Abstract
The localization of cell division protein FtsQ in Escherichia coli wild-type cells was studied by immunofluorescence microscopy with specific monoclonal antibodies. FtsQ could be localized to the division site in constricting cells. FtsQ could also localize to the division site in ftsQ1(Ts) cells grown at the permissive temperature. A hybrid protein in which the cytoplasmic domain and the transmembrane domain were derived from the γ form of penicillin-binding protein 1B and the periplasmic domain was derived from FtsQ was also able to localize to the division site. This result indicates that the periplasmic domain of FtsQ determines the localization of FtsQ, as has also been concluded by others for the periplasmic domain of FtsN. Noncentral FtsQ foci were found in the area of the cell where the nucleoid resides and were therefore assumed to represent sites where the FtsQ protein is synthesized and simultaneously inserted into the cytoplasmic membrane.
Many essential cell division proteins have been identified in Escherichia coli (27). Though these proteins have been expected to function at the site of division, it has taken many years to develop the means to localize such proteins. A technical breakthrough was the demonstration of the existence of the FtsZ ring by immunogold labeling in dividing cells (3). Subsequently, the FtsZ ring has been visualized by immunofluorescence (1, 26) and with a fusion protein of FtsZ and green fluorescent protein (28, 35). The latter approach has also been successful for FtsA (28), ZipA (16), and MinD-associated MinE (31). More recently, the cytoplasmic membrane proteins FtsN (2), penicillin-binding protein 3 (PBP 3) (FtsI) (43, 44), FtsK (48), and FtsW (43) were found to localize to the division site in constricting cells by immunofluorescence microscopy. The localization of all membrane-bound proteins, except ZipA and FtsW, occurred late in the division process and was dependent on the localization of both FtsZ and FtsA. The order of appearance of division proteins at the division site as determined by immunofluorescence microscopy was consistent with the results obtained by phenotypic analysis of the various temperature-sensitive mutants (36). The data suggest that the division proteins appear and function at the division site in the following order: MinE-FtsZ-FtsA-FtsK-PBP 3 (FtsI)-FtsN. ZipA might act either before or after FtsZ (16). It is not clear at what point FtsW localizes, but both the results of a genetic study (21) and FtsZ localization in ftsW(Ts) filaments (5) suggest that it acts early in cell division.
The FtsQ homolog DivIB in Bacillus subtilis, which occurs in about 5,000 copies per cell, has been shown to localize to the septum (18). FtsQ shares its membrane topology with PBP 3 (FtsI) (4), FtsN (9), and FtsL (15), which all harbor an N-terminal cytoplasmic domain and a C-terminal periplasmic domain (7). Based on morphological analysis of various temperature-sensitive mutants, it has been deduced that FtsQ, like PBP 3 (FtsI), acts after FtsZ does (36); however, its biological function is still unknown. Although the amount of FtsQ is very low (about 25 to 50 molecules of FtsQ exist per cell [7]) FtsQ could be localized to the site of division in constricting cells. It could also be found at noncentral positions. These positions occur in the vicinity of the nucleoid, and they might correlate with sites where FtsQ synthesis occurs.
MATERIALS AND METHODS
Bacterial strains and plasmids.
As wild-type strains, LMC500 [MC4100 (F− araD139 Δ(argF-lac)U169 deoC1 flbB5301 ptsF25 rbsR relA1 rpsL150) lysA (36)], JM101 (47), and B/rA (23) were used. As a temperature-sensitive ftsQ mutant and an ftsQ depletion strain, LMC531 [LMC500, ftsQ1(Ts)] (36) and VIP210 (12) were used, respectively. POP2136 (40) was used to overproduce β-galactosidase fusion proteins.
Plasmid pNB2 was obtained as follows. An EcoRI-PvuII fragment (967 bp) from pZAQ (8) containing the complete ftsQ gene was cloned into an EcoRI-HincII pUC18 vector, resulting in plasmid pNB1. An EcoRI-PstI fragment from pNB1 was cloned into pBTac1 (Boehringer, Mannheim, Germany) between the EcoRI and PstI sites, behind an inducible tac promoter, resulting in plasmid pNB2. Plasmid pREP4 (Qiagen, Chatsworth, Calif.) is a multicopy plasmid containing the lacI gene. To construct the ponB-ftsQ hybrid, a two-step PCR was carried out to fuse the two genes. In the first PCR, the ponB part, which codes for the amino-terminal domain of PBP 1Bγ, was amplified with the primers pH1b (5′-CCGAATTCATGCCGCGCAAAGGT-3′) and pH1bQ (5′-GCGTTGCGCATCTTCCATGAGATAAACGCCGTA-3′) and with plasmid pBS99 (6) as the template DNA. Primer pH1bQ partially overlapped the ftsQ sequence. In the second PCR, the ftsQ part, which codes for the periplasmic domain of FtsQ, was amplified with lm40 (5′-CCCAGTCACGACGTTGTAAAACG-3′) and the ponB PCR product as primers and with plasmid pNB1 as the template DNA. The obtained insert was digested with EcoRI and PstI and ligated into an EcoRI- and PstI-digested pBTac1 vector, resulting in pNB9.
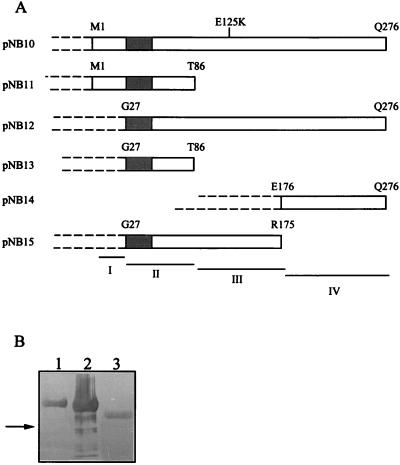
For the preparation of monoclonal antibodies (MAbs) (see also below), a β-galactosidase–FtsQ fusion protein was used. To obtain a lacZ-ftsQ fusion gene, an EcoRI-PvuII fragment (967 bp) from pZAQ was cloned in frame with the lacZ gene in an EcoRI- and SmaI-digested pEX2 vector (33), resulting in pNB10. For the epitope mapping, internal deletions (see Fig. 1A) were constructed in the lacZ-ftsQ fusion gene by the following procedure. A 620-bp KpnI fragment was deleted from pNB10, resulting in pNB11. From pNB1, an 870-bp BamHI-PstI fragment was subcloned into pEX2, resulting in pNB12. In pNB13, a 620-bp KpnI fragment was deleted from pNB12. From pNB12, a 550-bp SmaI-NruI fragment was deleted, resulting in pNB14. Deletion of a 428-bp NruI-PstI fragment from pNB12 resulted in pNB15. All constructs were sequenced with a T7 sequencing kit (Pharmacia, Uppsala, Sweden) with α-35S-dATP according to the dideoxy chain termination method of Sanger et al. (32).
