Abstract
The open reading frame immediately upstream of uspA is demonstrated to encode a 14-kDa protein which we named UspB (universal stress protein B) because of its general responsiveness to different starvation and stress conditions. UspB is predicted to be an integral membrane protein with at least one and perhaps two membrane-spanning domains. Overexpression of UspB causes cell death in stationary phase, whereas mutants of uspB are sensitive to exposure to ethanol but not heat in stationary phase. In contrast to uspA, stationary-phase induction of uspB requires the sigma factor ςS. The expression of uspB is modulated by H-NS, consistent with the role of H-NS in altering ςS levels. Our results demonstrate that a gene of the RpoS regulon is involved in the development of stationary-phase resistance to ethanol, in addition to the regulon’s previously known role in thermotolerance, osmotolerance, and oxidative stress resistance.
When Escherichia coli cells enter stationary phase due to a depletion of nutrients, a number of morphological and physiological changes occur. Stationary-phase cells accumulate storage compounds such as glycogen and polyphosphate, the cells become smaller and rounder, and the DNA condenses (20, 31). In addition, there are alterations in the composition of both the inner and outer membranes. The cytoplasmic membrane shrinks, and membrane phospholipids are degraded and used as a source of carbon and energy (11). There also are a number of changes in the fatty acid composition of the inner membrane and the protein composition of both the inner and outer membranes (11, 20). These changes and others result in stationary-phase resistance to a wide range of harmful environmental conditions, including high temperature, osmotic shock, ethanol, and oxidizing agents (18). The development of stationary-phase resistance and the typical morphological and physiological changes that occur in stationary phase are brought about by a programmed change in gene expression. Proteins made early in stationary phase are necessary for the survival of E. coli cells during long-term starvation (36), and much work has recently focused on the identification of the genes and proteins involved in stationary-phase physiology.
In E. coli, many of these characteristic features of stationary-phase cells are dependent on the alternative sigma factor ςS (RpoS). ςS is required for the induction of approximately 35 genes in stationary phase (17), and many of these are required for the development of stationary-phase resistance. Mutants of rpoS are sensitive to long-term starvation and fail to develop stationary-phase-induced thermotolerance and hydrogen peroxide resistance (23). In addition, genes under control of ςS have been found to be involved in osmoprotection and DNA repair and protection (3, 17). Thus, ςS-regulated genes play a central role in stationary-phase resistance to harmful environmental conditions.
In this report, we describe a new ςS-dependent gene which is involved in stationary-phase resistance to another stress condition, ethanol. We name this gene uspB (universal stress protein B) since it is induced by growth arrest in general and is situated immediately upstream of the divergently transcribed, stationary-phase-inducible uspA gene, which is involved in stationary-phase survival (32–34).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains used in this work are listed in Table 1. Cultures were grown aerobically in liquid LB or M9 (37) medium in Erlenmeyer flasks in a rotary shaker at 37°C. M9 medium included glucose (0.4%) and thiamine (10 mM). Nutrient limiting media contained 1/10 the normal amount of the limited nutrient: glucose, 0.04%; nitrogen, 1.9 mM; and phosphate, 6 mM. Phosphate limiting medium was supplemented with MOPS (morpholinepropanesulfonic acid; 1 mM) as a buffer. Defined medium with amino acids was M9 glucose medium containing all 20 amino acids (43). When appropriate, the media were supplemented with kanamycin (50 μg/ml), carbenicillin (50 μg/ml), tetracycline (20 μg/ml), chloramphenicol (30 μg/ml), or IPTG (isopropyl-β-d-thiogalactopyranoside; 5 mM). Testing for growth on other carbon sources was done on M9 minimal plates supplemented with 0.4% appropriate carbon source.
TABLE 1.
Bacterial strains and plasmids used
| E. coli strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| MC4100 | F−araD-139 Δ(argF-lac)U169 rpsL-150 relA1 flbB-5301 deoC-1 ptsF-25 rbsR | M. Givskov |
| AF607 | MC4100 ΔuspB::cam | This work |
| AF613 | MC4100/pTN38Xho | This work |
| AF620 | MC4100/pAF619/F′ lacIq Tn10 | This work |
| AF629 | MC4100 uspB::lacZ-Kmr | This work |
| AF633 | MC4100 λφ(PuspB-lacZ) | 15 |
| AF635 | AF633 ΔuspB::cam | This work |
| AF643 | AF633 fadR::Tn10 | This work |
| AF644 | AF633 uspA::Kmr | This work |
| AF680 | AF633 rpoS::Kmr | This work |
| AF800 | AF633 hns::cam | This work |
| KK147 | AF680/pMMkatF2 | This work |
| KK140 | AF633/pHNαβ | This work |
| KK139 | AF633/pKK223 | This work |
| KK138 | uspB131-lacZ/pHNαβ | This work |
| KK137 | uspB131-lacZ/pKK223 | This work |
| RW11 | fadR::Tn10 | C. C. DiRusso |
| TN4100 | MC4100 uspA::lacZ-Kmr | 14 |
| K4633 | recD | D. Friedman |
| TT379 | rpoS::Kmr | V. deLorenzo |
| BSN22 | hns::cam | B. E. Uhlin |
| pKOK5 | lacZ-Kmr operon fusion vector | 22 |
| pTN6093 | 2.3-kb KpnI/PstI fragment harboring uspA and uspB in pBluescript vector (Stratagene) | 32 |
| pAF602 | Fragment of pACYC184 containing Cmr replacing uspB gene in pTN6093 | This work |
| pTN38 | SalI-SalI fragment harboring uspB and pit in pBluescript | 32 |
| pTN38Xho | SalI-XhoI fragment harboring uspB in pBluescript | T. Nyström |
| pAF619 | SacII-XhoI fragment harboring uspB in pBluescript (Plac-uspB) | This work |
| pKK223 | Vector for pHNαβ | V. deLorenzo |
| pHNαβ | Ptac-himA Ptac-hip | V. deLorenzo |
| pMMkatF2 | rpoS plasmid | 28 |
General methods.
