Abstract
Sequencing of the vicinity of the staphylococcal pbp2 gene and transcriptional analysis by primer extension and promoter fusions were used to show that pbp2 is part of an operon that also includes a gene with high homology to prfA of Bacillus subtilis. Two distinct promoters were identified directing transcription of pbp2 either alone or together with prfA. It was recently reported that transposon inactivation of pbp2 causes a reduction in methicillin resistance, but complementation experiments were not fully successful. We now show that introduction of the intact pbp2 gene with its two newly identified promoters into the chromosome of the transposon mutant resulted in the full recovery of high-level methicillin resistance.
All strains of Staphylococcus aureus have four penicillin-binding proteins (PBPs) which are assumed to participate in the assembly of cell wall peptidoglycan. Of this native set of PBPs, PBP2 was reported to be a major peptidoglycan transpeptidase (5), since selective inhibition of this protein by ceftizoxime or cefotaxime led to the inhibition of peptidoglycan elongation (16) and to leakage of cytoplasmic contents due to cell lysis (5). Homology between the N-terminal half of S. aureus PBP2 and Escherichia coli PBP1A, a bifunctional protein, suggests that PBP2 also has a transglycosylase domain (13, 15).
Surprisingly, PBP2 also appears to have an important role in the expression of antibiotic resistance in methicillin-resistant S. aureus (MRSA) (18). MRSA has an additional PBP—PBP2A—which has a very low affinity for β-lactam antibiotics (8, 21) and has homology to monofunctional transpeptidase PBP3 of E. coli (6, 9, 13). In current models, PBP2A is assumed to take over the biosynthetic functions of normal PBPs in the presence of inhibitory concentrations of β-lactams. According to this model, normal PBPs no longer take part in the catalysis of cell wall synthesis in the presence of the antibiotic. It was therefore surprising to find that a mutant with a transposon insertion in pbp2 (RUSA 130) showed a massive reduction in methicillin resistance, despite its normal production of PBP2A, indicating that intact PBP2 is essential for the optimal expression of methicillin resistance in MRSA (18). As a possible explanation, we proposed that survival and growth in the presence of the antibiotic may require functional cooperation between the penicillin-insensitive transglycosylase domain of PBP2 and the transpeptidase domain of PBP2A or that effective functioning of PBP2A may require the presence of inactivated (acylated) PBP2, which serves as structural scaffolding (18). An additional possibility that could not be excluded was that a truncated PBP2 produced by the transposon-inactivated pbp2 gene may interfere with the function of PBP2A. This hypothesis was also suggested by the fact that attempts to recover the normal—high—level of antibiotic resistance by complementation with a plasmid-born pbp2 gene were only partially successful (18).
In an attempt to clarify the reasons for the lack of success in complementation, we proceeded to do more extensive sequencing in the vicinity of the pbp2 gene and also performed transcription analysis. This study showed that the pbp2 gene is part of an operon and can be transcribed alone or together with a newly identified PBP-related factor (PrfA), due to the presence of two distinct promoters. Introduction of a construct that contained the entire pbp2 operon into the chromosome of the pbp2 transposon mutant resulted in the full recovery of antibiotic resistance.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are described in Table 1. S. aureus strains were grown on tryptic soy broth (TSB; Difco Laboratories) with aeration as described previously (17). E. coli strains were grown in Luria-Bertani broth (Difco) with aeration. Antibiotics were used at the following concentrations: erythromycin, 10 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| S. aureus COL | Homogeneous Mcr (MIC, 1,600 μg/ml) | RU collection |
| S. aureus RN4220 | Mutant strain of 8325-4 that is r− | R. Novick |
| S. aureus RUSA130 | COLΩ703 (pbp2::Tn551) Emr heterogeneous Mcr (MIC, 12 μg/ml) | 4 |
| S. aureus RUSA130/pMGP19 | RUSA130 with pMGP19 plasmid containing prfA and pbp2 genes and promoters | This study |
| E. coli DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| pLC4 | Apr Cmr promoterless xylE gene | 20 |
| pro5/8 | pLC4 containing 678-bp fragment with promoter region upstream of pbp2 | This study |
| pro9/4 | pLC4 containing 311-bp fragment with promoter region upstream of prfA | This study |
| pro9/10 | pLC4 containing 967-bp fragment with two promoter regions | This study |
| pGC2 | E. coli-S. aureus shuttle vector, Apr Cmr | 25 |
| pSPT181 | E. coli-S. aureus shuttle vector, Apr Tcr | 10 |
| PSPT181cat | pSPT181 with cat gene | S. Wu |
| pMGP19 | pSPT181 with cat gene and 3.2-kb fragment with prfA and pbp2 | This study |
DNA methods.