FIG. 1.
(A) β-Galactosidase–FtsQ fusion proteins with internal deletions produced by overexpression of the corresponding constructs described in Materials and Methods. The four different antigenic domains are designated I to IV. The dark-gray region at the amino terminus represents the membrane-spanning sequence of FtsQ. The β-galactosidase part of the fusion protein is indicated by dashed lines. The amino acid substitution at position 125 (E125K) in the ftsQ1(Ts) mutant is indicated. (B) Reactivity of MAb 1-F7 with β-galactosidase–FtsQ fusion proteins with internal deletions. Lane 1, β-galactosidase–FtsQM1-Q276 (pNB10); lane 2, β-galactosidase–FtsQG27-Q276 (pNB12); lane 3, β-galactosidase–FtsQE176-Q276 (pNB14). Similar results were obtained with MAb 2-H1 and MAb 6-H5. The arrow indicates the position of β-galactosidase. Total cell extracts were applied to the gel.
Overproduction of FtsQ and PBP 1Bγ-FtsQ.
LMC500 harboring pNB2 and pREP4 (LMC1227) and JM101 harboring pNB2 (LMC1141) were used as FtsQ-overproducing strains. LMC500 harboring pNB9 and pREP4 (LMC1289) was used as the PBP 1Bγ-FtsQ-overproducing strain. Derivatives of LMC500 were grown to steady state in glucose minimal medium containing 6.33 g of K2HPO4 · 3H2O, 2.95 g of KH2PO4, 1.05 g of (NH4)2, 0.10 g of MgSO4 · 7H2O, 0.28 mg of FeSO4 · 7H2O, 7.1 mg of Ca(NO3)2 · 4H2O, 4 mg of thiamine, 4 g of glucose, and 50 μg of lysine per liter (pH 7.0) at 28°C. If required, ampicillin (200 μg/ml) and kanamycin (50 μg/ml) were added. To overproduce FtsQ and PBP 1Bγ-FtsQ, gene expression was induced for 1 h by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 450 nm of 0.1. JM101 harboring pNB2 was grown in TY medium (1% tryptone, 0.5% yeast extract, 3 mM NaOH, 0.5% NaCl) at 37°C. Gene expression was induced by the addition of 2 mM IPTG at an optical density at 600 nm of 0.3, and the cells were grown for an additional 3 h at 37°C.
Detection of FtsQ by Western blotting.
Electrophoresis and immunoblotting were performed as described by Laemmli (24) and Towbin et al. (37), respectively. The membrane was probed with FtsQ-specific MAbs or polyclonal antibodies. Washing steps were performed according to the ECL Western blotting protocol (Amersham, Little Chalfont, Buckinghamshire, England). The membrane was incubated with horseradish peroxidase-conjugated sheep anti-mouse or sheep anti-rabbit antibodies and developed with chemiluminescence reagents (Amersham).
Preparation of MAbs against FtsQ. (i) Expression of the lacZ-ftsQ fusion gene and isolation of the fusion protein.
Expression of the lacZ-ftsQ fusion gene from pNB10 was performed as described by Voskuil et al. (42). The 148-kDa fusion protein was isolated from a preparative sodium dodecyl sulfate–5.8% polyacrylamide gel after staining it in 300 mM CuCl2 and destaining it in distilled water. The excised fusion protein band of 148 kDa was washed three times for 20 min in 0.25 M EDTA–0.25 M Tris-HCl (pH 9.0) (25). The fusion protein was electroeluted overnight at 3 W in 0.3% Tris-HCl (wt/vol), 1.5% glycine (wt/vol), and 0.025% sodium dodecyl sulfate (wt/vol) according to the method described by Jacobs and Clad (20).
(ii) Immunization procedure.
BALB/c mice were immunized by injection with β-galactosidase–FtsQ fusion protein as described by Voskuil et al. (42). At day 0, 82 μg of protein in incomplete Freund’s adjuvant was injected. At day 66, 74 μg of protein in incomplete Freund’s adjuvant was injected. At day 79, 320 μg of protein in complete adjuvant was injected. At day 141, 150 μg of protein in phosphate-buffered saline (PBS) was injected. At day 322, 323 μg of protein in 150 μl of 0.15 M NaCl was injected. Three days later, antiserum was obtained, the lymphocytes were fused with NS1 myeloma cells, and the resulting hybridomas were grown in microtiter plates as described previously (22).
(iii) Screening and selection of hybridomas.
Screening of the hybridomas was performed in an enzyme-linked immunosorbent assay (ELISA) and by Western blotting. Cell envelopes were isolated from cells disrupted by sonication as described by Zijderveld et al. (49). A protein fraction enriched with cytoplasmic membrane proteins was obtained by incubating cell envelopes with sodium-lauryl sarcosinate according to the method of Filip et al. (14). Polystyrene microtiter plates with high binding capacity (Greiner, Nürtingen, Germany) were coated with 0.5 μg of protein fraction enriched with cytoplasmic membrane proteins of the FtsQ-overproducing strain LMC1141 and were incubated overnight at 4°C. Control ELISA plates were coated either with 0.25 μg of β-galactosidase (Sigma Chemical Co., St. Louis, Mo.) or with 0.5 μg of protein of cell-free lysate of ftsQ-depleted VIP210 cells (12). Incubation with hybridoma culture supernatant, washing steps, and colorimetric analysis of antibody binding were performed as described by Voskuil et al. (42). Hybridomas were selected on the basis of a strongly positive reaction in the ELISA with the fraction of the FtsQ-overproducing strain and a negative reaction with both β-galactosidase and the cell-free lysate of the ftsQ-depleted VIP210 cells. Reactivities of the hybridomas, which were positive in the ELISA, were also tested in a Western blot assay. Polyclonal antibodies against FtsQ (a gift from M. Vicente) were used as a positive control for the FtsQ bands on Western blots. The hybridomas positive by ELISA and/or Western blotting were cloned by the limiting-dilution technique at a density of 1 cell per well and were subcloned twice to a density of 0.3 cell per well (22).
(iv) Purification and determination of the classes and subclasses of the MAbs.
The cloned hybridomas were grown in bulk, and the MAbs were isolated from the culture supernatants by affinity chromatography on protein G matrix (Pharmacia). The immunoglobulin subclasses of the MAbs were determined by ELISA with subclass-specific antisera (MonoAb Screen ID kit; Zymed Laboratory Inc., San Francisco, Calif.). MAb 1-F7, MAb 2-H1, and MAb 6-H5 belonged to the immunoglobulin G1 subclass harboring κ light chains.