Plasmid DNA was purified by using Qiagen columns (Qiagen, Inc.) or a Wizard minipreparation kit (Promega, Inc.) according to the protocols provided by the manufacturers. DNA fragments were isolated after separation on an agarose gel, using the Gene-Clean Kit (Bio 101 Inc.) according to the manufacturer’s instructions. P1 transductions and plasmid transformations were performed as described by Miller (25) and Sambrook et al. (37). DNA sequencing was done by using Sequenase (U.S. Biochemical) according to the manufacturer’s instructions, and Southern blot analysis was done as described elsewhere (37). In vitro transcription/translation was done by using a kit from Promega according to the manufacturer’s instructions. Primer extension was performed as described elsewhere (37), using polynucleotide kinase to end label the primer (5′ CGACGGTGCTTATCATACCTC) with 32P and using Moloney murine leukemia virus reverse transcriptase for the extension. Template RNA was total cellular RNA isolated as described elsewhere (37) from strain MC4100. The product of the primer extension reaction was run on a sequencing gel (37), using a sequencing ladder as the marker.
Construction of λ-PuspB-lacZ lysogens and uspB::lacZ-Kmr insertional mutants.
A uspB::lacZ-Kmr (kanamycin resistance cassette) fusion was constructed by inserting the SmaI fragment of plasmid pKOK5 (22) containing lacZ-Kmr into the MscI site of uspB carried on plasmid pTN38 (SalI-uspB-SalI [32]). The MscI site is located approximately two-thirds into the uspB coding sequence. A fragment containing the uspB::lacZ-Kmr fusion was integrated into the chromosome of the E. coli recD strain K4633 by linear transformation and subsequently moved by P1 transduction into strain MC4100.
A second PuspB-lacZ transcriptional fusion has been previously described (15); AF633 contains the uspB promoter (up to the MscI site within the gene) fused to lacZ from pTL61T (24) and integrated into the λ attachment site by using λRS45 (39). A deletion of the upstream region in this fusion (uspB131-lacZ) was created by using a PCR primer binding to the putative integration host factor (IHF) binding site (Fig. 1A; 5′ ATATCTGCAGAGTGGTTAACCTTCTGG) with a primer within the lacZ gene and amplifying the uspB promoter from a plasmid carrying the fusion (AF631 [15]). This was cloned into pTL61T cut with PstI and recombined into λRS45 as described above. A single-copy lysogen was chosen for further studies.
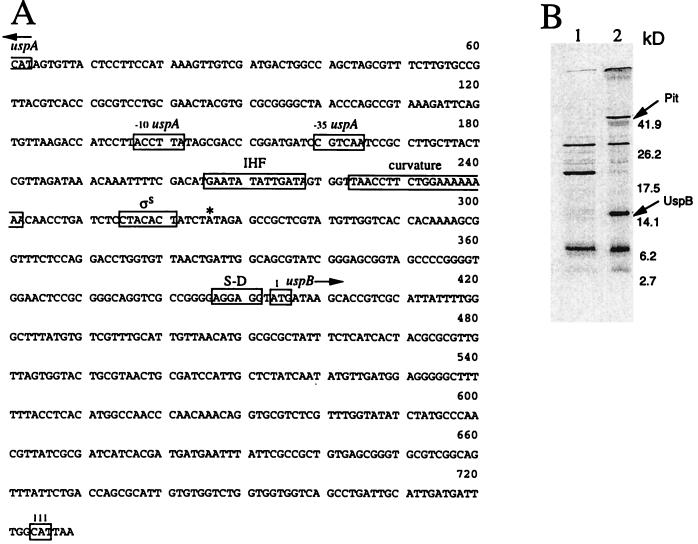
FIG. 1.
The uspB gene and its protein product. (A) Sequence of the uspB gene and upstream region. The putative uspB promoter (ςS) is indicated, as are the uspA promoter (−10, and −35), the putative IHF binding site, ribosome binding site (S-D), and intrinsic DNA curvature. The first and last (111) codons of the uspB ORF are boxed; ∗ indicates the first nucleotide (A) of the mRNA encoding uspB as determined by primer extension analysis. (B) Autoradiogram of in vitro transcription/translation products. Plasmid pBluescript (vector; lane 1) and pTN38 (lane 2) encoding uspB and pit were used in the assay, and protein was labeled with [35S]methionine. Extracts were prepared and run on a sodium dodecyl sulfate–15% polyacrylamide gel, and radioactive proteins were visualized by exposure to X-ray film (37). Sizes of the molecular weight markers used are indicated. Arrows indicate the positions of UspB and Pit.
