Abstract
Tight coordination of the spatial relationships between protein complexes is required for cellular function. In neuronal synapses, many proteins responsible for neurotransmission organize into subsynaptic nanoclusters whose trans-cellular alignment modulates synaptic signal propagation. However, the spatial relationships between these proteins and NMDA receptors (NMDARs), which are required for learning and memory, remain undefined. Here, we mapped the relationship of key NMDAR subunits to reference proteins in the active zone and postsynaptic density using multiplexed super-resolution DNA-PAINT microscopy. GluN2A and GluN2B subunits formed nanoclusters with diverse configurations that, surprisingly, were not localized near presynaptic vesicle release sites marked by Munc13–1. However, a subset of presynaptic sites was configured to maintain NMDAR activation: these were internally denser, aligned with abundant PSD-95, and associated closely with specific NMDAR nanodomains. This work reveals a new principle regulating NMDAR signaling and suggests that synaptic functional architecture depends on assembly of multiprotein nanodomains whose interior construction is conditional on trans-cellular relationships.
INTRODUCTION
Many cellular functions are performed by macromolecular protein ensembles that require nanoscale spatial relationships with neighboring ensembles to facilitate complex signaling. These critical relationships are required in healthy signaling and disrupted in disease across biological systems. An important case where this is clear is the neuronal synapse1–3. Synapses mediate highly complex intercellular signaling to respond extremely rapidly and with high fidelity to diverse stimuli, propagating information in the brain. Despite their small size (<500 nm), the spatial position of signaling events within the synapse can critically influence their effect on neuronal signaling4–7. Indeed, converging lines of evidence suggest the nanoscale positioning, relative to one another, of the protein ensembles that mediate synaptic signaling is a key determinant of local synapse function8.
A clear case of how nanoscale coordination of protein ensembles influences synaptic transmission is the regulation of ionotropic glutamate receptor activation. Presynaptic vesicle release machinery, postsynaptic receptors, and the scaffolds that position them each concentrate in <100 nm diameter subsynaptic regions of high protein density (nanoclusters, NCs)8. Alignment of NCs across the synapse from one another into the trans-synaptic “nanocolumn” plays a critical role in regulating synaptic strength by enriching AMPA receptors (AMPARs) directly across the synaptic cleft from release sites9,10. Indeed, perturbing this complex nanoscale relationship between AMPARs and release sites disrupts synaptic transmission1,11–13, most remarkably even if synaptic receptor content is not altered14, demonstrating the fundamental role of nanoscale protein contextual relationships in neuronal function.
Despite the clear importance of nanoscale context for AMPAR function, how synaptic NMDA receptors (NMDARs) are organized relative to release sites remains unanswered. NMDARs are required for learning and memory and disrupted in many neurological disorders15, highlighting the need to understand their regulation. Importantly, though, the molecular context of NMDARs is critical for determining synaptic function even beyond controlling their activation, as the receptors form large signaling super-complexes that are intimately involved in both driving plasticity and establishing molecular organization within synapses15–17. Indeed, NMDARs signal via both Ca2+ flux-dependent and independent mechanisms16,18, and are attached via their large extracellular domains and long C-terminal tails to myriad extra and intracellular signaling proteins by which they are presumed to organize subsynaptic signaling domains19–21. These roles for NMDARs also depend on receptor subunit composition: receptor kinetics and biophysics are subunit specific16, super-resolution microscopy has shown differences in subunit distribution within single synapses22,23, and GluN2 subunit interactions with scaffolds and signaling molecules are differentially mediated by subunit C-tails19,20,24. It is therefore critical to understand NMDAR subunit positioning and molecular context, as these key attributes will control receptor activation and may also indicate regions where NMDARs create their own unique environments to facilitate subsynaptic signaling. However, we have until recently lacked the tools to investigate multiple protein complex relationships simultaneously, and therefore lack basic maps of receptor context to aid these determinations.
To map the spatial relationships of endogenous NMDAR GluN2 subunits to key pre and postsynaptic nanodomains in synapses, we utilized the multiplexing capabilities of DNA Exchange-PAINT (Points Accumulation for Imaging in Nanoscale Topography). We targeted the relationships in cultured rat hippocampal neurons between the critical GluN2 subunits GluN2A and GluN2B15, the major NMDAR postsynaptic scaffold protein PSD-9525, and presynaptic release sites, as marked by the vesicle priming protein Munc13–126. This multiplexed mapping approach revealed nanodomain assembly principles in single synapses. Most surprisingly, we find that NMDARs are typically lacking from the immediate nanoscale region of the PSD directly across from presynaptic release sites. However, the protein content and context of individual presynaptic release sites in single synapses differed markedly from one another. Only a subset of Munc13–1 NCs were enriched across the synapse with high-density PSD-95, and these aligned NCs had significantly higher Munc13–1 protein density. Critically, these nanocolumnar release sites were dramatically more enriched with GluN2 subunits than non-nanocolumnar release sites in the same synapses, likely ensuring a higher probability of activation of specific subtypes and subsets of receptors following release from appropriate presynaptic fusion sites. These results demonstrate that NMDAR positioning and organization are governed by a trans-cellular assembly of multiprotein ensembles and suggest that overall synapse architecture may arise from local formation of subsynaptic domains with unique functional roles.
RESULTS
Mapping endogenous NMDAR organization with DNA-PAINT
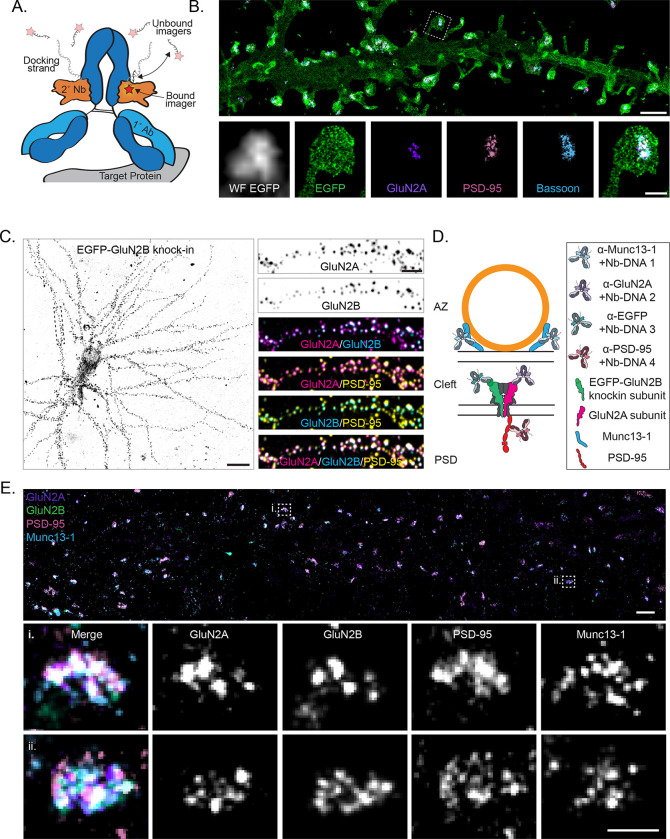
We took advantage of the high resolution and multiplexing flexibility of the single molecule localization microscopy (SMLM) technique DNA Exchange-PAINT27 to map the nanoscale relationships of multiple proteins at a single synapse. To facilitate multiplexing, we preincubated28,29 primary antibodies separately with secondary nanobodies conjugated to orthogonal DNA-PAINT docking strands, then combined them on the sample (Fig 1A, and see Materials and Methods). This method allows simple, antibody species-independent multiplexing and saturates the primary antibody with a defined number of DNA strands close to ideal for SMLM (1 per nanobody; 2 nanobodies per antibody). The use of small secondary nanobodies also significantly reduces linkage error compared to a secondary antibody28,29 and avoids the technical hurdles of direct primary antibody conjugation. NC-sized (40–100 nm) regions of high protein density can be observed within single synapses (~300–500 nm) in dendritic spines (~1 μm) in DNA Exchange-PAINT renderings of GluN2A, the pre- and post-synaptic scaffolds Bassoon and PSD-95, and a myristoylated-EGFP cell fill (Fig 1B), revealing the power of this technique to resolve the nanoscale organization and context of multiple proteins at a single synapse.
Figure 1.
Mapping endogenous NMDA receptor organization with DNA-PAINT. a) Schematic of a primary antibody labeled with a DNA-PAINT docking strand-conjugated secondary nanobody and imaged with fluorescent imager strands. Red star indicates fluorophore. b) (top) DNA-PAINT rendering (10 nm pixels) of myristoylated-EGFP cell fill, surface expressed GluN2A, PSD-95, and Bassoon demonstrating four-target, synaptic DNA-PAINT. Scale bar 2 μm. (bottom) Boxed region from top, including widefield image of myr-EGFP. Scale bar 500 nm. c) (left) Confocal image of EGFP-GluN2B CRISPR knock-in cell. Scale bar 20 μm. (right) Boxed region from left, showing surface expressed GluN2A, surface expressed GluN2B (EGFP knock-in), and PSD-95 labeling colocalized at synapses. Scale bar 4 μm. d) Schematic of four-target DNA-PAINT labeling of endogenous, surface expressed NMDAR subunits GluN2A and GluN2B with pre (Munc13–1) and postsynaptic (PSD-95) molecular context using primary antibodies preincubated with secondary nanobodies. This labeling scheme is used throughout the figures. e) (top) DNA-PAINT rendering (10 nm pixels) of endogenous, surface expressed GluN2A and GluN2B (EGFP knock-in), PSD-95, and Munc13–1. Scale bar 1 μm. (bottom) Zoom-in on two representative synapses showing nanoclusters of each protein and their co-organization. Scale bar 200 nm.
