influenza virus both in vitro and in vivo. This work broadens our understanding for the host-IAV interaction and provides new ideas for the development of anti-viral drugs, PLK3 may also become the next target for the treatment of influenza virus.
Supplementary Material
Acknowledgements
We thank Professor Wendy S Barclay (Imperial College, London) for helpful discussions and comments. We thank Professor Wenjun Ma (University of Missouri, USA) and Xiao Xiao (Huazhong Agricultural University, China) for critically proofreading the manuscript. This work was supported by the National Key Research and Development Program (2021YFD1800204), the National Natural Science Foundation of China (32025036 and 32060795), Fundamental Research Funds for the Central Universities (2662023PY005), Hubei Hongshan Laboratory (2022hszd005), the earmarked fund for CARS-41 and the Natural Science Foundation of Hubei Province (2021CFA016).
Funding Statement
This work was supported by National Key Research and Development Program of China: [Grant Number 2021YFD1800204]; National Natural Science Foundation of China: [Grant Number 32025036]; National Natural Science Foundation of China: [Grant Number 32060795]; Fundamental Research Funds for the Central Universities: [Grant Number 2662023PY005]; Hubei Hongshan Laboratory: [Grant Number 2022hszd005]; the earmarked fund for CARS-41 and the Natural Science Foundation of Hubei Province: [Grant Number 2021CFA016].
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Ma W, Richt JA.. Swine influenza vaccines: current status and future perspectives. Anim Health Res Rev. 2010 Jun;11(1):81–96. doi: 10.1017/S146625231000006X [DOI] [PubMed] [Google Scholar]
- 2.Torremorell M, Allerson M, Corzo C, et al. . Transmission of influenza A virus in pigs. Transbound Emerg Dis. 2012 Mar;59(1):68–84. doi: 10.1111/j.1865-1682.2011.01300.x [DOI] [PubMed] [Google Scholar]
- 3.Hao L, Sakurai A, Watanabe T, et al. . Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008 Aug 14;454(7206):890–893. doi: 10.1038/nature07151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brass AL, Huang IC, Benita Y, et al. . The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009 Dec 24;139(7):1243–1254. doi: 10.1016/j.cell.2009.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shapira SD, Gat-Viks I, Shum BO, et al. . A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009 Dec 24;139(7):1255–1267. doi: 10.1016/j.cell.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konig R, Stertz S, Zhou Y, et al. . Human host factors required for influenza virus replication. Nature. 2010 Feb 11;463(7282):813–817. doi: 10.1038/nature08699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlas A, Machuy N, Shin Y, et al. . Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010 Feb 11;463(7282):818–822. doi: 10.1038/nature08760 [DOI] [PubMed] [Google Scholar]
- 8.Ferrari M, Scalvini A, Losio MN, et al. . Establishment and characterization of two new pig cell lines for use in virological diagnostic laboratories. J Virol Methods. 2003 Feb;107(2):205–212. doi: 10.1016/S0166-0934(02)00236-7 [DOI] [PubMed] [Google Scholar]
- 9.Zhou H, Wang C, Yang Y, et al. . Pandemic (H1N1) 2009 virus in swine herds, People's Republic of China. Emerg Infect Dis. 2011 Sep;17(9):1757–1759. doi: 10.3201/eid1709.101916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmittgen TD, Livak KJ.. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann E, Neumann G, Kawaoka Y, et al. . A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA. 2000 May 23;97(11):6108–6113. doi: 10.1073/pnas.100133697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe T, Kawakami E, Shoemaker JE, et al. . Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe. 2014 Dec 10;16(6):795–805. doi: 10.1016/j.chom.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pohl MO, von Recum-Knepper J, Rodriguez-Frandsen A, et al. . Identification of polo-like kinases as potential novel drug targets for influenza A virus. Sci Rep. 2017 Aug 17;7(1):8629. doi: 10.1038/s41598-017-08942-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lansing TJ, McConnell RT, Duckett DR, et al. . In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol Cancer Ther. 2007 Feb;6(2):450–459. doi: 10.1158/1535-7163.MCT-06-0543 [DOI] [PubMed] [Google Scholar]
- 15.Turrell L, Hutchinson EC, Vreede FT, et al. . Regulation of influenza A virus nucleoprotein oligomerization by phosphorylation. J Virol. 2015 Jan 15;89(2):1452–1455. doi: 10.1128/JVI.02332-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turrell L, Lyall JW, Tiley LS, et al. . The role and assembly mechanism of nucleoprotein in influenza A virus ribonucleoprotein complexes. Nat Commun. 2013;4:1591. doi: 10.1038/ncomms2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salvi M, Trashi E, Cozza G, et al. . Investigation on PLK2 and PLK3 substrate recognition. Biochim Biophys Acta. 2012 Dec;1824(12):1366–1373. doi: 10.1016/j.bbapap.2012.07.003 [DOI] [PubMed] [Google Scholar]