Immunofluorescence procedure.
Derivatives of strain LMC500 were grown to steady state in glucose minimal medium. If required, ampicillin (200 μg/ml) and kanamycin (50 μg/ml) were added. Strain B/rA was grown in minimal medium with 0.08% l-alanine as the carbon source (23). For temperature shift experiments, a culture of strain LMC531 growing at 28°C was diluted in prewarmed medium at 42°C and grown for two mass doublings. VIP210 was grown at 28°C in minimal medium supplemented with 0.05% Casamino Acids and 34 μg of chloramphenicol per ml. To deplete cells of their ftsQ genes, the cells were diluted in prewarmed medium at 42°C and grown for four mass doublings.
Cells were fixed in 2.8% formaldehyde and 0.04% glutaraldehyde in growth medium at room temperature for 15 min. The cells were centrifuged at 8,000 × g for 5 min, washed three times in PBS (pH 7.2), and subsequently incubated in 0.1% Triton X-100 in PBS for 45 min at room temperature. The cells were washed three times in PBS and incubated in PBS containing 100 μg of lysozyme per ml and 5 mM EDTA for 45 min at room temperature. Finally, the cells were washed three times in PBS.
Prior to immunofluorescence staining, nonspecific binding sites were blocked by incubating the cells in 0.5% (wt/vol) blocking reagents (Boehringer) in PBS for 30 min at 37°C. As primary antibodies, either MAbs against FtsQ or PBP 1B (11) were used. The antibodies were diluted in blocking buffer, and incubation was carried out overnight at 37°C. The cells were washed three times with PBS containing 0.05% (vol/vol) Tween 20 (PBST). Incubation with secondary antibodies (goat anti-mouse antibody conjugated with Alexa546 [Molecular Probes Inc., Eugene, Oreg.]) diluted in blocking buffer was carried out for 30 min at 37°C. The cells were washed again three times in PBST. The nucleoids were stained with DAPI (4′,6-diamidino-2-phenylindole) at a final concentration of 0.5 μg/ml in H2O. The cells were washed once in H2O and resuspended in H2O.
Microscopic and image analyses.
Cells were immobilized on agarose slides as described by van Helvoort and Woldringh (39) and photographed with a cooled Princeton change-coupled device camera mounted on an Olympus BX-60 fluorescence microscope. Images were taken with the program IPlab Spectrum, version 3.0 (Signal Analytics Co., Vienna, Va.). In all experiments the cells were first photographed in the phase-contrast mode, next with a DAPI fluorescence filter (illuminated at 330 to 385 nm with a transmission range above 420 nm), and last with an Alexa filter (illuminated at 530 to 550 nm with a transmission range above 590 nm). The three photographs were stacked, and the length of each cell was measured in the phase-contrast image, the lengths and the positions of the nucleoids were measured in the DAPI image, and the foci were detected in the fluorescence image. Interactive measurements were taken as a “structured point collection” on a Macintosh 7200 computer with the public domain program Object-Image1.62n by Norbert Vischer (University of Amsterdam; simon.bio.uva.nl/object-image.html), which is based on NIH Image software (41).
RESULTS
Epitope mapping of MAbs against FtsQ.
To study the localization of FtsQ in situ, specific MAbs against FtsQ were produced. The selection of the hybridomas resulted in three MAbs, i.e., MAb 1-F7, MAb 2-H1, and MAb 6-H5. MAb 2-H1 and MAb 6-H5 reacted positively both in an ELISA with the protein fraction enriched with cytoplasmic membrane proteins of the FtsQ-overproducing strain LMC1141 and in a Western blot with the 31.5-kDa FtsQ band of the same fraction. MAb 1-F7 showed a strongly positive reaction in an ELISA but did not give a reaction in a Western blot (data not shown). The three antibodies did not show reactivity with β-galactosidase or with cell extracts from the ftsQ-depleted VIP210 cells (12), indicating that the antibodies specifically recognized the FtsQ protein (data not shown but see Fig. 2B, which shows that the FtsQ protein disappears in ftsQ-depleted cells).
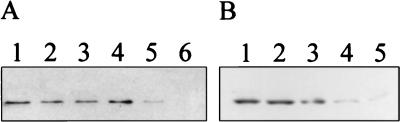
FIG. 2.
Detection of wild-type FtsQ in membranes of wild-type cells (LMC500), ftsQ1(Ts) cells (LMC531), and ftsQ-depleted cells (VIP210) disrupted with a French press. (A) Lane 1, LMC500 grown at 28°C; lanes 2 to 4, LMC500 grown at 42°C for 5, 30, and 60 min, respectively; lane 5, LMC531 grown at 28°C; lane 6, LMC531 grown at 42°C for 15 min. Each lane contains 40 μg of protein of the membrane fraction. (B) Lane 1, VIP210 grown at 28°C; lanes 2 to 5, VIP210 grown at 42°C for one mass doubling, two mass doublings, three mass doublings, and four mass doublings, respectively. Each lane contains 10 μg of protein of the membrane fraction. The proteins were detected with a mixture of MAbs against FtsQ.
To determine the epitopes on FtsQ, the reactivities of the antibodies against β-galactosidase–FtsQ fusion proteins harboring internal deletions in FtsQ were tested (Fig. 1). MAb 1-F7, MAb 2-H1, and MAb 6-H5 recognized β-galactosidase–FtsQM1-Q276, β-galactosidase–FtsQG27-Q276, and β-galactosidase–FtsQE176-Q276 both in Western blots (Fig. 1B) and in dot blots (data not shown). The three antibodies did not react with β-galactosidase–FtsQM1-T86, β-galactosidase–FtsQG27-T86, and β-galactosidase–FtsQG27-R175 (data not shown), indicating that the FtsQ epitopes are located in the last 100 amino acids of the protein (region IV).
Detection of FtsQ by Western blotting.
In the ftsQ1(Ts) mutant, a guanine-to-adenine transition at bp 397 has occurred (34). The transition mutation resulted in the substitution of a basic lysine residue for an acidic glutamate residue at amino acid 125 (FtsQE125K). When it was grown at the permissive temperature in minimal medium, the protein FtsQE125K could be detected on a Western blot although it was less abundant (Fig. 2A, lane 5) than FtsQ in wild-type cells (Fig. 2A, lane 1). After 15 min at the nonpermissive temperature, filaments with blunt constrictions were formed and FtsQE125K could no longer be detected with the MAbs (Fig. 2A, lane 6). However, the protein could be detected with polyclonal antibodies, raised in chicken against a histidine-tagged FtsQ protein, suggesting that the protein was not degraded at the nonpermissive temperature (data not shown). The amino acid substitution at position 125 might have affected folding of the protein and part of the C-terminal region of FtsQ might have been modified, resulting in a partial loss of the epitope region. Both FtsQE125K and wild-type FtsQ could be solubilized by treatment with Sarkosyl, indicating that the proteins were located in the cytoplasmic membrane (data not shown).