We aimed to measure the distribution of endogenous, surface expressed NMDAR subunits with both pre- and postsynaptic molecular context. To do this, we performed 2D DNA Exchange-PAINT of GluN2A, GluN2B, PSD-95 and Munc13–1 together at synapses in DIV21 rat primary hippocampal cultures. We labeled surface-expressed GluN2A (Fig S1A–C) and total PSD-95 and Munc13–1 with antibodies to avoid overexpression artifacts. As we were unsatisfied with commercially available antibodies targeting surface expressed GluN2B, we used ORANGE CRISPR30 to knock in EGFP to the extracellular domain of endogenous GluN2B, then labeled surface expressed EGFP-GluN2B with an anti-EGFP antibody (Fig 1C–D and S1D). We were routinely able to identify synapses containing all four proteins (Fig 1E), and only analyzed synapses containing EGFP-GluN2B localizations, as those without EGFP-GluN2B may either genuinely lack the subunit or could have accumulated an indel during genome editing and be GluN2B knock-outs. We further only selected synapses perpendicular (en face) to the optical axis for further analysis to best measure positional relationships between proteins with high fidelity (Fig S2). Together, these data demonstrate a workflow to map the distribution of surface-expressed, endogenous NMDAR subunit nanoclustering and position in the spatial context of other key pre and postsynaptic nanodomains.
Endogenous GluN2 subunits form diverse nanodomain types
In pyramidal neurons in hippocampus and neocortex, the dominantly expressed GluN2A and GluN2B subunits15 are each found in subsynaptic NCs that are postulated to control receptor subtype-specific activation and activity-regulated positioning22,23,31 relevant for signaling and plasticity. The characteristics and mutual relationships of these receptor-containing areas may shape their relationships with other synaptic constituents. Therefore, before investigating the spatial organization of NMDARs relative to other nanodomains, we first examined the nanoscale organization of GluN2A and GluN2B.
As expected23, both GluN2A and GluN2B formed small, tight NCs within the synapse that were readily discernible in local density heat maps (Fig 2A). To characterize these NCs, we first measured the normalized autocorrelation10 of each subunit (Fig 2B), which indicates the length scales over which the density of a protein correlates to itself, normalized to a uniform distribution at the average density. The autocorrelation thus assesses nanoclustering without needing to define NC boundaries. Both GluN2A and GluN2B showed autocorrelation magnitudes greater than one at short length scales that quickly decayed and plateaued after 35–45 nm, consistent with each protein forming NCs near that diameter. Both autocorrelations also plateaued below one, consistent with most of the protein being concentrated within NCs and sparse between, as was visually apparent (Fig 2A). This can be contrasted with the broader autocorrelation for PSD-95, reflecting the known larger size of its NCs and stronger presence of PSD-95 nearly continuously between NCs10,32–34.
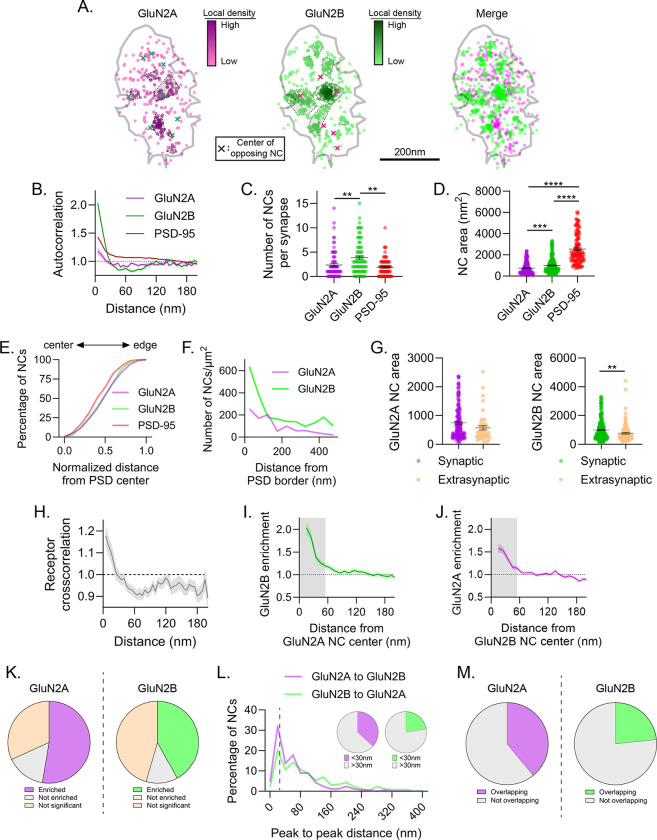
Figure 2.
Endogenous GluN2 subunits form diverse nanodomain types. a) Example synapse of DNA-PAINT localizations of GluN2A and GluN2B showing GluN2 NCs have diverse co-organization. Each point is a localization and its heat map codes normalized local density. NCs are indicated by gray-shaded, dash-bordered areas. Centers of the NCs of the opposing protein are indicated by colored x’s. Gray line indicates PSD border, defined by PSD-95 localizations (not shown). b-e) Characterization of GluN2 subunit subsynaptic organization. GluN2 subunit autocorrelations decayed faster than PSD-95 and plateaued below one, indicating small NCs with few localizations between them (b). Detected GluN2B NCs were more numerous than GluN2A or PSD-95 NCs (c), and both GluN2A and GluN2B NCs were smaller (d) and distributed slightly less centrally (e) than PSD-95 NCs. f-g) Characterization of extrasynaptic GluN2 subunit distribution. Extrasynaptic GluN2 tended to be within ~200 nm of the PSD edge (f) and formed clusters on average smaller than synaptic GluN2 NCs (g). h-m) Characterization of GluN2 subunit nanodomains. Cross-correlation (h) and cross-enrichment (i-j) indicated strong overlap of GluN2A and GluN2B densities at short distances. 52.6% of GluN2A and 42.0% of GluN2B NCs were significantly enriched with the opposite subunit (k), 36.7% of GluN2A and 22.6% of GluN2B NC peaks were located within 30 nm of an opposite GluN2 NC peak (l), and 39.0% of GluN2A and 23.4% of GluN2B NC areas spatially overlapped with the opposite subunit (m), indicating that a subset of GluN2 NCs are co-enriched at the synapse. Data in b and h-j are means ± SEM shading. Points in c are individual synapses and points in d and g individual NCs. Lines in c, d and g are means ± SEM. Data in l are shown as frequency histograms (20 nm bins), with dashed line indicating the division summarized in inset pie charts. *p<0.05, **p<0.01, ****p<0.0001
We measured the properties of individual NCs by identifying them directly using DBSCAN. We identified on average 2.3 ± 0.3 GluN2A and 3.8 ± 0.4 GluN2B NCs per synapse, with average areas of 748 ± 44 nm2 and 997 ± 44 nm2, respectively, consistent with the size of clustering suggested by the autocorrelation (~30–40 nm diameters if approximating a circular NC) (Figs 2C–D). These NCs were clearly different in size and number from the larger, less frequent PSD-95 NCs identified at the same synapses. GluN2 NCs were distributed throughout the radial extent of the synapse, compared to PSD-95 NCs that skewed slightly more central (Fig 2E). As has been reported also in tissue slices35, some synapses had centrally located NMDAR NCs, whereas others had prominent NCs positioned near the PSD edge. We also observed GluN2 NC-sized objects in extrasynaptic regions. These were largely concentrated within ~200 nm of the edge of the synapse (Fig 2F), consistent with recent single particle tracking data of expressed receptor subunits36, and on average were slightly smaller than their respective synaptic NCs (GluN2A: 569 ± 71 nm2, p=0.0679 vs synaptic; GluN2B: 763 ± 57 nm2, p=0.0026 vs synaptic; Fig 2G).
NMDARs of different GluN2 subunit compositions may form synaptic nanodomains in which separate diheteromeric receptor types (diheteromers) intermix, or they could represent triheteromeric GluN1/2A/2B receptors (triheteromers), or be a combination of both. Given the unique kinetics and interactors of each subunit, these mixed subunit nanodomains could have unique activation properties or downstream signaling pathways depending on their constituents and proximity to other complexes. We therefore assessed the nanoscale relationship of endogenous GluN2 subunits to one another. We first measured the relationship of receptor subunits using a normalized cross-correlation10, which, like the autocorrelation, indicates the spatial scales over which two protein distributions correlate. The magnitude of the cross-correlation of GluN2A and GluN2B was greater than one at length scales less than 30 nm (Fig 2H), similar to the predicted size of GluN2 NCs (Fig 2B,D), indicating that on average, GluN2A and GluN2B NCs are strongly spatially associated. Indeed, a cross-enrichment analysis10, which measures the density of one protein surrounding the peak density position of each NC of another protein (normalized to a randomized distribution), showed that each subunit was on average significantly enriched near the opposite subunit’s NC peak (Fig 2I–J). When the cross-enrichment curves were separated into those that statistically can be determined as enriched or de-enriched within the first 60 nm from the peak10, we found that 52.6% of GluN2A NCs were enriched with GluN2B and 42.0% of GluN2B NCs were enriched with GluN2A. By contrast, only 15.6% of GluN2A and 12.4% of GluN2B NCs were statistically de-enriched with the other subunit (Fig 2K). Consistent with this finding, 36.7% of GluN2A NC peaks could be found within 30 nm of the nearest GluN2B NC peak, and 22.6% of GluN2B NC peaks were within 30 nm of the nearest GluN2A peak (Fig 2L). Further, 39.0% and 23.4% of GluN2A and GluN2B NC areas, respectively, spatially overlapped (Fig 2M). These data together indicate that 30–50% of GluN2A and GluN2B NCs are closely spatially associated with one another at these synapses, a number strikingly similar to the predicted percentage of triheteromeric NMDARs at mature synapses37,38. These results suggest subtype-specific NMDAR trafficking mechanisms establish a diverse array of nanodomain types within single synapses, where the co-enriched population likely represents either triheteromers or mixed nanodomains of GluN2A and GluN2B diheteromers, while the remaining NCs likely represent nanodomains that accumulate a single diheteromer type.
Only a subpopulation of release sites is enriched with GluN2 subunits
NMDARs with different subunit compositions have different sensitivity to distance from the release site39, and their position within the synapse may affect what intracellular signaling cascades are activated after Ca2+ influx. To examine the relationship of GluN2 subunits to release sites, we detected Munc13–1 NCs, which are the best available immunostaining-based indicators of synaptic vesicle docking and fusion sites26. The Munc13–1 autocorrelation sharply decayed and plateaued below one around 45 nm, indicating small NCs with few localizations between (Fig 3A–B). This was supported by direct NC detection, which showed a wide range of NC numbers per synapse (up to 17, mean 6.9 ± 0.5 with area 850 ± 27 nm2, Fig 3C), consistent with previous reports26,40. In contrast to our expectations, GluN2A and GluN2B were each quite strongly de-enriched from Munc13–1 NC peaks (Fig 3D). Nearest the peak, GluN2A enrichment was only 0.76 ± 0.04, and GluN2B enrichment 0.58 ± 0.04, of randomly distributed receptor density, and their enrichments did not reach random until 95 and 155 nm from the Munc13–1 NC peak, respectively. Further, the GluN2 enrichment indices, or the average enrichment within 60 nm of the Munc13–1 NC peak10, was 0.88 ± 0.03 and 0.76 ± 0.03 for GluN2A and GluN2B, respectively, indicating a significant lack of both subunits immediately across the synapse from Munc13–1 NC peaks. This surprising observation provides direct evidence that NMDAR distribution within synapses is sensitive to presynaptic organization.