Routine DNA manipulations were performed by using standard methods (2, 22). All of the enzymes were purchased from either New England Biolabs or Boehringer Mannheim and used as recommended by the manufacturers. DNA sequencing was done at the Rockefeller University Protein/DNA Technology Center with the Taq fluorescent dye terminator sequencing method by using a PE/ABI 377 automated sequencer.
Inverted PCR.
COL chromosomal DNA was digested with EcoRI, purified with the Wizard DNA Clean-Up System (Promega), and ligated with T4 DNA ligase. The ligation mixture, after purification, was used as a template for a PCR with the GeneAmp PCR Reagent Kit with AmpliTaq DNA polymerase (Perkin Elmer) in accordance with the manufacturers’ instructions and by using 20 pmol each of primers PBP2P8 (CTTAGGCTGAGAAGATCCTT) and PBP2P9 (AGCTTGGCAATCAGTTAAGC). The following conditions were used: 94°C for 2 min; 35 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 2.5 min; and one final extension step of 72°C for 5 min. The 2,862-bp fragment was sequenced by primer walking, starting with the same primers used for the PCR.
Isolation of RNA and Northern blot hybridization.
Overnight cultures of S. aureus were diluted 1:50 in TSB and grown to mid-log phase (optical density at 620 nm [OD620], 0.7). The cells were pelleted and processed with an RNeasy Mini Kit (Qiagen) or with a FastRNA Blue isolation kit (Bio 101, Inc.) in combination with FastPrep FP120 (Bio 101 Savant) in accordance with the manufacturer’s recommendations. RNA (5 μg) was electrophoresed through a 1.2% agarose–0.66 M formaldehyde gel in morpholinepropanesulfonic acid running buffer (Sigma). Blotting of RNA onto Hybond N+ membranes (Amersham) was performed with the Turboblotter alkaline transfer system (Schleicher & Schuell). For the detection of specific transcripts, DNA probes were labelled by using the Gene Images random prime labelling module (Amersham Life Science) and hybridized under high-stringency conditions. The blots were subsequently washed and autoradiographed.
RT-PCR.
Reverse transcription (RT)-PCR was performed by using the GeneAmp RNA PCR kit (Perkin Elmer). COL RNA treated with DNase was used as the template. Random hexamers were used for the reverse transcriptase reaction, and a primer internal to prfA—PrfA1 (ATGTCAACTATCCTAAGCGG)— and a primer internal to pbp2—IP6 (TCTTAGCATCTTCCCACTGT)—were used in the PCR. The following conditions were used: 94°C for 2 min; 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 2 min; and one final extension step of 72°C for 5 min.
Constructions of promoter fusions.
Three fragments encompassing the region upstream of pbp2 were amplified by high-fidelity PCR with the GeneAmp XL PCR kit (Perkin Elmer), which includes rTth DNA polymerase XL. To further decrease the probability of errors during the PCR, a hot start and the following conditions were used: 94°C for 2 min, 20 cycles of 94°C for 30 s, and 55°C for 1.75 min; and one final extension step of 55°C for 5 min. Primer pairs pro4-pro9, pro5-pro8, and pro9-pro10 (Fig. 1) were used to amplify the three fragments that were subsequently cloned into pLC4.