The presence of FtsQ in the ftsQ depletion strain was also analyzed by Western blotting (Fig. 2B). The membrane fractions of filaments obtained by growth at the nonpermissive temperature for up to four doubling times were used. The amount of FtsQ decreased in time; however, a faint band was still visible after four doubling times (Fig. 2B, lane 5). Detection with polyclonal antibodies also showed a decrease in the amount of FtsQ (data not shown). This indicates that although the ftsQ gene is depleted, the protein itself is very stable.
Localization of FtsQ by immunofluorescence microscopy.
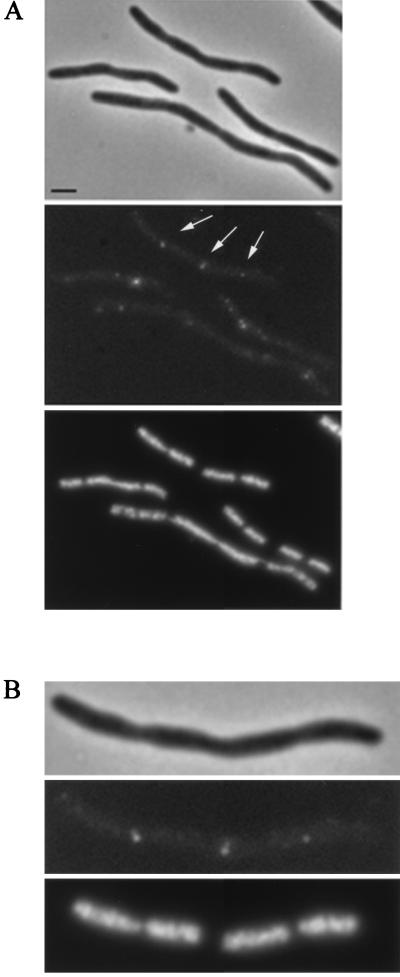
Although the amount of FtsQ protein in wild-type cells was very low (about 25 to 50 molecules exist per cell [7]), the FtsQ protein could be visualized by immunofluorescence microscopy with a mixture of the specific MAbs and a bright fluorophore (Alexa546) (Fig. 3 and 4). No fluorescence could be detected under conditions in which only the secondary antibodies were used (data not shown). The localization of FtsQ was studied in two different wild-type E. coli strains, i.e., a derivative of MC4100 and B/rA. The latter strain was chosen because the timing of its various cell cycle events is well known (19).
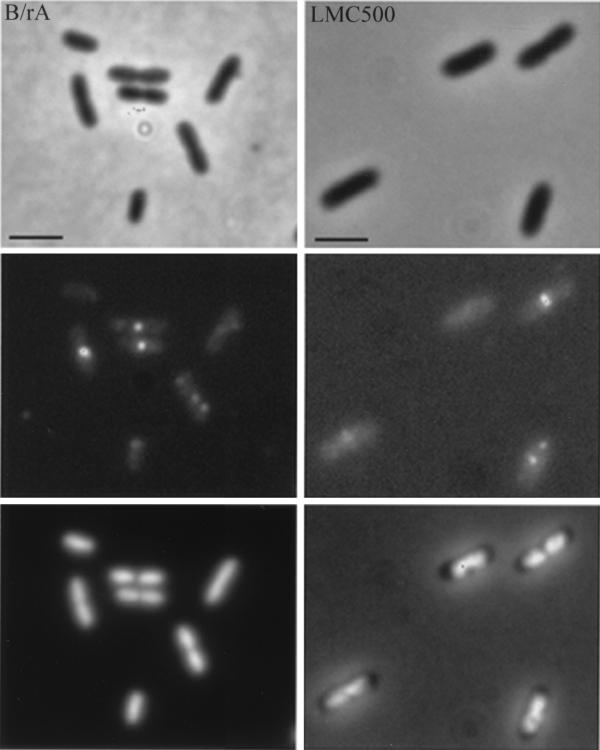
FIG. 3.
Localization of wild-type FtsQ by in situ immunofluorescence microscopy in B/rA (left) and LMC500 (right). The upper panels are phase-contrast images, the middle panels are immunofluorescence images, and the lower panels are DAPI images. Labeling was performed with a mixture of MAbs against FtsQ. Secondary antibodies were conjugated with Alexa. Scale bars, 2 μm.
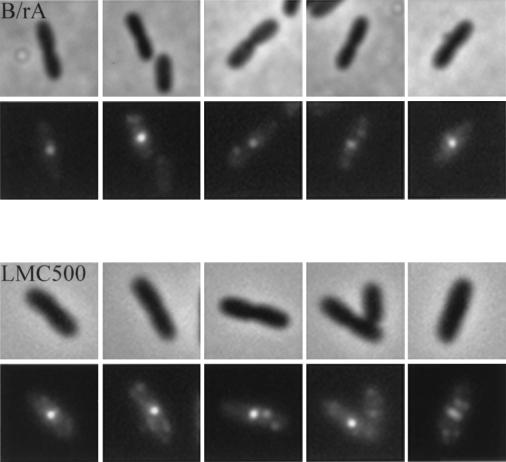
FIG. 4.
Localization of FtsQ in constricting cells of B/rA and LMC500. The upper panels are phase-contrast images, and the lower panels are immunofluorescence images. Labeling was performed with a mixture of MAbs against FtsQ. Secondary antibodies were conjugated with Alexa. The scale is identical to that used for Fig. 3.