Construction of the ΔuspB::cam mutant.
A deletion of uspB was constructed by replacing the SacII-XhoI fragment of pTN6093 containing the entire uspB coding sequence with a SacII-SalI fragment containing the chloramphenicol resistance gene (cam) from pACYC184 (8). The resulting plasmid (pAF602) was linearized by cutting with HindIII and PstI and transformed into K4633 (recD). P1 transduction was then used to move the ΔuspB::cam mutation to MC4100. The presence of this replacement in the new strain (AF607) was confirmed by Southern blot analysis using a probe upstream of the insertion and chromosomal DNA cut with PstI.
Construction of Plac-uspB fusion plasmid.
pAF619 is a high-copy-number plasmid (pBluescript vector) in which the uspB gene is placed downstream of the lac promoter. It was made by first cutting pTN38 (32) with XhoI and religating (pTN38Xho). pTN38Xho was then cut with SacII and religated; this deletes the uspB promoter and brings the uspB coding sequence downstream of the Plac promoter present in the vector sequence.
Measurements of β-galactosidase activity.
β-Galactosidase levels were measured as described by Miller (25) with modifications (1). Samples were centrifuged before they were measured spectrophotometrically to determine the optical density at 420 nm (OD420) (β-galactosidase). The β-galactosidase activity is expressed as follows: 1,000 × (OD420/[OD600 culture × reaction time × volume]). Duplicate measurements within an experiment gave less than 10% variation, and experimental values varied less than 20% between experiments. Shown in the figures are data from single representative experiments, but all experiments were repeated several times to ensure the reproducibility of results.
Computer analysis of sequence.
Homology searches were performed with both BLAST (4) and BLITZ (41) computer programs, and subsequent sequence alignments were performed with the Clustal-W program (42). For protein secondary structure analysis, we used a number of publicly available programs, including TM-pred, DAS, and SOSUI, which identify transmembrane domains (10, 19, 26), and Signal P (30), which identifies signal peptides. DNA curvature was analyzed with the BEND program (DNASTAR Inc.). Yersinia pestis sequence data were obtained from the Y. pestis Sequencing Group at the Sanger Centre and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/yp.
RESULTS
Identification of uspB.
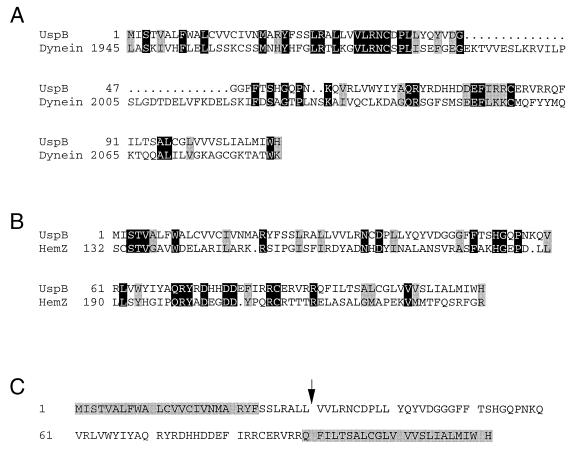
We had noted that plasmids carrying DNA from the region immediately upstream of uspA at 77 min on the chromosome were difficult to clone and unstable (see below). During our sequencing of this region, the entire sequence around 77 min was published by the E. coli sequencing project (40). Both sequencing efforts predicted the uspA upstream region to contain a gene encoding a 111-amino-acid protein (SwissProt accession no. P37632 [40]; Fig. 1A). We demonstrated that this putative gene, referred to as f111 (40) or yhiO (7) and now called uspB, indeed expressed a protein product of approximately 14 kDa by subjecting a plasmid containing uspB (pTN38 [32]) to in vitro transcription/translation analysis (Fig. 1B). Searching the database for genes similar to uspB revealed only one clear homologue. This open reading frame (ORF) was found in Y. pestis (contig 1401, bp 334 to 669) and is 86% identical to the predicted amino acid sequence of the E. coli UspB. Interestingly, approximately 700 bases upstream of the uspB-like sequence of Y. pestis, there is an ORF (contig 1401, bp 1379 to 1807) that is 89% identical to UspA transcribed in the opposite direction from the uspB homologue. Thus, the genetic organization of this locus appears to be conserved between E. coli and Y. pestis. The Y. pestis gene was the only clear homologue to UspB, but the UspB sequence does exhibit similarity to one region of dynein of Saccharomyces cerevisiae (Fig. 2A) as well as the ferrochelatase genes of E. coli (Fig. 2B) and Y. enterocolitica. The UspB protein sequence was examined by using a number of computer programs to analyze its putative structure (Materials and Methods); this analysis indicates that UspB contains two putative transmembrane domains. The first is at the N terminus and may be a signal peptide with a cleavage site located between amino acids 30 and 31, and the second is at the C terminus (Fig. 2C).
FIG. 2.
