Abstract
Extracellular vesicles (EVs) are membrane-bound nanoparticles with different types of cargo released by cells and postulated to mediate functions such as intercellular communications. Recent studies have shown that long non-coding RNAs (lncRNAs) or their fragments are present as cargo within EVs. LncRNAs are a heterogeneous group of RNA species with a length exceeding 200 nucleotides with diverse functions in cells based on their localization. While lncRNAs are known for their important functions in cellular regulation, their presence and role in EVs have only recently been explored. While certain studies have observed EV-lncRNAs to be tissue-and disease-specific, it remains to be determined whether or not this is a global observation. Nonetheless, these molecules have demonstrated promising potential to serve as new diagnostic and prognostic biomarkers. In this review, we critically evaluate the role of EV-derived lncRNAs in several prevalent diseases, including cancer, cardiovascular diseases, and neurodegenerative diseases, with a specific focus on their role as biomarkers.
Keywords: Extracellular vesicles, Long-noncoding RNA, Cardiovascular disease, Cancer, Neurological disease
1. Introduction
Fundamentally, all cells present in both eukaryotic and prokaryotic organisms communicate with each other through a plethora of intercellular communication strategies. Most cells harness this potential for rapid, effective, and precise biological cargo delivery by releasing nanosized, lipid bilayer membrane-enclosed packages called EVs into their surrounding environment. EVs can be retrieved from an array of biological fluids such as blood plasma, serum, urine, saliva, semen, breast milk, bile, ascitic, cerebrospinal, bronchoalveolar, and synovial fluid.1–4 EVs contain an intricate array of biomolecules not limited to but including protein, lipids, DNA, coding, and non-coding RNA (ncRNA). Owing to the phospholipid bilayer structure of EV membrane, fragile biomolecules, such as several RNA species that are otherwise prone to degradation, are sheltered from degradative enzymes, pH, and osmolar conditions that prevail in biofluids. In fact, EV contents can be transferred not only in a paracrine manner to the surrounding cells (horizontal transfer) but also to distant recipient tissues.5,6 Since EVs contain various populations, including certain functional RNA molecules, their role in genetically-encoded information exchange between cells has sparked in-depth scientific research in this direction and their impact at a cellular phenotypic level ought to be further explored.7–9 More importantly, the uptake of EV contents, specifically the transcriptomic cargo, could have profound implications for the recipient cells, as reflected by specific phenotypic alterations in their function or gene expression levels.10–13 Moreover, the cargo of vesicles is often subject to changes that are conferred upon different disease states and homeostatic perturbations and offer novel insights into the disease pathology. Although the EV cargo is composed of various molecules, non-coding RNAs have recently emerged as dynamic biomarkers.
There is an entire repertoire of RNAs, called ncRNAs, defying the central dogma of biology, that are transcribed from the genome but do rarely encode for proteins. Though their discovery began as early as 1955 through the discovery of ribosomal RNA (rRNA) as a part of the Ribonucleoprotein (RNP) complex, the discovery of newer ncRNAs continues until today.14 The advent of technological advancements such as next-generation sequencing (NGS) paved the way for identifying an increasing number of these non-protein-coding sequences that account for about 98% of the human genome.15,16 Thus, a considerable part of our transcriptome is composed only of ncRNAs. NcRNAs are divided into (a) infrastructural ncRNAs such as transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs) and (b) regulatory ncRNAs including micro RNAs, circular RNAs (circRNA), enhancer RNAs, piwi RNAs, lncRNAs, Promoter associated RNAs, pseudogenes, the majority of which are comprised of lncRNAs.17–19 LncRNAs are a heterogeneous group of ncRNAs with more than 200 nucleotides, transcribed by RNA polymerase II (Pol II), capped/methyl-guanylated at the 5′ ends (m7G), many with polyadenylated at the 3′ ends and spliced like mRNA, but with longer and fewer exons.20,21 Next generation sequencing (NGS) has helped strand-specific, and allele-specific identification of lncRNAs, and newer such methodologies are consistently developed for more precise identification of lncRNAs.22,23 LncRNAs, though initially mistaken as pervasive transcripts, are crucial elements in cell regulation. Due to the lack of a better classification system, for ease of understanding, lncRNAs have been artificially categorized on the basis of origin from genomic location, mode of action, and function. Briefly, based on the position with respect to the genome (Fig. 1), there are (1) sense lncRNAs that overlap with the nearest protein-coding genes and are transcribed from the same strand; (2) antisense lncRNAs that are transcribed from the opposite strand of proximate protein-coding genes; (3) intronic lncRNAs, transcribed from the introns of protein-coding genes on both sense and anti-sense strands; (4) bidirectional lncRNAs that are transcribed in the opposite direction of the nearest protein-coding gene and lie generally within 1000bases (5) intergenic lncRNAs transcribed from regions between two protein-coding genes and account for most of the lncRNAs. LncRNAs have two modes of action and can regulate gene transcription via cis-acting (same region) or trans-acting (distant region) mechanisms based on where they are transcribed and where they act. LncRNAs are versatile molecules that have numerous functions ranging from reproduction, development, maintenance of cellular homeostasis with metabolic re-programming and regulation, cellular differentiation, migration, waste management and in specific processes such as receptor–ligand signaling, survival, apoptosis, angiogenesis, wound healing and balancing immune response as represented in Fig. 1.24–29 They are masters of genome regulation vital to both transcription and translation while contributing significantly towards epigenetic modulations. Given their role in genomic regulation, lncRNAs are abundantly found in the nucleus. However, they are also found in the cytoplasm, and their function usually depends upon their localization.30 For instance, in the nucleus, they act as decoys, scaffolds, guides, repressors, enhancers, enzyme modulators, Barr bodies, etc.,31–33 and in the cytoplasm, they play the role of competing endogenous RNAs, small RNA precursors, and RNAs bound to proteins that regulate translation.32,34,35 More recently, lncRNAs and their fragments have been identified in EVs with important roles in intercellular communication as discussed later.
Fig. 1.
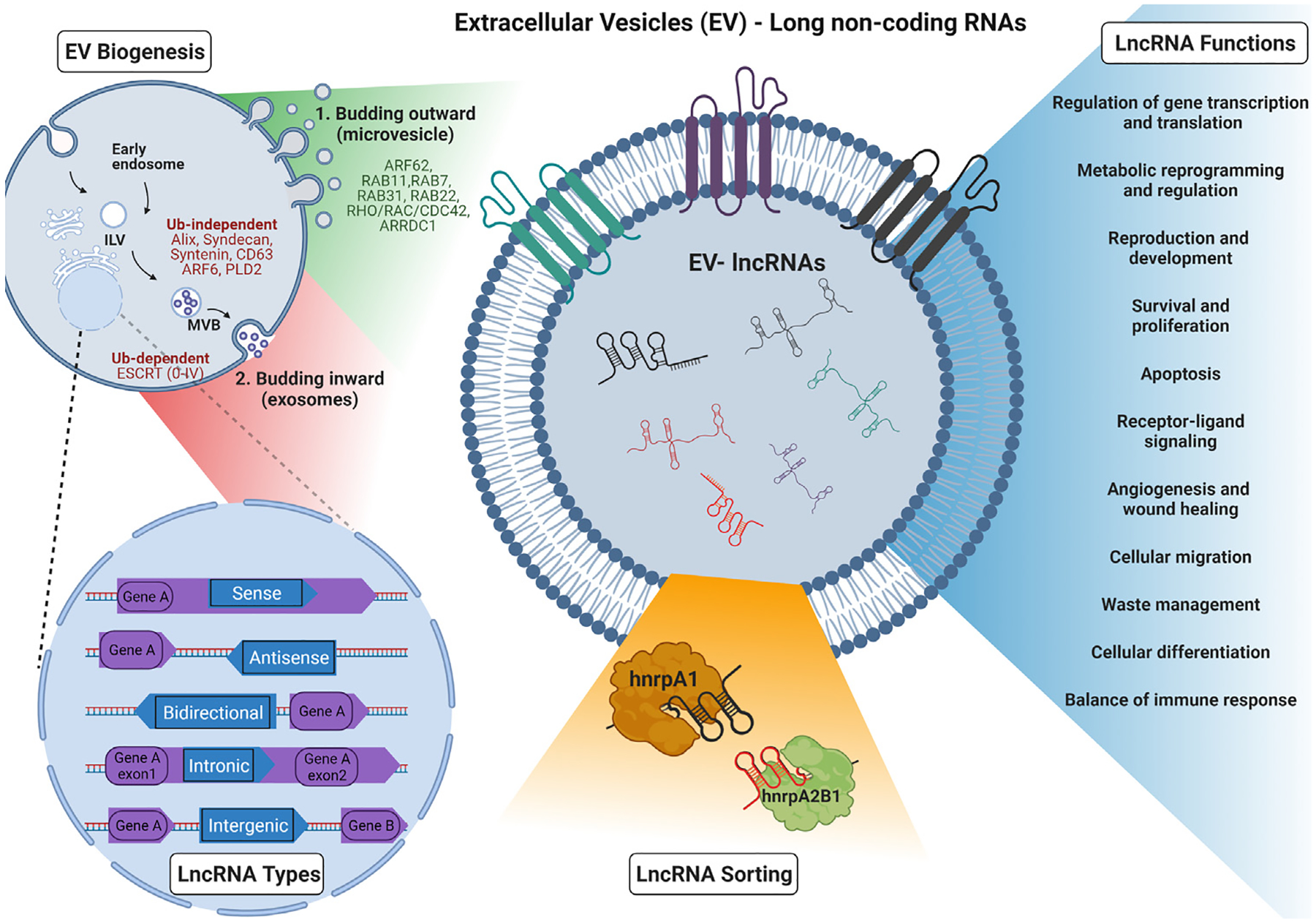
Extracellular vesicles-derived long non-coding RNAs — EV biogenesis, lncRNA types, sorting and function.
In fact, lncRNA gene expression is usually cell-type specific, tissue-specific, species-specific and in many cases, disease-specific.36–39 Accumulating evidence has demonstrated that these molecules have a central role in the pathogenesis of many diseases; the presence of tissue- or disease-related lncRNAs or their fragments in EVs serve as an intriguing type of biomarker for the early detection of disease as a ‘liquid biopsy’. However, the biological role of circulating lncRNAs and the pathways regulating their levels still remain elusive. Nonetheless, many studies have reported that lncRNAs packaged in EVs, play crucial roles in various systemic human diseases, such as cancer,40,41 cardiovascular,42,43 infectious,44,45 and neurodegenerative diseases.44,45 Notably, EVs exhibit tissue specificity through their unique transcriptomic profile and can be traced back to the tissue-of-origin from a mixture of tissues/biofluids using computational algorithms on their transcriptional data.46 Moreover, another mode of identifying the tissue-specificity of EVs is through proteomic methods such as mass spectrometry.47,48 Although there is much to be gleaned in this field, it is interesting and relevant to understand the process of EVs biogenesis from cells and why certain transcripts are selectively packaged into EVs, directing them as coded messages for specified cellular functions. Many recent studies converge on the idea that small RNAs, such as micro RNAs (miRNAs), and tRNA-derived small RNAs49 are selectively packaged in EVs.50 However, the same cannot be said for long RNAs in EVs, which may be more of a stochastic process, hence can provide a useful ‘biopsy’ that can faithfully represent cellular transcriptome.51
2. Sorting and packaging of lncRNAs into EVs
International Society for Extracellular Vesicles (ISEV) defines EVs as particles naturally released from the cell that are delimited by a lipid bilayer and cannot replicate, i.e., lack a functional nucleus.52 EVs are sized from 40 nm to more than 1000 nm and can be variously categorized into different subtypes according to their biogenetic origin, size, physical characteristics, surface, or cargo marker expression and/or cell of origin. Current classification systems categorize EVs based upon their biogenesis, release pathways, content, and vesicular size. To name a few, based on biogenetic origin, EVs can be subcategorized into exosomes (vesicles derived from endosomes) and ectosomes (vesicles deriving from plasma membrane) whereas considering biophysical characteristics, three main types can be inferred: exosomes, microvesicles, and apoptotic bodies.53–55 It is certainly noteworthy that newer non-vesicular nanoparticles that may carry specific cargo are being discovered in certain contexts such as exomeres, and supermeres.56–58 These are recently described nanoparticles typically less than 50 nm in size and characterized by absence of lipid membrane and unique lipid and protein composition. Exomeres show enrichment for heat shock protein 90 alpha family class B member 1(HSP90AB1) while supermeres are enriched in several proteins that are associated with diseases such as transforming growth factor beta-1 (TGFβ1-associated with colon cancer), amyloid precursor protein (APP-associated with Alzheimer’s disease), α-enolase and glypican-1 (associated with cancer), angiotensin-converting enzyme and angiotensin-converting enzyme 2 (ACE and ACE2 associated with cardiovascular disease and COVID-19).59,60
All currently described EV biogenesis pathways can be grouped into two main biosynthetic routes, both of which start with a membrane budding that encloses selectively sorted cargo in the lipid vesicle (Fig. 1). The first biogenesis pathway involves EVs that bud outward off the plasma membrane (e.g. ectosomes, sometimes referred to as microvesicles in literature),61 under the tight control of several proteins, such as ARF62, RAB11, RAB7, RAB31, RAB22, Rho/Rac/cdc42 and ARRDC1.62,63 The second biogenesis pathway starts with inward budding of endosomal membrane to form intralumenal vesicles (ILVs) within early endosomes that progressively mature into multivesicular bodies (MVBs) that, in turn, will fuse with the plasma membrane and be released (e.g. exosomes).64 For biomolecules to be sorted and transported into the ILVs, positive sorting signals are required. A positive sorting signal is a molecule which is added usually post-translationally and marks a specific cargo for a specific process, such as influx into an MVB in this instance. One such signal is the presence of a Ubiquitin (Ub) protein, and its addition is known as Ubiquitination.65 Recent research has shed light onto the sorting mechanisms that underlie the selective cargo incorporation into MVBs which can be further categorized as Ub-dependent and -independent. The Endosomal Sorting Complexes Required for Transport (ESCRTs) machinery is essential for Ub-dependent MVB biogenesis.66 The ESCRT system is comprised of a set of 4 interacting protein clusters, along with accessory proteins, that recognize ubiquitinated membrane proteins and promote their sorting into MVBs. ESCRT-0 (tumor susceptibility 101, TSG101) and ESCRT-I (Signal transducing adapter molecule 1) select cargo (TSG101 recognizes the Ub-tagged molecules)67 and trigger ESCRT-II/III recruitment and ILV formation, to complete vesicle formation after which a deubiquitinating enzyme removes the ubiquitin tag from the cargo proteins prior to final fission of the vesicle.68,69 Alix, Syndecan and Syntenin participate and interact within the ESCRT pathways prior to endosomal EVs release in Ub-independent MVB biogenesis. Syntenin binds syndecan to ALIX and aids in vesicle formation mediated by ESCRT-III.70 Selective cargo sorting of CD63 into vesicles and ALIX-Syntenin ILV formation pathway is regulated by the small GTPase ARF6 (ADP ribosylation factor 6) and the effector protein PLD2 (phospholipase D2).71 Another classification system which is based upon ESCRT involvement describes EVs as derived via an ESCRT-dependent or ESCRT-independent mechanism, however the pathways might be overlapping in some cases.72 Nonetheless, although in theory each EV sub-type stems from a distinct cellular compartment and exhibits distinct characteristics, recent evidence suggests there is considerable marker- and cargo-overlap between the different categories of EVs.73 Since defining a specific biogenetic pathway for an EV remains remarkably difficult; in this review, we use size to classify EVs and focus on small EVs (sEVs), that are <200 nm, while other EV categorizations are mentioned or discussed when relevant.
EVs carry an extensive repertoire of nucleic acids ranging from DNAs, mRNAs, and different types of ncRNAs such as rRNAs, tRNAs as well as their fragments, miRNAs, Y RNAs, snoRNA, snRNAs, piwi RNAs, circ RNAs, lncRNAs.74 It should be noted that there are other stable mechanisms of RNA transport in the plasma apart from EVs when bound to RNA binding proteins (RBPs) such as lipoproteins, and argonaute proteins.74 While most small RNAs are present in their full-length form in the EVs, long RNAs such as mRNAs and lncRNAs have been reported to be fragmented (although circ RNAs could remain intact in EVs).75,76 mRNA fragments have been reported to usually be 200 bases in length, with some extending even as long as 4 Kb.77 There have also been reports of lncRNAs loaded in EVs to have a mean size of 700 nucleotides.78 Considering the tissue-specific EV transcriptomics as mentioned earlier,79 one might infer that the loading of the lncRNAs or their fragments has a specific mechanism. Several studies probing into the mechanism of loading of miRNAs into EVs concluded that the overexpression of miRNAs in cells also correspondingly increases their levels in the EVs.80 While it is natural to expect lncRNAs that are abundant in cells to also be abundant in EVs, as observed in a previous study where there was a high correlation between EV-RNAs and cellular RNAs upon stress,51 there have been reports of the opposite being observed.81 Here, the authors report a selective abundance of certain lncRNAs in EVs even though they were less expressed in the parent cell line suggesting that these lncRNAs were distinctively loaded onto the EVs.81 Although this avenue is still being explored, there are a few studies that have provided an insight into this process.82 A heterogeneous ribonucleoprotein A2B1 (hnRNPA2B1), also as m6a reader, has been reported to be involved in the sorting of several lncRNAs, mostly in cancers, based on particular motifs such as GGAG-containing.83–85 In fact, an EV-lncRNA called lymph node metastasis-associated transcript 2 (LNMAT2) that has been reported to be involved in bladder cancer for promoting lymphangiogenesis, was selectively loaded onto EVs after binding with hnRNPA2B1.86 This RNA-RBP complex was also involved in the functionality of LNMAT2 when transported into recipient cells by locating to nucleus, further mediating H3 lysine 4 trimethylation to activate the PROX1 gene, thereby contributing towards metastatic phenotype. Similarly, in a study by Chen et al, the lncRNA EV-mediated LN-associated transcript 1 (ELNAT1) has been reported to be selectively loaded onto the EVs of bladder cancer cells through the ESCRT pathway when the RBP heterogeneous ribonucleoprotein A1 (hnRNPA1) residue underwent small ubiquitin-like modifier (SUMO) binding or SUMOylation at 113 lysine residue.87 It is pertinent to mention that the hnRNP family of proteins are important RBPs, primarily present in the nucleus, are involved in various cellular processes ranging from pre-mRNA or RNA processing to RNA splicing, RNA packaging, and RNA modifications. One or more members of this family are likely to be involved in the sorting of lncRNAs in EVs, and this warrants further investigation.88,89 Another study found that the RBPs YB-1 and NSUN2 could bind specifically to selected motifs, namely, ACCAGCCU, CAGUGAGC and UAAUCCCA in mRNAs and lncRNAs and selectively direct them to EVs.90 However, the selective lncRNA loading onto EVs warrants extensive research to precisely understand how and why certain lncRNAs are found in specific disease states and whether the RBP loading system holds true for most EV-lncRNA transport should be further explored.