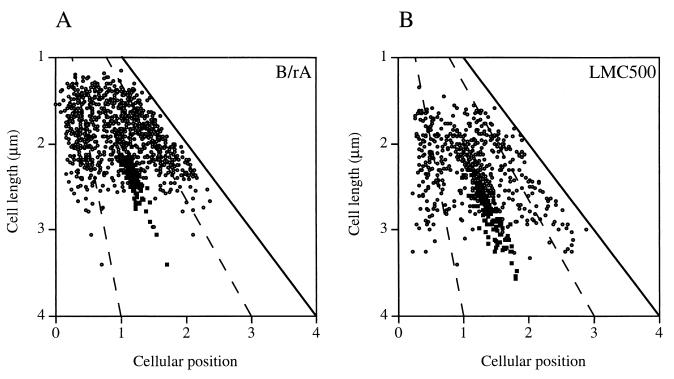
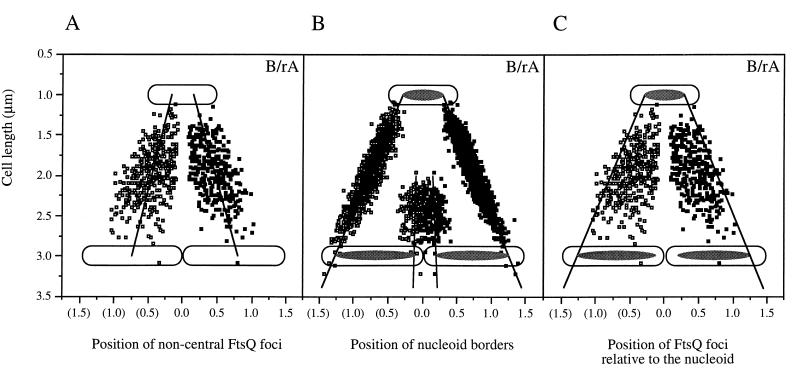
In both strains FtsQ could be visualized as foci (Fig. 3). Cells were analyzed according to cell length, length and position of the nucleoid, and fluorescent-labeling pattern with the image analysis program Object-Image (41) (Table 1). According to the fluorescent-labeling pattern, four different classes of cells could be distinguished, i.e., cells without foci, cells containing a single focus, cells containing two foci, and cells with more than two foci. However, unlike in LMC500, cells with more than two foci were not observed in B/rA. In Fig. 5 the position of FtsQ in cells with one or two foci is shown. The FtsQ foci that localized at midcell in constricting cells could clearly be distinguished from the asymmetrically localized foci in these cells. In nonconstricting cells (short cells) FtsQ foci could also be found at midcell. In both strains the average cell length of nonconstricting cells without foci was smaller than the average cell length of cells with one or more foci (for B/rA, 1.68 and 1.87 μm, respectively, and for LMC500, 1.99 and 2.13 μm, respectively). Nonconstricting cells with one asymmetrically localized fluorescent spot were somewhat smaller than cells in which the spot was localized at midcell (for B/rA, 1.76 and 1.83 μm, respectively, and for LMC500, 2.02 and 2.27 μm, respectively). These data suggest that FtsQ might be localized to the division site before a constriction is visible, similar to what was found for DivIB of B. subtilis (18). Alternatively, the FtsQ spot found at midcell might represent a site where FtsQ synthesis occurs (see below). A regression line with a significant coefficient was calculated both for FtsQ foci, with relative positions between 0 and 0.45, and for FtsQ foci, with relative positions between 0.55 and 1 (Fig. 6A). In small cells, i.e., with cell lengths between 0.9 and 1.8 μm for B/rA and between 1.30 and 2.5 μm for LMC500, the FtsQ foci were on average localized at one-third and two-third positions in the cell, whereas in longer cells the spots were on average localized at one-quarter and three-quarter positions. To better determine the positions of foci not located at midcell, we determined the positions of borders of the DAPI-stained nucleoids in relation to the length of the cell (Fig. 6B and C). From these data it was clear that the FtsQ foci were confined to that area of the cell where the nucleoid is located.
TABLE 1.
Parameters of length distributionsa
| Strain | t (min)b | No. of cells measured | Lavgc (μm) | CV%d | % of cells constricted | Lavgce (μm) | CV% |
|---|---|---|---|---|---|---|---|
| B/rA | 138 | 1,859 | 1.81 | 20.2 | 12.2 | 2.39 | 8.4 |
| LMC500 | 72 | 1,308 | 2.20 | 19.5 | 17.4 | 2.84 | 9.9 |
The parameters of length distributions of the total cell populations of both strains are similar to the data described by Koppes et al. (23).
Doubling times (t) were obtained with alanine in the case of B/rA and with glucose in the case of LMC500 as the carbon sources.
Lavg, average cell length.
CV, coefficient of correlation.
Lavgc, average cell length of cells showing constriction.
FIG. 5.
Position of FtsQ in B/rA cells (n = 1859) and LMC500 cells (n = 1308) with one or two foci depending on cell length. Dashed lines represent one-quarter and three-quarter positions in the cell, and the solid line represents cell length. Asymmetrically localized FtsQ foci in nonconstricting and constricting cells and foci at midcell in nonconstricting cells are indicated as open circles; FtsQ foci at midcell in constricting cells are indicated as filled squares.
FIG. 6.
(A) Positions of FtsQ foci with relative positions between 0 and 0.45 (filled symbols) and between 0.55 and 1 (open symbols) in B/rA cells. Representative 1- and 3-μm cells are schematically drawn to scale. Regression lines with a significant coefficient were calculated. Relative positions of foci are plotted relative to cell length values and against cell length relative to midcell. For FtsQ foci with relative positions between 0 and 0.45, the equation of the regression line is y = 0.323 · x − 0.186 (r = 0.513). For FtsQ foci with relative positions between 0.55 and 1, the equation of the regression line is y = 0.301 · x − 0.137 (r = 0.440). (B) Positions of the nucleoid borders as a function of cell length in B/rA. The method described above was also used to determine the correlation between the positions of the nucleoid borders and cell length. The equations of the regression lines are y = 0.485 · x − 0.206 (r = 0.937) and y = 0.488 · x − 0.196 (r = 0.915). (C) Combination of the data from panels A and B.
In constricting cells, as in nonconstricting cells, different fluorescent-labeling patterns could be observed. Unlike those of classes of nonconstricting cells, the average cell lengths of the different classes of constricting cells were similar. For DivIB of B. subtilis different labeling patterns in constricting cells could be correlated with cell length. A two-dot pattern was the earliest stage of localization, a line was the second stage, and a center dot was the last stage of the division process. However, FtsQ could be visualized only as one clear spot at the site of division (Fig. 4). The fact that 5,000 molecules of DivIB are present per cell compared with 50 molecules of FtsQ per cell might explain the difference in labeling patterns. Furthermore, the amino acid sequences of both proteins are similar but the percentage of homology is low, suggesting that DivIB and FtsQ are not identical but that they might be functionally related.
Only 50% of the constricting cells contained a central FtsQ spot in both LMC500 and B/rA. This finding might indicate that FtsQ disappears from the division site before septation of the two daughter cells is completed or, alternatively, that FtsQ can be visualized by immunofluorescence microscopy only if a sufficient number of molecules are grouped together.
Localization of FtsQ in an ftsQ(Ts) strain and in an ftsQ depletion strain.