The UspB protein alignments to other known proteins and putative transmembrane regions. (A and B) Comparison of UspB with the heavy chain of dynein (A) from S. cerevisiae (SwissProt accession no. Z21877 [12]) or with ferrochelatase (HemZ; SwissProt accession no. P23871 [27]) from E. coli (B). Black shading indicates identical amino acids, gray shading indicates similar amino acids, and dots represent gaps. Alignment was done as described in Materials and Methods. (C) Predicted transmembrane domains of UspB are indicated by shaded amino acids, and a possible signal peptide cleavage site is indicated by the arrow.
uspB is induced in stationary phase.
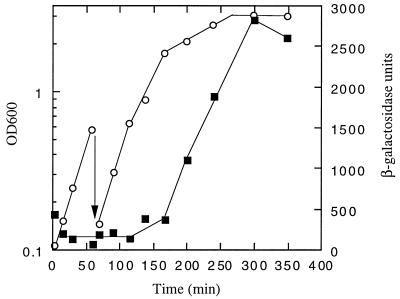
The expression pattern of uspB was examined by using two transcriptional gene fusions to lacZ. The first, in strain AF629, has a uspB::lacZ fusion linked to Kmr in the normal uspB chromosomal site. The second (AF633) has the same fusion junction, but the fusion was recombined into λ phage and integrated at the λatt site. These two gene fusions gave identical patterns of expression, though AF633 always had approximately threefold-higher levels of expression. Expression from each of the fusion strains was examined during growth in LB medium. As shown in Fig. 3 (AF633), uspB is expressed at a very low level during log-phase growth and induced approximately 50-fold as cells enter stationary phase.
FIG. 3.
PuspB-lacZ expression. AF633 (λφPuspB-lacZ) was grown in LB medium aerobically at 37°C. Cell density (open circles) and β-galactosidase activity (filled squares) were measured. The arrow indicates dilution of the culture.
uspB is induced by carbon, phosphate, and nitrogen starvation as well as osmotic shock and oxidative stress.
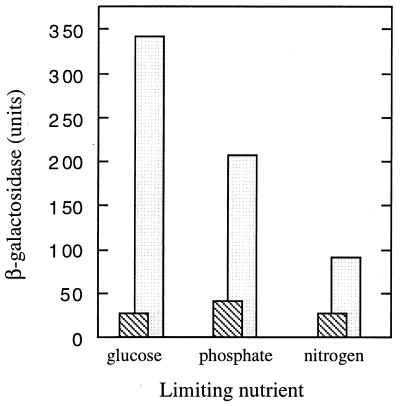
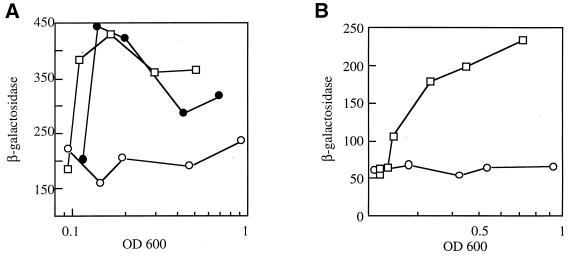
To examine the nature of the signal required to induce uspB, the β-galactosidase levels of the fusion strains were examined in minimal medium limited for various nutrients. Limiting growth by glucose starvation gave the highest level of induction, but both phosphate and nitrogen limitation also induced the fusion (Fig. 4). It should be noted that the log-phase levels of expression of the PuspB-lac fusions were higher in minimal medium than in LB (e.g., compare control cultures in Fig. 5A and B); however, PuspB-lacZ expression was not proportional to growth rate, as there was no correlation between growth rates in minimal medium containing different carbon sources and expression of the fusion (data not shown). In addition to starvation conditions, other stresses induce the PuspB-lacZ fusions. The addition of NaCl or sucrose (osmotic shock) to exponentially growing cells induced expression twofold (Fig. 5A), and the addition of H2O2 (1.5 mM) gave a fivefold induction of PuspB-lacZ (Fig. 5B). The addition of ethanol (4%) to exponentially growing cells also induced the fusion, albeit poorly (threefold), and lower concentrations failed to induce at all, indicating that a significant decrease in growth rate (as in the entry to stationary phase) may be needed to induce the fusion.
FIG. 4.
Expression of uspB::lacZ-Kmr fusion during starvation for various nutrients. Cells (AF629) were grown in minimal medium limited for one nutrient as indicated, and β-galactosidase was measured during exponential growth (striped bars) and 16 h after growth was arrested (open bars), which was the maximal β-galactosidase activity measured.
FIG. 5.
Expression from the uspB promoter is induced by osmotic stress and hydrogen peroxide. (A) Expression of PuspB-lacZ during osmotic stress. Cells (AF633) growing exponentially in glucose minimal medium were diluted to an OD600 of 0.08 in the same medium (no addition; open circles), medium containing 0.464 M sucrose (closed circles), or medium containing 0.3 M NaCl (open squares), and the levels of β-galactosidase were determined. The cell growth rate was inhibited approximately 50% by the addition of NaCl and 10% by addition of sucrose. (B) Expression of PuspB-lacZ after exposure to hydrogen peroxide. Cells (AF633) growing in LB medium were diluted to an OD600 of 0.2 in the same medium (circles) or LB containing 1.5 mM H2O2 (squares). Growth was inhibited 50% by the addition of H2O2.
uspB induction is dependent on ςS and modulated by IHF and H-NS.