Having summarized the biogenesis and sorting of EV-lncRNA, we briefly describe the most popularly accepted methods to study EVs and EV-derived lncRNAs. EVs are usually isolated using ultracentrifugation, differential ultracentrifugation, size exclusion chromatography, and commercially available EV-isolation kits. EVs, after isolation, are first examined for their sizes using particle analyzers such as nano particle tracking analysis (NTA), Spectradyne; the EVs could be visualized using Transmission Electron microscopy or quantitative single molecule localization microscopy (qSMLM),91 and further characterized qualitatively using western blot assay for the presence of tetraspanins such CD9, CD63, CD81 and cargo proteins such as Alix, syntenin. LncRNAs present in EVs are confirmed usually using RT-PCR or digital PCR. Interestingly, detection of biofluid EV-derived lncRNA has been well optimized by long RNA-sequencing techniques.92 Their presence can also be verified using Northern blot, Fluorescence in Situ Hybridization, Molecular beacons, and microarray techniques.
The omnipresence of EVs in human tissue and biological fluids, coupled with unique transcriptional cargo provided by lncRNAs along with their combinatorial distinct tissue-specificity render EV-derived lncRNAs as ideal biomarkers for human theranostics. Here, we explore the role of these EV-lncRNAs in the context of clinical biomarkers in some of the most lethal diseases wherein early indicators have a critical life-saving function.
3. Cancer
Cancer is an extremely heterogeneous disease with more than 200 types in the human body,93,94 with increasing prevalence and mortality.95 This accentuates the need for important prognostic and diagnostic biomarkers that aid in the better clinical management of this disease. In fact, nearly 90% of cancer mortality is attributed to the metastatic disease as opposed to the primary tumor itself,96,97 largely owing to tumor microenvironment-driven intercellular communication in its progression.98 Interestingly, cancer cells may use EVs as carriers of oncogenic protein for their spread and metastasis, demonstrating the significance of EVs in the tumor milieu.99,100 Specifically, PD-L1 in tumor-derived EVs contributes to immune evasion, which is a critical hallmark of cancer.101 Thus, inhibitors of EV release or fusion are considered a potential therapy for many cancers,102,103 although the specific role of any EV-lncRNAs in this process is undetermined. Further studies to investigate if inhibition of EV-mediated lncRNA transfer to recipient cells from malignant cells can impact cancer progression are needed. However, this is complicated by certain unique features of cancer EVs. Firstly, EVs secreted by tumor cells are much greater in number compared to normal cells.104 Secondly, tumor-derived EVs are comprised of highly diverse populations (e.g., “oncosomes” that are quite specific for cancers.57) Here, we will discuss EV-lncRNAs based on their potential to serve either as diagnostic or prognostic biomarkers in the most lethal cancer types (specifically in lung cancer, colorectal cancer, and breast cancer), prioritized by their functional role in the cellular context, such as tumor progression, immune evasion, and chemoresistance, specifically. However, it is important to mention that the functional roles have mostly been assigned due to the cellular role of these lncRNAs, and there have been very few studies that have characterized the EV-derived lncRNA role.
Lung Cancer is one of the cancers with the highest mortality in men and women globally, with non-small cell lung cancer (NSCLC) accounting for most of it.105 NSCLC is usually diagnosed at later stages with an extremely poor prognosis,106 necessitating the identification of good early diagnostic markers with proper prognostic markers and therapeutic targets. Colorectal cancer (CRC) is the second most common cause of cancer-related mortality, after lung cancer, causing almost 935,000 deaths.107 Currently, the most commonly used biomarker is carcinoembryonic antigen (CEA) but with sub-optimal sensitivity and specificity. Therefore, given the very poor 5-year survival rate of CRC patients, better diagnostic markers that can detect disease at an early stage are still a necessity.107–109 Breast cancer (BC) is the most common cancer among women globally, accounting for 12% of all new cancer cases worldwide.109–111 While Cancer antigen 153 (CA153) and CEA are the commonly used biomarkers, there remains a need for better BC diagnostic and prognostic markers for BC. In each of these cancers, a few promising EV-lncRNAs biomarkers have been identified recently with important roles. However, we will try and limit the scope of discussion to those EV-lncRNAs (Fig. 2, Table 1) that have been validated in patient cohorts.
Fig. 2.
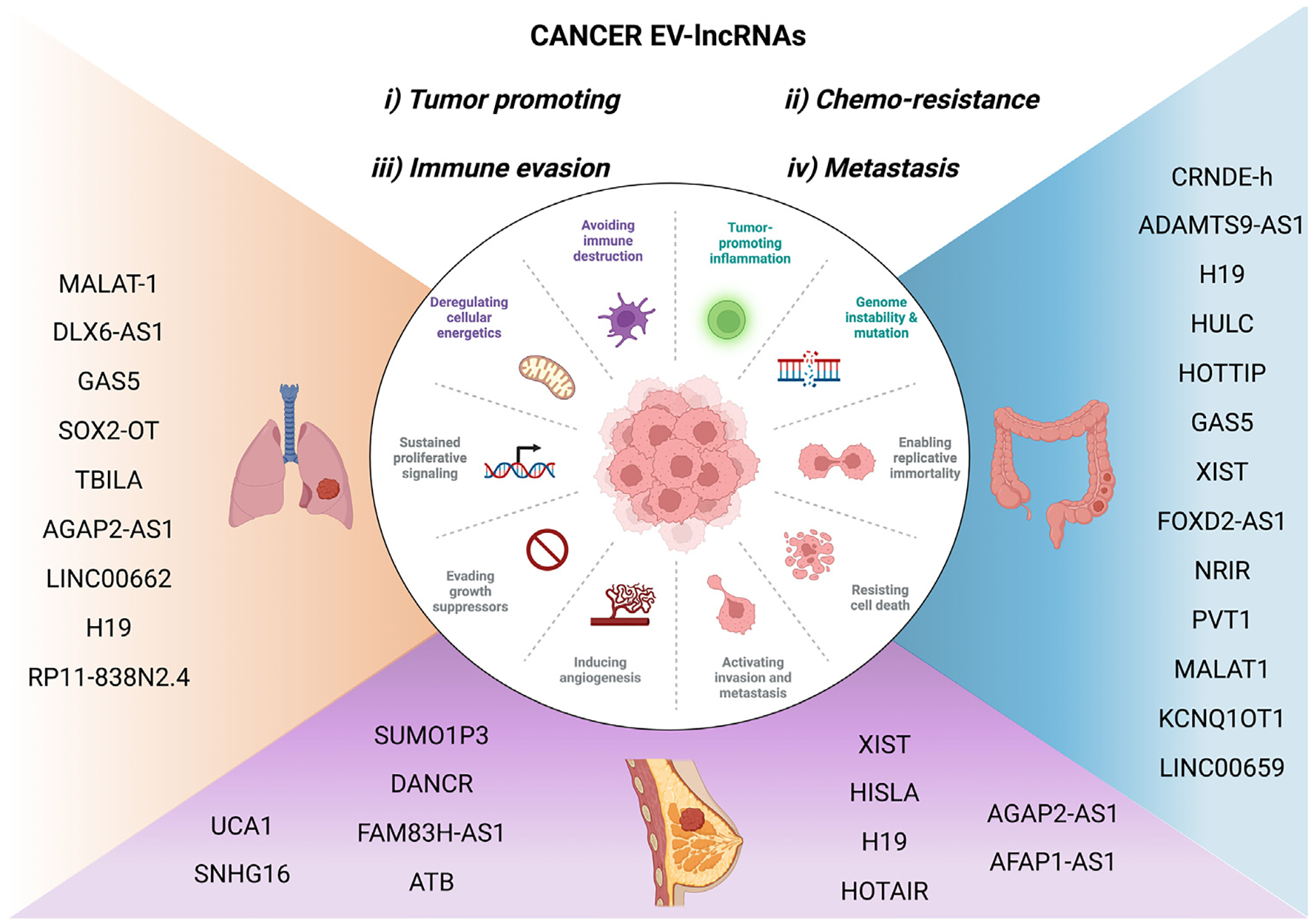
EV-lncRNAs in cancer.
Table 1.
EV lncRNAs in Cancer.
| Cancer type | lncRNA | Expression | Function | EV isolation method | Biofluid | Type of study | Validated in (number) | AOC curve | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Non-small cell lung cancer (NSCLC) | MALAT-1 | Upregulated | Oncogene | ExoQuick TC | Serum | qPCR, cell culture validated | Control (30) and NSCLC (77) | 0.703 | Ref. 112 |
| MALAT-1 | Upregulated | oncogene | ExoQuick precipitation kit | Serum | qPCR, cell culture validated | Control (52) and NSCLC (52) | NA | Ref. 113 | |
| DLX6-AS1 | Upregulated | oncogene | ExoQuick precipitation kit | Serum | qPCR | Control (64) and NSCLC (72) | 0.806 | Ref. 114 | |
| LINC00662 | Upregulated | oncogene | ExoQuick precipitation kit | Plasma | qPCR, cell culture validated | Control (50) and NSCLC (50) | NA | Ref. 115 | |
| UFC1 | Upregulated | oncogene | NA | Serum | qPCR, cell culture validated, in vivo | Control (40) and NSCLC (54) | 0.794 | Ref. 116 | |
| SOX2OT | Upregulated | Bone metastasis | Ultracentrifugation | Plasma | qPCR, cell culture validated | NSLC(142) and BoM | NA | Ref. 117 | |
| H19 | Upregulated | drug resistance | ExoQuick precipitation kit | serum | qPCR, cell culture validated | Responding(28) and nonresponding 30) | 0.799 | Ref. 118 | |
| RP11-838N2.4 | Upregulated | drug resistance | ExoQuick precipitation kit | serum | qPCR, cell culture validated | Responding(43) and nonresponding (35) | 0.754 | Ref. 119 | |
| HOTTIP | Upregulated | Prognostic marker | ExoQuick precipitation kit | serum | qPCR | Relapsed (7) and recovered (11) post-surgery and therapy | 0.935 | Ref. 120 | |
| GAS5 | Downregulated | Diagnostic marker | Exosome Isolation Kit | Control (40) and NSCLC (64) | 0.857 | Ref. 121 | |||
| TBILA, AGAP2-AS1 | Upregulated | Diagnostic marker | ExoQuick precipitation kit | Serum | qPCR | Control (100) and NSCLC (100) | 0.775,0.734 | Ref. 122 | |
| Lung Squamous Cell Carcinoma (LSCC) | SOX2OT | Upregulated | oncogene | Ultracentrifugation | Plasma | qPCR, cell culture validated | Control (79) and LSCC (72) | 0.815 | Ref. 123 |
| Colorectal Cancer (CRC) | UCA1 | Downregulated | Diagnostic marker | ExoQuick TC | Serum | qPCR | Control (20) and CRC (20) | 0.718 | Ref. 124 |
| CRNDE-h | Upregulated | Diagnostic marker | ExoQuick TC | Serum | qPCR | Control (80) and CRC (148) | 0.892 | Ref. 125 | |
| LINC00659 | Upregulated | Oncogene | Ultracentrifugation | CAF Evs | qPCR | NF (8) and CAF (8) | Ref. 126 | ||
| ADAMTS9-AS1 | Upregulated | Diagnostic marker | Ultracentrifugation | qPCR | Control (130) and CRC (130) | 0.855 | Ref. 127 | ||
| HOTTIP | Upregulated | Diagnostic marker | Thermofisher Exosome Isolation Kit | qPCR | Control (52) and CRC (52) | 0.75 | Ref. 128 | ||
| CCAL | Upregulated | Oncogene | CAF Evs | qPCR | NF (15) and CAF (15) | Ref. 129 | |||
| H19 | Upregulated | Oncogene | ultracentrifugation | CAF Evs | qPCR | NF (10) and CAF (10) | Ref. 130 | ||
| GAS5 | Downregulated | Diagnostic and prognostic, tumor supressor | ultracentrifugation | Plasma | qPCR | Control (178) and CRC (153) | 0.961 | Ref. 131 | |
| XIST | Upregulated | Diagnostic and prognostic | ultracentrifugation | Serum | qPCR | Control (41) and CRC (94) | 0.864 | Ref. 132 | |
| Breast Cancer (BC) | H19 | Upregulated | Diagnostic marker | NA | Plasma | qPCR | Control (96) and BC (102) | 0.81 | Ref. 133 |
| H19 | Upregulated | drug resistance | ExoQuick TC | serum | qPCR | Responding(36) and non responding (46) | 0.752 | Ref. 134 | |
| XIST | Upregulated | Diagnostic, prognostic, recurrence | Exosome Isolation Kit | serum | qPCR | Control(50) and TNBS(91) | 0.888 | Ref. 135 | |
| HOTAIR | Upregulated | diagnostic, prognostic, drug-resistance | ExoQuick TC | serum | qPCR | Control(15), BC(15) | 0.916 | Ref. 136 | |
| DANCR | Upregulated | diagnostic, prognostic, drug-resistance | Ultracentrifugation | serum | qPCR | Control(105), Benign breast disease(70), BC(120) | 0.88 | Ref. 137 | |
| H19 | Upregulated | diagnostic, prognostic, drug-resistance | Exosome Extraction Reagent | serum | qPCR | Control(50), Benign breast disease(50), BC(50) | 0.87 | Ref. 138 | |
| AGAP2-AS1 | Upregulated | drug resistance | ExoQuick TC | serum | qPCR, cell culture validated, in vivo | Her2+ BC-Responding(45) and non responding (45) | 0.784 | Ref. 139 | |
| MALAT1 | Upregulated | oncogene | ultracentrifugation | serum | qPCR, cell culture validated, in vivo | BC (80) | NA | Ref. 140 | |
| Triple Negative Breast Cancer (TNBC) | SUMO1P3 | Upregulated | prognostic marker | ultracentrifugation | serum | qPCR | Control(30) and BC(90, stage I-30,II-30,III-30) | Ref. 141 |
(i). Markers of Tumor-progression
Metastasis associated lung adenocarcinoma transcript 1 (MA LAT-1), a well-established lncRNA for its pivotal role as a promising prognostic marker in lung cancer,142 was also identified by Zhang et al. in 2017 to be expressed abundantly in serum EVs of NSCLC patients with an AUC of 0.703.143 In this study, serum EV-derived MALAT-1, analyzed using qPCR, was also positively associated with tumor stage and lymphatic metastasis. Further functional studies suggested that EV-associated MALAT-1 could stimulate tumor growth and migration while suppressing apoptosis in vitro in lung cancer cell lines as corroborated in another study.144 Further, its role in lung cells was examined in vitro and in vivo using knockdown and overexpression models to reveal that MALAT-1 acted as competing endogenous RNA to miR-515-5p, which in turn regulated the Eukaryotic Translation Elongation Factor 2 (EEF2) expression to aid in tumor progression. In another study, two serum EV lncRNAs TGF-β Induced lncRNA (TBILA) and ArfGAP with GTPase Domain, Ankyrin Repeat And PH Domain 1 (AGAP2-AS1) were detected to be increased in NSCLC patients compared to controls and individually had an AUC of 0.775 and 0.734 respectively.145 When these were combined with the commonly used serum tumor biomarker Cyfra21-1, the specific diagnostic accuracy for NSCLC patients enhanced considerably (AUC = 0.853). Interestingly, MALAT1 and AGAP2-AS1 were found to be elevated in EVs of smokers and NSCLC patients when compared to controls, indicating that these lncRNAs should be probed for their role as early diagnostic markers in larger cohorts. In BC, EV-derived MALAT1 was observed to be overexpressed in BC patients, correlated with poorer prognosis and clinical outcomes. This was brought about by possibly promoting cell proliferation, as observed by in vitro and in vivo experiments.146 Further, MALAT1 has been reported to be secreted by CRC tumor cells to mediate disease progression using their competing endogenous RNA (ceRNA) function by sequestering miR-26a/26b and regulating Fucosyltransferase 4 (FUT4) fucosylation to activate the PI3K/AKT pathway.147 While this study specifically proposes a role mediated by EV-derived MALAT-1, the validation on patient samples is indirect and only associative.