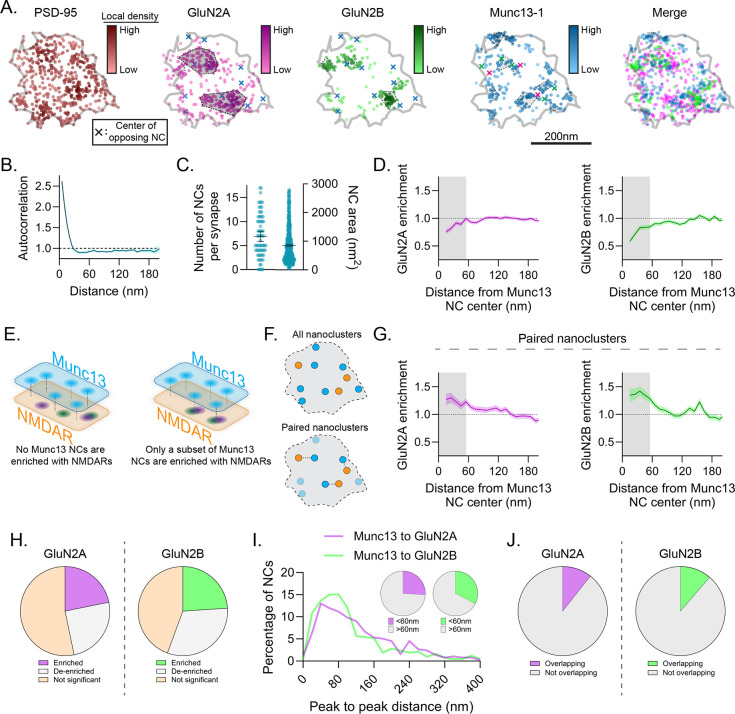
Figure 3.
Only a subpopulation of release sites is enriched with GluN2 subunits. a) Example synapse of DNA-PAINT localizations of PSD-95, GluN2A, GluN2B, and Munc13–1 showing arrangement of receptor subunits relative to release sites. Markers as described in Fig 2a. b-c) Characterization of Munc13–1 NCs. Munc13–1 autocorrelation decayed rapidly and plateaued below one (b), consistent with its many small NCs (c). d) Munc13–1 NCs were, on average, de-enriched with GluN2A (left), and GluN2B (right) at distances <55 nm (shading). e-j) A subset of Munc13–1 NCs were enriched with GluN2 subunits. Schematic indicates possible configurations of GluN2 and Munc13–1 NCs (e). Schematic of NC pairing to reveal stereotyped distances of closely-associated NCs (f). Paired Munc13–1 NCs were enriched with GluN2A and GluN2B within 55 nm of their center (g). 21.8% and 24.0% of Munc13–1 NCs were statistically enriched with GluN2A or GluN2B (h), 26.0% and 32.7% of Munc13–1 NCs had a GluN2A or GluN2B NC peak within 60 nm (i), and 10.6% and 11.3% of Munc13–1 NCs spatially overlapped with GluN2A or GluN2B NCs (j), respectively. Data in b, d and g are means ± SEM shading. Points in c (left) are synapses and (right) NCs, with lines at mean ± SEM. Data in i are shown as frequency histograms (20 nm bins), with dashed line indicating the division summarized in inset pie charts.
The offset from release sites was intriguing given that it would appear to decrease the likelihood of NMDAR activation, particularly of receptors containing GluN2B, which was on average more strongly de-enriched from Munc13–1 NCs and also has a stronger predicted distance-dependence for activation due to its slower glutamate binding39. However, having observed diversity in the spatial relationship between GluN2 subunits, we asked whether the relationship of GluN2 subunits to Munc13–1 NCs was a result of a systematic de-enrichment of receptors around all Munc13–1 NCs, or if there existed a subset of release sites enriched with GluN2 subunits (Fig 3E). Such diversity would suggest a tight interplay of NMDAR position and active zone structure.
As a first step, we paired mutually nearest Munc13–1 and GluN2 NCs and measured their cross-enrichment (Fig 3F). If GluN2 density were systematically de-enriched from Munc13–1 NCs, then it would remain de-enriched even after selecting for nearest pairs. However, we observed that paired Munc13–1 NCs were significantly cross-enriched with both subunits, consistent with there being a subpopulation of release sites enriched with NMDARs (Fig 3G). Indeed, even without pairing NCs, 21.8% and 24.0% of Munc13–1 NCs were statistically enriched with GluN2A and GluN2B, respectively, despite a slightly larger population being statistically de-enriched (25.0% for GluN2A and 31.6% for GluN2B, Fig 3H). Further, 26.0% and 32.7% of Munc13–1 NC peaks had a GluN2A or GluN2B NC peak, respectively, within 60 nm (Fig 3I), and 10.6% and 11.3% of Munc13–1 NCs showed spatial overlap with GluN2A and GluN2B, respectively (Fig 3J). Intriguingly, while 33.1% of the release sites enriched with GluN2 subunits were specifically enriched with both GluN2A and GluN2B, just 6.9% were enriched with only one subunit and de-enriched with the other, suggesting triheteromer or mixed diheteromer nanodomains may be specifically enriched closer to release sites.
We expect the diversity of this organization to impact receptor activation, particularly for GluN2B-containing receptors. We estimated the likelihood of NMDAR opening in response to release at a given Munc13–1 NC based on distance-dependent open probabilities from Santucci et al.39, comparing release from Munc13–1 NCs either near (median distance in first quartile; GluN2A = 37 nm, GluN2B = 35 nm) or far (median distance in third quartile; GluN2A = 246 nm; GluN2B = 227 nm) from their nearest receptor NC. With these parameters, the probability of a GluN2B diheteromer opening is reduced by nearly 70% for receptors far from release sites vs those nearby, and by about 10% for GluN2A diheteromers (Popen: GluN2B near = 0.49 vs GluN2B far = 0.15; GluN2A near = 0.76 vs GluN2A far = 0.69). Note that triheteromer distance-dependence to release is unknown as the glutamate-dependence of their opening rates has not been tested similarly, but as NMDAR opening requires glutamate binding at both GluN2 subunits41, they are likely dominated by the slow GluN2B glutamate binding rate and may therefore be affected similarly to GluN2B diheteromers. These results are all consistent with the presence of a subpopulation of Munc13–1 NCs that are enriched with both GluN2A and GluN2B, amongst a larger population of putative release sites that lack NMDARs.
A subset of structurally unique Munc13–1 NCs is enriched with PSD-95 and in the nanocolumn
Our results suggest there may be unique, trans-cellular molecular contexts for some release sites that may influence receptor positioning. To resolve this, we examined the relative trans-synaptic enrichment of PSD-95 near Munc13–1 NCs (Fig 4A). PSD-95 anchors receptors within the synapse25 and is a central component of the trans-synaptic nanocolumn9,10,14. However, there are approximately 3.5 times as many Munc13–1 NCs (Fig 3C) as PSD-95 NCs (Fig 2C), suggesting an architectural diversity that may be important for the control of receptor subsynaptic positioning. When all NCs were analyzed, Munc13–1 was weakly enriched on average across from PSD-95 NC centers and PSD-95 was essentially randomly distributed across from Munc13–1 NCs, consistent with the large numerical mismatch (Fig 4B). However, this average did not reflect a systematic or consistent offset between Munc13–1 and PSD-95, as identifying mutually paired NCs to even the numerical imbalance revealed strong, bidirectional enrichment (Fig 4C).
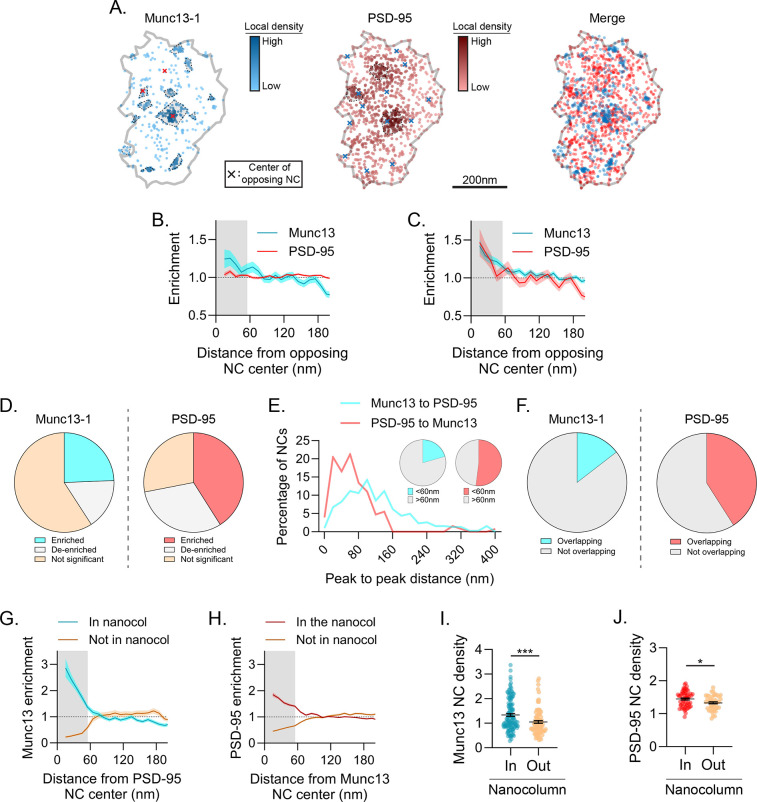
Figure 4.