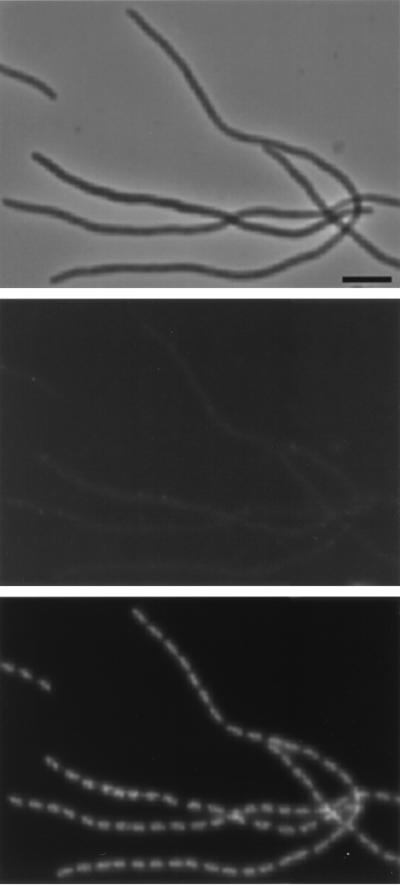
To demonstrate that the immunofluorescence microscopy results of FtsQ in wild-type cells specifically located FtsQ, we also studied the temperature-sensitive ftsQ1(Ts) mutant and the ftsQ depletion strain VIP210 (12). In ftsQ1(Ts) cells grown at the permissive temperature, FtsQ could localize to the division site even in cells without a visible constriction (data not shown). Taschner et al. (36) showed that the constriction period in these cells was longer than that in wild-type cells. Thus, FtsQ could be observed at the constriction site early in the division process and also appeared more persistent at this location than in wild-type cells. Although the ftsQ1(Ts) mutant protein FtsQE125K could not be detected with the MAbs in membranes of ftsQ1(Ts) filaments by Western blotting (Fig. 2), fluorescent foci were observed by immunofluorescence microscopy (Fig. 7). Some of these foci were found at potential division sites, indicating that the protein is still able to localize to the division site but that it is inactive in the division process. In wild-type cells, an average of 0.60 foci per μm of cell length was calculated. For the ftsQ1(Ts) mutant, an average of 0.34 foci per μm was calculated for cells grown at the permissive temperature and 0.18 foci per μm was calculated for cells grown for two mass doublings at the nonpermissive temperature. This decreasing number of foci per micrometer is in line with the Western blot data (Fig. 2) which show that the FtsQ protein was less abundant in ftsQ1(Ts) cells and could not be detected in ftsQ1(Ts) filaments.
FIG. 7.
Labeling of FtsQ in the ftsQ1(Ts) mutant. (A) Filaments obtained after growth at the nonpermissive temperature for two mass doublings. The upper panels are phase-contrast images, the middle panels are immunofluorescence images, and the lower panels are DAPI images. Labeling was performed with a mixture of MAbs against FtsQ. Secondary antibodies were conjugated with Alexa. The arrows indicate fluorescent label at (potential) constriction sites. Scale bar, 2 μm. (B) Filament shown in more detail.
In the ftsQ depletion strain, FtsQ could also localize to the division site in cells grown at the permissive temperature (data not shown). In filaments obtained after growth at the nonpermissive temperature for four doubling times, no fluorescent spots could be detected (Fig. 8). We do not know why there are no fluorescent spots in the ftsQ-depleted filaments, even though by Western blotting the FtsQ protein could still be detected after four doubling times. Remarkably, similar observations have been made by Weiss et al. (44) with an ftsI temperature-sensitive mutant and an ftsI depletion strain.
FIG. 8.
Labeling of FtsQ in the ftsQ depletion strain. Cells grown at the nonpermissive temperature for four mass doublings were labeled with a mixture of MAbs against FtsQ. Secondary antibodies were conjugated with Alexa. The upper panels are phase-contrast images, the middle panels are immunofluorescence images, and the lower panels are DAPI images. Scale bar, 4 μm.
Localization of a hybrid protein of PBP 1B and FtsQ by immunofluorescence microscopy.
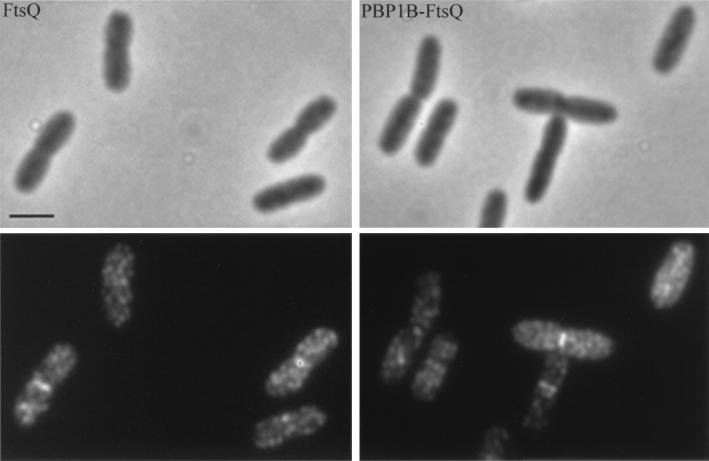
To determine which domain of FtsQ is needed for its localization at the cell center, a hybrid protein in which the cytoplasmic domain and the transmembrane domain were derived from the γ form of PBP 1B (amino acids M46 to L87) and the periplasmic domain was derived from FtsQ (amino acids M49 to Q276) was used. PBP 1B is a bitopic cytoplasmic membrane protein with a membrane topology (10, 13) similar to that of FtsQ and is involved in peptidoglycan synthesis during cell elongation (45). Both the PBP 1Bγ-FtsQ hybrid protein and the wild-type FtsQ protein were overproduced in a wild-type strain, and their localizations were determined by in situ labeling with MAbs against FtsQ.
As shown in Fig. 9, label could be found at midcell. The PBP 1Bγ-FtsQ hybrid protein, as well as overproduced wild-type FtsQ, could also be found as foci in parts of the cell other than at the constriction. The PBP 1Bγ-FtsQ hybrid protein could restore cell division in the ftsQ1(Ts) mutant when the mutant was grown at the nonpermissive temperature. Furthermore, an FtsQ protein whose carboxy terminus was truncated by more than 80% was not functional in vivo (data not shown). This indicates that the carboxy-terminal periplasmic domain of FtsQ is important for its function in cell division. The images of the hybrid protein are quite different from those obtained after overproduced PBP 1B was labeled with MAbs against PBP 1B alone in the sense that in the latter case no preferential label was at midcell (data not shown). This result indicates that the periplasmic domain of FtsQ determines the localization of FtsQ, as has also been concluded for the periplasmic domain of FtsN (2).
FIG. 9.