Serum EV-derived lncRNA DLX6 antisense RNA 1 (DLX6-AS1) was found to be elevated in 72 NSCLC patients compared to their 64 healthy controls and could be a promising diagnostic marker with AUC of 0.806.148 Not only was this lncRNA elevated in lung cancer patients in TCGA datasets, but more importantly, DLX6-AS1 levels in serum dynamically decreased post-operatively after tumor excision, suggesting that the enrichment of this lncRNA could be tumor-associated. In contrast, a serum EV-derived lncRNA growth arrest specific 5 (GAS5) was observed to be downregulated in NSCLC patients with respect to control patients with an AUC of 0.857 and when it was combined with Carcinoembryonic Antigen (CEA, a conventional tumor marker) the AUC went up to 0.929 making this a very promising biomarker.149 Another study has implicated low EV-GAS5 to mechanistically promote angiogenesis in the tumor supported by in vivo analysis.150
(ii). Markers of Chemoresistance
Given that drug resistance is a major factor that deters the effectiveness of cancer therapy, EV-lncRNAs such as H19151 and RP11-838N2.4152 have been observed to play a role specifically with erlotinib resistance having an AUC of 0.799 and 0.754 respectively as observed in NSCLC patient serum samples. EV-derived H19 has been recently implicated in gefitinib resistance of NSCLC as well.153 An EV lncRNA HOXA Distal Transcript Antisense RNA (HOTTIP) (oncogenic) was found to be a prognostic marker that can predict the relapse (AUC = 0.935, for 1st post-operative sample) by serially checking for this lncRNA in the serum of NSCLC patients from pre-surgical to post-surgical state (between 3–6 months).154
We now know that the tumor milieu is composed of cancer-associated fibroblasts (CAFs) and tumor-associated macrophages (TAMs) that form important components in aiding the cancer development, progression, metastasis, and drug resistance. This aspect was highlighted in a recent study wherein lncRNA Long Intergenic Non-Protein Coding RNA 659 (LINC00659) was significantly upregulated in EVs derived from CAF rather than normal fibroblasts (NF) from CRC patients.155 This lncRNA was observed to be transferred to CRC cells, where it acted as a sponge for miR-342-3p to increase ANXA2 expression and promote cancer progression. In a similar mechanism, CAF-secreted colorectal cancer-associated lncRNA (CCAL) was observed to promote oxaliplatin (Oxa) resistance in CRC cells.156
Drug resistance and cancer recurrence are the principal sources of poor survival in breast cancer, and several EV-lncRNAs have been identified to play a role in promoting drug resistance. While circulating plasma-derived lncRNA H19 was reported to be a promising biomarker (AUC = 0.81) of BC,157 serum EV-derived H19 was significantly elevated in BC patients compared to benign breast disease and control with an AUC of 0.87 as opposed to CEA153(0.822) and CEA (0.811).158 In this cohort, H19 also acted as a prognostic marker as it was associated significantly with Estrogen Receptor (ER), Progesterone Receptor (PR), human epidermal growth factor receptor-2 (Her-2), tumor node metastasis (TNM) stages, lymph node metastasis (LNM), and distant metastasis. Also, serum EV-derived H19 was observed to be elevated in non-responding patients (46) as opposed to responsive patients (36) after Doxorubicin (DOX) therapy with an AUC of 0.752 as evidenced by in vitro experiments as well.159 Another study,160 substantiated the predictive role of H19 after neo-adjuvant chemotherapy suggesting this lncRNA should be more carefully evaluated for drug resistance in larger cohorts. Serum EV-derived Small ubiquitin-like modifier 1 pseudogene 3 (SUMO1P3) was significantly elevated in Triple-Negative BC (TNBC), a subtype of BC.161 More importantly, this lncRNA also significantly decreased in chemosensitive patients after therapy, and high EV SUMO1P3 was associated with poor prognosis. Serum EV-associated lncRNA AGAP2-AS1 is another trastuzumab resistance-predicting biomarker with an AUC of 0.784 and in cells, works through the ELAV like protein 1 (ELAVL1)/AGAP2-AS1/ATG10 axis to induce autophagy that in turn promotes drug resistance.139 Moreover, another lncRNA actin filament-associated protein 1-antisense RNA 1 (AFAP1-AS1) was identified in serum-derived EVs of trastuzumab non-responders.162 Han et al. demonstrated using in vitro and in vivo methods that trastuzumab-resistant tumor cells secrete AFAP1-AS1 taken up by the drug-sensitive cells where it binds to AU-binding factor 1 (AUF1) protein to promote the translation of er-BB2 receptor tyrosine kinase 2 (ERBB2) gene further conferring resistance in these cells. EV-derived urothelial cancer associated 1 (UCA1) has been reported to confer tamoxifen resistance in BC cells in vitro.163 In 2018, Barbagallo et al. examined the expression of certain lncRNAs in CRC cell-line and biopsies along with EVs in cell-culture medium and serum samples of the same patients.164 Based on the extent of expression, their analysis yielded a set of lncRNAs differentially expressed at the cellular level and EV level. Interestingly, for several lncRNAs including UCA1, and Taurine Up-Regulated 1 (TUG1 ) — were identified to have an inverse correlation with respect to expression, i.e., while UCA1 was upregulated in CRC tissues, it was downregulated in the EVs with respect to control. TUG1 followed the inverse pattern where it was more in EVs and downregulated in CRC tissues. Mechanistically, this suggests a selective retention or expulsion of the EV-lncRNA based on its tumor promoting or tumor suppressing role. However, the diagnostic efficacy estimated using the AUC curve for UCA1 was 0.719 while with TUG1, it was not significant. But on combining TUG1 and UCA1, their AUC went up to 0.814. In this study, UCA1 was found to aid in CRC progression by either binding to 3’ UTR of mRNAs (ANLN, BIRC5, IPO7, KIF2 A, and KIF23) to stabilize them or acting as ceRNA to miRNAs that degrade these mRNAs. Interestingly, a 2021 study165 corroborated the downregulation of circulating plasms UCA1 in 76 CRC patients with respect to 20 healthy controls. They confirmed that UCA1 levels helped identify BRAF mutation status in CRC patients. In another study,166 EV-derived UCA1 was found to be enriched in CRC patient tissues using microarray, and mechanistically in cells, it regulated the miR-143/MYO6 axis by acting as a ceRNA. While EV-UCA1 has been reported to be elevated in Controls compared to CRC patients, Yang et al. showed that EV-UCA1 levels increase upon cetuximab-resistant CRC as evidenced both in vitro and in CRC patients under cetuximab therapy.167
(iii). Markers of Immune-evasion
Another tumor-promoting lncRNA Ubiquitin-Fold Modifier- Conjugating Enzyme 1 (UFC1), was identified to be upregulated in both serum EVs and NSCLC tumor tissues of patients with an AUC of 0.812 in serum and 0.794 in serum EVs with a positive correlation with age and tumor infiltration.168 Using both in vitro and in vivo analysis, EV-transmitted UFC1 was noted to aid in cancer progression by binding to enhancer Enhancer of Zeste 2 Polycomb Repressive Complex 2 Subunit (EZH2) to promote epigenetic silencing of Phosphatase And Tensin Homolog (PTEN).168 Colorectal neoplasia differentially expressed-h (CRNDE-h) was a lncRNA that was identified in colorectal tissues. Its expression is both tissue-specific and age-specific where its expression is mainly during early development and minimally in adult tissues. In 2016, Liu et al.examined the serum EV-derived CRNDE-h expression in CRC patients (n = 148) and found that its expression was significantly upregulated when compared to controls (n = 80).169 Its biomarker potential (AUC = 0.892) was better than the traditional CEA biomarker (AUC = 0.688), with their combination being even better (AUC = 0.913). Given that T helper 17 cells (Th17) aid in tumor progression, Sun et al. showed that CRC tumor EV-derived CRNDE-h lncRNA is transmitted to CD4+ Tcells to increase the Th17 population and positively contribute toward CRC spread as seen in vitro and in vivo.170 In this study, CRNDE-h levels observed in the serum of CRC patients were noted to be positively correlated with Th17 cell proportion in tumor-infiltrating T cells.
EV-lncRNAs KCNQ1 Opposite Strand/Antisense Transcript 1 (KCNQ1OT1) secreted by CRC tumor cells acted as a ceRNA on the miR-30a-5p/USP22 axis by regulating PDL-1 ubiquitination thereby inhibiting CD8+ T cell s.171 This autocrine effect mediates immune escape by CRC tumor cells and aids in CRC progression. One of the main limitations of this study is the indirect correlation demonstrated on the patient CRC tissues.
Tumor-infiltrating lymphocytes (TILs) have long been associated with the prognosis of BC, and depending upon which subtype forms the majority of TILs, their function could either be tumor-promoting or tumor-suppressing.172 Ni et al. identified that CD73+ Vδ1 T cells were the most common regulatory T cells (Tregs) in BC (validated in 40 BC patient samples) and were also induced by them.173 BC tumor cells secreted EVs that contained Small nucleolar RNA host gene 16 (SNHG16) that were taken up by Vδ1 T cells in the tumor.173 In the EV recipient cells, SNHG16 acted as a miR-16-5p sponge that resulted in the increase of SMAD5 mRNA causing upregulation of the TGF beta pathway that further enhanced CD73 expression and activation of these Tregs.173 This process was hypothesized to promoted immunosuppression that in turn aided in cancer progression.
(iv). Markers of metastasis
Plasma EV-derived Growth Arrest Specific 5 (GAS5) was analyzed between control (178) and CRC (153) patients, could distinguish between the two groups with an AUC of 0.964 and was observed to be associated with the TNM stage, LNM, local recurrence rate and distant metastasis rate. Interestingly, expression levels of its target miR-221 was also expressed in tissues, and plasma/EV level was associated with tumor size, TNM stage, LNM, local recurrence rate, and distant metastasis rate.174 Strikingly, plasma EV-derived lncRNA SOX2 Overlapping Transcript (SOX2OT) was specifically elevated in NSCLC patients with bone metastasis with increased expression correlating with a poorer prognosis. Mechanistically, SOX2OT acts as a ceRNA for miR-194-5p consequently increasing the expression of Rac Family Small GTPase 1 (RAC1) mRNAs as well as elevating the expression TGF-β, Parathyroid hormone-related protein (PTHrP) and receptor activator of nuclear factor-κB ligand (RANKL) observed in bone lesions.175 Importantly, these studies show a pro-metastatic role for EV-derived SOX2OT on osteoclasts cells and mouse models. It is worth mentioning that plasma EV SOX2OT can also be used as a promising biomarker for Lung Squamous Cell Carcinoma with an AUC of 0.815.176
Apart from drug resistance, distal metastasis is a common cause of BC mortality, and the brain is one of the most common secondary sites for BC metastasis. EV-derived lncRNA GS1-600G8.5 has been reported using in vitro and in vivo studies to be secreted by brain metastasis of BC cells, and it appears to compromise the integrity of the Blood-Brain Barrier further facilitating secondary tumors in the brain.177 Serum-derived FAM83H antisense RNA 1 (FAM83H-AS1) and lncRNA activated by TGF β (lncRNA ATB) were evaluated in 90 BC patients for their biomarker role. LncRNA ATB was found to be a more promising diagnostic given that the AUC values were 0.744 and 0.906.178 FAM83H-AS1 was observed to be a prognostic marker and showed a significant correlation with large tumor size, TNM stages, and LNM. Serum EV-associated X-inactive specific transcript (XIST) was found to be elevated in patients with TNBC (AUC = 0.888) and dynamically and significantly reduced after tumor resection.179 However, in patients showing recurrence, this lncRNA showed a marked increase and was also associated with a poorer prognosis. HOX Transcript Antisense RNA (HOTAIR), identified by Tang et al. in serum-derived EVs was markedly elevated in BC patient samples compared to controls with an AUC of 0.916 as opposed to 0.7378 for CA153.180 Interestingly, HOTAIR levels decreased post-surgery suggesting that they vary dynamically in concordance with tumor mass; HOTAIR could also act as a prognostic marker as its high expression was associated with poor overall and disease-free survival and non-responsiveness to therapy. The biomarker role of HOTAIR in BC was confirmed by Wang et al in another study where it was noted to be correlated with ErbB2/HER2 positivity (n = 23).181 Another serum EV-lncRNA plasmacytoma variant translocation 1 (PVT1) was found to be elevated in colon cancer patients and the cellular PVT1 through miR-152-3p/VEGFA axis promoted stemness and metastasis of cancer.182
Although this was an attempt to categorize the EV-lncRNAs roughly based on their functional contribution toward certain lethal cancers, many of them have multiple roles. For example, EV-lncRNAs such as MALAT-1, and UCA-1 not only acted as early diagnostic markers but may also be involved in immune evasion, metastasis, and chemoresistance. Given their prominent role in cancer, it is certainly crucial to validate their detection and function in several patient cohorts. Another aspect worth pondering is that since many of these EV-lncRNAs are expressed in more than one type of cancer, could there be a common EV-lncRNA that could act as an early cancer detection biomarker. However, one must bear in mind that many of these are also expressed in other disease conditions, raising questions about their specificity for particular diseases. It is possible that these are secreted due to a particular cellular dysfunction that coincides with multiple disease conditions and, unfortunately, cannot solely play a role of a biomarker of a particular disease. An alternate would be to find a signature of EV-lncRNAs that comprise more than one candidate that can uniquely identify or mark a disease. In fact, if a discovery cohort spanning multiple cancer types carefully assess common EV-lncRNAs across different cancers, a panel of specific set of EV-lncRNA signature that can act as a pan-cancer early detection biomarker is a possibility. However, it is imperative that this is objectively assessed in multiple and large patient cohorts to qualify as a disease biomarker for clinical applications in the future.
4. Cardiovascular diseases
Cardiovascular diseases (CVD) have an ever-increasing contribution to world-wide mortality and healthcare costs.183–185 Clinical biomarkers are an integral part of the diagnostic and prognostic cardiovascular workup for many CVDs, including atherosclerosis (AS), coronary artery disease (CAD), acute myocardial infarction (AMI), and heart failure (HF). However, even though several protein biomarkers for CVD (such as Creatine Kinase-MB, Natriuretic peptides, N-terminal fragment, and high-sensitivity cardiac troponin) have been incorporated into cardiovascular risk assessment scoring systems, specific and sensitive circulating biomarkers that can serve in CVD diagnosis, prognosis, and treatment response prediction are still the need of the hour. Recent studies in the cardiovascular field have shown that dysregulated lncRNAs are associated with different kinds of CVDs, as part of the underlying pathophysiologic pathways, revealing their potential as novel therapeutic targets, probes, and biomarkers of disease. Lately, extracellular lncRNAs which are incorporated in sEVs, have attracted vast research traction mainly due to their tissue-specificity and their detectability from all sorts of human biofluids.186 EV-lncRNAs (Fig. 3, Table 2) show great potential in enhancing or even replacing the traditionally used clinical biomarkers in the future.
Fig. 3.
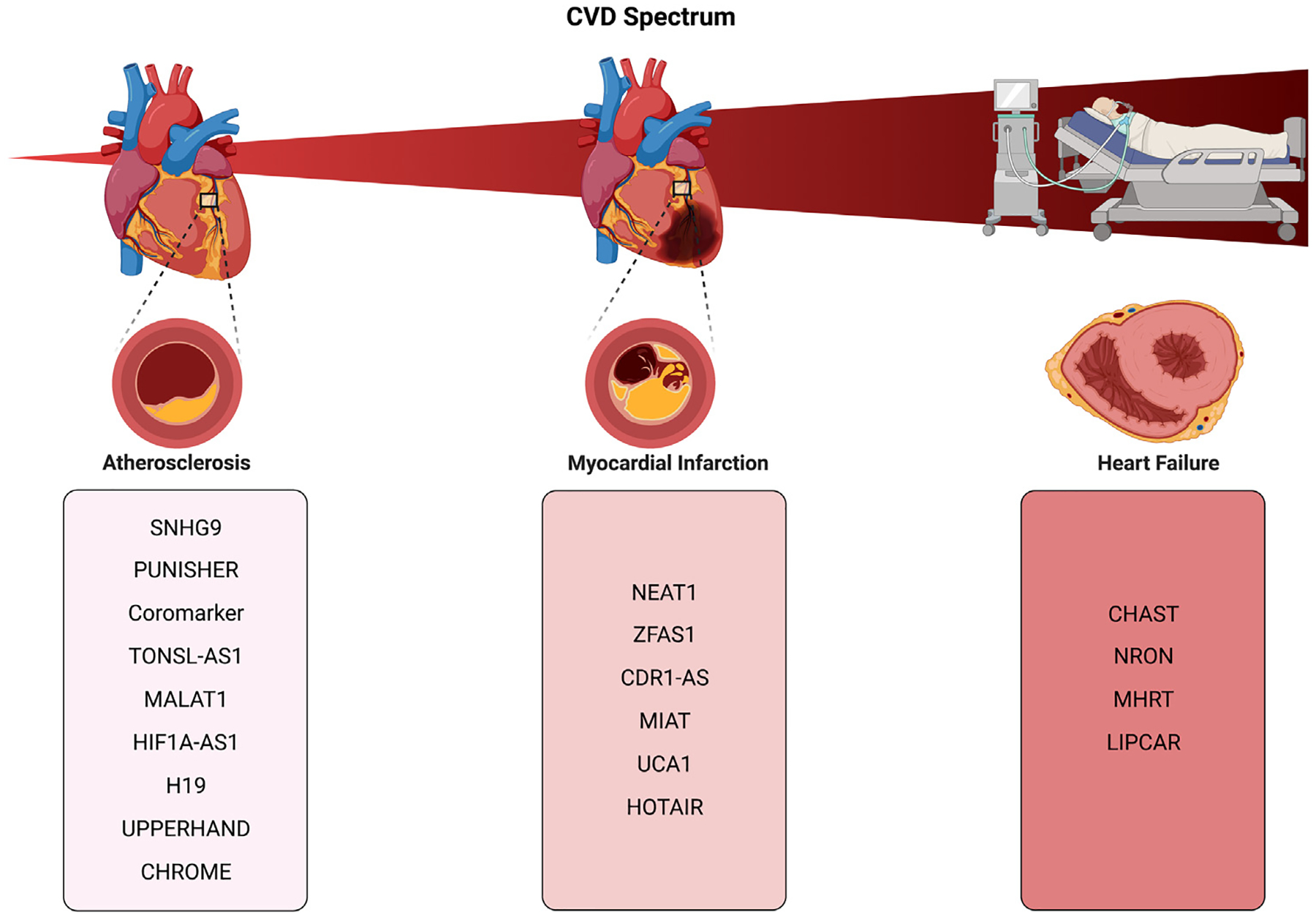
EV-lncRNAs in cardiovascular diseases.
Table 2.