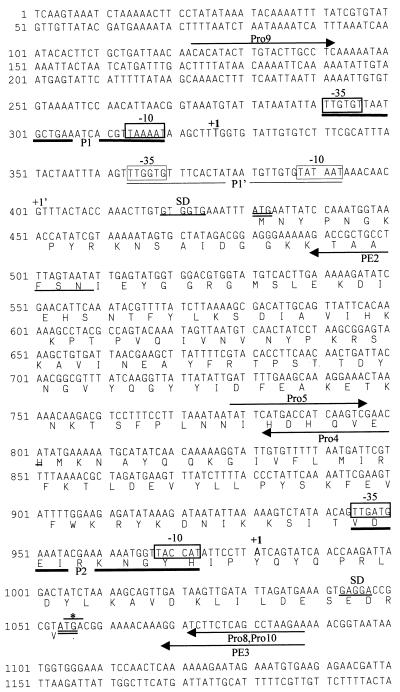
FIG. 1.
Nucleotide sequence of the region upstream of pbp2. Putative promoter regions are highlighted by boxed sequences and labelled −10 and −35. The promoters are designated P1 and P2. P1’ corresponds to a weaker signal in the primer extension analysis. Putative ribosome-binding sites are underlined and labelled SD. The 5′ end of the RNA determined by primer extension is labelled +1. Start codons are in boldface and double underlined. The prfA stop codon is indicated by an asterisk. Primers are indicated by arrows. The deduced amino acid sequence of prfA is aligned under the DNA sequence.
Enzyme assays.
Catechol 2,3-dioxygenase assays were performed essentially as previously described (23), except for the lysis of bacteria, which was done by using glass beads and FastPrep 120 (Bio 101 Savant) in 100 mM phosphate buffer (pH 7.5) containing 10% acetone. Volumes of 20 to 3 ml corresponding to different OD620 values were serially removed from cultures growing in TSB. Assays contained 100 μl of extract, and the reactions were conducted at room temperature for 5 min with OD375 readings taken at 30-s intervals. One milliunit corresponds to the formation at room temperature of 1 nmol of 2-hydroxymuconic semialdehyde per min. Specific activity is reported in milliunits per milligram of total protein. Protein concentrations were measured by using the bicinchoninic acid protein assay reagent kit (Pierce).
Primer extension analysis.
Primer extension analysis was performed by using primers PE2 and PE3 (Fig. 1), which were end labelled with [γ-32P]ATP and purified with Sephadex G-25 spin columns (Boehringer Mannheim). RNA from COL (50 μg) or RN4220pro9/10 (10 μg) was hybridized with the appropriate primer at 65°C for 90 min and slowly cooled to room temperature. RT was carried out by using SuperScript RT (Gibco BRL) at 42°C for 90 min, and the reaction mixture was heated at 65°C for 10 min to inactivate the enzyme. The reaction product was incubated with RNase H (3 U) at 37°C for 30 min, ethanol precipitated, resuspended in 10 μl of Sequenase stop solution, denatured, and applied to a 6% sequencing gel. Sequencing reaction mixtures prepared by using the T7 Sequenase Kit vs2.0 (Amersham Life Sciences) primed by an oligonucleotide identical to that used for primer extension were also applied to the gel.
Complementation of mutant RUSA130.
A 3.2-kb fragment was amplified from the COL chromosome by using primers PBP2B (CGGGATCCCACATACTTGTACTTGCCTC) and PBP2P7P (AACTGCAGTCCCACCATAAAAGATGAAG) by high-fidelity PCR under the same conditions as for the construction of promoter fusions, except for an extension time of 5 min. This fragment was cloned into shuttle vector pSPT181 digested with BamHI and PstI. A chloramphenicol resistance marker amplified by high-fidelity PCR from pGC2 with primers cat1 (TTCCCCGGGACCAGTCATACCAATAAC) and cat2 (TTCCCCGGGCTCAACGTCAATAAAGCA) was cloned into the AvaI site of the polylinker.
S. aureus RN4220 was used as the recipient for electroporation of the shuttle plasmid, which was performed as previously described (25). The plasmids were subsequently introduced into RUSA130 by transduction using phage 80α (17).
Population analysis profiles were performed by plating 10-μl samples of 100, 10−1, 10−2, 10−4, 10−5, and 10−6 dilutions of an overnight culture on plates containing methicillin (0, 0.75, 1.5, 3, 6, 12, 25, 50, 100, 200, 400, and 800 μg/ml). The plates were then tipped onto their sides (90° angle), and the spots were allowed to migrate in parallel tracks across the agar surface to the opposite side of each plate, and the plates were incubated at 30°C for 48 h (11).