A subset of structurally unique Munc13–1 NCs is enriched with PSD-95 and in the nanocolumn. a) Example synapse of DNA-PAINT localizations of Munc13–1 and PSD-95, showing variable position of Munc13–1 NCs relative to PSD-95 density. Markers as in Fig 2a. b-f) A subset of Munc13–1 NCs is enriched with PSD-95. On average Munc13–1 and PSD-95 were weakly enriched with one another (b), but after pairing (as in 3g) both were significantly enriched (c). When subsetting the data, 24.4% of Munc13–1 and 41.0% of PSD-95 NCs were statistically enriched with the other protein (d), 20.6% of Munc13–1 and 52.0% of PSD-95 NCs had a NC peak of the other protein within 60 nm (e), and 14.5% of Munc13–1 and 40.9% of PSD-95 NCs were spatially overlapped (f). g-h) Cross-enrichments of PSD-95 NCs with Munc13–1 (g) or vice versa (h) demonstrate strong cross-enrichment of these proteins when subset by the data in 4d. i-j) Munc13–1 (i) and PSD-95 (j) NCs were denser when in the nanocolumn than when outside it. Data in b-c and g-h are means ± SEM shading. Points in i and j are NCs with lines at mean ± SEM. ****p<0.0001, *p<0.05.
This subpopulation of Munc13–1 NC sites closely associated with PSD-95 was also evident in other measures. When we tested each NC for whether it was enriched with the other protein (within 60 nm), we found 24.4% of Munc13–1 NCs (~1.5–2 per synapse, on average) were enriched with PSD-95 and 41.0% of PSD-95 NCs (~1 per synapse, on average) were enriched with Munc13–1 (Fig 4D). Additionally, 20.6% of Munc13–1 NCs and 52.0% of PSD-95 NCs had a nearest NC peak of the opposite protein within 60 nm (Fig 4E), indicating a subpopulation of closely associated NCs. This fraction was similar to the proportion of each NC that spatially overlaid one another (14.5% for Munc13 NCs and 40.9% for PSD-95 NCs, Fig 4F). These results together suggest that some Munc13–1 NCs have a privileged location closely associated with PSD-95 across the synapse. Indeed, this can be clearly seen in their cross-enrichment profiles (Fig 4G–H) when subset by the data in Fig 4D. We designate these mutually co-enriched PSD-95 and Munc13–1 densities as nanocolumnar, which allowed us to make further conditional comparisons based on subsetting the data by nanocolumn status. Intriguingly, Munc13–1 NCs in the nanocolumn had a higher Munc13–1 density within 60 nm of their peak than those outside the nanocolumn (27.6% higher; 1.34 ± 0.06 inside vs 1.05 ± 0.06 outside, p=0.0009; Fig 4I), which may reflect a difference in priming characteristics of vesicles at these nanocolumnar release sites. PSD-95 peak NC density was also significantly higher in the nanocolumn, though not to the same magnitude as Munc13–1 (9.0% higher; 1.45 ± 0.03 inside vs 1.33 ± 0.03 outside, p=0.0152; Fig 4J). Together, these results suggest the nanocolumn represents a complex, macromolecular context that endows a privileged subset of release sites with high Munc13–1 NC density to spatially associate their neurotransmitter release most closely with PSD-95 NCs.
Subunit-specific NMDAR nanodomains are organized with distinct trans-synaptic molecular contexts
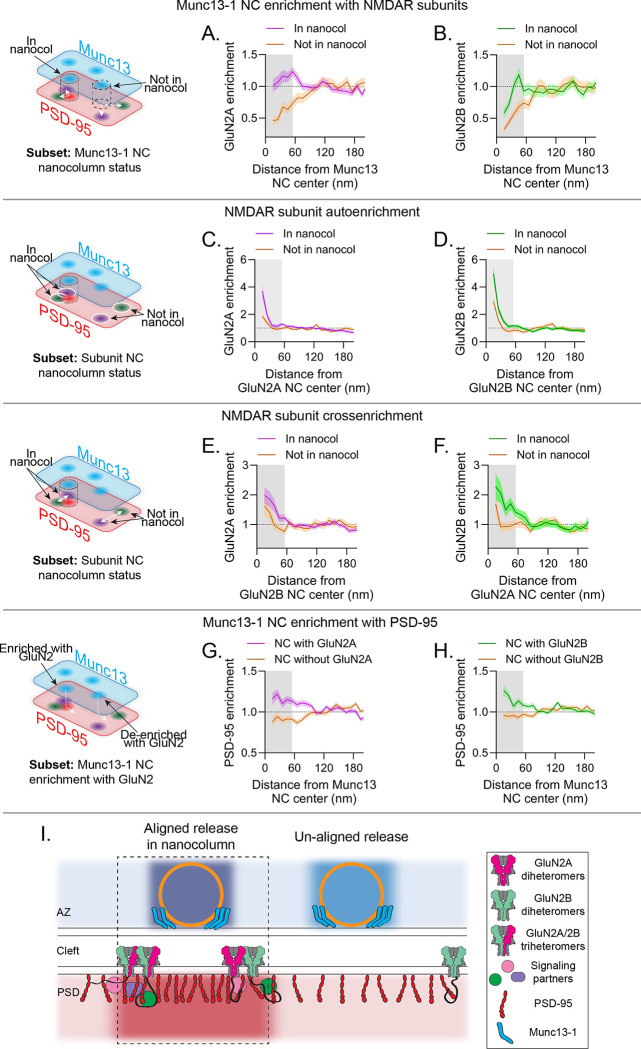
With this approach in hand to molecularly identify subsets of Munc13–1 NCs, we hypothesized that nanocolumn organization may determine NMDAR positioning with respect to release sites. To test this, we made further conditional comparisons, assigning each Munc13–1 NC as being within the nanocolumn (enriched with PSD-95) or outside it (de-enriched with PSD-95), then measured GluN2 enrichment to these subsets. GluN2A was strongly de-enriched from Munc13–1 NCs outside the nanocolumn, considerably more so than to Munc13–1 NCs overall (enrichment index (EI): 0.60 ± 0.07 to Munc13–1 NC outside the nanocolumn vs 0.88 ± 0.03 to all Munc13–1 NCs (Fig 3D); p=0.0004). However, GluN2A enrichment near Munc13–1 NCs within the nanocolumn was entirely rescued from de-enrichment, reaching a maximum enrichment of 1.23 at 55 nm (EI: 1.13 ± 0.69 to Munc13–1 NCs inside the nanocolumn vs 0.60 ± 0.07 to Munc13–1 NCs outside the nanocolumn, p<0.0001) (Fig 5A, S3A). Similar to GluN2A, GluN2B was strongly de-enriched from Munc13–1 NCs outside the nanocolumn (EI: GluN2B: 0.55 ± 0.06 vs GluN2A: 0.60 ± 0.07 to Munc13–1 NCs outside the nanocolumn, p=0.56), but like GluN2A was significantly more enriched to Munc13–1 NCs in the nanocolumn (EI: 0.89 ± 0.07 to Munc13–1 NCs inside the nanocolumn vs 0.55 ± 0.06 to Munc13–1 NCs outside the nanocolumn, p=0.0327) (Fig 5B, S3B). These data indicate that NMDA receptors are generally positioned away from release sites but are significantly more associated with release sites in the nanocolumn.
Figure 5.
Subunit-specific NMDAR nanodomains are organized with distinct trans-synaptic molecular contexts. a-h) In each row, the schematic indicates the conditional comparison being made, and each column shows the measurements made with respect to GluN2A (a,c,e,g) and GluN2B (b,d,f,h). Each panel shows the cross-enrichment plot, where data are means ± SEM shading. a-b) Munc13–1 NCs in the nanocolumn were significantly more enriched with both GluN2A and GluN2B than Munc13–1 NCs outside the nanocolumn. c-d) GluN2A and GluN2B NCs in the nanocolumn were denser than those outside the nanocolumn. e-f) GluN2A and GluN2B NCs were more cross-enriched with one another when in the nanocolumn. g-h) Munc13–1 NC enrichment with GluN2A and GluN2B can predict Munc13–1 NC enrichment with PSD-95. i) Proposed model: NMDAR distribution in synapses is governed by nanodomains with distinct trans-synaptic molecular contexts. Active zones contain molecularly diverse release sites likely with differing vesicle priming and release properties and whose functional impact depends in part on differential transsynaptic alignment. Receptor nanodomains near nanocolumn release sites contain GluN2A and GluN2B subunits; NMDARs accumulated near release sites will be activated more efficiently and potentially transduce unique signaling due to the presence of a mixed population of intracellular C-termini.
We pursued the molecular identity of these locations more fully by analyzing the characteristics of receptor NCs depending on their molecular context. Interestingly, both GluN2A and GluN2B NCs within the nanocolumn were denser compared to those outside the nanocolumn (Fig 5C–D, S3C–D), suggesting these receptor nanodomains may contain more receptors, or that the receptors are more tightly clustered near the nanocolumn. Because the enhanced accumulation was apparent for both subunits, we considered the possibility that the nanocolumn comprises a subdomain of specific receptor nanodomain types. We examined the cross-enrichments of each subunit to the other and found that GluN2A enrichment around GluN2B NCs was nearly 29% higher in the nanocolumn than out (Fig 5E, S3E; EI: 1.58 ± 0.17 inside nanocolumn vs 1.13 ± 0.17 outside of nanocolumn, p=0.0741). Even more strikingly, GluN2B enrichment around GluN2A NCs was enhanced almost 38% (Fig 5F, S3F; EI: 1.79 ± 0.22 inside nanocolumn vs 1.11 ± 0.15 outside of nanocolumn, p=0.0257). These observations suggest more abundant closely positioned subunits of each type within the nanocolumn. In further support of this, none of the nanocolumnar Munc13–1 NCs that were statistically enriched with any subunit were also de-enriched with the other. Instead, 34.6% of the nanocolumnar Munc13–1 NCs enriched with any subunit were significantly enriched with both subunits, with the remainder significantly enriched with one subunit and at least neutral with the other. This reveals a preferential positioning of enlarged, heterogeneous NMDAR nanodomains near release sites aligned with PSD-95, and more generally indicates that receptor subsynaptic organizational characteristics are dependent on trans-synaptic context.