EV lncRNAs in CVD.
| lncRNA | Disease | Status | Function | Origin | Study design | Validation | References |
|---|---|---|---|---|---|---|---|
| SNHG9 | CAD, Obesity, PAD | reduced | TRADD gene suppression | plasma, Adipose-derived stem cells (ADSCs) and Human umbilical vein endothelial cells (HUVECs)-derived EVs | Cohort study; Control group normal-weight (n = 32), obese-only (n = 17); obese with endothelial dysfunction (DB, n = 18; CAD, n = 23; PAD, n = 16; HBP, n = 15) | Ref. 187 | |
| PUNISHER | CAD | increased | Modulates VEGFA activity | plasma s Evs | Cohort study; Total cohort n = 221; lncRNA profiling cohort (CAD; n = 3, controls; n = 3) Validation cohort (CAD; n = 30, controls; n = 30) | (CAD; n = 30, controls; n = 30) | Ref. 188 |
| Coromarker | CAD | increased | Unknown | plasma, monocytes | Cohort study; CAD group (n = 221); non-CAD control group (n = 187) | CAD group (n = 221) non-CAD control group (n = 187) | Ref. 189 |
| TONSL-AS1 | CAD | reduced | upregulates BCL-2 | Plasma | Cohort study; CAD group (n = 60); non-CAD control group (n = 60) | Ref. 190 | |
| MALAT1 | CAD | increased | Induction of M2 phenotype in macrophages, p38/AKT signaling pathway activation, NLRP3 modulation, miR-497 suppression | serum, arterial tissue, endothelial cells, HUVEC-derived Evs | Cohort study; CAD group (n = 20) | Refs. 191–193 | |
| HIF1A-AS1 | CAD | increased | Induced by Brahma-related gene 1 (BRG1), promotes apoptosis and inhibits proliferation of VSMCs | plasma s Evs | Cohort study; AS group (n = 65); non-AS group (n = 68) | Refs. 194–197 | |
| H19 | CAD | increased | Regulated by IGF-2, suppresses SOCS1 expression by sponging miR-19a, promotes cardiomyocyte senesence | plasma | Cohort study; CAD group (n = 300) ; control group (n = 180) | Refs. 198–202 | |
| Upperhand | CAD | increased | Regulates Hand2 expression | whole blood | Cohort study; Total cohort n = 324; Discovery cohort: microarray analysis (CAD; n = 6, controls; n = 6), | Validation cohort 1; (CAD; n = 30, controls; n = 30) Validation cohort 2; (CAD; n = 137, controls; n = 115) | Refs. 203, 204 |
| CHROME | CAD | increased | Regulates d miRNAs miR-27b, miR-33a, miR-33b and miR-128 | plasma | Cohort study; CAD group (n = 14); control group (n = 33) | Ref. 205 | |
| NEAT1 | STEMI | increased | sponges miR-495-3p/indirectly activates MAPK6 pathway | plasma s Evs | Cohort study; Control group (n = 27); UA group (n = 24); STEMI group (n = 47) | Refs. 206–208 | |
| ZFAS1 | AMI/HF | reduced | Modulates SERCA2a protein/induces mitochondria-mediated apoptosis via cytosolic Ca2+ overload | whole blood | Cohort study; AMI group (n = 103); non-AMI group (n = 149); control group (n = 95) | Refs. 209–212 | |
| CDR1AS | AMI | increased | Sponges miR-7a/upregulates PARP,SP1 | whole blood | Cohort study; AMI group (n = 103); non-AMI group (n = 149); control group (n = 95) | Refs. 209, 213 | |
| MIAT | AMI | increased | sponges miR-22-3p/upregulates DAPK2 | Plasma | Cohort study; STEMI group (n = 58); UA control group (n = 50) | Refs. 214–216 | |
| UCA1 | AMI | increased | sponges miR-873/XIAP upregulation/AMPK phosphorylation and increased level of the antiapoptotic protein BCL2 | plasma s Evs | Cohort study; AMI group (n = 26, ccu patients); control group (n = 26) | Ref. 217 | |
| HOTAIR | AMI | Decreased | sponges miR-126-5p | Plasma | Cohort study; AMI group (n = 50); control group (n = 50) | Refs. 218, 219 | |
| CHAST | AMI/HF | increased | suppresses Pleckstrin homology domain-containing protein family M member 1 | Plasma | Cohort study; AMI group (n = 53); control group (n = 90) | Refs. 220, 221 | |
| NRON | HF | increased | upregulates HIF1a | plasma | Cohort study; HF group (n = 72) non-HF control group (n = 60) | Refs. 222, 223 | |
| MHRT | HF | increased | antagonizes the function of Brg1/suppresses miR-3185/reduces myocardin expression through KLF4 modulation | plasma | Cohort study; HF group (n = 72) non-HF control group (n = 60) | Refs. 222, 224–226 | |
| LIPCAR | STEMI/HF | increased | Unknown | plasma | HF Cohort study 1; MI and LV remodeling group n = 246 ; HF Cohort study 2; STEMI group (n = 46); control group (n = 40); HF Cohort study 3; non-HF post-AMI group (n = 68); HF post-AMI group (n = 59), 30 control group (n = 30) HF Cohort study 4; HF group (n = 967) | validation cohort 1; chronic HF group (n = 344); validation cohort 2; chronic HF group 2 (n = 198); STEMI | Refs. 227–230 |
(i). Atherosclerotic cardiovascular disease (ASCVD)
Atherosclerotic CAD culminating in acute coronary syndrome or myocardial infarction is a central contributor to CVD morbidity and mortality, with about 20% of patients with myocardial infarction having fatal outcomes within the first month of the event.231 Endothelial injury and inflammation are considered the initial events in the development of AS and CAD. EV-lncRNAs have been shown to exert regulatory effects and thus actively participate in endothelium homeostasis.232 EV-lncRNA, PUNISHER (also known as AGAP2-AS1) was shown to act on the endothelial cells after being loaded on sEVs via the RNA-binding protein (RBP), heterogeneous nuclear ribonucleoprotein K (hnRNPK), and modulate the expression of the proangiogenic protein vascular endothelial growth factor A (VEGFA).233 PUNISHER was found to be significantly upregulated in the plasma of patients with stable CAD compared to controls while studies on human coronary artery cells showed that pro-atherogenic factors such as oxLDL or TNF-α caused the upregulation of PUNISHER levels.188 Endothelial activation, monocyte attachment, and transmigration are hallmarks in the development of CAD.234 EV-lncRNAs have been reported to take part in the communication between ECs and immune cells.235 Monocyte-derived EV-lncRNA Coromarker (or AC100865.1) was shown to be upregulated in EVs isolated from plasma of CAD patients compared to controls. In the validation cohort of 221 stable CAD patients and 187 non-CAD controls, Coromarker exhibited 78.05% sensitivity and 86.49% specificity in discriminating CAD.189 H19, which is under the regulation of insulin growth factors, promotes senescence in cardiomyocytes, and is normally expressed during embryologic development and downregulated later in adult life198–200; however, in patients with CAD, re-expression of lncRNA H19 is observed as indicated by its high levels both in plasma and peripheral blood mononuclear cells (PBMC).201,202 Thus, PBMCs could react to certain pathologic stimuli by releasing H19 to the periphery. In another CAD cohort of 137 patients, circulating lncRNA Upperhand (also known as HAND2-AS1), known to regulate Hand2 expression during heart development, was significantly upregulated compared with 115 non-CAD controls and exhibited high sensitivity and specificity in diagnosing CAD (0.737 and 0.652 respectively).203,204 It should be noted that even though H19 and Upperhand levels were detected in human plasma and whole blood, respectively, none of these studies’ protocols included separate EV isolation steps and thus, whether these lncRNAs are indeed harbored within EVs remained unconfirmed. Furthermore, it has been demonstrated that PUNISHER, H19, and Upperhand are also involved in other pathophysiological processes such as human cancer. These confounding associations which could diminish their diagnostic and prognostic biomarker potential have remained unexplored in the current CVD studies.
(ii). Acute myocardial infarction (AMI)
ASCVD is driven by lipid accumulation in the arterial wall, inflammation, and vascular injury that lead to coronary arterial stenosis, plaque formation, and AMI.236 EV-lncRNAs are involved in all aspects of this atherothrombotic spectrum.237 Currently, lncRNA urothelial carcinoma associated 1 (UCA1) is one of the very few lncRNA molecules that have been studied in human plasma-derived EVs and are associated with AMI in patient cohorts. In fact, the extracellular levels of UCA1 were significantly higher in AMI patients compared to healthy volunteers, while the AUC for AMI prediction was 0.82, indicating that human plasma extracellular UCA1 has the potential to be a non-invasive biomarker for the diagnosis of AMI.217 However, attention needs to be drawn to the confounding potential of certain cancers, such as colorectal cancer, that can also increase circulating UCA1 levels in the serum of patients.238 Function-wise, UCA1 was found to suppress miR-873, which in turn increases X-Linked Inhibitor Of Apoptosis (XIAP) levels and promotes apoptosis via the AMPK phosphorylation pathway.217 Zinc finger antisense 1 (ZFAS1) along with Cdr1 antisense (CDR1AS) were significantly different in whole blood samples of AMI and controls. Interestingly, the logistic regression analysis showed that the combination of reduced ZFAS1 levels and increased CDR1AS improved the predictive value for AMI.209 Cellular CDR1AS upregulates two apoptotic genes, poly (ADP-ribose) polymerase (PARP) and Specificity protein 1 (SP1), by sponging miR-7a, in myocardial cells, during MI injury.213 Cellular ZFAS1 has a dual mechanism of action on cardiomyocytes — inhibiting the sarcoplasmic reticulum Ca2+ ATPase 2a (SERCA2a) after AMI210 and activating the mitochondrial apoptosis pathway.211 Although the combination of circulating ZFAS1 and CDR1AS was shown to exhibit a diagnostic and prognostic value for AMI, there are still a lot of obstacles to overcome to be clinically utilized. First, similar to several other lncRNA markers, both ZFAS1 and CDR1AS were found to be upregulated in cancer patients,239,240 suggesting that cancer is a confounder and might hamper ZFAS1 and CDR1AS diagnostic specificity. Secondly, two separate independent studies showed that, circulating ZFAS1 levels were associated with HF212 and type 2 diabetes (T2D)241 limiting its diagnostic utility in AMI, given that T2D is strongly associated with AS and AMI.242 It is also worth noting that both ZFAS1 and CDR1AS were detected as circulating in whole blood, and the question of whether can be found in the EV compartment was left unanswered. In another cohort of 58 STEMI patients and 50 UA controls, plasma levels of pro-fibrotic EV-lncRNA myocardial infarction associated transcript (MIAT) were shown to be elevated in patients with AMI compared to controls. MIAT was positively associated with cardiac dysfunction enzymes, creatine kinase-MB and cardiac troponin T (cTnT). After ROC analysis was performed, it revealed that MIAT had the same diagnostic value as cTnT.214 Mechanistically, MIAT exhibits a pro-apoptotic effect in cardiomyocytes by sponging miR-22-3p and, in turn, upregulating the death-associated protein kinase 2 (DAPK2).215 Although MIAT is purported in many studies as a specific biomarker for AMI, in another study MIAT was found to be upregulated in the plasma and EVs of patients with atrial fibrillation (Afib). Notably, none of those patients had a prior history of AMI.216 Moreover, another study found that MIAT levels are also higher in patients with multiple sclerosis (MS), a neurodegenerative disorder.243 These data suggest that MIAT levels could fluctuate responding not only to ischemic stimuli and cardiac injury, but also to other diseases. Nevertheless, to answer the question of whether MIAT can be used clinically as a biomarker for AMI necessitates larger trials that include both AMI and Afib patients as well as neurologic diseases to be assessed as possible confounding factors. Finally, EV-lncRNA HOTAIR, which acts by sponging miR-126-5p, is another case of EV-lncRNA that showed utility as a biomarker in CVD as its levels were found to be markedly elevated in blood samples of patients with CAD and AMI218,219. However, numerous other conditions, especially neoplastic disorders,244 have also been shown to significantly increase its levels in plasma and, therefore hinder the potential of HOTAIR to be a specific biomarker for CVD. Thus, further larger studies are needed to assess HOTAIR specificity for diagnosing AMI. Notably for all these studies, comparisons need to be made to the current gold standard biomarker for MI diagnosis (high sensitivity cardiac troponin T, cTnT) to determine if any of these EV-lncRNAs can add to the diagnostic or prognostic performance of cTnT. Finally, given that myocardial infarction is an acute condition, timely detection, diagnosis, and treatment are of the essence. However, due to the unpredictable nature of such an event data on changes in EV-lncRNA levels immediately following an MI are limited. Additionally current biomarkers of acute coronary syndrome such as high sensitivity troponin are excellent biomarkers, suggesting that the role of EV-lncRNAs as diagnostic markers may be limited, although their role as prognostic markers of post-MI sequelae need to be established. Further research is needed to determine the dynamics of EV-lncRNAs expression and their association with post-MI outcomes.
(iii). Heart Failure (HF)
In the aftermath of AMI; fibrosis and myocardial scarring lead to ventricular remodeling, which, in turn, triggers arrhythmia and HF.245 EV-lncRNAs have been shown to be key players in cardiac tissue remodeling and HF development and thus can be used for diagnostic and prognostic purposes in both chronic and acute setting.76 Recently, novel EV-lncRNAs with mechanisms that converge on the nuclear factor of activated T-cells (NFAT) pathway have been implicated in HF. NFAT is a molecule with pivotal functions in regulating intracellular Ca2+ homeostasis in HF and pathological hypertrophy.246,247 The expression levels of lncRNA cardiac hypertrophy-associated transcript (CHAST), which is a transcriptional target of NFAT, were increased acutely and significantly in patients with AMI, and correlated with cardiac systolic function. CHAST was found to be an independent predictor of cardiac contractile function and thus could serve as a clinically useful biomarker for cardiac remodeling.220 Furthermore, CHAST has been implicated in cardiac hypertrophy pathways where it was found to suppress Pleckstrin homology domain-containing protein family M member 1, inhibit cardiomyocyte autophagy, and thus promote hypertrophy.221 Although more studies are needed, to date, CHAST has not been associated with other diseases besides cardiac hypertrophy and HF, signifying its promising potential for clinical utility in the future. Other EV-lncRNAs implicated in the NFAT pathway, namely., non-coding repressor of NFAT (NRON) along with myosin heavy chain related to RNA transcription (MHRT) were found to be significantly upregulated in plasma of patients with HF compared to non-HF controls. In fact, ROC analysis showed the area under the curve for NRON and MHRT to be 0.865 and 0.702 respectively. Regression analyses identified NRON and MHRT as independent predictors of HF, demonstrating that both can serve as HF diagnostic biomarkers.222 However, it should be noted that NRON was also found to be upregulated in the plasma of patients with MS.243 Mechanistically speaking, animal studies have demonstrated cellular lncRNA NRON to be involved in cardiac hypertrophy and myocardial ischemia via hypoxia inducible factor 1a (HIF-1a) modulation.223,248 On the other hand, lncRNA MHRT has been found to exert somewhat ambiguous effects on myocardial tissue, either alleviating cardiac hypertrophy, in some studies224,225 or promoting cardiac fibrosis after MI through miR-3185.226 Finally, another circulating lncRNA long intergenic noncoding RNA predicting cardiac remodeling (LIPCAR,also known as uc022bqs.1) was shown, in two separate studies, to be a predictor for adverse outcomes both in patients with AMI and chronic HF, since its levels were associated with LV cardiac remodeling and a higher risk for cardiovascular death.227,228 In par with those results, several studies found circulating LIPCAR levels to be significantly elevated in patients with HF and showed a positive correlation with NYHA class, NT-proBNP, and high-sensitivity cTnT while negatively associated with left ventricular ejection fraction (LVEF). Furthermore, after ROC analysis, LIPCAR was demonstrated to have a better diagnostic value (AUC: 0.985) of HF following AMI during hospitalization than NT-proBNP (AUC: 0.714).229,230 Although its place in AMI or HF diagnosis remains questionable, LIPCAR may have excellent prognostic and predictive value for AMI and HF patients in a clinical setting, and thus, further clinical studies are warranted.
5. Neurodegenerative diseases
Advanced age inevitably leads to the loss of neuronal structure and function, which is the hallmark of neurodegenerative diseases (NDD). NDD (Alzheimer’s disease; AD, Parkinson’s disease; PD and motor neuron diseases including amyotrophic lateral sclerosis; ALS) collectively affected almost 6.0 million individuals in the U.S. between 2016–2017. These diseases were responsible for almost 273,000 deaths and 3.0 million disability-adjusted life years in 2016.249–251 Clinical biomarkers for early detection and diagnosis of NDDs are currently lacking. Therefore, it is paramount that reliable markers for diagnosis, prognosis, and monitoring disease activity be discovered.
All cells that comprise the central nervous system, including neurons, astroglia, microglia, and oligodendrocytes, release EVs.252–255 Studies investigating the potential of EV ncRNAs in NDD diagnosis have largely focused on the miRNA cargo of EVs and have shown promising potential.256 On the other hand, lncRNAs have been shown to play major roles in the physiological development of the human brain, exhibit brain region- and cell-type-specificity, suggesting that their perturbations might be central to NDD pathophysiology.257 Thus, these characteristics deem EV-lncRNAs as potential circulating diagnostic biomarkers. Hereafter, we review the potential applications of circulating lncRNAs as biomarkers and probes of NDDs in clinical practice.