Nucleotide sequence accession number.
The complete nucleotide sequence determined in this study is available in the EMBL and GenBank databases under accession no. Y17795.
RESULTS
Sequencing of the region upstream of the pbp2 gene.
A fragment of 2,862 bp, which included the region upstream of the pbp2 gene of strain COL, was obtained by inverted PCR and sequenced by primer walking. The sequence of the pbp2 gene from COL was obtained by using primers based on the published sequence of S. aureus SRM705 (15) (accession no. X62288). The fragment upstream of pbp2 is presented in Fig. 1.
Computer analysis identified an open reading frame (ORF) of 627 bp upstream of pbp2. In fact, the last four nucleotides of this ORF overlap the pbp2 coding sequence. The deduced amino acid sequence of this ORF was compared to sequences of known polypeptides in the EMBL and GenBank databases by using the Gapped Blast algorithm (1). Significant homology (52% identity) was found with the protein encoded by gene prfA (PBP-related factor A) from Bacillus subtilis (19), which is located upstream of the gene that encodes PBP1 (the B. subtilis homologue of S. aureus PBP2). This protein has also been identified in Streptococcus oralis and Streptococcus pneumoniae (12).
Analysis of the transcription of pbp2.
To determine if the prfA and pbp2 genes were transcribed together, a Northern blot of total RNA of COL was hybridized with a probe internal to pbp2 (nucleotides 330 to 716 of the pbp2 sequence). The appearance of two bands (data not shown), one with a molecular size of 2.1 kb, corresponding to the size expected for the pbp2 transcript, and another with a molecular size of 2.9 kb, corresponding to the size of a transcript with the two ORFs, suggests that the pbp2 gene can be transcribed either alone or together with prfA.
A third band, corresponding to a molecular size of 1.6 kb and located just above the rRNA band, was occasionally observed in some of the Northern blot hybridizations. The size of this band is smaller than that of the pbp2 gene, and it probably corresponds to degradation products retained in this region.
RT-PCR was performed by using one primer internal to prfA and another internal to pbp2. The successful amplification of a 1.2-kb fragment clearly indicates that the two ORFs can be transcribed together.
These results suggested the existence of two promoters, one upstream of prfA (promoter 1) that would direct the transcription of the 2.9-kb transcript and another upstream of pbp2 (promoter 2) that would direct the transcription of the 2.1-kb transcript.
We amplified, by high-fidelity PCR, three fragments: one containing 678 bp (from nucleotide 133 to nucleotide 802 in Fig. 1) and including the region where we expected promoter 1 to be; a second, 311-bp fragment (from nucleotide 777 to nucleotide 1088 in Fig. 1) including the region of promoter 2; and a third, 967-bp fragment (from nucleotide 133 to nucleotide 1088 in Fig. 1) including both promoter regions. The fragments were cloned into pLC4, and the inserts were sequenced. This plasmid has a promoterless xylE gene, and production of catechol 2,3-dioxygenase in S. aureus is dependent on the introduction of a promoter, upstream of xylE, that is functional in the gram-positive host (26).
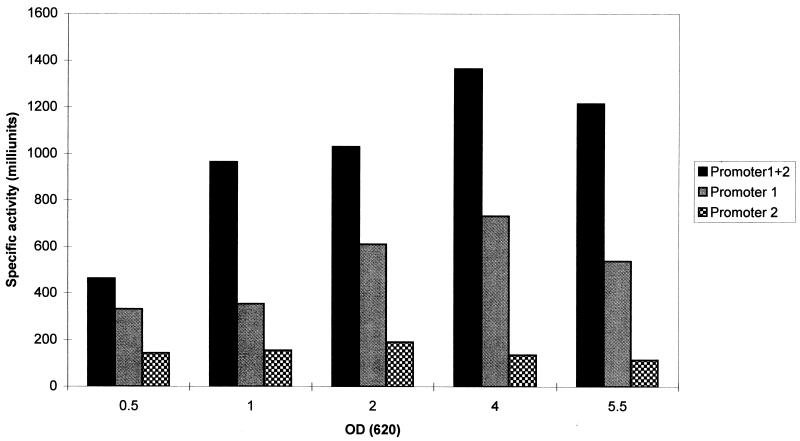
Determination of the specific activity of catechol 2,3-deoxygenase (Fig. 2) indicated that promoter 1 generates higher levels of catechol 2,3-dioxygenase activity than promoter 2 and that the activity of both promoters was found to decrease with the end of the exponential growth phase.