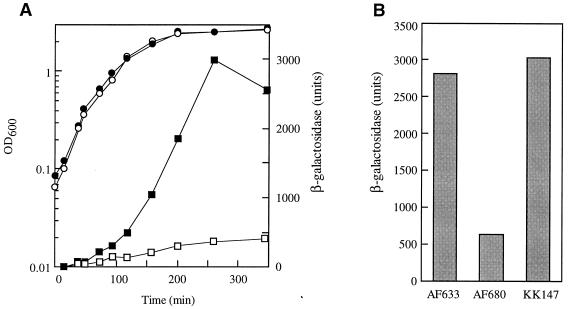
As a first step to determine the mode of regulation of uspB, a number of mutations in global regulators were introduced into the PuspB-lacZ fusion strains. FadR, which, in part, regulates uspA expression (15), has no effect on uspB expression. Mutations in the gene encoding adenylate cyclase (cya) or cyclic AMP repressor protein (crp) had only a small (negative) effect on uspB expression. However, a mutation in the sigma factor (ςS, encoded by rpoS) known to regulate a number of stationary-phase-induced genes (17) abolished induction of uspB in both LB (Fig. 6A) and glucose-limited medium (data not shown). In addition, transformation of the rpoS mutant strain with the plasmid carrying rpoS was able to restore induction of PuspB-lacZ (Fig. 6B). In the sequence upstream of the proposed uspB start site there is a sequence very similar to the consensus sequence for ςS recognition proposed by Espinoso-Urgel and coworkers (13); the consensus is in two parts, a −10 sequence (CTATACT) and an intrinsic DNA curvature upstream of this sequence. Both of these components are found upstream of the uspB coding sequence (Fig. 1A). Primer extension analysis was used to demonstrate that there is a single transcriptional start site upstream of uspB located 5 bases downstream from the 3′ end of the −10 sequence (Fig. 1A). This spacing is similar to that observed for other ςS-dependent promoters (13).
FIG. 6.
Transcription from the uspB promoter requires ςS. (A) Expression of PuspB-lacZ in an ςS mutant. Growth (circles) and β-galactosidase activities (squares) from strains AF633 (filled symbols) and AF680 (rpoS::Kmr; open symbols) are shown. Cells were grown in LB medium aerobically at 37°C. (B) Complementation of PuspB-lacZ expression in an rpoS mutant by a plasmid carrying rpoS. Cells were grown overnight in LB medium aerobically at 37°C, and samples for β-galactosidase activity measurements were taken. Strains used are AF633 (PuspB-lacZ), AF680 (AF633 rpoS::Kmr), and KK147 (AF680/pMMkatF2).
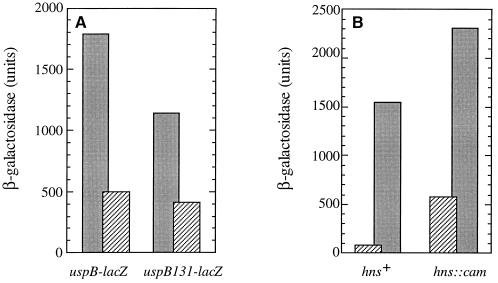
In addition to the consensus ςS promoter, there is a sequence resembling an IHF binding site (29) situated almost exactly between the promoters of uspA and uspB (Fig. 1A). We have shown previously that this site has no role in uspA regulation (15), and so we wanted to test if IHF and this site are involved in uspB expression. A plasmid encoding both subunits of IHF under Ptac control was introduced into the fusion strain. As shown in Fig. 7A, overexpression of IHF by the addition of IPTG in strain KK140 reduced the β-galactosidase levels in stationary phase about threefold compared to the control strain. However, a deletion of the putative IHF binding sequence did not alter the effect of IHF overproduction on PuspB-lacZ expression (Fig. 7A), although it is clear that deletion of this upstream region did reduce uspB expression. Thus, it appears that the IHF overproduction effect on uspB expression is most likely indirect but that there are other sequences within or upstream of the IHF consensus important for uspB expression.
FIG. 7.
uspB promoter activity is modulated by IHF and H-NS. (A) β-Galactosidase levels in stationary-phase cells overexpressing IHF. KK139 (PuspB-lacZ/pKK223), KK140 (PuspB-lacZ/pHNαβ), and the upstream deletion mutants KK137 (uspB131-lacZ/pKK223) and KK138 (uspB131-lacZ/pHNαβ) were grown overnight in LB medium containing IPTG and carbenicillin. β-Galactosidase activity was assayed as described in Materials and Methods. Black bars are measurements from strains carrying pKK223 (vector), and striped bars are measurements from those carrying pHNαβ. (B) Effect of an hns mutation on PuspB-lacZ expression. Cells were grown in defined medium (M9 glucose plus amino acids), and β-galactosidase activity was measured during exponential growth (OD600 of <1.0; hatched bars) or 2 h after entry into stationary phase (black bars). These values represent the minimum and maximum expression observed. Strains are AF633 (PuspB-lacZ) and AF800 (AF633 hns::cam).