(i). Alzheimer’s Disease
Alzheimer’s disease (AD) is the most common form of dementia worldwide and accounts for 60%–80% of all dementia cases.258 AD is characterized by progressive cognitive decline and memory deficits. Anatomical manifestations include brain volume loss mainly due to hippocampal degeneration, whereas pathological examination of brain tissue reveals intracellular neurofibrillary tangles rich in hyperphosphorylated Tau and extracellular amyloid-β (Aβ) deposition leading to senile plaque formation. Aβ deposition is associated with neurodegeneration, inflammation, and neuronal cell death in AD.259 β-secretase-1 (BACE1) cleaves amyloid precursor protein (APP) to generate the amyloid-β 1–42 and amyloid-β 1–40, one of the inciting events in AD pathophysiology. The antisense transcript of BACE1 (lncRNA BACE1-AS) was shown to upregulate BACE1 and increase Aβ expression in patients with AD through a feed-forward regulatory mechanism. BACE1-AS was also elevated in the brain tissue of patients with AD and APP transgenic mice.260 Circulating BACE1-AS, which acts by sponging miR-214-3p, was elevated in a small cohort of 35 AD patients.261 In another study with 72 AD participants, circulating BACE1-AS levels in sEVs were significantly higher compared to 62 controls. ROC curve analysis demonstrated that the AUC for AD prediction was 0.761, the sensitivity was 87.5%, and the specificity was 61.3%. In the same study, when BACE1-AS expression data was combined with the volume and thickness of the right entorhinal cortex measured by MRI, the specificity and sensitivity were raised to 96.15 and 90.91%, respectively, suggesting that BACE1-AS levels combined with imaging data may serve as a potential diagnostic biomarker for AD.262 Towards the purpose of biomarker discovery in neurological diseases, EVs have successfully been isolated from cerebrospinal fluid (CSF).263 In a study that enrolled 120 AD patients, 120 PD patients and 120 healthy controls lncRNA MALAT1 was measured along with miR-125b, FOXQ1, PTGS2, and CDK5 in both CSF and plasma by RT-qPCR. MALAT1 was found to be downregulated both in plasma and CSF of AD patients compared to PD patients and controls. In fact, MALAT1 was able to differentiate AD patients from PD patients (AUC: 0.892) and controls (AUC: 0.830). In AD patients, correlations with mini-mental state exam (MMSE) score, and proteins Aβ42, t-tau and p-tau showed that MALAT1 and FOXQ1, in plasma and CSF, correlated with decreased disease severity, while miR-125b, PTGS2 and CDK5 correlated with increased disease severity.264 On the other hand, serious specificity concerns arise when considering the influence of cancer265 and other prevalent conditions on MALAT1 levels. In that regard, large cohorts are needed to investigate the associations between MALAT1 and confounding factors such as sex, age, and systemic diseases.266
(ii). Parkinson’s disease
PD is the second most common neurodegenerative disorder characterized by progressive motor dysfunction in older individuals, often accompanied by cognitive abnormalities267 with around 2%–3% prevalence in individuals aged more than 65.268 The most common feature of PD is the degeneration of dopaminergic neurons in the mid-brain area called substantia nigra (SN), which consequently causes striatal dopaminergic deficiency.268 This is caused by the formation of Lewy bodies in the cytoplasm of these dopaminergic neurons attributed to the aggregation of α-synuclein leading to clinical symptoms usually associated with motor functions such as bradykinesia, resting tremors, rigidity, and motor failure accompanied by cognitive decline and even dementia.269,270 However, since many of these symptoms are found in other neurodegenerative diseases, they are often misdiagnosed because the exact mechanism of PD is still poorly understood. Therefore, identifying appropriate diagnostic biomarkers for this disease is an unmet need, and EV-lncRNAs could offer a promising biomarker role. One of the first studies to explore the EV-lncRNA role was done by Wang et al. wherein they compared the transcriptome of plasma-derived EVs obtained through ultracentrifugation in age and sex-matched 7 PD patients along with 7 healthy controls using RNA-sequencing.271 On applying log2 fold-change ± 1.5 and p ≤.05, they identified 24 downregulated and 15 upregulated lncRNAs in the PD group. They further chose to explore lnc-Makorin Ring Finger Protein 2-42:1 (lnc-MKRN2-42:1), downregulated in PD patients, by bioinformatically identifying its mRNA targets, such as EIF4E, MKNK1, BTD, and TMEM78. This was in turn validated in 32 PD patients along with 12 healthy controls using qPCR, with some of them yielding promising results. Although this study is not an extensive characterization for identifying EV-lncRNA biomarkers for PD, it offers a glimpse of the approach. Certainly, further exploration, along with validation in larger cohorts, could help ascertain useful biomarkers. More interestingly, Zou et al. pursued a more focused approach toward biomarker identification.272 Based on PD pathology, α-synuclein aberrant aggregation plays a pivotal role in the development of this disease. Certain studies have observed the release of α-synuclein in EVs in mouse models of PD273 and further, EVs derived from brain tissue of dementia seem to be able to contribute to the formation of Lewy bodies demonstrated by in vitro and in vivo models.274 To further probe the mechanism of such EVs, Zou et al. performed a microarray analysis in neuronal-derived EVs with L1 cell adhesion molecules (L1CAM) isolated from blood using antibody-based capture, that was also positive for α-synuclein. Based on this, Long Intergenic- POU class 3 homeobox 3 (Linc-POU3F3), known for its role in the autophagic lysosomal pathway, was selected for further analysis.272 Upon comparing the levels of plasma-EV derived Linc-POU3F3 in 93 PD patients and 85 controls, this lncRNA was upregulated in PD patients, while α-synuclein was also elevated in these EVs. β-Glucocerebrosidase (GCase) has been reported to be associated with α-synuclein proteostasis and correspondingly, the GCase activity was shown to be significantly downregulated in PD patient plasma. Each of these lncRNAs exhibited good diagnostic capabilities for PD, with Linc-POU3F3 having an AUC of 0.763, for α-synuclein was 0.616, GCase activity 0.684, and all these three combined had an AUC of 0.824 with a sensitivity of 71%, and a specificity of 83% at a cutoff of 0.66. Additionally, Linc-POU3F3 levels correlated with disease severity, however, its biomarker role has to be further researched to be translated at a clinical level. In another approach, Elkouris et al. probed the role of lncRNA in PD patient samples by examining bioinformatically genes proximal to lncRNAs (less than 0.5 kilobases) and closely involved with PD, such as GBA1, SNCA (PARK4), UCH-L1 (PARK5), PINK1 (PARK6), DJ-1 (PARK7), LRRK2 (PARK8), and MAPT.275 When they examined the expression of these lncRNAs, namely., SNCA-AS1, AK127687, UCHL1-AS1, PINK1-AS1, AX747125, GBAP1, and MAPT-AS1 in substantia nigra of healthy and PD patients, all of them except GBAP1 were significantly downregulated in PD. These lncRNAs were then assessed in PBMC (Peripheral Blood Mononuclear Cells) and in the EVs derived from CSF using ultracentrifugation. Four out of the six lncRNAs, α-Synuclein Antisense transcript 1 (SNCA-AS1), Microtubule Associated Protein Tau Antisense transcript 1 (MAPT-AS1), AK127687, and AX747125, were detected in the CSF EVs. However, a more in-depth analysis is required to gauge their biomarker role. The exploration of EV-lncRNAs in PD is still in nascent stages, and further studies are required to identify a suitable clinical biomarker for this disease.
(iii). Amyotrophic Lateral Sclerosis
Amyotrophic Lateral Sclerosis (ALS) is an adult-onset, progressive neurodegenerative disorder characterized by loss of motor neurons in the cerebral cortex with fatal outcomes. Since the pathogenesis of ALS is still unclear, any insight and clinical biomarkers are urgently required. Although several studies have explored the role of lncRNAs in ALS, few studies have investigated the role of EV-derived lncRNAs. In a recent study by Sproviero et al. transcriptomic analysis was performed on plasma-derived small (sEVs) and large EVs (lEVs) isolated using ultracentrifugation from patients with neurodegenerative diseases such as AD (n = 6), PD (n = 9), Sporadic ALS (n = 6), frontotemporal dementia/FTD ( n = 9)276 as well as control patients (n = 6). Interestingly, the EVs from ALS patients were larger in size than those from other NDD’s. Furthermore, the number of EVs in ALS and FTD patients was higher than in controls. Moreover, this study reported that sEVs had a significantly higher number of differentially expressed long RNAs in ALS than lEVs. In addition, this study failed to identify DEGs in AD and PD; however, several targets were found to be differentially expressed between ALS and FTD, including 9 lncRNAs that were upregulated and 4 that were downregulated. While a major limitation of this study is that it does not validate its results through experimental methods such as qPCR. It is evident that further research is needed to identify appropriate EV-lncRNA biomarkers for ALS.
Compared to cancer and cardiovascular diseases, there are fewer EV-lncRNAs that have been identified and studied in the context of NDDs (Table 3, Fig. 4). Certainly, extensive research should be conducted before these EV-lncRNAs can be utilized as biomarkers for NDDs in clinical practice.
Table 3.
EV lncRNAs in NDD.
| NDD type | lncRNA | Expression | Source | EV isolation method | Patient Cohort | RNA validation method | AUC | Reference |
|---|---|---|---|---|---|---|---|---|
| Alzheimer’s Disease (AD) | BACE1-AS | Upregulated | Plasma | NA | 35 | qPCR | Ref. 260 | |
| BACE1-AS | Upregulated | Plasma | ExoQuick Precipitation | 72 | qPCR | 0.761 | Ref. 261 | |
| MALAT1 | Downregulated | PLasma | NA | 120 AD, 120 PD, 120 normal | qPCR | 0.83 | Ref. 263 | |
| Parkinson’s disease (PD) | lnc-MKRN2–42:1 | Downregulated | Plasma | Ultracentrifugation | Discovery cohort (7 Healthy;7PD), Validation cohort (32 PD; 12 healthy) | qPCR | NA | Ref. 270 |
| Linc-POU3F3 | Upregulated | Plasma | LICAM Pulldown | 93 PD patients and 85 controls | qPCR | 0.763 | Ref. 271 |
Fig. 4.
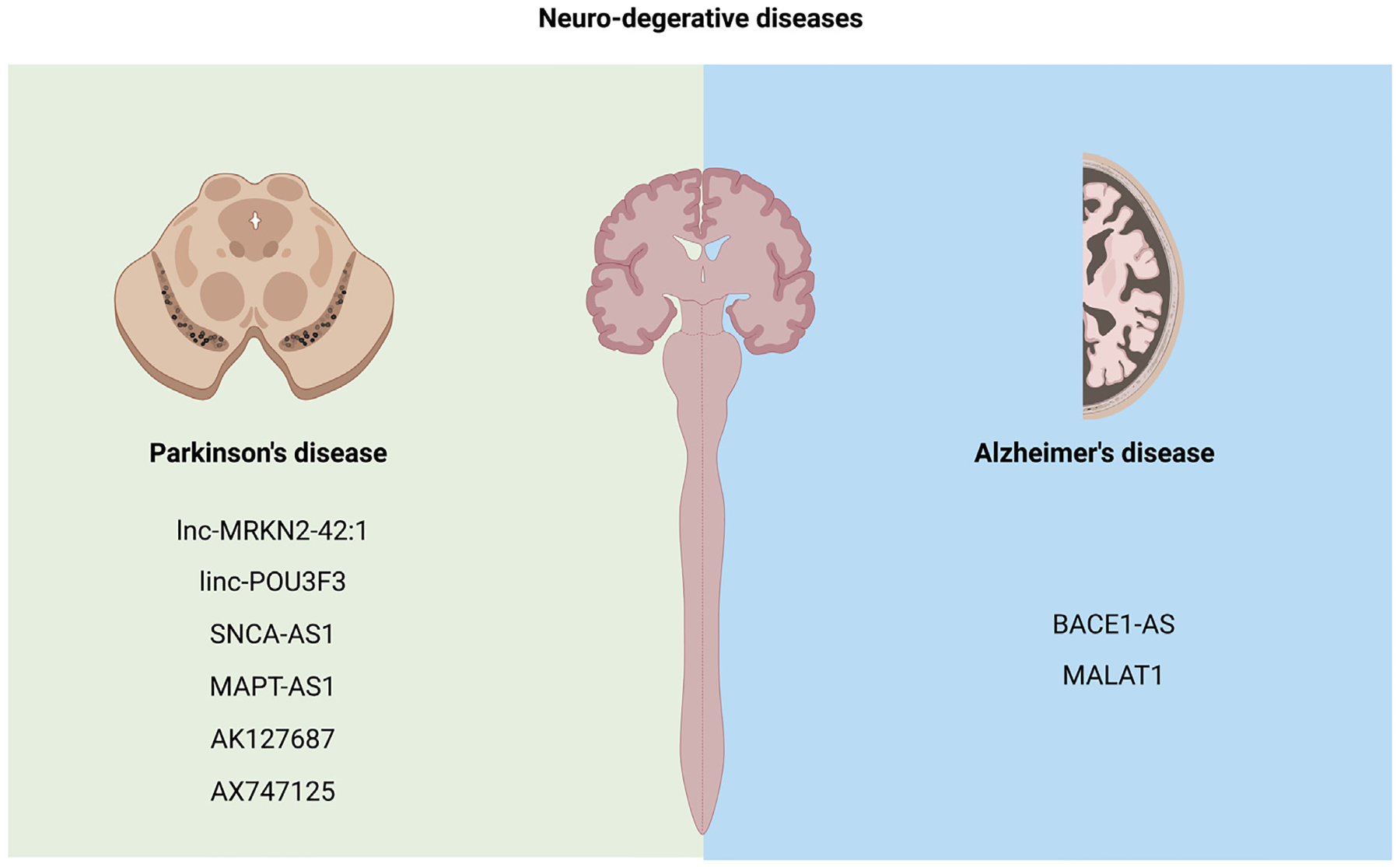
EV-lncRNAs in neuro-degenerative diseases.
6. Challenges in EV-lncRNA research
Given that both research on EVs and lncRNAs are ever-expanding with room for greater clarity concerning their classification, function, and myriad roles in normal physiology and pathogenesis, extracellular RNA research is perhaps doubly challenged by the limitations of each of these fields. Firstly, the EV isolation method used in each case has significant implications on the outcome of the downstream analysis.277 Hence, the lack of standardized isolation protocols poses a challenge in comparing findings made between different research groups. The commercially available isolation kits diminish but do not eliminate the reproducibility problem. Secondly, identifying specific markers for EVs is still challenging, despite extensive research done in this direction. Newer methodologies that not only characterize but also visualize intact EVs more clearly, such as super-resolution microscopy and better classification systems, are still the need of the hour. Third, the annotation of lncRNAs is currently insufficient. While many large-scale efforts to refine current annotations are on the way, annotating the profiled lncRNAs is based on several different databases which might have implications on biomarker validation efforts between different groups. Fourth, while RNA-sequencing (RNA-seq) has replaced microarray technology due to a wider range of RNA quantification and higher throughput capabilities, there is no single RNA-seq method that allows simultaneous profiling of all RNA species. Depending on the RNA-seq technology used, different RNA biotypes can be profiled, however the sequencing method needs to be aligned with the experimental needs, aims and design. Moreover, profiling lncRNA expression in biofluids presents additional challenges. In human plasma, extracellular lncRNAs are usually found in fragments, which in turn necessitates a thorough library preparation procedure. It is still unclear whether the long RNA species is present as fragments or full-length in EVs. Especially in the case of cellular stress, lncRNAs could be packaged in EVs to prevent any toxic build-up to improve cell-viability and consequently be fragmented. Interestingly, in some cases, cells subjected to stress preferentially increase the expression of a certain lncRNA fragment in the EVs, suggesting that the fragmentation itself is not a random process and seems to follow a specific mechanism (unpublished observations). While there are emerging reports of fragments in EVs, it is unclear if the fragmentation is a way of clearance from the cell or if the fragments themselves have a functional role.278 In fact, the functionality of lncRNA fragments is only recently explored, and its specific role in EVs is currently unknown. Most reports focus only on the functionality of the lncRNAs packaged in EVs, but have not critically evaluated the nature or the function of the EV-lncRNA itself. Finally, studying the mechanism of long RNA packaging in EVs is a huge challenge given the different types of s-EVs and a possible differential cargo. Therefore, most studies have focused on small RNA packaging in EVs, with very few exploring the mechanism of long RNA packaging in EVs. Given that this is a huge unmet need in the field, hopefully, future studies focus on studying this aspect of EV-lncRNAs.
7. Conclusions and future perspectives
While this review offers a glimpse of what lies ahead for EV-lncRNA research in diseases, one can conclude that EV-derived lncRNAs and their fragments have a dynamic expression and are highly sensitive to physiological changes aiding in their promising biomarker role, sometimes even outperforming the conventional protein biomarkers. However, in light of these challenges, we should focus on identifying and understanding more precisely how these EV-lncRNAs function in diseases, how they change in response to disease progression or therapy, and what associations with clinical outcomes in order to have a more targeted approach to theranostics and prognosis. Finally, probing disease-related EV-lncRNAs may provide insight into novel druggable pathways and targets in the cells of origin.
Acknowledgments
SD was funded by NIH, USA grant R35HL150807 and R01DK133847. PG is funded by an AHA post-doctoral award, USA and GL is funded by NIH DK133847 and an AHA Career Development Award, USA.
Disclosures
SD is a founder and has equity in Switch Therapeutics and Thryv Therapeutics, has received research funding from Abbott and Bristol–Myers–Squibb, and consulted with Thryv Therapeutics and Boston Scientific. None of these entities are relevant to the material in this manuscript.
Footnotes
CRediT authorship contribution statement
Michail Spanos: Literature review, Generated the initial draft, Generated all the art and table, Involved in the editing and final production of the manuscript. Priyanka Gokulnath: Literature review, Generated the initial draft, Generated all the art and table, Involved in the editing and final production of the manuscript. Emeli Chatterjee: Review & editing of manuscript. Guoping Li: Review & editing of manuscript. Dimitrios Varrias: Review & editing of manuscript. Saumya Das: Formulate the initial design of the manuscript, Editing, Final approval, Funding for the efforts of the authors.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Saumya Das reports financial support was provided by National Institutes of Health.