FIG. 2.
XylE activity in constructs with promoter 1 (plasmid pro9/4), promoter 2 (plasmid pro5/8), and both promoters (plasmid pro9/10) during the growth cycle. An OD620 nm of 4 corresponds to the end of the exponential growth phase. Values are presented in milliunits per milligram of cellular protein. The control strain with the vector pLC4 had no XylE activity.
Determination of transcription initiation sites.
Primer extension analysis was performed to determine the transcription start site corresponding to each promoter by using primers specific for the prfA and pbp2 transcripts. RNAs prepared from both COL (data not shown) and RN4220pro9/10 (Fig. 3) (i.e., the strain containing a plasmid with an insert encompassing both promoter regions) were used as templates for the RT reaction.
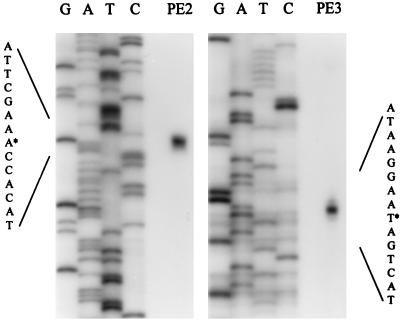
FIG. 3.
Mapping of the 5′ ends of two pbp2 transcripts by primer extension. The sequence encompassing the transcription start site (marked by asterisks) is enlarged. PE2 and PE3 were the primers used for the study of promoters 1 and 2, respectively.
Based on this analysis, it was determined that the transcript that includes prfA initiates at a thymine residue (start point of promoter P1 in Fig. 1) located 104 bp upstream of the prfA start codon. Nevertheless, a fainter band was consistently observed at a quanine residue 74 bp downstream (start point of promoter P1’ in Fig. 1). The pbp2 transcript initiates at an adenine residue located 73 bp upstream of the pbp2 start codon and therefore in the prfA coding sequence.
Recovery of methicillin resistance in mutant RUSA130.
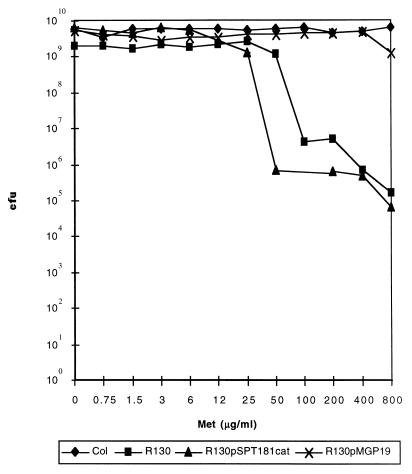
A 3.2-kb fragment containing the complete sequences of prfA and pbp2, as well as the promoter regions, was amplified by high-fidelity PCR and cloned into pSPT181. Plasmid pMGP19 was first introduced into RN4220 by electroporation and afterwards into RUSA130 by transduction. This plasmid is incompatible with the tetracycline resistance-encoding plasmid present in COL and in the RUSA130 mutant. Therefore, strain RUSA130 with pMGP19 had the plasmid integrated into the chromosome by a Campbell-type mechanism and also retained the transposon-inactivated copy of pbp2—as confirmed by Southern blotting, followed by hybridization with a probe specific for pbp2 (data not shown). Figure 4 shows that RUSA130/pMGP19 fully recovered the high-level methicillin resistance of parental strain COL.
FIG. 4.
Population analysis profiles of parental strain COL, mutant RUSA130, mutant RUSA130 with an intact copy and an inactivated copy of pbp2 in the chromosome (RUSA130/pMGP19), and the mutant with the vector pSPT181cat.