Mutations in hns have been shown to increase ςS levels in log phase (45); thus we tested whether the PuspB-lacZ fusion was affected by introduction of an hns::cam mutation. As shown in Fig. 7B, PuspB-lacZ expression was increased sevenfold in log phase and twofold in stationary phase in the hns mutant compared to the wild-type derivative but still exhibited growth phase regulation. A mutation in hns was unable to overcome the effects of an rpoS mutation (data not shown) as has been reported for some rpoS-dependent genes (5, 6). Finally, uspB does not appear to be autoregulated: a deletion of uspB (see below) does not affect the expression of PuspB-lacZ. In addition, an insertion mutation in uspA does not affect the expression of uspB, and a deletion of uspB has only a small effect on uspA expression.
Overexpression of UspB is detrimental to cell viability.
Early in our investigations of uspB, we observed that plasmids containing the entire uspB gene were extremely unstable. For example, overnight cultures of a strain carrying one of these plasmids bearing uspB (pTN38Xho) contained only 1 of 104 cells that were still resistant to the selectable marker (carbenicillin). Most cells had lost the plasmid and persisted in the culture only because of the degradation of carbenicillin during growth of the culture. In addition, those cells which were still carbenicillin resistant were found to have rearrangements of the plasmids. We suspect that these strains had lost part of the uspB gene, but this was not examined further. Deletion of the uspB gene in these plasmids (e.g., pAF602) eliminated the instability phenotype. To confirm that UspB overproduction caused the instability observed in these experiments, we constructed a gene fusion (Plac-uspB) which allowed us to control the production of UspB (see Materials and Methods). A plasmid carrying this fusion was stable in a strain carrying lacIq (AF620) and in the absence of the inducer, IPTG. Growth and starvation of AF620 in the presence, but not in the absence of IPTG caused a reduction in cells carrying the plasmid to only 15% after 16 h of starvation.
A uspB mutant is sensitive to ethanol stress in stationary phase.
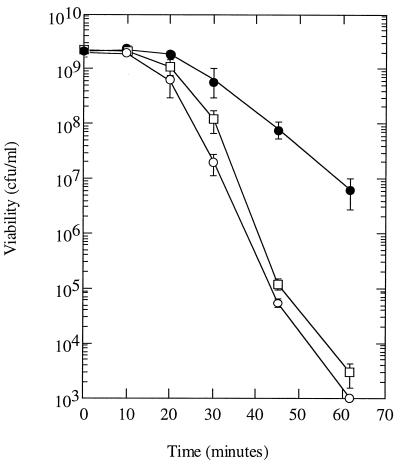
The strain carrying a deletion of uspB (AF607) or an insertion mutation (uspB::lacZ-Kmr; AF629) was compared to its parent strain with respect to the ability to grow and survive in a number of environmental conditions. Most conditions yielded no difference between the mutant and parent strains; specifically, the following were tested: growth in minimal medium and LB, survival in glucose starvation and LB stationary phase, recovery from long-term starvation (up to several weeks), survival during osmotic, heat (52 and 55°C), and oxidative (H2O2) stress in stationary phase, growth on various carbon sources (lactose, succinate, ribose, glycerol, acetate, and glucose), growth under anaerobiosis, and survival in stationary phase after growth in LB containing glucose (a condition which acidifies the medium greatly). The only difference observed between the strains was during exposure to ethanol. During exponential growth, the mutant and parent were equally sensitive to either 4 or 10% ethanol. In stationary phase, both the parent and uspB mutant became more resistant to ethanol exposure (4% had no effect on viability), but the mutant failed to develop the same high degree of resistance (to 10% ethanol) as the parent (Fig. 8). Higher concentrations of ethanol killed both the wild type and mutant too quickly for us to determine if there was a difference between their rate of killing. Thus, we conclude that uspB mutants are unable to fully develop stationary-phase-induced resistance to ethanol.
FIG. 8.
Survival of uspB mutants after exposure to ethanol during stationary phase. Cells were grown overnight in LB medium and then mixed with ethanol to 10%. Cultures were incubated with shaking at 37°C, and samples were taken periodically to measure CFU on LB plates at 30°C after appropriate dilution in M9 medium lacking glucose. The strains used were MC4100 (wild type; filled circles), AF607 (ΔuspB::cam; open squares), and AF629 (uspB::lacZ-Kmr; open circles). The error bars represent 1 standard deviation.
DISCUSSION
The uspB gene appears to be primarily regulated by the alternative sigma factor ςS. This alternative sigma factor is involved in the induction of at least 35 different genes during stationary phase, many of which are involved in the development of the resistance to harmful environmental conditions during starvation or stationary phase (reviewed in reference 17). In particular, thermotolerance, osmotic shock, and oxidative stress resistance in stationary phase are mediated in part by ςS-dependent genes. Our results demonstrate that another stationary-phase resistance, that to ethanol, is also mediated at least in part by an ςS-regulated gene.