References
- 1.Yáñez Mó M, Siljander PRM, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:1–60. 10.3402/jev.v4.27066, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Höög JL, Lötvall J. Diversity of extracellular vesicles in human ejaculates revealed by cryo-electron microscopy. J Extracell Vesicles. 2015:4. 10.3402/jev.v4.28680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and Saliva is concentrated in exosomes. PLoS One. 2012:7. 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monguió-Tortajada M, Morón-Font M, Gámez-Valero A, Carreras-Planella L, Borràs FE, Franquesa M. Extracellular-vesicle isolation from different biological fluids by size-exclusion chromatography. Curr Protoc Stem Cell Biol. 2019:49. 10.1002/cpsc.82. [DOI] [PubMed] [Google Scholar]
- 5.Gutiérrez-Vázquez C, Villarroya-Beltri C, Mittelbrunn M, Sánchez-Madrid F. Transfer of extracellular vesicles during immune cell–cell interactions. Immunol Rev. 2013:251. 10.1111/imr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawamura Y, Sanchez Calle A, Yamamoto Y, Sato TA, Ochiya T. Extracellular vesicles mediate the horizontal transfer of an active LINE-1 retrotransposon. J Extracell Vesicles. 2019:8. 10.1080/20013078.2019.1643214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008:105. 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of mirna for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014:3. 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ban JJ, Lee M, Im W, Kim M. Low pH increases the yield of exosome isolation. Biochem Biophys Res Commun. 2015:461. 10.1016/j.bbrc.2015.03.172. [DOI] [PubMed] [Google Scholar]
- 10.Lai CP, Kim EY, Badr CE, et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nature Commun. 2015:6. 10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, Flores-Bellver M, Pan J, et al. Human retinal organoids release extracellular vesicles that regulate gene expression in target human retinal progenitor cells. Sci Rep. 2021:11. 10.1038/s41598-021-00542-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Jin X, Liang J, et al. Extracellular vesicles derived from ODN-stimulated macrophages transfer and activate Cdc42 in recipient cells and thereby increase cellular permissiveness to EV uptake. Sci Adv. 2019:5. 10.1126/sciadv.aav1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee E, Rodosthenous RS, Kujala V, et al. Circulating extracellular vesicles in human cardiorenal syndrome promote renal injury. medRxiv; 2023. 10.1101/2023.02.07.23285599, 2023.02.07.23285599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morillon A Long non-coding RNA. first. London, UK: Elsevier Ltd; 2018. [Google Scholar]
- 15.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. 10.1146/annurev-biochem-051410-092902, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattick JS. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. 10.1093/embo-reports/kve230, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarroux J, Morillon A, Pinskaya M. Long non coding RNA biology. Adv Exp Med Biol. 2017;1008:1–46. 10.1007/978-981-10-5203-3. [DOI] [PubMed] [Google Scholar]
- 18.Mattick JS, Makunin Iv. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No):17–29. [DOI] [PubMed] [Google Scholar]
- 19.Geisler S, Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhat SA, Ahmad SM, Mumtaz PT, et al. Long non-coding RNAs: Mechanism of action and functional utility. Noncoding RNA Res. 2016;1:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das S, Shah R, Dimmeler S, et al. Noncoding RNAs in cardiovascular disease: Current knowledge, tools and technologies for investigation, and future directions: A scientific statement from the American heart association. Circ Genom Precis Med. 2020:350–372. 10.1161/HCG.0000000000000062, [DOI] [PubMed] [Google Scholar]
- 22.Marguerat S, Bähler J. RNA-seq: From technology to biology. Cell Mol Life Sci. 2010;67:569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hon CC, Ramilowski JA, Harshbarger J, et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, Zhou L, Yu F, Zhang Y, Li P, Wang K. The functional roles of exosomal long non-coding RNAs in cancer. Cell Mol Life Sci. 2019;76(11):2059–2076. 10.1007/S00018-019-03018-3, 2019, 76, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor DH, Chu ETJ, Spektor R, Soloway PD. Long non-coding RNA regulation of reproduction and development. Mol Reprod Dev. 2015;82:932–956. 10.1002/MRD.22581, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miguel V, Lamas S, Espinosa-Diez C. Role of non-coding-RNAs in response to environmental stressors and consequences on human health. Redox Biol. 2020:37. 10.1016/J.REDOX.2020.101580, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nadhan R, Isidoro C, Song YS, Dhanasekaran DN. Signaling by LncRNAs: Structure, cellular homeostasis, and disease pathology. Cells. 2022:11. 10.3390/CELLS11162517, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bocchetti M, Scrima M, Melisi F, et al. LncRNAs and immunity: Coding the immune system with noncoding Oligonucleotides. Int J Mol Sci. 2021;22:1–30. 10.3390/IJMS22041741, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang N, Zhang X, Gu X, Li X, Shang L. Progress in understanding the role of lncRNA in programmed cell death. Cell Death Discov. 2021;7(1):1–11. 10.1038/s41420-021-00407-1, 2021, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlevaro-Fita J, Johnson R. Molecular cell review global positioning system: Understanding long noncoding RNAs through subcellular localization. Mol Cell. 2019;73:869–883. 10.1016/j.molcel.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Guh CY, Hsieh YH, Chu HP. Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. J Biomed Sci. 2020;27:1–14. 10.1186/S12929-020-00640-3/FIGURES/7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2020;22(2):96–118. 10.1038/s41580-020-00315-9, 2020, 22, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sas-Nowosielska H, Magalska A. Long noncoding RNAs—Crucial players organizing the landscape of the neuronal nucleus. Int J Mol Sci. 2021:22. 10.3390/IJMS22073478, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. 10.1007/978-981-10-5203-3_1/COVER, [DOI] [PubMed] [Google Scholar]
- 35.Rashid F, Shah A, Shan G. Long non-coding RNAs in the cytoplasm. Genom Proteom Bioinform. 2016;14:73–80. 10.1016/J.GPB.2016.03.005, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grote P, Wittler L, Hendrix D, et al. The tissue-specific lncRNA fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. 10.1016/j.devcel.2012.12.012, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chishima T, Iwakiri J, Hamada M. Identification of transposable elements contributing to tissue-specific expression of long non-coding RNAs. Genes. 2018;9:23. 10.3390/GENES9010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amin V, Harris RA, Onuchic V, et al. Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs. Nature Commun. 2015;6(1):1–10. 10.1038/ncomms7370, 2015, 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Goede OM, Nachun DC, Ferraro NM, et al. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell. 2021;184:2633–2648e19. 10.1016/J.CELL.2021.03.050/ATTACHMENT/8951D696-BB59-4013-8539-C8420C43EBB1/MMC8.PDF, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen F, Wang N, Tan HY, Guo W, Zhang C, Feng Y. The functional roles of exosomes-derived long non-coding RNA in human cancer, vol. 20. 2019:583–592. 10.1080/15384047.2018.1564562, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang M, Zhou L, Yu F, Zhang Y, Li P, Wang K. The functional roles of exosomal long non-coding RNAs in cancer. Cell Mol Life Sci. 2019;76(11):2059–2076. 10.1007/S00018-019-03018-3, 2019, 76, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan Z, Huang W. New developments in exosomal lncRNAs in cardiovascular diseases. Front Cardiovasc Med. 2021;8:750. 10.3389/FCVM.2021.709169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Y Exosomal lncRNAs from mesenchymal stem cells as the novel modulators to cardiovascular disease. Stem Cell Res Ther. 2020;11:1–2. 10.1186/S13287-020-01812-6/METRICS, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C, Ni YQ, Xu H, et al. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct Target Ther. 2021;6(1):1–31. 10.1038/s41392-021-00779-x, 2021, 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dolati S, Shakouri SK, Dolatkhah N, Yousefi M, Jadidi-Niaragh F, Sanaie S. The role of exosomal non-coding RNAs in aging-related diseases. BioFactors. 2021;47:292–310. 10.1002/BIOF.1715, [DOI] [PubMed] [Google Scholar]
- 46.Li Y, He X, Li Q, et al. EV-origin: Enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile. Comput Struct Biotechnol J. 2020:18. 10.1016/j.csbj.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rontogianni S, Synadaki E, Li B, et al. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun Biol. 2019;2(1):1–13. 10.1038/s42003-019-0570-8, 2019, 2, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muraoka S, Hirano M, Isoyama J, Nagayama S, Tomonaga T, Adachi J. Comprehensive proteomic profiling of plasma and serum phosphatidylserine-positive extracellular vesicles reveals tissue-specific proteins. IScience. 2022;25:104012. 10.1016/j.isci.2022.104012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li G, Manning AC, Bagi A, et al. Distinct stress-dependent signatures of cellular and extracellular tRNA-derived small RNAs. Adv Sci. 2022;9:2200829. 10.1002/ADVS.202200829, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barman B, Sung BH, Krystofiak E, et al. VAP-a and its binding partner CERT drive biogenesis of RNA-containing extracellular vesicles at ER membrane contact sites. Dev Cell. 2022;57:974–994e8. 10.1016/J.DEVCEL.2022.03.012, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cherbonneau F, Li G, Gokulnath P, et al. TRACE-seq: A transgenic system for unbiased and non-invasive transcriptome profiling of living cells. IScience. 2022;25:103806. 10.1016/J.ISCI.2022.103806, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Théry C, Witwer KW, Aikawa E, et al. (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. Minimal Inform Stud Extracell Vesicl J Extracell Vesicl. 2018;2018:7. 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;(1979):367. 10.1126/science.aau6977, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meldolesi J Extracellular vesicles (exosomes and ectosomes) play key roles in the pathology of brain diseases. Mol Biomed. 2021:2. 10.1186/s43556-021-00040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doyle L, Wang M. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019;8:727. 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q, Jeppesen DK, Higginbotham JN, et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat Cell Biol. 2021;23:1240–1254. 10.1038/s41556-021-00805-8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rak J, Guha A. Extracellular vesicles - vehicles that spread cancer genes. BioEssays. 2012;34:489–497. 10.1002/bies.201100169, [DOI] [PubMed] [Google Scholar]
- 58.Anand S, Samuel M, Mathivanan S. Exomeres: A new member of extracellular vesicles family. Subcell Biochem. 2021;97:89–97. 10.1007/978-3-030-67171-6_5, [DOI] [PubMed] [Google Scholar]
- 59.Winter D, Becker T, Clancy JW, Boomgarden AC. Profiling and promise of supermeres. Nat Cell Biol. 2021;22:1216–1223. 10.1038/s41556-021-00801-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Q, Jeppesen DK, Higginbotham JN, et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nature Cell Biol. 2022;23(12):1240–1254. 10.1038/s41556-021-00805-8, 2021, 23, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci U S A. 2012:109. 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc Natl Acad Sci U S A. 2012:109. 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han QF, Li WJ, Hu KS, et al. Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Molecular Cancer. 2022;21(1):1–26. 10.1186/S12943-022-01671-0, 2022, 21, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foot N, Henshall T, Kumar S. Ubiquitination and the regulation of membrane proteins. Physiol Rev. 2017;97:253–281. 10.1152/PHYSREV.00012.2016/ASSET/IMAGES/LARGE/Z9J0011727930004.JPEG, [DOI] [PubMed] [Google Scholar]
- 66.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. 10.1038/NATURE07961, [DOI] [PubMed] [Google Scholar]
- 67.Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP. Ubiquitin recognition by the human TSG101 protein. Mol Cell. 2004;13:783–789. 10.1016/S1097-2765(04)00129-7, [DOI] [PubMed] [Google Scholar]
- 68.Mizuno E, Kobayashi K, Yamamoto A, Kitamura N, Komada M. A deubiquitinating enzyme UBPY regulates the level of protein ubiquitination on endosomes. Traffic. 2006;7:1017–1031. 10.1111/J.1600-0854.2006.00452.X, [DOI] [PubMed] [Google Scholar]
- 69.Li J, Chai QY, Liu CH. The ubiquitin system: A critical regulator of innate immunity and pathogen-host interactions. Cell Mol Immunol. 2016;13:560–576. 10.1038/CMI.2016.40, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baietti MF, Zhang Z, Mortier E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012:14. 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 71.Ghossoub R, Lembo F, Rubio A, et al. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nature Commun. 2014:5. 10.1038/ncomms4477. [DOI] [PubMed] [Google Scholar]
- 72.Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 2017;27:172–188. 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mathieu M, Névo N, Jouve M, et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nature Commun. 2021:12. 10.1038/s41467-021-24384-2, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Janas T, Janas MM, Sapoń K, Janas T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015;589:1391–1398. 10.1016/J.FEBSLET.2015.04.036, [DOI] [PubMed] [Google Scholar]
- 75.Prieto-Vila M, Yoshioka Y, Ochiya T. Biological functions driven by mRNAs carried by extracellular vesicles in cancer. Front Cell Dev Biol. 2021:9. 10.3389/FCELL.2021.620498, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gokulnath P, Spanos M, Lehmann HI, et al. Plasma extracellular vesicle transcriptome as a dynamic liquid biopsy in acute heart failure. medRxiv; 2023. 10.1101/2023.02.17.23285936, 2023.02.17.23285936. [DOI] [Google Scholar]
- 77.Batagov AO, Kurochkin Iv. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3′ -untranslated regions. Biol Direct. 2013;8:12. 10.1186/1745-6150-8-12, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kossinova OA, Gopanenko Av, Tamkovich SN, et al. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim Biophys Acta Proteins Proteom. 2017;1865:664–673. 10.1016/J.BBAPAP.2017.03.010, [DOI] [PubMed] [Google Scholar]
- 79.Veziroglu EM, Mias GI. Characterizing extracellular vesicles and their diverse RNA contents. Front Genet. 2020;11:1–30. 10.3389/fgene.2020.00700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Squadrito ML, Baer C, Burdet F, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. 10.1016/J.CELREP.2014.07.035, [DOI] [PubMed] [Google Scholar]
- 81.Gezer U, Özgür E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38:1076–1079. 10.1002/CBIN.10301, [DOI] [PubMed] [Google Scholar]
- 82.Zhang R, Xia Y, Wang Z, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490:406–414. 10.1016/J.BBRC.2017.06.055, [DOI] [PubMed] [Google Scholar]
- 83.Qiu Y, Li P, Zhang Z, Wu M. Insights into exosomal non-coding RNAs sorting mechanism and clinical application. Front Oncol. 2021:11. 10.3389/FONC.2021.664904, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen T, Gu C, Xue C, et al. LncRNA-uc002mbe.2 interacting with hnrnpa2b1 mediates AKT deactivation and p21 up-regulation induced by trichostatin in liver cancer cells. Front Pharmacol. 2017:8. 10.3389/fphar.2017.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Grady T, Njock M-S, Lion M, et al. Sorting and packaging of RNA into extracellular vesicles shape intracellular transcript levels. BMC Biol. 2022;20:1–21. 10.1186/S12915-022-01277-4/TABLES/1, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Corrado C, Barreca MM, Zichittella C, Alessandro R, Conigliaro A. Molecular mediators of RNA loading into extracellular vesicles. Cells. 2021;10:3355. 10.3390/CELLS10123355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen C, Zheng H, Luo Y, et al. Sumoylation promotes extracellular vesicle–mediated transmission of lncRNA ELNAT1 and lymph node metastasis in bladder cancer. J Clin Invest. 2021:131. 10.1172/JCI146431, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jean-Philippe J, Paz S, Caputi M. hnRNP A1: The Swiss army knife of gene expression. Int J Mol Sci. 2013;14:18999. 10.3390/IJMS140918999, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen C, Huang L, Sun Q, Yu Z, Wang X, Bu L. HNRNPA2B1 demonstrates diagnostic and prognostic values based on pan-cancer analyses. Comput Math Methods Med. 2022:2022. 10.1155/2022/9867660, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kossinova OA, Gopanenko AV, Tamkovich SN, et al. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim Biophys Acta Proteins Proteom. 2017;1865:664–673. 10.1016/J.BBAPAP.2017.03.010, [DOI] [PubMed] [Google Scholar]
- 91.Lennon KM, Saftics A, Abuelreich S, et al. Cardiac troponin T in extracellular vesicles as a novel biomarker in human cardiovascular disease. Clin Transl Med. 2022:12. 10.1002/CTM2.979, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rodosthenous RS, Hutchins E, Reiman R, et al. Profiling extracellular long RNA transcriptome in human plasma and extracellular vesicles for biomarker discovery. IScience. 2020:23. 10.1016/J.ISCI.2020.101182, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. 10.1016/J.CELL.2016.11.037, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hassanpour SH, Dehghani M. Review of cancer from perspective of molecular. J Cancer Res Pract. 2017;4:127–129. 10.1016/j.jcrpr.2017.07.001. [DOI] [Google Scholar]
- 95.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017:66. 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 96.Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: A Hallmark of cancer revisited. Signal Transduct Target Ther. 2020;5(1):1–17. 10.1038/s41392-020-0134-x, 2020, 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. 10.1016/J.CELL.2016.11.037, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trosko JE, Chang CC, Upham BL, Tai MH. Ignored Hallmarks of carcinogenesis: Stem cells and cell-cell communication. Ann N Y Acad Sci. 2004;1028:192–201. 10.1196/ANNALS.1322.023, [DOI] [PubMed] [Google Scholar]
- 99.Wan Z, Gao X, Dong Y, et al. Exosome-mediated cell–cell communication in tumor progression. Am J Cancer Res. 2018;8:1661. [PMC free article] [PubMed] [Google Scholar]
- 100.O’Neill CP, Gilligan KE, Dwyer RM. Role of extracellular vesicles (EVs) in cell stress response and resistance to cancer therapy. Cancers (Basel). 2019;11:136. 10.3390/cancers11020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560(7718):382–386. 10.1038/s41586-018-0392-8, 2018, 560, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang H, Lu J, Liu J, Zhang G, Lu A. Advances in the discovery of exosome inhibitors in cancer. J Enzyme Inhib Med Chem. 2020;35:1322–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Datta A, Kim H, McGee L, et al. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: A drug repurposing strategy for advanced cancer. Sci Rep. 2018;8(1):1–13. 10.1038/s41598-018-26411-7, 2018, 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bebelman MP, Janssen E, Pegtel DM, Crudden C. The forces driving cancer extracellular vesicle secretion. Neoplasia (United States). 2021;23:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Entezari M, Ghanbarirad M, Taheriazam A, et al. Long non-coding RNAs and exosomal lncRNAs: Potential functions in lung cancer progression, drug resistance and tumor microenvironment remodeling. Biomed Pharmacother. 2022;150:112963. 10.1016/J.BIOPHA.2022.112963, [DOI] [PubMed] [Google Scholar]
- 106.Zhang R, Xia Y, Wang Z, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490:406–414. 10.1016/J.BBRC.2017.06.055, [DOI] [PubMed] [Google Scholar]
- 107.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017:66. 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 108.Editors GAC. Non-coding RNAs in colorectal cancer. 2016.