Finally, we found the parallelized nanoimaging approach with DNA-PAINT further revealed that measuring the multiprotein context allows deeper and more accurate prediction of the organizational determinants of critical molecules. Notably, the presence of GluN2 density near a Munc13–1 NC was predictive of the postsynaptic environment around the release site: Munc13–1 NCs that were enriched with either GluN2A or GluN2B were on average also enriched with PSD-95, while those de-enriched with GluN2A or GluN2B were also de-enriched with PSD-95 (Fig 5G–H, S3G-H; EI of Munc13–1 NCs with PSD-95: 1.15 ± 0.04 with GluN2A vs 0.91 ± 0.03 without GluN2A, p<0.0001; 1.15 ± 0.03 with GluN2B vs 0.95 ± 0.03 without GluN2B, p<0.0001). Together, these observations reveal that synaptic molecular architecture depends on the assembly of multiprotein nanodomains whose interior construction is conditional on their trans-cellular relationships.
DISCUSSION
We leveraged the high resolution and multiplexing capabilities of DNA Exchange-PAINT to map the nano-organization of GluN2A and GluN2B with respect to the key release site and scaffold proteins Munc13–1 and PSD-95. Based on four-target super-resolution imaging and new analysis of the mutually conditional distributions of these proteins, we propose a model (Fig 5I) showing the unique distribution of NMDARs within the synapse, particularly with respect to release sites. GluN2 NCs were only well aligned to release sites enriched with PSD-95 i.e. located at the nanocolumn, and otherwise were de-enriched from release sites. This multi-protein relationship was strong enough that Munc13–1 NC enrichment with PSD-95 could be predicted by Munc13–1 enrichment with NMDAR subunits. Further, Munc13–1 NCs outnumbered PSD-95 and receptor NCs, varied substantially in their interior density of Munc13–1, and some Munc13–1 NCs were quite far (100s of nm) from the nearest NMDAR NC, suggesting variability in NMDAR responses within a single synapse will depend on which release site is activated. This spatial segregation raises the possibility that independent fusion of vesicles at different release sites are likely to activate unique ratios of GluN2A or GluN2B-containing receptors, expanding the computational potential of the synapse.
A major question therefore is whether the structurally definable subsets of Munc13–1 NCs we have observed are functionally distinct. One possibility is they have different preferred release modes (synchronous vs asynchronous and spontaneous vs evoked), which are suggested to be spatially segregated at the presynapse42–46. Release mode diversity is critically associated with NMDAR function, as functionally distinct pools of NMDARs respond more to spontaneous vs evoked release47,48 and activate unique downstream signaling cascades49,50. NMDARs may also be preferentially activated by asynchronous release51, though the biological sequelae specific to this remain to be determined. Molecular determinants establishing release site preference for one or more different release modes have not been defined, but our data raise the possibility that specific trans-cellular molecular contexts may be involved.
Structurally distinct Munc13–1 NCs may also quantitatively tune action potential-evoked release. The number of Munc13–1 NCs correlates strongly with the number of evoked release sites26,52, and our data along with recent results indicate that release sites within a single synapse have diverse molecular properties6,52–54. We found that Munc13–1 density was higher at nanocolumnar NCs than at those outside the nanocolumn, and increased Munc13–1 density is positively correlated with vesicle priming and release probability52,55. This suggests Munc13–1 NCs at the nanocolumn may support higher Pr there than at other synaptic sites, which in turn would further regulate its frequency-dependent engagement during paired action potentials or sustained activity. It is tempting to speculate that the trans-synaptic molecular context of a release site coordinately confers unique properties to evoked release and the associated receptor pool.
While some modeling based on the high affinity of NMDARs suggests a minimal effect of nanodomain organization on NMDAR activation56, taking the subunit-specific kinetic behavior into consideration suggests NMDAR activation is sensitive to release site position, especially for receptors containing GluN2B39. Because so few NMDARs are activated during typical responses57, this organization could help promote the relatively dependable activation of NMDARs during basal release by allowing nanocolumnar release sites to efficiently activate at least the small population of NMDARs nearby. Furthermore, it will be important to explore whether the remaining population of NMDAR subunits that do not align with Munc13–1 NCs instead are anchored at other specific synaptic subdomains to maintain distinct functions or organize proteins there, or perhaps represent a mobile pool of diffusing receptors. Further analysis of how NMDAR positioning is conditional on additional proteins should prove helpful in establishing the mechanisms that determine their distribution.
Even when in the nanocolumn, the position of GluN2 peak enrichment was still offset relative to the presynaptic NC center (55 and 45 nm for GluN2A and GluN2B respectively, Fig 5), well beyond the expected linkage error in our primary antibody/secondary nanobody system28,29. This offset is small enough that those nearest receptors will be activated at close to their maximum probability (which is still predicted to be fairly low, especially for GluN2B), but this distinctive organization may carry several other functions. Indeed, NMDARs act as synaptic signaling hubs16,58 that scaffold diverse downstream signaling molecules through their long C-terminal tails20. It’s possible that NMDAR maximal activation is balanced to allow for many or large binding partners to fit near scaffold nanodomains that concentrate further downstream signaling proteins. For example, the holoenzyme of the major GluN2 intracellular binding partner CaMKII is ~20 nm in diameter59, a size that could be disruptive to fit into the dense PSD-95 NC environment. Another potential reason for this shift could be to allow space for AMPARs to access maximal activation, as suggested by Hruska et al.22, which is important not only because of their biophysical properties60, but also because their C-tails and auxiliary proteins, such as the PSD-95-anchoring stargazin/TARPγ2, are targets for CaMKII phosphorylation61. This model appears consistent with biochemical experiments showing that while TARPγ2, PSD-95, GluN2B C-tail, and CaMKII form phase condensates together, the highest concentrations of TARPγ2 and PSD-95 are spatially separated from those of GluN2B C-tail and CaMKII62. In either case, this offset organization seems likely to be facilitated by the large size of GluN2 C-tails (~660 amino acids), which could result in the receptor channel and the extracellular domains mapped here being localized laterally quite far from their PSD-95-anchoring C-terminus25. While the structures of GluN2 C-tails remain unsolved and are presumably flexible in neurons63, high-resolution, multiplexed mapping of receptor intracellular and extracellular domains with their interacting proteins will provide additional insight to this organization.
We observed significant co-enrichment of GluN2A and GluN2B subunit nanodomains, and their selective enrichment with a subset of release sites in the nanocolumn. Previous work has observed minimal overlap between subunit nanodomains22,23. This may arise from differences in culture age, expression levels, surface vs total staining, or the imaging modality, though we note that we accomplished our mapping without subunit overexpression. Nevertheless, the key insight we add is that we have mapped the overlapping GluN2 nanodomains simultaneously and in the molecular context of two key synaptic proteins, which revealed their enrichment to the nanocolumn and suggests their relative importance in the synapse, regardless of whether they are the majority receptor population. This subunit co-enrichment specifically across from nanocolumnar release sites suggests particular synaptic subregions (nanodomains) may facilitate activation of specific NMDAR subtypes. This in turn may facilitate transduction of sparse signals. Ca2+ is quickly buffered by calmodulin after entry into the dendritic spine64,65, and as only a few of the estimated 10–20 NMDARs per synapse are activated per stimulus57, there may also be postsynaptic hotspots of Ca2+ influx. Beyond Ca2+-dependent signaling, NMDARs also pass non-ionotropic signals via effector proteins such as PP1 that interact directly with the receptor C-tail18. Therefore, positioning NMDAR signaling partners within the proposed NMDAR functional nanodomains could facilitate their downstream activation.
Co-enriched GluN2 subunit nanodomains could be constructed from either mixed populations of diheteromers or of triheteromers, which could confer unique signaling properties to these nanodomains. Triheteromers make up ~50% of synaptic NMDARs in mature synapses37,38 and have unique kinetic properties compared to diheteromers66,67. Although delineating the trafficking of triheteromers has not yet been feasible, it is notable that a triheteromer carrying dual GluN2 C-tails likely engages in a broader range of interactions than either diheteromer carrying only one type. We observed a slightly higher enrichment of nanocolumnar release sites with GluN2A vs GluN2B, which could come about due to preferential binding of GluN2A over GluN2B to PSD-9519,68. However, there is still a significant portion of GluN2A outside the nanocolumn, suggesting other mechanisms are at play. For example, a receptor carrying GluN2A and GluN2B C-tails together could create an avidity effect that increases the range of conformational possibilities with PSD-95, or perhaps the combination of GluN2A and GluN2B binding with other MAGUKs, such as PSD-93 or SAP102, could create a binding environment that gathers both subunits. This could have implications for LTP, as triheteromers have fast, GluN2A-like kinetics and could bring the receptor to PSD-95 nanodomains, while the GluN2B C-tail recruits interactors required for LTP like CaMKII20. In fact, when GluN2B diheteromers, but not triheteromers, are blocked, LTP remains intact, but a complete GluN2B subunit deletion ablates LTP69,70, suggesting a specific role of triheteromers that might be facilitated by their position relative to release sites. This suggests a nanoscale signaling complex where the very precise spatial combination of the NMDAR coincidence detection mechanism that gates Ca2+ influx is combined with the ability to interact with downstream LTP effectors and concentrated near high Pr release sites. In the future, direct visualization of NMDARs of specific molecular compositions in the context of release sites and other proteins will help clarify the specific roles each receptor subtype plays at the synapse in neurotransmission and plasticity.
Our observations of NMDAR molecular context we believe help illuminate general rules by which synapses are assembled. We suggest that a critical level at which synaptic function is established is through assembly of specific nanodomain configurations from available cell type-specific components, rather than from following an overall synapse-wide scheme such as a center-surround architecture. Several observations support this idea. Here, we document variability in molecular characteristics of presynaptic release sites within a single active zone and show that NMDAR subsynaptic distribution is dependent on highly local transcellular context. Other recent work has shown that postsynapses contain diverse scaffold molecules beyond PSD-95 that are organized in unique and developmentally regulated NCs10,71,72. Synaptic nanoclustering and trans-synaptic alignment are conserved in evolution and observed across several synapse types40,73,74, the detailed characteristics of which depend on cleft-resident synaptic organizing complexes1,9,11,14. Further, the specific complement of these proteins differs across cell types and may individualize the nano-organization even of the same proteins at different excitatory synapse types40, or related ones at inhibitory synapses73. The ability to assemble a range of nanoscale protein relationships substantially broadens the functional range of a synapse. Given that assembly and regulation of multi-protein ensembles at the nanoscale level is a ubiquitous requirement for diverse cell functions, the power of DNA-PAINT super-resolution microscopy to provide high-resolution multiplexed protein localization will be critical for analysis of how conditional distribution features sculpt these complex relationships.