- 18.Mondal A, Potts GK, Dawson AR, et al. . Phosphorylation at the homotypic interface regulates nucleoprotein oligomerization and assembly of the influenza virus replication machinery. PLoS Pathog. 2015 Apr;11(4):e1004826. doi: 10.1371/journal.ppat.1004826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bortz E, Westera L, Maamary J, et al. . Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. mBio. 2011;2(4):e00151–11. doi: 10.1128/mBio.00151-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer D, Molawi K, Martinez-Sobrido L, et al. . Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J Proteome Res. 2007 Feb;6(2):672–682. doi: 10.1021/pr060432u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagata K, Kawaguchi A, Naito T.. Host factors for replication and transcription of the influenza virus genome. Rev Med Virol. 2008 Jul-Aug;18(4):247–260. doi: 10.1002/rmv.575 [DOI] [PubMed] [Google Scholar]
- 22.Song S, Lee KS.. A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis. J Cell Biol. 2001 Feb 5;152(3):451–469. doi: 10.1083/jcb.152.3.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Carcer G, Escobar B, Higuero AM, et al. . Plk5, a polo box domain-only protein with specific roles in neuron differentiation and glioblastoma suppression. Mol Cell Biol. 2011 Mar;31(6):1225–1239. doi: 10.1128/MCB.00607-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YC, Su WC, Huang JY, et al. . Polo-like kinase 1 is involved in hepatitis C virus replication by hyperphosphorylating NS5A. J Virol. 2010 Aug;84(16):7983–7993. doi: 10.1128/JVI.00068-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colicino EG, Hehnly H.. Regulating a key mitotic regulator, polo-like kinase 1 (PLK1). Cytoskeleton. 2018 Nov;75(11):481–494. doi: 10.1002/cm.21504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diab A, Foca A, Fusil F, et al. . Polo-like-kinase 1 is a proviral host factor for hepatitis B virus replication. Hepatology. 2017 Dec;66(6):1750–1765. doi: 10.1002/hep.29236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi F, Chen Q, Chen H, et al. . WAC promotes polo-like kinase 1 activation for timely mitotic entry. Cell Rep. 2018 Jul 17;24(3):546–556. doi: 10.1016/j.celrep.2018.06.087 [DOI] [PubMed] [Google Scholar]
- 28.Sun D, Luthra P, Li Z, et al. . PLK1 down-regulates parainfluenza virus 5 gene expression. PLoS Pathog. 2009 Jul;5(7):e1000525. doi: 10.1371/journal.ppat.1000525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu J, Zhang T, Cheng J, et al. . Hepatitis B virus X protein-regulated expression of Plk1. Chinese Journal of Hepatology. 2016 Jan;24(1):46–50. [DOI] [PubMed] [Google Scholar]
- 30.Matthew EM, Yen TJ, Dicker DT, et al. . Replication stress, defective S-phase checkpoint and increased death in Plk2-deficient human cancer cells. Cell Cycle. 2007 Oct 15;6(20):2571–2578. doi: 10.4161/cc.6.20.5079 [DOI] [PubMed] [Google Scholar]
- 31.Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, et al. . SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005 Dec 20;15(24):2199–2207. doi: 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- 32.Habedanck R, Stierhof YD, Wilkinson CJ, et al. . The polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005 Nov;7(11):1140–1146. doi: 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- 33.Treffers EE, Tas A, Scholte FE, et al. . Temporal SILAC-based quantitative proteomics identifies host factors involved in chikungunya virus replication. Proteomics. 2015 Jul;15(13):2267–2280. doi: 10.1002/pmic.201400581 [DOI] [PubMed] [Google Scholar]
- 34.Pickar A, Zengel J, Xu P, et al. . Mumps virus nucleoprotein enhances phosphorylation of the phosphoprotein by polo-like kinase 1. J Virol. 2016 Feb 1;90(3):1588–1598. doi: 10.1128/JVI.02160-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallina A, Simoncini L, Garbelli S, et al. . Polo-like kinase 1 as a target for human cytomegalovirus pp65 lower matrix protein. J Virol. 1999 Feb;73(2):1468–1478. doi: 10.1128/JVI.73.2.1468-1478.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helmke C, Becker S, Strebhardt K.. The role of Plk3 in oncogenesis. Oncogene. 2016 Jan 14;35(2):135–147. doi: 10.1038/onc.2015.105 [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Beauchemin M, Bertrand R.. Bcl-xL phosphorylation at Ser49 by polo kinase 3 during cell cycle progression and checkpoints. Cell Signal. 2011 Dec;23(12):2030–2038. doi: 10.1016/j.cellsig.2011.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Gonzalez S, Dai W, et al. . Effect of hypoxia-regulated polo-like kinase 3 (Plk3) on human limbal stem cell differentiation. J Biol Chem. 2016 Aug 5;291(32):16519–16529. doi: 10.1074/jbc.M116.725747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu D, Yao Y, Lu L, et al. . Plk3 functions as an essential component of the hypoxia regulatory pathway by direct phosphorylation of HIF-1alpha. J Biol Chem. 2010 Dec 10;285(50):38944–38950. doi: 10.1074/jbc.M110.160325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui L, Mahesutihan M, Zheng W, et al. . CDC25B promotes influenza A virus replication by regulating the phosphorylation of nucleoprotein. Virology. 2018 Dec;525:40–47. doi: 10.1016/j.virol.2018.09.005 [DOI] [PubMed] [Google Scholar]