Localization of FtsQ in FtsQ-overproducing cells (left) and in PBP 1Bγ-FtsQ-overproducing cells (right). The upper panels are phase-contrast images, and the lower panels are immunofluorescence images. Labeling was performed with a mixture of MAbs against FtsQ. Secondary antibodies were conjugated with Alexa. Scale bar, 2 μm.
DISCUSSION
In a pioneering experiment, Maddock and Shapiro (29) demonstrated that chemotaxis proteins localized as bright fluorescent foci to the cell poles. Importantly, the authors corroborated this result by immunogold electron microscopy, and they could thus show that fluorescent foci are markers of genuine cellular components. Similarly, results of immunofluorescence microscopy of FtsZ correlated well with the original finding of the FtsZ ring by immunogold electron microscopy (3). Of course, electron microscopy has a higher resolution than light microscopy, and this is reflected in the precision of labeling. For instance, electron microscopy can reveal whether markers are associated with the inner or the outer membrane. However, the brightness of fluorescence signals makes detection much easier, and in this sense light microscopy surpasses electron microscopy. Thus, it is not surprising that localization of FtsI (PBP 3) to the cell center has been achieved by fluorescence microscopy (43, 44) and not by electron microscopy.
For the localization of the low-abundance membrane-bound cell division protein FtsQ by immunofluorescence microscopy, we used MAbs as primary antibodies. To assess the specificity of FtsQ immunostaining, we carried out different tests. (i) Epitope mapping with the aid of truncated fusion proteins and immunoblot detection allowed us to localize the epitopes between amino acids 176 and 276, and β-galactosidase was clearly not recognized. (ii) No bright foci were observed in cells labeled with secondary antibody only. (iii) Fluorescent label decreased after depletion of FtsQ.
Location of FtsQ at midcell.
FtsQ localized to the division site in most but not all constricting cells. This indicates either that FtsQ disappears from the constriction site before separation of the two daughter cells or that FtsQ is localized early in division but that it can be labeled only if a sufficient number of molecules are grouped closely together. The latter option seems more likely since the FtsQ homolog DivIB of B. subtilis was found to localize early to the division site, even before a visible septation could be observed, and remained localized throughout division (18). In the ftsQ1(Ts) mutant the glutamate residue at position 125 is replaced by a lysine residue. This amino acid is located in a highly conserved domain of FtsQ in various gram-negative and gram-positive bacteria. In this mutant, FtsQ localized to the division site at the permissive temperature. In filaments in which cell division was inhibited, FtsQ foci occurred occasionally at the abortive division site (Fig. 7). This finding raises the possibility that the localization of FtsQ is independent of its function in the division process. For the localization of DivIB and FtsN, the external and periplasmic C-terminal domains, respectively, are required. For DivIB it has been suggested that this domain is involved in interactions with other membrane-bound division proteins (17). A MalG-FtsN fusion protein could localize to the division site and was also found to be functional in cell division (2, 9). Possible candidates for interaction with FtsN are PBP 3 and/or FtsQ, because the activities of both proteins are required for FtsN localization (2). The PBP 1Bγ-FtsQ hybrid protein was also found to localize to the division site. Because localization of the hybrid protein was studied under overproducing conditions, one might argue that localization at midcell is not specific and that this would be caused by crowding of the protein in a region highly composed of membrane. However, PBP 1B itself was not found localized to the division site when the protein was overproduced. Therefore, it can be concluded that the periplasmic domain of FtsQ determines its localization, similar to what was found for FtsN and DivIB.
Location of FtsQ at other sites.
As pointed out above, fluorescence microscopy allows visualization of clusters of proteins. So what can be the significance of foci not located at the site of division? It is known that cytoplasmic membrane proteins are inserted cotranslationally into the membrane (46). Also with FtsQ there is evidence that translation and membrane insertion are coupled (38). It is therefore likely that foci not located at midcell represent groups of newly synthesized FtsQ molecules. In line with this interpretation, LMC500 cells contained more fluorescent foci than B/rA cells. Since the doubling time of strain LMC500 is shorter than the doubling time of B/rA, LMC500 cells probably contain more than one chromosome equivalent, which might result in multiple FtsQ synthesis sites and therefore in more FtsQ foci.
Is the arrangement of the nucleoid spatially related to the membrane insertion of FtsQ? The phenomenon of cotranscriptional-cotranslational insertion of membrane proteins implies that the nucleoid is linked through mRNA and ribosomes to the cytoplasmic membrane (reference 30 and references therein). Our measurements on the cellular nucleoid positions (Fig. 5) reveal that the noncentral FtsQ foci are confined to this cellular area, which suggests that FtsQ becomes inserted in the vicinity of the nucleoid. An interesting speculation is that the foci might occur where the division and cell wall gene cluster resides. To corroborate this intriguing possibility, it will be necessary to combine gene and protein localization studies.
ACKNOWLEDGMENTS
This work was supported by the Life Sciences Foundation (grant 805-33-221P), which is subsidized by the Netherlands Organization for Scientific Research.
We thank Jannet van Leeuwen and Sjoukje Kuijper for their assistance in the production of the MAbs against FtsQ. We thank Tanneke den Blaauwen and Martine Nguyen-Distèche for critically reading the manuscript and Marco Roos for help with statistical analysis.