- 109.Liu L, Meng T, Yang XH, et al. Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark. 2018;22:283–299. 10.3233/CBM-171011, [DOI] [PubMed] [Google Scholar]
- 110.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics 2022. CA Cancer J Clin. 2022;72:7–33. 10.3322/CAAC.21708, [DOI] [PubMed] [Google Scholar]
- 111.Iacoviello L, Bonaccio M, de Gaetano G, Donati MB. Epidemiology of breast cancer, a paradigm of the common soil hypothesis. Semin Cancer Biol. 2021:72. [DOI] [PubMed] [Google Scholar]
- 112.Zhang R, Xia Y, Wang Z, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490:406–414. 10.1016/J.BBRC.2017.06.055, [DOI] [PubMed] [Google Scholar]
- 113.Rong F, Liu L, Zou C, Zeng J, Xu Y. MALAT1 promotes cell tumorigenicity through regulating miR-515–5p/EEF2 axis in non-small cell lung cancer. Cancer Manag Res. 2020;12:7691. 10.2147/CMAR.S242425, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang X, Guo H, Bao Y, Yu H, Xie D, Wang X. Exosomal long non-coding RNA dlx6-as1 as a potential diagnostic biomarker for non-small cell lung cancer. Oncol Lett. 2019;18:5197–5204. 10.3892/OL.2019.10892/HTML. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lv X, Lian Y, Liu Z, Jianguang X, Zhang D, Yin X. Exosomal long non-coding RNA LINC00662 promotes non-small cell lung cancer progression by miR-320d/E2F1 axis. Aging (Albany NY). 2021;13:6010. 10.18632/AGING.202522, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zang X, Gu J, Zhang J, et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020;11(4):1–13. 10.1038/s41419-020-2409-0, 11, 2020, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ni J, Zhang X, Li J, et al. Tumour-derived exosomal lncRNA-SOX2OT promotes bone metastasis of non-small cell lung cancer by targeting the miRNA-194–5p/RAC1 signalling axis in osteoclasts. Cell Death Dis. 2021;12(7):1–10. 10.1038/s41419-021-03928-w, 12, 2021, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pan R, Zhou H. Exosomal transfer of lncRNA H19 promotes erlotinib resistance in non-small cell lung cancer via miR-615–3p/ATG7 axis. Cancer Manag Res. 2020;12:4283. 10.2147/CMAR.S241095, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang W, Cai X, Yu J, Lu X, Qian Q, Qian W. Exosome-mediated transfer of lncRNA RP11–838n2.4 promotes erlotinib resistance in non-small cell lung cancer. Int J Oncol. 2018;53:527–538. 10.3892/IJO.2018.4412, [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120.Han B, Marrades RM, Viñolas N, et al. Monitoring HOTTIP levels on extracellular vesicles for predicting recurrence in surgical non-small cell lung cancer patients. Transl Oncol. 2021;14. 10.1016/j.tranon.2021.101144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li C, Lv Y, Shao C, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234:20721–20727. 10.1002/JCP.28678, [DOI] [PubMed] [Google Scholar]
- 122.Tao Y, Tang Y, Yang Z, et al. Exploration of serum exosomal LncRNA TBILA and AGAP2-AS1 as promising biomarkers for diagnosis of non-small cell lung cancer. Int J Biol Sci. 2020;16:471. 10.7150/IJBS.39123, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Teng Y, Kang H, Chu Y. Identification of an exosomal long noncoding RNA sox2-ot in plasma as a promising biomarker for lung squamous cell carcinoma. Genet Test Mol Biomarkers. 2019;23:235–240. 10.1089/GTMB.2018.0103/ASSET/IMAGES/LARGE/FIGURE5.JPEG, [DOI] [PubMed] [Google Scholar]
- 124.Barbagallo C, Brex D, Caponnetto A, et al. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Ther Nucleic Acids. 2018;12:229–241. 10.1016/J.OMTN.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551–85563. 10.18632/oncotarget.13465, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhou L, Li J, Tang Y, Yang M. Exosomal LncRNA LINC00659 transferred from cancer-associated fibroblasts promotes colorectal cancer cell progression via miR-342–3p/ANXA2 axis. J Transl Med. 2021;19:1–14. 10.1186/s12967-020-02648-7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li N, Li J, Mi Q, et al. Long non-coding RNA ADAMTS9-AS1 suppresses colorectal cancer by inhibiting the Wnt/β-catenin signalling pathway and is a potential diagnostic biomarker. J Cell Mol Med. 2020;24:11318–11329. 10.1111/JCMM.15713, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oehme F, Krahl S, Gyorffy B, et al. Low level of exosomal long non-coding RNA HOTTIP is a prognostic biomarker in colorectal cancer. RNA Biol. 2019;16:1339–1345. 10.1080/15476286.2019.1637697/SUPPL_FILE/KRNB_A_1637697_SM3834.ZIP, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Deng X, Ruan H, Zhang X, et al. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int J Cancer. 2020;146:1700–1716. 10.1002/IJC.32608, [DOI] [PubMed] [Google Scholar]
- 130.Ren J, Ding L, Zhang D, et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8. 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu L, Meng T, Yang XH, et al. Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark. 2018;22:283–299. 10.3233/CBM-171011, [DOI] [PubMed] [Google Scholar]
- 132.Yu J, Dong W, Liang J. Extracellular vesicle-transported long non-coding RNA (LncRNA) X inactive-specific transcript (XIST) in serum is a potential novel biomarker for colorectal cancer diagnosis. Med Sci Monit. 2020;26:e924448–1. 10.12659/MSM.924448, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang K, Luo Z, Zhang Y, et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomarkers. 2016;17:187–194. 10.3233/CBM-160630, [DOI] [PubMed] [Google Scholar]
- 134.Wang X, Pei X, Guo G, et al. Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J Cell Physiol. 2020;235:6896–6904. 10.1002/JCP.29585, [DOI] [PubMed] [Google Scholar]
- 135.Lan F, Zhang X, Li H, Yue X, Sun Q. Serum exosomal lncRNA XIST is a potential non-invasive biomarker to diagnose recurrence of triple-negative breast cancer. J Cell Mol Med. 2021;25:7602–7607. 10.1111/JCMM.16009, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tang S, Zheng K, Tang Y, Li Z, Zou T, Liu D. Overexpression of serum exosomal HOTAIR is correlated with poor survival and poor response to chemotherapy in breast cancer patients. J Biosci. 2019;44(2):1–8. 10.1007/S12038-019-9861-Y, 44, 2019, [DOI] [PubMed] [Google Scholar]
- 137.Shi W, Jin X, Wang Y, Zhang Q, Yang L. High serum exosomal long non-coding RNA DANCR expression confers poor prognosis in patients with breast cancer. J Clin Lab Anal. 2022;36:e24186. 10.1002/JCLA.24186, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhong G, Wang K, Li J, Xiao S, Wei W, Liu J. Determination of serum exosomal H19 as a noninvasive biomarker for breast cancer diagnosis. Onco Targets Ther. 2020;13:2563. 10.2147/OTT.S243601, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Qian X, Qu H, Zhang F, et al. Exosomal long noncoding RNA AGAP2-AS1 regulates trastuzumab resistance via inducing autophagy in breast cancer. Am J Cancer Res. 2021;11:1962. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Zhang P, Zhou H, Lu K, Lu Y, Wang Y, Feng T. Exosome-mediated delivery of MALATI induces cell proliferation in breast cancer. Onco Targets Ther. 2018;11. 10.2147/OTT.S155134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Na-er A, Xu YY, Liu YH, Gan YJ. Upregulation of serum exosomal SUMO1P3 predicts unfavorable prognosis in triple negative breast cancer. Eur Rev Med Pharmacol Sci. 2021;25:154–160. 10.26355/eurrev_202101_24379, [DOI] [PubMed] [Google Scholar]
- 142.Sun Y, Ma L. New insights into long non-coding RNA MALAT1 in cancer and metastasis. Cancers (Basel). 2019:11. 10.3390/CANCERS11020216, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang R, Xia Y, Wang Z, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490:406–414. 10.1016/J.BBRC.2017.06.055, [DOI] [PubMed] [Google Scholar]
- 144.Rong F, Liu L, Zou C, Zeng J, Xu Y. Malat1 promotes cell tumorigenicity through regulating mir-515–5p/eef2 axis in non-small cell lung cancer. Cancer Manag Res. 2020;12:7691–7701. 10.2147/CMAR.S242425, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tao Y, Tang Y, Yang Z, et al. Exploration of serum exosomal LncRNA TBILA and AGAP2-AS1 as promising biomarkers for diagnosis of non-small cell lung cancer. Int J Biol Sci. 2020;16:471. 10.7150/IJBS.39123, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang YL, Liu LC, Hung Y, et al. Long non-coding RNA HOTAIR in circulatory exosomes is correlated with ErbB2/HER2 positivity in breast cancer. Breast. 2019;46:64–69. 10.1016/j.breast.2019.05.003, [DOI] [PubMed] [Google Scholar]
- 147.Xu J, Xiao Y, Liu B, et al. Exosomal MALAT1 sponges miR-26a/26b to promote the invasion and metastasis of colorectal cancer via FUT4 enhanced fucosylation and PI3K/Akt pathway. J Exper Clin Cancer Res. 2020;39:1–15. 10.1186/S13046-020-01562-6/FIGURES/6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang X, Guo H, Bao Y, Yu H, Xie D, Wang X. Exosomal long non-coding RNA dlx6-as1 as a potential diagnostic biomarker for non-small cell lung cancer. Oncol Lett. 2019;18:5197–5204. 10.3892/OL.2019.10892/HTML. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li C, Lv Y, Shao C, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234:20721–20727. 10.1002/JCP.28678, [DOI] [PubMed] [Google Scholar]
- 150.Li C, Lv Y, Shao C, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234:20721–20727. 10.1002/JCP.28678, [DOI] [PubMed] [Google Scholar]
- 151.Pan R, Zhou H. Exosomal transfer of lncRNA H19 promotes erlotinib resistance in non-small cell lung cancer via miR-615–3p/ATG7 axis. Cancer Manag Res. 2020;12:4283. 10.2147/CMAR.S241095, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang W, Cai X, Yu J, Lu X, Qian Q, Qian W. Exosome-mediated transfer of lncRNA RP11–838n2.4 promotes erlotinib resistance in non-small cell lung cancer. Int J Oncol. 2018;53:527–538. 10.3892/IJO.2018.4412, [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 153.Lei Y, Guo W, Chen B, Chen L, Gong J, Li W. Tumor-released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non-small cell lung cancer. Oncol Rep. 2018;40:3438. 10.3892/OR.2018.6762, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Han B, Marrades RM, Viñolas N, et al. Monitoring HOTTIP levels on extracellular vesicles for predicting recurrence in surgical non-small cell lung cancer patients. Transl Oncol. 2021;14:101144. 10.1016/J.TRANON.2021.101144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhou L, Li J, Tang Y, Yang M. Exosomal LncRNA LINC00659 transferred from cancer-associated fibroblasts promotes colorectal cancer cell progression via miR-342–3p/ANXA2 axis. J Transl Med. 2021;19:1–14. 10.1186/s12967-020-02648-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Deng X, Ruan H, Zhang X, et al. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int J Cancer. 2020;146:1700–1716. 10.1002/IJC.32608, [DOI] [PubMed] [Google Scholar]
- 157.Zhang K, Luo Z, Zhang Y, et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. 2016;17:187–194. 10.3233/CBM-160630, [DOI] [PubMed] [Google Scholar]
- 158.Zhong G, Wang K, Li J, Xiao S, Wei W, Liu J. Determination of Serum exosomal H19 as a noninvasive biomarker for breast cancer diagnosis. Onco Targets Ther. 2020;13:2563. 10.2147/OTT.S243601, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang X, Pei X, Guo G, et al. Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J Cell Physiol. 2020;235:6896–6904. 10.1002/JCP.29585, [DOI] [PubMed] [Google Scholar]
- 160.Özgür E, Ferhatoǧlu F, Sen F, Saip P, Gezer U. Circulating lncRNA H19 may be a useful marker of response to neoadjuvant chemotherapy in breast cancer. Cancer Biomark. 2020;27:11–17. 10.3233/CBM-190085, [DOI] [PubMed] [Google Scholar]
- 161.Na-er A, Xu YY, Liu YH, Gan YJ. Upregulation of serum exosomal SUMO1p3 predicts unfavorable prognosis in triple negative breast cancer. Eur Rev Med Pharmacol Sci. 2021;25:154–160. 10.26355/eurrev_202101_24379, [DOI] [PubMed] [Google Scholar]
- 162.Han M, Gu Y, Lu P, et al. Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol Cancer. 2020;19:1–18. 10.1186/S12943-020-1145-5/FIGURES/9, [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 163.Xu CG, Yang MF, Ren YQ, Wu CH, Wang LQ. Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol Sci. 2016;20:4362–4368. [PubMed] [Google Scholar]
- 164.Barbagallo C, Brex D, Caponnetto A, et al. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Ther Nucleic Acids. 2018;12:229–241. 10.1016/J.OMTN.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Maeda T, Konishi H, Takaki W, et al. Significance of plasma UCA1 for predicting colorectal cancer and BRAF mutation status. Anticancer Res. 2021;41:1761–1769. 10.21873/ANTICANRES.14941, [DOI] [PubMed] [Google Scholar]
- 166.Luan Y, Li X, Luan Y, et al. Circulating lncRNA UCA1 promotes malignancy of colorectal cancer via the miR-143/MYO6 axis. Mol Ther Nucleic Acids. 2020;19:790. 10.1016/J.OMTN.2019.12.009, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang YN, Zhang R, Du JW, et al. Predictive role of UCA1-containing exosomes in cetuximab-resistant colorectal cancer. Cancer Cell Int. 2018;18:164. 10.1186/S12935-018-0660-6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zang X, Gu J, Zhang J, et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020;11(4):1–13. 10.1038/s41419-020-2409-0, 2020, 11, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551–85563. 10.18632/oncotarget.13465, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sun J, Jia H, Bao X, et al. Tumor exosome promotes Th17 cell differentiation by transmitting the lncRNA CRNDE-h in colorectal cancer. Cell Death Dis. 2021;12(1):1–14. 10.1038/s41419-020-03376-y, 2021, 12, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xian D, Niu L, Zeng J, Wang L. LncRNA KCNQ1OT1 secreted by tumor cell-derived exosomes mediates immune escape in colorectal cancer by regulating PD-L1 ubiquitination via MiR-30a-5p/USP22. Front Cell Dev Biol. 2021:9. 10.3389/fcell.2021.653808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stanton SE, Disis ML. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J Immunother Cancer. 2016;4:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ni C, Fang QQ, Chen WZ, et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+γ δ1 treg cells. Signal Transduct Target Ther. 2020;5(1):1–14. 10.1038/s41392-020-0129-7, 2020, 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu L, Meng T, Yang XH, et al. Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark. 2018;22:283–299. 10.3233/CBM-171011, [DOI] [PubMed] [Google Scholar]
- 175.Ni J, Zhang X, Li J, et al. Tumour-derived exosomal lncRNA-SOX2OT promotes bone metastasis of non-small cell lung cancer by targeting the miRNA-194–5p/RAC1 signalling axis in osteoclasts. Cell Death Dis. 2021;12(7):1–10. 10.1038/s41419-021-03928-w, 2021, 12, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Teng Y, Kang H, Chu Y. Identification of an exosomal long noncoding RNA sox2-ot in plasma as a promising biomarker for lung squamous cell carcinoma. Genet Test Mol Biomark. 2019;23:235–240. 10.1089/GTMB.2018.0103/ASSET/IMAGES/LARGE/FIGURE5.JPEG, [DOI] [PubMed] [Google Scholar]
- 177.Lu Y, Chen L, Li L, Cao Y. Exosomes derived from brain metastatic breast cancer cells destroy the blood-brain barrier by carrying lncRNA GS1–600g8.5. Biomed Res Int. 2020:2020. 10.1155/2020/7461727, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.El-Ashmawy NE, Hussien FZ, El-Feky OA, Hamouda SM, Al-Ashmawy GM. Serum LncRNA-ATB and FAM83H-AS1 as diagnostic/prognostic non-invasive biomarkers for breast cancer. Life Sci. 2020;259:118193. 10.1016/J.LFS.2020.118193, [DOI] [PubMed] [Google Scholar]
- 179.Lan F, Zhang X, Li H, Yue X, Sun Q. Serum exosomal lncRNA XIST is a potential non-invasive biomarker to diagnose recurrence of triple-negative breast cancer. J Cell Mol Med. 2021;25:7602–7607. 10.1111/JCMM.16009, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tang S, Zheng K, Tang Y, Li Z, Zou T, Liu D. Overexpression of serum exosomal HOTAIR is correlated with poor survival and poor response to chemotherapy in breast cancer patients. J Biosci. 2019;44(2):1–8. 10.1007/S12038-019-9861-Y, 2019, 44, [DOI] [PubMed] [Google Scholar]
- 181.Wang YL, Liu LC, Hung Y, et al. Long non-coding RNA HOTAIR in circulatory exosomes is correlated with ErbB2/HER2 positivity in breast cancer. Breast. 2019;46:64–69. 10.1016/j.breast.2019.05.003, [DOI] [PubMed] [Google Scholar]
- 182.Lai SW, Chen MY, Bamodu OA, et al. Exosomal lncRNA PVT1/VEGFA axis promotes colon cancer metastasis and stemness by downregulation of tumor suppressor miR-152–3p. Oxid Med Cell Longev. 2021:2021. 10.1155/2021/9959807, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Roth GA, Mensah GA, Johnson CO, et al. Global Burden of cardiovascular diseases and risk factors, 1990–2019: Update from the GBD 2019 study. J Am Coll Cardiol. 2020:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Townsend N, Kazakiewicz D, Lucy Wright F, et al. Epidemiology of cardiovascular disease in Europe. Nat Rev Cardiol. 2022;19:133–143. 10.1038/s41569-021-00607-3. [DOI] [PubMed] [Google Scholar]
- 185.Lee Y-TH, Fang J, Schieb L, Park S, Casper M, Gillespie C. Prevalence and trends of coronary heart disease in the United States, 2011 to 2018. JAMA Cardiol. 2022;7:459. 10.1001/jamacardio.2021.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Everaert C, Helsmoortel H, Decock A, et al. Performance assessment of total RNA sequencing of human biofluids and extracellular vesicles. Sci Rep. 2019:9. 10.1038/s41598-019-53892-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Song Y, Li H, Ren X, Li H, Feng C. SNHG9, delivered by adipocyte-derived exosomes, alleviates inflammation and apoptosis of endothelial cells through suppressing TRADD expression. Eur J Pharmacol. 2020;872. 10.1016/J.EJPHAR.2020.172977, [DOI] [PubMed] [Google Scholar]
- 188.Hosen MR, Li Q, Liu Y, et al. CAD increases the long noncoding RNA PUNISHER in small extracellular vesicles and regulates endothelial cell function via vesicular shuttling. Mol Ther Nucleic Acids. 2021:25. 10.1016/j.omtn.2021.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yang Y, Cai Y, Wu G, et al. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin Sci. 2015:129. 10.1042/CS20150121. [DOI] [PubMed] [Google Scholar]
- 190.Wu L, Tan G, Li X, et al. LncRNA TONSL-AS1 participates in coronary artery disease by interacting with miR-197. Microvasc Res. 2021;136. 10.1016/j.mvr.2021.104152. [DOI] [PubMed] [Google Scholar]
- 191.Li H, Zhu X, Hu L, Li Q, Ma J, Yan J. Loss of exosomal MALAT1 from oxLDL-treated vascular endothelial cells induces maturation of dendritic cells in atherosclerosis development. Cell Cycle. 2019;18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 192.Huang C, Han J, Wu Y, et al. Exosomal MALAT1 derived from oxidized low-density lipoprotein-treated endothelial cells promotes M2 macrophage polarization. Mol Med Rep. 2018;18. 10.3892/mmr.2018.8982. [DOI] [PubMed] [Google Scholar]
- 193.Shaath H, Vishnubalaji R, Elkord E, Alajez NM. Single-cell transcriptome analysis highlights a role for neutrophils and inflammatory macrophages in the pathogenesis of severe COVID-19. Cells. 2020;9. 10.3390/cells9112374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang Y, Liang J, Xu J, et al. Circulating exosomes and exosomal lncRNA HIF1A-AS1 in atherosclerosis. Int J Clin Exp Pathol. 2017;10. [PMC free article] [PubMed] [Google Scholar]