Methods
DNA constructs:
pORANGE GFP-Grin2b KI (Addgene plasmid #131487), pFUGW spCas9 (Addgene plasmid #131506) and pFUGW mCherry-KASH (Addgene plasmid #131505) were gifts from Harold MacGillavry. psPAX2 (Addgene plasmid #12260) and pMD2.G (Addgene plasmid #12259) were gifts from Didier Trono. LentiCRISPRv2GFP (LCV2) was a gift from David Feldser (Addgene plasmid #82416). GFP-LRRTM2 knockdown/rescue was previously described14. SEP-GluN2A and SEP-GluN2B were gifts of Andres Barria. Rat GluN1–1a pcDNA3.1+ was a gift of Gabriela Popescu. pFUGW ORANGE GFP-Grin2b KI was made by subcloning the U6 promoter-sgRNA-GFP-Grin2b donor cassette from pORANGE into the PacI restriction site of pFUGW mCherry-KASH with NEBuilder HiFi assembly, then subsequently removing mCherry-KASH with NEB Q5 site-directed mutagenesis. pFSW myr(Fyn)-EGFP-LDLRct75 was made by subcloning a synthetic double-stranded DNA fragment of the promoter and ORF (Twist Bioscience) into the PacI and XbaI sites of pFW (pFUGW with the ubiquitin promoter-EGFP removed by NEB Q5 mutagenesis) with restriction/ligation. LCV2 Grin1 KO was created by ligating annealed oligos for the previously described gRNA targeting GRIN176 into the BsmBI sites of LCV2. DNA constructs are detailed Supplementary Table 1.
Lentivirus:
HEK293T cells (ATCC CRL-3216) were maintained in DMEM + 10% FBS and penicillin/streptomycin at 37˚C and 5% CO2. For lentiviral production, cells were plated at 5×106 cells/10 cm plate and transfected 12–24h later with 6 μg of either pFUGW ORANGE GFP-Grin2B KI, pFUGW spCas9 or LCV2 Grin1 KO + 4 μg psPAX2 + 2 μg pMD2.G using PEI for 4–6 hours. After 48h, the media was harvested, debris removed by centrifugation at 1000 RPM for 5 min and 0.45 μm PES filtering, and single use aliquots were frozen at −80°C for long term storage without further concentration. Titers were ~105 IFU/mL and routinely infected 90% or more of the cells on the coverslip at the volumes used.
Neuron and HEK culture:
All animal procedures were approved by the University of Maryland Animal Use and Care committee. Dissociated hippocampal cultures were prepared from E18 Sprague-Dawley rats of both sexes as described previously40 and plated on poly-L-lysine-coated coverslips (#1.5, 18 mm, Warner) at a density of 30,000 cells/coverslip. For most experiments, neurons were infected with 100–150 μl each of pFUGW ORANGE GFP-Grin2b KI and pFUGW spCas9 lentivirus at DIV4–6 and fixed at DIV20–21. For DNA-PAINT of dendritic spines, neurons were transfected with 1 μg of pFSW myr(Fyn)-EGFP-LDLRct at DIV14–16 with Lipofectamine 2000 per manufacturer’s instructions, and fixed at DIV20–21. For testing anti-GluN2A specificity, neurons were infected with 100 μl LCV2 Grin1 KO lentivirus at DIV5 and fixed at DIV21. For SEP-GluN2 overexpression tests, HEK cells were plated on poly-L-lysine and fibronectin-coated 18 mm coverslips at a density of 100,000 cells/coverslips, transfected 24h later with 250 ng SEP-GluN2A or GluN2B + 125 ng GluN1–1a + 125 ng mCherry-C1 with Lipofectamine 2000, then maintained for 24h in fresh media with 150 μM APV + 11.25 μM MK-801 before fixation.
Antibody conjugation and preincubation:
Primary antibodies are detailed in Supplementary Table 2, and secondary reagents in Supplementary Table 3. Donkey anti-rabbit IgG was conjugated with Cy3B as previously described40. Secondary single domain antibodies (sdAbs) for DNA-PAINT were custom-made by Massive Photonics and each carried one of four oligonucleotide docking strands optimized for DNA-PAINT77. To stain multiple targets in the same sample with antibodies form the same species, we preincubated28,29,40,53,71 primary antibodies with 2.5-fold molar excess of the appropriate species secondary sdAb labeled with DNA-PAINT docking strands for 20 minutes at room temperature (RT) in either PBS + 100 mM glycine (PBS/Gly) for fixed staining or ACSF (10 mM HEPES pH7.4, 139 mM NaCl, 2.5 mM KCl, 10 mM glucose, 2 mM MgCl2, 2 mM CaCl2) for live staining, then bound excess sdAb by adding 2-fold molar excess of Fc fragment (Jackson Immunoresearch) of the same species as the secondary sdAb for a further 20 minutes at RT. Preincubated antibodies for a given incubation were then pooled together and diluted to their final concentrations in PBS/Gly or ACSF for use in immunostaining.
Immunostaining:
For DNA-PAINT experiments, DIV20–21 neurons were removed from culture media and placed in ACSF containing primary antibodies preincubated with sdAbs conjugated to DNA-PAINT docking strands. Neurons were incubated in the antibody mixture at 16°C for 60 min and then transferred to fixative (2% PFA in 10mM MES (pH6.8), 138mM KCl, 3mM MgCl2, 2mM EGTA, 320mM sucrose) for 15 min at room temperature. Following fixation, neurons were washed in PBS/Gly 3 × 5 min at RT, permeabilized with 0.3% Triton X-100 for 20 min at RT, and blocked with 10% donkey serum in PBS/Gly + 0.2% Triton X-100 for 45 min at RT. Neurons were then incubated overnight at 4°C with sdAb-preincubated primary antibodies diluted in 50% blocking buffer. Neurons were washed 3 × 5 min in PBS/Gly, postfixed in PBS containing 4% PFA and 4% sucrose for 15 min at RT, and finally washed 3 × 5 min with PBS/Gly. For confocal experiments, cells were stained using the protocol above, but without preincubating primaries. Secondary antibodies diluted in PBS/Gly were applied for 1h at RT after washing off overnight primaries. Cells were washed 3 × 5 min in PBS/Gly before postfixing as above. For detailed use of antibodies in each experiment, see Supplementary Table 3.
Confocal microscopy and analysis:
Confocal images were acquired on a Nikon TI2 inverted microscope equipped with an Andor Dragonfly spinning disk confocal, a Plan Apo λD 60x/1.42 NA oil immersion objective, and a Plan Apo 20x/0.75 NA air objective. Excitation light (405/488/561/640) was supplied by an Andor ILE and reflected to the sample through a 405/488/561/638 quadband polychroic (Chroma), and emission light was passed through the confocal unit and appropriate emission filters (ET525/50, ET600/50 (Chroma) or Em01-R442/647 (Semrock)) to a Zyla 4.2+ sCMOS camera (Andor). Neurons were imaged at 50% laser power and 200 ms exposure, and Z-stacks were acquired using a piezo fitted in a Nikon stage. Z-stacks were converted to maximum intensity projections using FIJI78. The mouse anti-EGFP dilution series was analyzed with a custom FIJI macro that first background subtracted the 1st percentile pixel intensity from each channel, then thresholded the GFP-LRRTM2 signal to create a mask and measured the mean EGFP and anti-EGFP intensities inside the ROI.
Single-molecule microscopy:
DNA-PAINT images were acquired on an Olympus IX81 inverted microscope with an Olympus 100x/1.49 NA TIRF oil immersion objective. Excitation light (405/488/561) from an Andor ALC and a Toptica iBeam Smart (640) was reflected to the sample through a 405/488/561/638 quadband polychroic (Chroma) at an incident angle greater than the critical angle to achieve Highly Inclined and Laminated Optical (HILO) illumination. Emission light was passed through an adaptive optics device (MicAO, Imagine Optic), which corrected aberrations present in the point-spread function, followed by a DV2 image splitter (Photometrics) equipped with a T640lpxr dichroic and ET655lp single band (far-red) and 59004m dual band (red and green) emission filters to allow identification of GFP-Grin2b KI cells with the 488 nm laser followed by simultaneous collection of red and far-red emissions during DNA-PAINT imaging. Emission was finally collected on an iXon+ 897 EM-CCD camera (Andor). Z stability was maintained by the Olympus ZDC2 feedback positioning system. The microscope, ALC, and camera were controlled by iQ3 (Andor), the Toptica laser by TOPAS iBeam Smart GUI, and the Micao by separate Imagine Optic software. An additional arc lamp provided epifluorescence illumination for identifying GFP-Grin2b KI cells. The microscope was contained inside an insulated box with temperature control to minimize sample drift.
90 nm gold nanoparticles (Cytodiagnostics) were added at a 1:3 dilution for 10 minutes before imaging to act as fiducials for drift and chromatic correction. EGFP-Grin2b KI cells were identified based on GFP-Booster AF488 staining and selected to have similar AF488 intensity and cell morphology across experiments. Four targets were imaged in two exchange rounds. Cy3B and Atto643 DNA-PAINT imager strands (Massive Photonics) (one each) were diluted into imaging buffer (1x PBS pH7.4 + 500 mM NaCl + oxygen scavengers (PCA/PCD/Trolox))27 to the indicated concentrations and added to the sample. Drift was allowed to settle for 10 minutes, then 50,000 frames were acquired with 50 ms exposure. Output laser power at the objective on the Olympus setup was ~27 mW for the 640 nm laser and ~18 mW for the 561 nm laser, yielding power densities of ~3.3 and ~2.2 and kW/cm2, respectively. After acquisition, the imagers were removed by gently exchanging the imaging buffer with 20 mL exchange buffer (1x PBS pH7.4), then replacing the exchange buffer with fresh imaging buffer containing the next set of imager strands. After letting drift settle 10 minutes, the second round of imaging was acquired as before. TetraSpeck beads (100 nm; Invitrogen) were immobilized on separate coverslips prepared with poly-L-lysine as for the cultured neurons, and 8–10 fields of beads were imaged for 100 frames at 50 ms exposure each and used to generate transforms to correct chromatic aberrations between the two channels.