DISCUSSION
In previous studies of pbp2, it was suggested that the −10 region of the pbp2 promoter (annotated in the GenBank database, accession no. L25426 [7]) was located in the region corresponding to nucleotides 1027 to 1032 of Fig. 1. Our data shows that this is unlikely and that, in fact, not one but two promoters direct the transcription of pbp2 (promoters 1 and 2 in Fig. 1), neither one of which coincides with the previously suggested promoter.
The lack of success in recovering high-level antibiotic resistance in the complementation experiments described before (18) may have been caused by the use of pbp2 without a correct promoter. By using the pbp2 operon as defined by the results described here, it was possible to recover the high, parental level of methicillin resistance in pbp2 transposon mutant RUSA130. Construct RUSA130/pMGP19 contained single copies of both the truncated and normal forms of the pbp2 gene on the chromosome, each preceded by native promoters. The possibility could not be previously excluded that the truncated allele of PBP2 present in RUSA130 might interfere with the function of PBP2A and thus cause the reduction of methicillin resistance in strain RUSA130 (18). However, the recovery of high methicillin resistance in RUSA130/pMGP19 makes it unlikely that the truncated allele could have a dominant negative effect on the activity of PBP2A. The reappearance of parental-level methicillin resistance in this construct also provides final proof of the importance of functional PBP2 in the expression of resistance to methicillin.
The results described here indicate that transcription of pbp2 can occur together with that of prfA, a gene located immediately upstream of pbp2 with an overlap of four nucleotides, and the two genes therefore constitute an operon. However, pbp2 can also be transcribed alone. It is conceivable that changes in the preferential use of the two promoters may occur under specific physiological conditions, for instance, in the presence of antibiotics. However, analysis of the activities of the two promoters through the growth cycle did not show any striking change of promoter usage: promoter 1 activity was always higher, and the expression of pbp2 from both promoters declined as the bacteria entered stationary phase.
The function of the protein encoded by prfA is unknown. In B. subtilis, the combined effects of mutations in the PBP1 (ponA) and prfA genes on the bacterial growth rate were more dramatic than the effect of either one of the individual mutations, suggesting involvement of PrfA with the function of PBP1 (19), a homologue of S. aureus PBP2. The existence of two promoters in B. subtilis has also been suggested, although attempts to determine their positions by primer extension were inconclusive (19). The fact that the two genes are part of an operon in at least two different organisms reinforces the hypothesis that their functions may be related. PrfA may modulate the activity of the staphylococcal pbp2 promoter, although the presence of prfA in a multicopy plasmid (pro9/10) did not significantly affect the transcription directed by the pbp2 promoters in the catechol 2,3-deoxygenase assay. It is possible that this protein interacts with PBP2, as a part of a multienzyme complex responsible for the catalysis of cell wall synthesis in a manner similar to the one proposed for E. coli (3). Studies are in progress to test this possibility.
Overexpression of PBP2 was observed in vancomycin- and teicoplanin-resistant S. aureus (14, 24). The identification of pbp2 promoter regions, as well as the characterization of the PrfA protein, may be relevant to the understanding of the mechanisms of resistance to both β-lactams and glycopeptides.
ACKNOWLEDGMENTS
Partial support for this work was provided by contracts PRAXIS XXI 2/2.1/BIA/349/94 and PRAXIS XXI 2/2.1/BIO/1154/95 (Portugal) awarded to H. de Lencastre and by the Aaron Diamond Foundation and the Bodman Foundation to A. Tomasz. Mariana Pinho was supported by grant PRAXIS XXI/BD/9079/96.