- 195.He Q, Tan J, Yu B, Shi W, Liang K. Long noncoding RNA HIF1A-AS1A reduces apoptosis of vascular smooth muscle cells: Implications for the pathogenesis of thoracoabdominal aorta aneurysm. Pharmazie. 2015;70. 10.1691/ph.2015.4830. [DOI] [PubMed] [Google Scholar]
- 196.Wang J, Chen L, Li H, et al. Clopidogrel reduces apoptosis and promotes proliferation of human vascular endothelial cells induced by palmitic acid via suppression of the long non-coding RNA HIF1A-AS1 in vitro. Mol Cell Biochem. 2015;404. 10.1007/s11010-015-2379-1. [DOI] [PubMed] [Google Scholar]
- 197.Wang S, Zhang X, Yuan Y, et al. BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding RNA HIF1A-AS1 in vitro. Eur J Cardio-Thoracic Surg. 2015;47. 10.1093/ejcts/ezu215. [DOI] [PubMed] [Google Scholar]
- 198.Gabory A, Jammes H, Dandolo L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. BioEssays. 2010:32. [DOI] [PubMed] [Google Scholar]
- 199.Han DKM, Khaing ZZ, Pollock RA, Haudenschild CC, Liau G. H19, a marker of developmental transition, is reexpressed in human atherosclerotic plaques and is regulated by the insulin family of growth factors in cultured rabbit smooth muscle cells. J Clin Invest. 1996:97. 10.1172/JCI118543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhuang Y, Li T, Xiao H, et al. LncRNA-H19 drives cardiomyocyte senescence by targeting miR-19a/socs1/p53 axis. Front Pharmacol. 2021:12. 10.3389/fphar.2021.631835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bitarafan S, Yari M, Broum MA, et al. Association of increased levels of lncRNA H19 in PBMCs with risk of coronary artery disease. Cell J. 2019:20. 10.22074/cellj.2019.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhang Z, Gao W, Long QQ, et al. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci Rep. 2017:7. 10.1038/s41598-017-07611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li X, Zhao Z, Gao C, et al. Identification of a peripheral blood long non-coding RNA (upperhand) as a potential diagnostic marker of coronary artery disease. Cardiol J. 2018:25. 10.5603/CJ.a2017.0133. [DOI] [PubMed] [Google Scholar]
- 205.Hennessy EJ, van Solingen C, Scacalossi KR, et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primates. Nat Metab. 2019;1. 10.1038/s42255-018-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen Z, Yan Y, Wu J, Qi C, Liu J, Wang J. Expression level and diagnostic value of exosomal NEAT1/mir-204/MMP-9 in acute ST-segment elevation myocardial infarction. IUBMB Life. 2020;72. 10.1002/iub.2376. [DOI] [PubMed] [Google Scholar]
- 207.Luo M, Sun Q, Zhao H, Tao J, Yan D. Long noncoding RNA NEAT1 sponges miR-495-3p to enhance myocardial ischemia-reperfusion injury via MAPK6 activation. J Cell Physiol. 2020;235. 10.1002/jcp.28791. [DOI] [PubMed] [Google Scholar]
- 208.Wang L, Xia JW, Ke ZP, Zhang BH. Blockade of NEAT1 represses inflammation response and lipid uptake via modulating miR-342–3p in human macrophages THP-1 cells. J Cell Physiol. 2019;234. 10.1002/jcp.27340. [DOI] [PubMed] [Google Scholar]
- 209.Zhang Y, Sun L, Xuan L, et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep. 2016:6. 10.1038/srep22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhang Y, Jiao L, Sun L, et al. LncRNA ZFAS1 as a SERCA2a inhibitor to cause intracellular Ca 2+ overload and contractile dysfunction in a mouse model of myocardial infarction. Circ Res. 2018;122:1354–1368. 10.1161/CIRCRESAHA.117.312117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jiao L, Li M, Shao Y, et al. lncRNA-ZFAS1 induces mitochondria-mediated apoptosis by causing cytosolic Ca2+ overload in myocardial infarction mice model. Cell Death Dis. 2019:10. 10.1038/s41419-019-2136-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chang G, Zhang W, Zhang M, Ding G. Clinical value of circulating ZFAS1 and miR-590–3p in the diagnosis and prognosis of chronic heart failure. Cardiovasc Toxicol. 2021:21. 10.1007/s12012-021-09678-7. [DOI] [PubMed] [Google Scholar]
- 213.Geng H-H, Li R, Su Y-M, et al. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11:e0151753. 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Azat M, Huojiahemaiti X, Gao R, Peng P. Long noncoding RNA MIAT: A potential role in the diagnosis and mediation of acute myocardial infarction. Mol Med Rep. 2019:20. 10.3892/mmr.2019.10768. [DOI] [PubMed] [Google Scholar]
- 215.Zhou X, Zhang W, Jin M, Chen J, Xu W, Kong X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017:8. 10.1038/cddis.2017.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Chen Y, Chen X, Li H, et al. Serum extracellular vesicles containing MIAT induces atrial fibrosis, inflammation and oxidative stress to promote atrial remodeling and atrial fibrillation via blockade of miR-485–5p-mediated CXCL10 inhibition. Clin Transl Med. 2021:11. 10.1002/ctm2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sun L, Zhu W, Zhao P, et al. Long noncoding RNA UCA1 from hypoxia-conditioned hMSC-derived exosomes: A novel molecular target for cardioprotection through miR-873–5p/XIAP axis. Cell Death Dis. 2020:11. 10.1038/s41419-020-02783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Liu L, Cui S, Wan T, et al. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote glioma progression by sponging miR-126–5p. J Cell Physiol. 2018:233. 10.1002/jcp.26432. [DOI] [PubMed] [Google Scholar]
- 219.Gao L, Liu Y, Guo S, et al. Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell Physiol Biochem. 2017:44. 10.1159/000485588. [DOI] [PubMed] [Google Scholar]
- 220.Wang X, Wang L, Ma Z, et al. Early expressed circulating long noncoding RNA CHAST is associated with cardiac contractile function in patients with acute myocardial infarction. Int J Cardiol. 2020:302. 10.1016/j.ijcard.2019.12.058. [DOI] [PubMed] [Google Scholar]
- 221.Viereck J, Kumarswamy R, Foinquinos A, et al. Long noncoding RNA chast promotes cardiac remodeling - supplementary materials. Sci Transl Med. 2016:8. [DOI] [PubMed] [Google Scholar]
- 222.Xuan L, Sun L, Zhang Y, et al. Circulating long non-coding RNAs NRON and MHRT as novel predictive biomarkers of heart failure. J Cell Mol Med. 2017:21. 10.1111/jcmm.13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hoepfner J, Leonardy J, Lu D, et al. The long non-coding RNA NRON promotes the development of cardiac hypertrophy in the murine heart. Mol Ther. 2022:30. 10.1016/j.ymthe.2021.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Han P, Li W, Lin CH, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014:514. 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Xu Y, Luo Y, Liang C, Zhang T. LncRNA-Mhrt regulates cardiac hypertrophy by modulating the miR-145a-5p/KLF4/myocardin axis. J Mol Cell Cardiol. 2020:139. 10.1016/j.yjmcc.2019.12.013. [DOI] [PubMed] [Google Scholar]
- 226.Lang M, Ou D, Liu Z, Li Y, Zhang X, Zhang F. LncRNA MHRT promotes cardiac fibrosis via miR-3185 pathway following myocardial infarction. Int Heart J. 2021:62. 10.1536/ihj.20-298. [DOI] [PubMed] [Google Scholar]
- 227.Kumarswamy R, Bauters C, Volkmann I, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014:114. 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 228.Li M, Wang LF, Yang XC, et al. Circulating long noncoding RNA LIPCAR acts as a novel biomarker in patients with ST-segment elevation myocardial infarction. Med Sci Monit. 2018:24. 10.12659/MSM.909348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Meessen JMTA, Bär C, di Dona FM, et al. LIPCAR is increased in chronic symptomatic HF patients. a sub-study of the GISSI-HF trial. Clin Chem. 2021:67. 10.1093/clinchem/hvab197. [DOI] [PubMed] [Google Scholar]
- 230.Yan L, Zhang Y, Wang M, Wang L, Zhang W, Ge ZR. Circulating LIPCAR is a potential biomarker of heart failure in patients post-acute myocardial infarction. Exp Biol Med. 2021:246. 10.1177/15353702211036055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ketchum ES, Dickstein K, Kjekshus J, et al. The seattle post myocardial infarction model (SPIM): Prediction of mortality after acute myocardial infarction with left ventricular dysfunction. Eur Heart J Acute Cardiovasc Care. 2014:3. 10.1177/2048872613502283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Golia E Adipose tissue and vascular inflammation in coronary artery disease. World J Cardiol. 2014:6. 10.4330/wjc.v6.i7.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hosen MR, Li Q, Liu Y, et al. CAD increases the long noncoding RNA PUNISHER in small extracellular vesicles and regulates endothelial cell function via vesicular shuttling. Mol Ther Nucleic Acids. 2021;25:388–405. 10.1016/j.omtn.2021.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Arnold KA, Blair JE, Paul JD, Shah AP, Nathan S, Alenghat FJ. Monocyte and macrophage subtypes as paired cell biomarkers for coronary artery disease. Exp Physiol. 2019:104. 10.1113/EP087827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Chen D, Lu T, Tan J, Li H, Wang Q, Wei L. Long non-coding RNAs as communicators and mediators between the tumor microenvironment and cancer cells. Front Oncol. 2019:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Palasubramaniam J, Wang X, Peter K. Myocardial infarction—From atherosclerosis to thrombosis. Arterioscler Thromb Vasc Biol. 2019:39. 10.1161/atvbaha.119.312578. [DOI] [PubMed] [Google Scholar]
- 237.Fang Y, Xu Y, Wang R, et al. Recent advances on the roles of LncRNAs in cardiovascular disease. J Cell Mol Med. 2020:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wang M, Zhang Z, Pan D, et al. Circulating lncRNA UCA1 and lncRNA PGM5-AS1 act as potential diagnostic biomarkers for early-stage colorectal cancer. Biosci Rep. 2021:41. 10.1042/BSR20211115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jian F, Yangyang R, Wei X, et al. The prognostic and predictive significance of circRNA CDR1as in tumor progression. Front Oncol. 2021:10. 10.3389/fonc.2020.549982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.He A, He S, Li X, Zhou L. ZFAS1: A novel vital oncogenic lncRNA in multiple human cancers. Cell Prolif. 2019:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Rezaeinejad F, Mirzaei A, Khalvati B, Sabz G, Alipoor B. Circulating expression levels of CircHIPK3 and CDR1as circular-RNAs in type 2 diabetes patients. Mol Biol Rep. 2022:49. 10.1007/s11033-021-06850-8. [DOI] [PubMed] [Google Scholar]
- 242.Lago RM, Nesto RW. Type 2 diabetes and coronary heart disease: Focus on myocardial infarction. Curr Diab Rep. 2009:9. [DOI] [PubMed] [Google Scholar]
- 243.Amiri M, Mokhtari MJ, Bayat M, et al. Expression and diagnostic values of MIAT, H19, and NRON long non-coding RNAs in multiple sclerosis patients. Egypt J Med Human Genet. 2022:23. 10.1186/s43042-022-00260-6. [DOI] [Google Scholar]
- 244.Hajjari M, Salavaty A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Gerber Y, Weston SA, Enriquez-Sarano M, et al. Atherosclerotic burden and heart failure after myocardial infarction. JAMA Cardiol. 2016:1. 10.1001/jamacardio.2016.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Dirkx E, Gladka MM, Philippen LE, et al. Nfat and miR-25 cooperate to reactivate the transcription factor Hand2 in heart failure. Nat Cell Biol. 2013:15. 10.1038/ncb2866. [DOI] [PubMed] [Google Scholar]
- 247.Wilkins BJ, De Windt LJ, Bueno OF, et al. Targeted disruption of NFATc3, but not NFATc4, reveals an intrinsic defect in calcineurin-mediated cardiac hypertrophic growth. Mol Cell Biol. 2002:22. 10.1128/mcb.22.21.7603-7613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Deng X, Liu Y, Xu Z, Wang Z. lncRNA nuclear factor of activated t cells knockdown alleviates hypoxia/reoxygenation-induced cardiomyocyte apoptosis by upregulating HIF-1α expression. J Cardiovasc Pharmacol. 2022:79. 10.1097/FJC.0000000000001198. [DOI] [PubMed] [Google Scholar]
- 249.Logroscino G, Piccininni M, Marin B, et al. Global, regional, and national burden of motor neuron diseases 1990–2016: A systematic analysis for the global burden of disease study 2016. Lancet Neurol. 2018:17. 10.1016/S1474-4422(18)30404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Nichols E, Szoeke CEI, Vollset SE, et al. Global, regional, and national burden of Alzheimer’s disease and other dementias, 1990–2016: A systematic analysis for the global burden of disease study 2016. Lancet Neurol. 2019:18. 10.1016/S1474-4422(18)30403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ray Dorsey E, Elbaz A, Nichols E, et al. Global, regional, and national burden of Parkinson’s disease, 1990–2016: A systematic analysis for the global burden of disease study 2016. Lancet Neurol. 2018:17. 10.1016/S1474-4422(18)30295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Fauré J, Lachenal G, Court M, et al. Exosomes are released by cultured cortical neurones. Mol Cell Neurosci. 2006:31. 10.1016/j.mcn.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 253.Tytell M. Release of heat shock proteins (Hsps) and the effects of extracellular Hsps on neural cells and tissues. Int J Hyperth. 2005:21. 10.1080/02656730500041921. [DOI] [PubMed] [Google Scholar]
- 254.Potolicchio I, Carven GJ, Xu X, et al. Proteomic analysis of microglia-derived exosomes: Metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J Immunol. 2005:175. 10.4049/jimmunol.175.4.2237. [DOI] [PubMed] [Google Scholar]
- 255.Krämer-Albers EM, Bretz N, Tenzer S, et al. Oligodendrocytes secrete exosomes containing major Myelin and stress-protective proteins: Trophic support for Axons? Proteom Clin Appl. 2007:1. 10.1002/prca.200700522. [DOI] [PubMed] [Google Scholar]
- 256.Sproviero D, Gagliardi S, Zucca S, et al. Different mirna profiles in plasma derived small and large extracellular vesicles from patients with neurodegenerative diseases. Int J Mol Sci. 2021:22. 10.3390/ijms22052737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Clark BS, Blackshaw S. Understanding the role of lncRNAs in nervous system development. Adv Exper Med Biol. 2017;1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Sosa-Ortiz AL, Acosta-Castillo I, Prince MJ. Epidemiology of dementias and Alzheimer’s disease. Arch Med Res. 2012:43. [DOI] [PubMed] [Google Scholar]
- 259.Murphy MP, Levine H. Alzheimer’s disease and the amyloid-β peptide. J Alzheimer’s Dis. 2010:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat Med. 2008:14. 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.He W, Chi S, Jin X, et al. Long non-coding RNA BACE1-AS modulates isoflurane-induced neurotoxicity to Alzheimer’s disease through sponging miR-214–3p. Neurochem Res. 2020;45:2324–2335. 10.1007/s11064-020-03091-2. [DOI] [PubMed] [Google Scholar]
- 262.Wang D, Wang P, Bian X, et al. Elevated plasma levels of exosomal BACE1-as combined with the volume and thickness of the right entorhinal cortex may serve as a biomarker for the detection of Alzheimer’s disease. Mol Med Rep. 2020;22:227–238. 10.3892/mmr.2020.11118, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Welton JL, Loveless S, Stone T, von Ruhland C, Robertson NP, Clayton A. Cerebrospinal fluid extracellular vesicle enrichment for protein biomarker discovery in neurological disease; Multiple sclerosis. J Extracell Vesicles. 2017:6. 10.1080/20013078.2017.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zhuang J, Cai P, Chen Z, et al. Long noncoding RNA MALAT1 and its target microRNA-125b are potential biomarkers for Alzheimer’s disease management via interactions with FOXQ1, PTGS2 and CDK5. Am J Transl Res. 2020:12. [PMC free article] [PubMed] [Google Scholar]
- 265.Sun Y, Ma L. New insights into long non-coding RNA MALAT1 in cancer and metastasis. Cancers (Basel). 2019:11. 10.3390/CANCERS11020216, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ruan L, Mendhe B, Parker E, et al. Long non-coding RNA MALAT1 is depleted with age in skeletal muscle in vivo and MALAT1 silencing increases expression of TGF-β1 in vitro. Front Physiol. 2022:12. 10.3389/FPHYS.2021.742004, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Stanton SE, Disis ML. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J Immunother Cancer. 2016;4:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Xin C, Liu J. Long non-coding RNAs in Parkinson’s disease. Neurochem Res. 2021;46:1031–1042. 10.1007/S11064-021-03230-3/TABLES/2, [DOI] [PubMed] [Google Scholar]
- 269.Rezaei O, Nateghinia S, Estiar MA, Taheri M, Ghafouri-Fard S. Assessment of the role of non-coding RNAs in the pathophysiology of Parkinson’s disease. Eur J Pharmacol. 2021;896:173914. 10.1016/J.EJPHAR.2021.173914, [DOI] [PubMed] [Google Scholar]
- 270.Elkouris M, Kouroupi G, Vourvoukelis A, et al. Long non-coding RNAs associated with neurodegeneration-linked genes are reduced in Parkinson’s disease patients. Front Cell Neurosci. 2019;13:1–13. 10.3389/FNCEL.2019.00058/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Wang Q, Han CL, Wang KL, et al. Integrated analysis of exosomal lncRNA and mRNA expression profiles reveals the involvement of lnc-MKRN2–42:1 in the pathogenesis of Parkinson’s disease. CNS Neurosci Ther. 2020;26:527–537. 10.1111/CNS.13277, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Zou J, Guo Y, Wei L, Yu F, Yu B, Xu A. Long noncoding RNA POU3F3 and α-synuclein in plasma L1CAM exosomes combined with β-glucocerebrosidase activity: Potential predictors of Parkinson’s disease. Neurotherapeutics. 2020;17:1104–1119. 10.1007/S13311-020-00842-5/FIGURES/5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Papadopoulos VE, Nikolopoulou G, Antoniadou I, et al. Modulation of β-glucocerebrosidase increases α-synuclein secretion and exosome release in mouse models of Parkinson’s disease. Hum Mol Genet. 2018;27:1696–1710. 10.1093/HMG/DDY075, [DOI] [PubMed] [Google Scholar]
- 274.Ngolab J, Trinh I, Rockenstein E, et al. Brain-derived exosomes from dementia with lewy bodies propagate α-synuclein pathology. Acta Neuropathol Commun. 2017;5:46. 10.1186/S40478-017-0445-5/FIGURES/5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Elkouris M, Kouroupi G, Vourvoukelis A, et al. Long non-coding RNAs associated with neurodegeneration-linked genes are reduced in Parkinson’s disease patients. Front Cell Neurosci. 2019;13:1–13. 10.3389/FNCEL.2019.00058/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Sproviero D, Gagliardi S, Zucca S, et al. Extracellular vesicles derived from plasma of patients with neurodegenerative disease have common transcriptomic profiling. Front Aging Neurosci. 2022;14:785741. 10.3389/FNAGI.2022.785741/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Clos-Sansalvador M, Monguió-Tortajada M, Roura S, Franquesa M, Borràs FE. Commonly used methods for extracellular vesicles’ enrichment: Implications in downstream analyses and use. Eur J Cell Biol. 2022;101:151227. 10.1016/j.ejcb.2022.151227. [DOI] [PubMed] [Google Scholar]
- 278.Husna AA, Rahman MM, Chen HW, Hasan MN, Nakagawa T, Miura N. Long non-coding RNA and transfer RNA-derived small fragments in exosomes are potential biomarkers for canine oral melanoma. Vet Comp Oncol. 2022;20:653–663. 10.1111/VCO.12818, [DOI] [PubMed] [Google Scholar]