- 41.Perwitasari O, Torrecilhas AC, Yan X, et al. . Targeting cell division cycle 25 homolog B to regulate influenza virus replication. J Virol. 2013 Dec;87(24):13775–13784. doi: 10.1128/JVI.01509-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bahassi el M, Hennigan RF, Myer DL, et al. . Cdc25C phosphorylation on serine 191 by Plk3 promotes its nuclear translocation. Oncogene. 2004 Apr 8;23(15):2658–2663. doi: 10.1038/sj.onc.1207425 [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Huang F, Tan L, et al. . Host protein moloney leukemia virus 10 (MOV10) acts as a restriction factor of influenza A virus by inhibiting the nuclear import of the viral nucleoprotein. J Virol. 2016 Apr;90(8):3966–3980. doi: 10.1128/JVI.03137-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chutiwitoonchai N, Aida Y.. NXT1, a novel influenza A NP binding protein, promotes the nuclear export of NP via a CRM1-dependent pathway. Viruses. 2016 Jul 28;8(8):e209. doi: 10.3390/v8080209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo W, Zhang J, Liang L, et al. . Phospholipid scramblase 1 interacts with influenza A virus NP, impairing its nuclear import and thereby suppressing virus replication. PLoS Pathog. 2018 Jan;14(1):e1006851. doi: 10.1371/journal.ppat.1006851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moisy D, Avilov SV, Jacob Y, et al. . HMGB1 protein binds to influenza virus nucleoprotein and promotes viral replication. J Virol. 2012 Sep;86(17):9122–9133. doi: 10.1128/JVI.00789-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Jiang L, Wang G, et al. . Influenza A virus use of BinCARD1 to facilitate the binding of viral NP to importin alpha7 is counteracted by TBK1-p62 axis-mediated autophagy. Cell Mol Immunol. 2022 Oct;19(10):1168–1184. doi: 10.1038/s41423-022-00906-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mondal A, Dawson AR, Potts GK, et al. . Influenza virus recruits host protein kinase C to control assembly and activity of its replication machinery. eLife. 2017 Jul 31;6:e26910. doi: 10.7554/eLife.26910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Portela A, Digard P.. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol. 2002 Apr;83(Pt 4):723–734. doi: 10.1099/0022-1317-83-4-723 [DOI] [PubMed] [Google Scholar]
- 50.Ng AK, Wang JH, Shaw PC.. Structure and sequence analysis of influenza A virus nucleoprotein. science in China series C. Life Sci. 2009 May;52(5):439–449. [DOI] [PubMed] [Google Scholar]
- 51.Gabriel G, Herwig A, Klenk HD.. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 2008 Feb 8;4(2):e11. doi: 10.1371/journal.ppat.0040011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biswas SK, Boutz PL, Nayak DP.. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J Virol. 1998 Jul;72(7):5493–5501. doi: 10.1128/JVI.72.7.5493-5501.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ortega J, Martin-Benito J, Zurcher T, et al. . Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J Virol. 2000 Jan;74(1):156–163. doi: 10.1128/JVI.74.1.156-163.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang C, Liu X, Gao Q, et al. . The nucleolar protein LYAR facilitates ribonucleoprotein assembly of influenza A virus. J Virol. 2018 Dec 1;92(23):e01042–18. doi: 10.1128/JVI.01042-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hutchinson EC, Denham EM, Thomas B, et al. . Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Pathog. 2012;8(11):e1002993. doi: 10.1371/journal.ppat.1002993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng W, Li J, Wang S, et al. . Phosphorylation controls the nuclear-cytoplasmic shuttling of influenza A virus nucleoprotein. J Virol. 2015 Jun;89(11):5822–5834. doi: 10.1128/JVI.00015-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui L, Zheng W, Li M, et al. . Phosphorylation status of tyrosine 78 residue regulates the nuclear export and ubiquitination of influenza A virus nucleoprotein. Front Microbiol. 2019;10:1816. doi: 10.3389/fmicb.2019.01816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Sun L, Zheng W, et al. . Phosphorylation and dephosphorylation of threonine 188 in nucleoprotein is crucial for the replication of influenza A virus. Virology. 2018 Jul;520:30–38. doi: 10.1016/j.virol.2018.05.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.