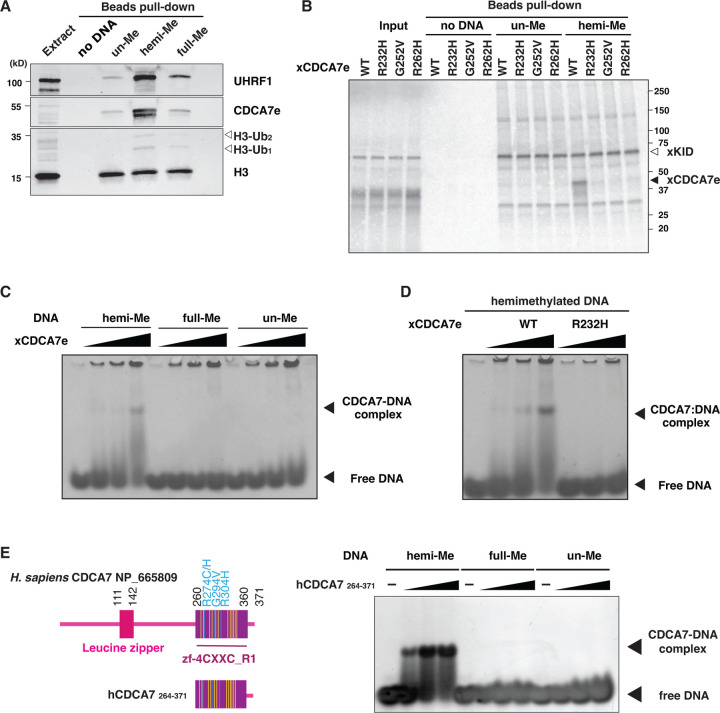
Fig. 2. CDCA7 selectively binds hemimethylated DNA.
(A) Magnetic beads coupled with pBluescript DNA with unmethylated CpGs (un-Me), hemimethylated CpGs (hemi-Me), or fully methylated CpGs (full-Me), were incubated with interphase Xenopus egg extracts. Beads were collected after 60 min and analyzed by western blotting. (B) 35S-labeled X. laevis CDCA7e proteins (wildtype or with the indicated ICF3-patient associated mutation) were incubated with control beads, or beads conjugated 200 bp unmethylated or hemimethylated DNA (Table S1). 35S-labeled Xkid (85), a nonspecific DNA-binding protein, was used as a loading control. Autoradiography of 35S-labeled proteins in input and beads fraction is shown. (C) and (D) Native gel electrophoresis mobility shift assay (EMSA) using recombinant X. laevis CDCA7eWT and CDCA7eR232H. (E) Left: schematic of H. sapiens CDCA7 (isoform 2 NP_665809). Positions of the zf-4CXXC_R1 domain (purple), three ICF3-patient mutations (cyan), and conserved cysteine residues (yellow) are shown. Right: EMSA assay using the purified zf-4CXXC_R1 domain (aa 264–371) of H. sapiens CDCA7. For C-E, double-stranded DNA oligonucleotides with an unmethylated, hemimethylated or fully-methylated CpG used for protein binding were visualized.