Single molecule processing:
Images were processed in batch with a combination of FIJI, Picasso v0.4.11 (https://github.com/jungmannlab/picasso), and custom MATLAB scripts similar to our previous work40,53,71. The analysis pipeline is described in detail in Supplemental Note 1 and Supplementary Figure 2. In brief, images were localized and drift corrected in Picasso, and chromatic aberrations between channels corrected with a custom MATLAB script. Localizations were subsequently filtered to remove spurious detections and linked to combine localizations persisting for more than one frame. Clusters of synaptic proteins were identified with DBSCAN and removed if they displayed kinetic properties of non-specific imager binding79. Finally, high-confidence synapses were picked by manually inspecting for the presence of the other imaged proteins in sufficient density for analysis and to confirm that the kept cluster was a synapse. Synapses were kept based on disc like shapes, overlap of pre- and post-synaptic proteins, a size range of ~100 – 800 nm diameter, and their position near a dendrite, then scored as “en face”, “side view” or “intermediate”, ie, somewhere between en face and side. Synapses were then further filtered for en face by removing those with a long/short axis ratio >2, then validated independently by three expert raters. In some cases, super-resolution images were rendered using the FIJI ThunderSTORM plugin’s80 average shifted histogram method with 10 nm pixels (magnification 16) for ease of visualization. Otherwise, localizations are plotted as heat maps of local density, calculated for each localization as the number of localizations within 2x epsilon (see below) for that protein and normalizing to the maximum value per synapse.
Single-molecule analyses:
Synapse analyses:
Protein autocorrelations (AC) and cross-correlations (CC) were determined using custom MATLAB functions as previously described71,81 with 5 nm render pixels and a max shift radius of 500 nm for AC and 5 nm render pixels and max shift radius of 250 nm for the CC. NCs were detected using DBSCAN with the following parameters: GluN2B epsilon = 16 nm, minpts = 11; GluN2A epsilon = 16 nm, minpts = 8; Munc13–1 epsilon = 17.6 nm, minpts = 9. DBSCAN parameters for PSD-95 were set per synapse to normalize for density variations between synapses, where epsilon was 5x standard deviations greater than the mean minimal distance of 50 randomizations of PSD-95 localization positions within the same space, and minpts was 5x standard deviations greater than the mean number of points within that epsilon. NC areas were determined using the MATLAB alphaShape function with ‘HoleThreshold’ set to suppress all interior holes and an alpha radius of 150 nm, followed by the area function. NCs with fewer than five localizations or with large outlier areas due to erroneous grouping by DBSCAN were removed from analysis (area maximums identified by GraphPad Prism’s ROUT method at Q = 0.1%: GluN2A = 2430.7 μm2, GluN2B = 3376.6 μm2, PSD-95 = 6202.9 μm2, Munc13–1 = 2818.6 μm2). To calculate NC position relative to PSD center and edge, PSD-95 localizations at each synapse were fit to an ellipse (modifying an approach by Nima Moshtagh (2007); Minimum Volume Enclosing Ellipsoid v1.2.0.0, https://www.mathworks.com/matlabcentral/fileexchange/9542-minimum-volume-enclosing-ellipsoid, MATLAB Central File Exchange, retrieved May 4, 2023) that was used to derive a concentric ellipse that included the center of a NC of interest on its perimeter. The ratio of the area of these two ellipses yields a two-dimensional measure of how close to the center or to the edge a given NC is located, the square root of which results in a linear representation, where 0 is a NC at the center of the synapse, 1 is a NC on the edge of the synapse, and 0.5 is a NC that is positioned halfway between center and edge. The distance of extransynaptic NCs to the PSD border was calculated from a vector connecting the extrasynaptic NC and PSD centroids by subtracting the distance between the PSD centroid and border (determined using MATLAB’s intersect function) from the distance between the PSD and NC centroids.
Nanocluster analyses:
NC peak-to-peak distances were determined as the linear distance (MATLAB pdist2) from the peak of a NC to the nearest peak of another protein NC. NC pairing was performed by, for each NC, identifying based on peak-to-peak distances whether there existed a NC of a second protein that was mutually nearest to it. Receptor activation was calculated from Munc13–1 NC peak-to-GluN2 NC-peak median 1st and 4th quartile distances using curves fit to the distance-dependent open probabilities () of GluN2A and GluN2B39, with a linear equation for GluN2A ; m = −0.000314; b = 0.7681; R2 = 0.88) or a one-phase exponential decay for GluN2B ; plateau = 0.05748; K = 0.008038; R2 = 0.99). NC overlap was determined using the separation index as described previously53, which normalizes the distance between NCs to the sum of their radii, resulting in a measure ranging from 0 (perfect overlap) to 1 (perfectly adjacent) and greater (no overlap).
Cross-enrichments:
Cross-enrichments (CE) were determined as described53,71,81. In brief, the peak density of one protein NC was used as the reference point, and the distance of all localizations of a second protein, or of a modified randomized synapse of that protein with 300x more localizations to avoid bins with zero localizations40,53, were determined to this point. CE is calculated as the number of localizations with distances to the reference position in 10 nm bins normalized to the same for the randomized synapse, with the 300x density factor divided out. CEs can be noisy due to the randomization resulting in a value that disproportionately represents the true density within a distance bin. To retain the information in that bin but minimize this noisiness, CEs were smoothed by using the MATLAB isoutlier function and its ‘quartiles’ method to detect outliers per distance bin and replacing the outlier values with the largest, non-outlier value in that bin. Auto-enrichments were calculated the same way except the distance of localizations of one protein was measured in reference to the peak density of its own NC. NC density was calculated as the mean of the first 60 nm of the auto-enrichment.
Conditional comparisons:
Nanoclusters were determined to be “enriched” or “not enriched” if the enrichment index (average of the first 60 nm of the CE) to the real data was greater than or less than 1.96x standard deviations from the mean of the enrichment index to a mean randomized synapse, respectively. Nanoclusters were then subset into these groups and their CE with another protein plotted. For example, a Munc13–1 NC can be conditioned on its nanocolumn status, ie, within the nanocolumn (Munc13–1 NC enriched with PSD-95) or outside the nanocolumn (Munc13–1 NC de-enriched with PSD-95), then the CE of those Munc13–1 NC groups with GluN2 subunits compared.
Statistics:
A total of 74 high-confidence, en face synapses from 8 neurons over 6 independent cultures were measured in this study. All statistical comparisons were performed in GraphPad Prism 10. Data were tested for normality using a Kolmogorov-Smirnov test. All normally distributed datasets in this work met the assumptions of homoscedasticity (F test), and differences between groups were tested using two-tailed unpaired t-tests. For non-parametric data, differences between groups were tested using two-tailed Mann-Whitney tests for two groups or a Kruskal-Wallis test, followed by post-hoc Dunn’s multiple comparisons tests, for greater than two groups.
Supplementary Material
Funding:
MCA – F31MH124283
ADL – F32MH119687
PAD – F31MH117920
TAB – R37MH080046, R01MH11982
REFERENCES
- 1.Fukata Y. et al. LGI1–ADAM22–MAGUK configures transsynaptic nanoalignment for synaptic transmission and epilepsy prevention. Proc. Natl. Acad. Sci. 118, e2022580118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu W. et al. Nanoscale reorganisation of synaptic proteins in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 49, e12924 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Zieger H. L. & Choquet D. Nanoscale synapse organization and dysfunction in neurodevelopmental disorders. Neurobiol. Dis. 158, 105453 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Choquet D. & Opazo P. The role of AMPAR lateral diffusion in memory. Semin. Cell Dev. Biol. 125, 76–83 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Eggermann E., Bucurenciu I., Goswami S. P. & Jonas P. Nanodomain coupling between Ca2+ channels and sensors of exocytosis at fast mammalian synapses. Nat. Rev. Neurosci. 13, 7–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gou X.-Z., Ramsey A. M. & Tang A.-H. Re-examination of the determinants of synaptic strength from the perspective of superresolution imaging. Curr. Opin. Neurobiol. 74, 102540 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Guzikowski N. J. & Kavalali E. T. Nano-Organization at the Synapse: Segregation of Distinct Forms of Neurotransmission. Front. Synaptic Neurosci. 13, 796498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biederer T., Kaeser P. S. & Blanpied T. A. Transcellular Nanoalignment of Synaptic Function. Neuron 96, 680–696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lloyd B. A., Han Y., Roth R., Zhang B. & Aoto J. Neurexin-3 subsynaptic densities are spatially distinct from Neurexin-1 and essential for excitatory synapse nanoscale organization in the hippocampus. Nat. Commun. 14, 4706 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang A.-H. et al. A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 536, 210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas K. T. et al. Pre-post synaptic alignment through neuroligin-1 tunes synaptic transmission efficiency. eLife 7, e31755 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Y. et al. Neuroligin-3 confines AMPA receptors into nanoclusters, thereby controlling synaptic strength at the calyx of Held synapses. Sci. Adv. 8, eabo4173 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinnen B. L. et al. Optogenetic Control of Synaptic Composition and Function. Neuron 93, 646–660.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsey A. M. et al. Subsynaptic positioning of AMPARs by LRRTM2 controls synaptic strength. Sci. Adv. 7, eabf3126 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paoletti P., Bellone C. & Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 14, 383–400 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Hansen K. B., Yi F., Perszyk R. E., Menniti F. S. & Traynelis S. F. NMDA Receptors in the Central Nervous System. in NMDA Receptors (eds. Burnashev N. & Szepetowski P.) vol. 1677 1–80 (Springer; New York, 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosokawa T. & Liu P.-W. Regulation of the Stability and Localization of Post-synaptic Membrane Proteins by Liquid-Liquid Phase Separation. Front. Physiol. 12, 795757 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dore K. et al. Unconventional NMDA Receptor Signaling. J. Neurosci. Off. J. Soc. Neurosci. 37, 10800–10807 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardoni F. & Di Luca M. Protein-protein interactions at the NMDA receptor complex: From synaptic retention to synaptonuclear protein messengers. Neuropharmacology 190, 108551 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Hardingham G. NMDA receptor C-terminal signaling in development, plasticity, and disease. F1000Research 8, 1547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petit-Pedrol M. & Groc L. Regulation of membrane NMDA receptors by dynamics and protein interactions. J. Cell Biol. 220, e202006101 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hruska M., Cain R. E. & Dalva M. B. Nanoscale rules governing the organization of glutamate receptors in spine synapses are subunit specific. Nat. Commun. 