REFERENCES
- 1.Addinal S G, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinal S G, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 3.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 4.Bowler L D, Spratt B G. Membrane topology of penicillin-binding protein 3 of Escherichia coli. Mol Microbiol. 1989;3:1277–1286. doi: 10.1111/j.1365-2958.1989.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 5.Boyle D S, Khattar M M, Addinall S G, Lutkenhaus J, Donachie W D. ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol. 1997;24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 6.Broome-Smith J K, Edelman A, Yousif S, Spratt B G. The nucleotide sequences of ponA and ponB genes encoding penicillin-binding proteins 1A and 1B of Escherichia coli. Eur J Biochem. 1985;147:437–446. doi: 10.1111/j.1432-1033.1985.tb08768.x. [DOI] [PubMed] [Google Scholar]
- 7.Carson M J, Barondess J, Beckwith J. The FtsQ protein of Escherichia coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J Bacteriol. 1991;173:2187–2195. doi: 10.1128/jb.173.7.2187-2195.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corton J C, Ward J E J, Lutkenhaus J. Analysis of cell division gene ftsZ (sulB) from gram-negative and gram-positive bacteria. J Bacteriol. 1987;169:1–7. doi: 10.1128/jb.169.1.1-7.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai K, Xu Y, Lutkenhaus J. Topological characterization of the essential Escherichia coli cell division protein FtsN. J Bacteriol. 1996;178:1328–1334. doi: 10.1128/jb.178.5.1328-1334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Blaauwen T, Nanninga N. Topology of penicillin-binding protein 1b of Escherichia coli and topography of four antigenic determinants studied by immunocolabeling electron microscopy. J Bacteriol. 1990;172:71–79. doi: 10.1128/jb.172.1.71-79.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.den Blaauwen T, Wientjes F B, Kolk A H J, Spratt B G, Nanninga N. Preparation and characterization of monoclonal antibodies against native membrane-bound penicillin-binding protein 1B of Escherichia coli. J Bacteriol. 1989;171:1394–1401. doi: 10.1128/jb.171.3.1394-1401.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dopazo A, Palacios P, Sanchez M, Pla J, Vicente M. An amino-proximal domain required for the localization of FtsQ in the cytoplasmic membrane, and for its biological function in Escherichia coli. Mol Microbiol. 1992;6:715–722. doi: 10.1111/j.1365-2958.1992.tb01520.x. [DOI] [PubMed] [Google Scholar]
- 13.Edelman A, Bowler L, Broome-Smith J K, Spratt B G. Use of β-lactamase fusion vector to investigate the organization of the penicillin-binding protein 1B in the cytoplasmic membrane of Escherichia coli. Mol Microbiol. 1987;1:101–106. doi: 10.1111/j.1365-2958.1987.tb00533.x. [DOI] [PubMed] [Google Scholar]
- 14.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzmán L, Barondess J, Beckwith J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 16.Hale C A, de Boer P A J. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:1–20. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 17.Harry E J, Stewart S B J, Wake R G. Characterization of mutations in divIB of Bacillus subtilis and cellular localization of the DivIB protein. Mol Microbiol. 1993;7:611–621. doi: 10.1111/j.1365-2958.1993.tb01152.x. [DOI] [PubMed] [Google Scholar]
- 18.Harry E J, Wake R G. The membrane-bound cell division protein DivIB is localized to the division site in Bacillus subtilis. Mol Microbiol. 1997;25:275–283. doi: 10.1046/j.1365-2958.1997.4581822.x. [DOI] [PubMed] [Google Scholar]
- 19.Helmstetter C E. Timing of synthetic activities in the cell cycle. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1627–1639. [Google Scholar]
- 20.Jacobs E, Clad A. Electroelution of fixed and stained membrane proteins from preparative SDS-polyacrylamide gels into a membrane trap. Anal Biochem. 1986;154:583–589. doi: 10.1016/0003-2697(86)90033-3. [DOI] [PubMed] [Google Scholar]
- 21.Khattar M M, Begg K J, Donachie W D. Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J Bacteriol. 1994;176:7140–7147. doi: 10.1128/jb.176.23.7140-7147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolk A H J, Ho M L, Klatser P R, Eggelte T A, Kuijper S, de Jonge S, van Leeuwen J. Production and characterization of monoclonal antibodies to Mycobacterium tuberculosis, M. bovis (BCG) and M. leprae. Clin Exp Immunol. 1984;58:511–521. [PMC free article] [PubMed] [Google Scholar]
- 23.Koppes L J H, Woldringh C L, Nanninga N. Size variations and correlation of different cell cycle events in slow-growing Escherichia coli. J Bacteriol. 1978;134:423–433. doi: 10.1128/jb.134.2.423-433.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli E K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee C, Levin A, Branton D. Copper staining: a five-minute protein stain for sodium dodecyl sulfate-polyacrylamide gels. Anal Biochem. 1987;166:308–312. doi: 10.1016/0003-2697(87)90579-3. [DOI] [PubMed] [Google Scholar]
- 26.Levin A P, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 27.Lutkenhaus J, Mukherjee A. Cell division. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1615–1626. [Google Scholar]
- 28.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 30.Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raskin D M, de Boer A J. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanley K K, Luzio J P. Construction of a new family of high efficiency bacterial expression vectors: identification of cDNA clones coding for human liver proteins. EMBO J. 1984;3:1429–1434. doi: 10.1002/j.1460-2075.1984.tb01988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storts D R, Markovitz A. A novel rho promoter::Tn10 mutation suppresses an ftsQ1(Ts) missense mutation in an essential Escherichia coli cell division gene by a mechanism not involving polarity suppression. J Bacteriol. 1991;173:655–663. doi: 10.1128/jb.173.2.655-663.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Q, Margolin W. FtsZ dynamics during the division cycle of live Escherichia coli cells. J Bacteriol. 1998;180:2050–2056. doi: 10.1128/jb.180.8.2050-2056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taschner P E M, Huls P G, Pas E, Woldringh C L. Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J Bacteriol. 1988;170:1533–1540. doi: 10.1128/jb.170.4.1533-1540.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valent Q A, Scotti P A, High S, de Gier J-W L, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Helvoort J M L M, Woldringh C L. Nucleoid partitioning in Escherichia coli during steady-state growth and upon recovery from chloramphenicol treatment. Mol Microbiol. 1994;13:577–583. doi: 10.1111/j.1365-2958.1994.tb00452.x. [DOI] [PubMed] [Google Scholar]
- 40.Vidal-Ingigliardi D, Raiboud O. The mac promoters: functional hybrid promoters activated by the malT product and repression by the lacI product. Nucleic Acids Res. 1985;13:1164–1174. doi: 10.1093/nar/13.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vischer N O E. Object-Image: an interactive image analysis program using structured point collection. Binary. 1994;6:160–166. [Google Scholar]
- 42.Voskuil J L A, Westerbeek C A M, Wu C, Kolk A H J, Nanninga N. Epitope mapping of Escherichia coli cell division protein FtsZ with monoclonal antibodies. J Bacteriol. 1994;176:1886–1893. doi: 10.1128/jb.176.7.1886-1893.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Khattar M K, Donachie W D, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weiss D S, Pogliano K, Carson M, Guzmán L-M, Fraipont C, Nguyen-Distèche M, Losick R, Beckwith J. Localization of the Escherichia coli cell division protein FtsI (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- 45.Wientjes F B, Nanninga N. On the role of the high-molecular-weight penicillin-binding proteins in the cell cycle of Escherichia coli. Res Microbiol. 1991;142:333–344. doi: 10.1016/0923-2508(91)90049-g. [DOI] [PubMed] [Google Scholar]
- 46.Wolin S L. From the elephant to E. coli: SRP-dependent protein targeting. Cell. 1994;77:787–790. doi: 10.1016/0092-8674(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 47.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 48.Yu X, Tran A H, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zijderveld C A L, Aarsman M E G, den Blaauwen T, Nanninga N. Penicillin-binding protein 1B of Escherichia coli exists in dimeric forms. J Bacteriol. 1991;173:5740–5746. doi: 10.1128/jb.173.18.5740-5746.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]