We thank Ambrose Cheung, who kindly provided plasmids pSPT181 and pLC4, and Shangwei Wu for plasmid pSPT181cat and helpful discussions.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1996. [Google Scholar]
- 3.Ayala J A, Garrido T, de Pedro M A, Vicente M. Molecular biology of bacterial separation. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science; 1994. pp. 73–101. [Google Scholar]
- 4.de Lencastre H, Tomasz A. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2590–2598. doi: 10.1128/aac.38.11.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Georgopapadakou N H, Dix B A, Mauriz Y R. Possible physiological functions of penicillin-binding proteins in Staphylococcus aureus. Antimicrob Agents Chemother. 1986;29:333–336. doi: 10.1128/aac.29.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goffin C, Fraipont C, Ayala J, Terrak M, Nguyen-Distèche M, Ghuysen J-M. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J Bacteriol. 1996;178:5402–5409. doi: 10.1128/jb.178.18.5402-5409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hackbarth C J, Kocagoz T, Kocagoz S, Chambers H F. Point mutations in Staphylococcus aureus PBP 2 gene affect penicillin-binding kinetics and are associated with resistance. Antimicrob Agents Chemother. 1995;39:103–106. doi: 10.1128/aac.39.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartman B J, Tomasz A. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holtje J V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jett B D, Hatter K L, Huycke M M, Gilmore M S. Simplified agar plate method for quantifying viable bacteria. Biotechniques. 1997;23:648–650. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 12.Martin C, Briese T, Hakenbeck R. Nucleotide sequences of genes encoding penicillin-binding proteins from Streptococcus pneumoniae and Streptococcus oralis with high homology to Escherichia coli penicillin-binding proteins 1a and 1b. J Bacteriol. 1992;174:4517–4523. doi: 10.1128/jb.174.13.4517-4523.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreira B, Boyle-Vavra S, deJonge B L, Daum R S. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:1788–1793. doi: 10.1128/aac.41.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami K, Fujimura T, Doi M. Nucleotide sequence of the structural gene for the penicillin-binding protein 2 of Staphylococcus aureus and the presence of a homologous gene in other staphylococci. FEMS Microbiol Lett. 1994;117:131–136. doi: 10.1111/j.1574-6968.1994.tb06754.x. [DOI] [PubMed] [Google Scholar]
- 16.Okonogi K, Noji Y, Nakao M, Imada A. The possible physiological roles of penicillin-binding proteins of methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J Infect Chemother. 1995;1:50–58. [Google Scholar]
- 17.Oshida T, Tomasz A. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J Bacteriol. 1992;174:4952–4959. doi: 10.1128/jb.174.15.4952-4959.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinho M G, Ludovice A M, Wu S, de Lencastre H. Massive reduction in methicillin resistance by transposon inactivation of the normal PBP2 in a methicillin-resistant strain of Staphylococcus aureus. Microb Drug Resist. 1997;3:409–413. doi: 10.1089/mdr.1997.3.409. [DOI] [PubMed] [Google Scholar]
- 19.Popham D L, Setlow P. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. J Bacteriol. 1995;177:326–335. doi: 10.1128/jb.177.2.326-335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ray C, Hay R E, Carter H L, Moran C P., Jr Mutations that affect utilization of a promoter in stationary-phase Bacillus subtilis. J Bacteriol. 1985;163:610–614. doi: 10.1128/jb.163.2.610-614.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynolds P E, Brown D F. Penicillin-binding proteins of β-lactam-resistant strains of Staphylococcus aureus. Effect of growth conditions. FEBS Lett. 1985;192:28–32. doi: 10.1016/0014-5793(85)80036-3. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Sheehan B J, Foster T J, Dorman C J, Park S, Stewart G S. Osmotic and growth-phase dependent regulation of the eta gene of Staphylococcus aureus: a role for DNA supercoiling. Mol Gen Genet. 1992;232:49–57. doi: 10.1007/BF00299136. [DOI] [PubMed] [Google Scholar]
- 24.Shlaes D M, Shlaes J H, Vincent S, Etter L, Fey P D, Goering R V. Teicoplanin-resistant Staphylococcus aureus expresses a novel membrane protein and increases expression of penicillin-binding protein 2 complex. Antimicrob Agents Chemother. 1993;37:2432–2437. doi: 10.1128/aac.37.11.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S, de Lencastre H, Sali A, Tomasz A. A phosphoglucomutase-like gene essential for the optimal expression of methicillin resistance in Staphylococcus aureus: molecular cloning and DNA sequencing. Microb Drug Resist. 1996;2:277–286. doi: 10.1089/mdr.1996.2.277. [DOI] [PubMed] [Google Scholar]
- 26.Zukowski M M, Gaffney D F, Speck D, Kauffmann M, Findeli A, Wisecup A, Lecocq J P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci USA. 1983;80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]