13, 920 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellermayer B. et al. Differential Nanoscale Topography and Functional Role of GluN2-NMDA Receptor Subtypes at Glutamatergic Synapses. Neuron 100, 106–119.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Sun Y. et al. The differences between GluN2A and GluN2B signaling in the brain. J. Neurosci. Res. 96, 1430–1443 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Niethammer M., Kim E. & Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J. Neurosci. 16, 2157–2163 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamoto H. et al. Synaptic weight set by Munc13–1 supramolecular assemblies. Nat. Neurosci. 21, 41–49 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Schnitzbauer J., Strauss M. T., Schlichthaerle T., Schueder F. & Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 12, 1198–1228 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Pleiner T., Bates M. & Görlich D. A toolbox of anti–mouse and anti–rabbit IgG secondary nanobodies. J. Cell Biol. 217, 1143–1154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sograte-Idrissi S. et al. Circumvention of common labelling artefacts using secondary nanobodies. Nanoscale 12, 10226–10239 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Willems J. et al. ORANGE: A CRISPR/Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. PLOS Biol. 18, e3000665 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dupuis J. P. et al. Surface dynamics of GluN2B-NMDA receptors controls plasticity of maturing glutamate synapses. EMBO J. 33, 842–861 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broadhead M. J. et al. PSD95 nanoclusters are postsynaptic building blocks in hippocampus circuits. Sci. Rep. 6, 24626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gürth C.-M. et al. Neuronal activity modulates the incorporation of newly translated PSD-95 into a robust structure as revealed by STED and MINFLUX. 10.1101/2023.10.18.562700 (2023) doi: 10.1101/2023.10.18.562700. [DOI] [Google Scholar]
- 34.MacGillavry H. D., Song Y., Raghavachari S. & Blanpied T. A. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron 78, 615–622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dani A., Huang B., Bergan J., Dulac C. & Zhuang X. Superresolution Imaging of Chemical Synapses in the Brain. Neuron 68, 843–856 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kortus S. et al. Subunit-Dependent Surface Mobility and Localization of NMDA Receptors in Hippocampal Neurons Measured Using Nanobody Probes. J. Neurosci. 43, 4755–4774 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Hallaq R. A., Conrads T. P., Veenstra T. D. & Wenthold R. J. NMDA Di-Heteromeric Receptor Populations and Associated Proteins in Rat Hippocampus. J. Neurosci. 27, 8334–8343 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rauner C. & Köhr G. Triheteromeric NR1/NR2A/NR2B Receptors Constitute the Major N-Methyl-d-aspartate Receptor Population in Adult Hippocampal Synapses. J. Biol. Chem. 286, 7558–7566 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santucci D. M. & Raghavachari S. The Effects of NR2 Subunit-Dependent NMDA Receptor Kinetics on Synaptic Transmission and CaMKII Activation. PLoS Comput. Biol. 4, e1000208 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dharmasri P. A., Levy A. D. & Blanpied T. A. Differential nanoscale organization of excitatory synapses onto excitatory vs inhibitory neurons. 10.1101/2023.09.06.556279 (2023) doi: 10.1101/2023.09.06.556279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen K. B. et al. Structure, function, and allosteric modulation of NMDA receptors. J. Gen. Physiol. 150, 1081–1105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaeser P. S. & Regehr W. G. Molecular Mechanisms for Synchronous, Asynchronous, and Spontaneous Neurotransmitter Release. Annu. Rev. Physiol. 76, 333–363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kusick G. F. et al. Synaptic vesicles transiently dock to refill release sites. Nat. Neurosci. 23, 1329–1338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melom J. E., Akbergenova Y., Gavornik J. P. & Littleton J. T. Spontaneous and Evoked Release Are Independently Regulated at Individual Active Zones. J. Neurosci. 33, 17253–17263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendonça P. R. F. et al. Asynchronous glutamate release is enhanced in low release efficacy synapses and dispersed across the active zone. Nat. Commun. 13, 3497 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C. S., Chanaday N. L., Monteggia L. M. & Kavalali E. T. Probing the segregation of evoked and spontaneous neurotransmission via photobleaching and recovery of a fluorescent glutamate sensor. eLife 11, e76008 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atasoy D. et al. Spontaneous and Evoked Glutamate Release Activates Two Populations of NMDA Receptors with Limited Overlap. J. Neurosci. 28, 10151–10166 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reese A. L. & Kavalali E. T. Single synapse evaluation of the postsynaptic NMDA receptors targeted by evoked and spontaneous neurotransmission. eLife 5, e21170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutton M. A., Wall N. R., Aakalu G. N. & Schuman E. M. Regulation of Dendritic Protein Synthesis by Miniature Synaptic Events. Science 304, 1979–1983 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Sutton M. A., Taylor A. M., Ito H. T., Pham A. & Schuman E. M. Postsynaptic Decoding of Neural Activity: eEF2 as a Biochemical Sensor Coupling Miniature Synaptic Transmission to Local Protein Synthesis. Neuron 55, 648–661 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Li S. et al. Asynchronous release sites align with NMDA receptors in mouse hippocampal synapses. Nat. Commun. 12, 677 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karlocai M. R. et al. Variability in the Munc13–1 content of excitatory release sites. eLife 10, e67468 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emperador-Melero J. et al. Molecular definition of distinct active zone protein machineries for Ca 2 + channel clustering and synaptic vesicle priming. 10.1101/2023.10.27.564439 (2023) doi: 10.1101/2023.10.27.564439. [DOI] [PubMed] [Google Scholar]
- 54.Maschi D. & Klyachko V. A. Spatiotemporal dynamics of multi-vesicular release is determined by heterogeneity of release sites within central synapses. eLife 9, e55210 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aldahabi M. et al. Different priming states of synaptic vesicles underlie distinct release probabilities at hippocampal excitatory synapses. Neuron 110, 4144–4161.e7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goncalves J. et al. Nanoscale co-organization and coactivation of AMPAR, NMDAR, and mGluR at excitatory synapses. Proc. Natl. Acad. Sci. 117, 14503–14511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nimchinsky E. A., Yasuda R., Oertner T. G. & Svoboda K. The Number of Glutamate Receptors Opened by Synaptic Stimulation in Single Hippocampal Spines. J. Neurosci. 24, 2054–2064 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan X., Jin W. Y. & Wang Y. T. The NMDA receptor complex: a multifunctional machine at the glutamatergic synapse. Front. Cell. Neurosci. 8, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Myers J. B. et al. The CaMKII holoenzyme structure in activation-competent conformations. Nat. Commun. 8, 15742 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raghavachari S. & Lisman J. E. Properties of Quantal Transmission at CA1 Synapses. J. Neurophysiol. 92, 2456–2467 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Lisman J., Yasuda R. & Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 13, 169–182 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hosokawa T. et al. CaMKII activation persistently segregates postsynaptic proteins via liquid phase separation. Nat. Neurosci. 24, 777–785 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Warnet X. L., Bakke Krog H., Sevillano-Quispe O. G., Poulsen H. & Kjaergaard M. The C-terminal domains of the NMDA receptor: How intrinsically disordered tails affect signalling, plasticity and disease. Eur. J. Neurosci. 54, 6713–6739 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Faas G. C., Raghavachari S., Lisman J. E. & Mody I. Calmodulin as a direct detector of Ca2+ signals. Nat. Neurosci. 14, 301–304 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sabatini B. L., Oertner T. G. & Svoboda K. The life cycle of Ca(2+) ions in dendritic spines. Neuron 33, 439–452 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Hansen K. B., Ogden K. K., Yuan H. & Traynelis S. F. Distinct Functional and Pharmacological Properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA Receptors. Neuron 81, 1084–1096 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stroebel D., Carvalho S., Grand T., Zhu S. & Paoletti P. Controlling NMDA Receptor Subunit Composition Using Ectopic Retention Signals. J. Neurosci. 34, 16630–16636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sans N. et al. A Developmental Change in NMDA Receptor-Associated Proteins at Hippocampal Synapses. J. Neurosci. 20, 1260–1271 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delaney A. J., Sedlak P. L., Autuori E., Power J. M. & Sah P. Synaptic NMDA receptors in basolateral amygdala principal neurons are triheteromeric proteins: physiological role of GluN2B subunits. J. Neurophysiol. 109, 1391–1402 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Foster K. A. et al. Distinct Roles of NR2A and NR2B Cytoplasmic Tails in Long-Term Potentiation. J. Neurosci. 30, 2676–2685 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Metzbower S. R., Dharmasri P. A., Levy A. D., Anderson M. C. & Blanpied T. A. Distinct SAP102 and PSD-95 nano-organization defines multiple types of synaptic scaffold protein domains at single synapses. 10.1101/2023.09.12.557372 (2023) doi: 10.1101/2023.09.12.557372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun S.-Y. et al. Correlative Assembly of Subsynaptic Nanoscale Organizations During Development. Front. Synaptic Neurosci. 14, 748184 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crosby K. C. et al. Nanoscale Subsynaptic Domains Underlie the Organization of the Inhibitory Synapse. Cell Rep. 26, 3284–3297.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muttathukunnel P., Frei P., Perry S., Dickman D. & Müller M. Rapid homeostatic modulation of transsynaptic nanocolumn rings. Proc. Natl. Acad. Sci. U. S. A. 119, e2119044119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kameda H. et al. Targeting green fluorescent protein to dendritic membrane in central neurons. Neurosci. Res. 61, 79–91 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Incontro S., Asensio C. S., Edwards R. H. & Nicoll R. A. Efficient, Complete Deletion of Synaptic Proteins using CRISPR. Neuron 83, 1051–1057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strauss S. & Jungmann R. Up to 100-fold speed-up and multiplexing in optimized DNA-PAINT. Nat. Methods 17, 789–791 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun C. et al. The prevalence and specificity of local protein synthesis during neuronal synaptic plasticity. Sci. Adv. 7, eabj0790 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ovesný M., Křížek P., Borkovec J., Švindrych Z. & Hagen G. M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen J.-H., Blanpied T. A. & Tang A.-H. Quantitative Analysis of Trans-synaptic Protein Alignment. 10.1101/573063 (2019) doi: 10.1101/573063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.