The fact that mutants lacking the uspB gene product are sensitive to ethanol only in stationary phase and not in exponential growth allows us to speculate about its role in stationary-phase physiology. Ethanol is known to have at least two effects on cells. The first is an effect on protein folding and/or denaturation. It is well known that ethanol induces a heat shock response in E. coli and that this response is signalled by an increase in unfolded proteins (reviewed in reference 16). Two mechanisms of heat shock response induction exist (reviewed in reference 16). The first operates during relatively mild heat shock (42°C) or ethanol exposure (4%) and is dependent on the alternative sigma factor ς32. This response is triggered by an increased level of unfolded proteins in the cytoplasm. The second mechanism of heat shock protein induction operates during severe heat shock (>50°C) or ethanol exposure (10%) and is mediated by ςE. The ςE pathway is thought to be triggered by the presence of unfolded or misfolded outer membrane porins in the periplasm (9). If UspB were involved in either of these pathways, we would expect that the mutants would be deficient in survival both during ethanol exposure and during heat treatment. This is not the case; we could find no evidence of increased sensitivity to high temperature in our mutants. Thus, we consider it unlikely that the defect in uspB mutants is related to ethanol-induced protein denaturation.
The second effect of ethanol exposure is on the membrane. Cells exposed to lethal concentrations of ethanol lyse, presumably because of effects on the lipids of the inner and outer membranes. These effects include disruption of membrane organization by intercalation of the hydrophobic tail and dehydration mediated by hydrogen bonding with the hydroxyl portion of the molecule (21). Although both heat shock and ethanol alter the E. coli membrane, there is evidence that the two stresses cause different alterations (38). Thus, resistance to this effect of ethanol could be mediated by a pathway separate from the heat shock-induced pathway.
Why are uspB mutants sensitive to ethanol treatment only in stationary phase? This is not completely unexpected since uspB appears to be expressed only in stationary phase (and is not induced well by ethanol in exponential phase), but in addition, ethanol effects in log phase and stationary phase differ in at least one respect. In log phase, in LB medium, ethanol-inflicted cell death is caused by lysis of cells which is dependent on growth, indicating that the concentrations of ethanol used do not completely disrupt the membrane integrity; instead, it is thought that lysis results from an inhibition of peptidoglycan cross-linking (21). In contrast, stationary-phase cells are by definition nongrowing and thus insensitive to the low concentrations of ethanol used to kill log-phase cells. Thus, uspB mutants in stationary phase may be more resistant to ethanol than log-phase cells simply because they are not growing, but fail to achieve the increased tolerance of a wild-type cell. This increased tolerance could be due to changes in lipid composition (reviewed in references 11 and 20) or peptidoglycan cross-linking (35, 44) that is known to occur in stationary phase. UspB contains two putative membrane-spanning domains, and thus the UspB protein is most likely an integral membrane protein. It is tempting to speculate that UspB may play a role in sensing or mediating needed alterations in membrane composition during stationary phase. Finally, it should be noted that UspA, encoded upstream of uspB, is also implicated in membrane function during stationary phase because it is regulated, in part, by FadR, the global regulator of fatty acid synthesis and degradation (14). Thus, these two universal stress proteins, regulated by different control circuits, may both be involved in the alterations of membrane composition during stationary phase.
ACKNOWLEDGMENTS
Victor deLorenzo is gratefully acknowledged for analyzing DNA curvature; Mårten Hammar, Concetta DiRusso, Michael Givskov, David Friedman, Bernt Eric Uhlen, and Victor deLorenzo provided strains essential to this work.
This work was funded by a grant from the Swedish Natural Science Research Council.
REFERENCES
- 1.Albertson N H, Nyström T. Effects of starvation for exogenous carbon on functional mRNA stability and the rate of peptide chain elongation in Escherichia coli. FEMS Microbiol Lett. 1994;117:181–188. doi: 10.1111/j.1574-6968.1994.tb06762.x. [DOI] [PubMed] [Google Scholar]
- 2.Aldea M, Garrido T, Pla J, Vicente M. Division genes in Escherichia coli are expressed coordinately to cell septum requirements by gearbox promoters. EMBO J. 1990;9:3787–3794. doi: 10.1002/j.1460-2075.1990.tb07592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almirion M, Link A J, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnquist A, Olsen A, Normark S. ςS-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by ς70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol. 1994;13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x. [DOI] [PubMed] [Google Scholar]
- 6.Barth M, Marschall C, Muffler A, Fischer D, Hengge-Aronis R. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation in sigma S-dependent genes in Escherichia coli. J Bacteriol. 1995;177:3455–3464. doi: 10.1128/jb.177.12.3455-3464.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 8.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vechicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly L, De Las Penas A, Alba B M, Gross C A. The response to extracytoplasmic stress in Escherichia coli is controlled by partially overlapping pathways. Genes Dev. 1997;11:2012–2021. doi: 10.1101/gad.11.15.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in procaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 11.DiRusso C C, Nyström T. The fats of Escherichia coli during infancy and old age: regulation by global regulators, alarmones and lipid intermediates. Mol Microbiol. 1998;27:1–8. doi: 10.1046/j.1365-2958.1998.00645.x. [DOI] [PubMed] [Google Scholar]
- 12.Eshel D, Urrestarazu L A, Vissers S, Jauniaux J-C, van Vliet-Reedijk J C, Planta R J, Gibbons I R. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinoso-Urgel M, Chamizo C, Tormo A. A consensus structure for ςS-dependent promoters. Mol Microbiol. 1996;21:657–659. doi: 10.1111/j.1365-2958.1996.tb02573.x. [DOI] [PubMed] [Google Scholar]
- 14.Farewell A, Diez A A, DiRusso C C, Nyström T. Role of the Escherichia coli FadR regulator in stasis survival and growth-phase dependent expression of the uspA, fad, and fab genes. J Bacteriol. 1996;178:6443–6450. doi: 10.1128/jb.178.22.6443-6450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farewell A, Kvint K, Nyström T. Negative regulation by RpoS: a case of sigma factor competition. Mol Microbiol. 1998;29:1039–1052. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 16.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 17.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 18.Hengge-Aronis R. The role of rpoS in early stationary phase gene regulation in Escherichia coli K12. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 171–194. [Google Scholar]
- 19.Hofmann K, Stoffel W. TMbase-a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 20.Huisman G W, Siegele D A, Zambrano M M, Kolter R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1672–1682. [Google Scholar]
- 21.Ingram L O, Vreelan N S. Differential effects of ethanol and hexanol on the Escherichia coli cell envelope. J Bacteriol. 1980;144:481–488. doi: 10.1128/jb.144.2.481-488.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 23.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 24.Linn T, St. Pierre R. Improved vector for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Mitaku, S., S. Boon-Chang, and T. Hirokawa. A theoretical method to distinguish between membrane and soluble proteins by physiochemical approach. Department of Biotechnology, Tokyo University of Agriculture and Technology, Tokyo, Japan.
- 27.Miyamota K, Nakahigashi K, Nishimura K, Inokuchi H. Isolation and characterization of visible light-sensitive mutants of Escherichia coli K12. J Mol Biol. 1991;219:393–398. doi: 10.1016/0022-2836(91)90180-e. [DOI] [PubMed] [Google Scholar]
- 28.Mulvey M R, Sorby P A, Triggs-Raine B L, Loewen P C. Cloning and physical characterization of katE and katF required for catalase HPII expression in Escherichia coli. Gene. 1988;73:337–345. doi: 10.1016/0378-1119(88)90498-2. [DOI] [PubMed] [Google Scholar]
- 29.Nash H. The HU and IHF proteins: accessory factors for complex protein-DNA assemblies. In: Lin E C C, Simon A, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes Co.; 1996. pp. 149–179. [Google Scholar]
- 30.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Nyström T. The trials and tribulations of growth arrest. Trends Microbiol. 1995;3:131–136. doi: 10.1016/s0966-842x(00)88901-5. [DOI] [PubMed] [Google Scholar]
- 32.Nyström T, Neidhardt F C. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol Microbiol. 1992;6:3187–3198. doi: 10.1111/j.1365-2958.1992.tb01774.x. [DOI] [PubMed] [Google Scholar]
- 33.Nyström T, Neidhardt F C. Isolation and properties of a mutant of Escherichia coli with an insertional inactivation of the uspA gene, which encodes a universal stress protein. J Bacteriol. 1993;175:3949–3956. doi: 10.1128/jb.175.13.3949-3956.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyström T, Neidhardt F C. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol Microbiol. 1994;11:537–544. doi: 10.1111/j.1365-2958.1994.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 35.Pisabarro A G, de Pedro M A, Vazquez D. Structural modifications in the peptidoglycan of Escherichia coli associated with changes in the state of growth of the culture. J Bacteriol. 1985;161:238–242. doi: 10.1128/jb.161.1.238-242.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reeve C A, Amy P S, Matin A. Role of protein synthesis in the survival of carbon-starved Escherichia coli K-12. J Bacteriol. 1984;160:1041–1046. doi: 10.1128/jb.160.3.1041-1046.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Sanchez R A, Charlier J E. Effect of stress conditions on Escherichia coli outer and inner membranes, separated by Sephacryl S-500 chromatography. Anal Biochem. 1989;179:202–205. doi: 10.1016/0003-2697(89)90226-1. [DOI] [PubMed] [Google Scholar]
- 39.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 40.Sofia H J, Burland V, Daniels D L, Plunkett G, Blattner F R. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. Nucleic Acids Res. 1994;22:2576–2586. doi: 10.1093/nar/22.13.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturrock S S, Collins J F. MPsrch version 1.3. Edinburgh, United Kingdom: Biocomputing Research Unit, University of Edinburgh; 1993. [Google Scholar]
- 42.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wanner B L, Kodaira R, Neidhardt F C. Physiological regulation of a decontrolled lac operon. J Bacteriol. 1977;130:212–222. doi: 10.1128/jb.130.1.212-222.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wensink J, Gilden N, Witholt B. Attachment of lipoprotein to the murein of Escherichia coli. Eur J Biochem. 1982;122:587–590. doi: 10.1111/j.1432-1033.1982.tb06479.x. [DOI] [PubMed] [Google Scholar]
- 45.Yamashino, Ueguchi C, Mizuno T. Quantitative control of the stationary phase-specific sigma factor, ςS, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J. 1995;14:594–602. doi: 10.1002/j.1460-2075.1995.tb07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]