Abstract
Background
The patient selection for optimal adjuvant therapy in gastrointestinal stromal tumors (GISTs) is provided by nomogram based on tumor size, mitotic index, tumor location, and tumor rupture. Although mutational status is not currently used to risk assessment, tumor genotype showed a prognostic influence on natural history and tumor relapse. Innovative measures, such as KIT/PDGFRA-mutant-specific variant allele frequency (VAF) levels detection from next-generation sequencing (NGS), may act as a surrogate of tumor burden and correlate with prognosis and overall survival of patients with GIST, helping the choice for adjuvant treatment.
Patients and Methods
This was a multicenter, hospital-based, retrospective/prospective cohort study to investigate the prognostic role of KIT or PDGFRA-VAF of GIST in patients with radically resected localized disease. In the current manuscript, we present the results from the retrospective phase of the study.
Results
Two-hundred (200) patients with GIST between 2015 and 2022 afferent to 6 Italian Oncologic Centers in the EURACAN Network were included in the study. The receiver operating characteristic (ROC) curves analysis was used to classify “low” vs. “high” VAF values, further normalized on neoplastic cellularity (nVAF). When RFS between the low and high nVAF groups were compared, patients with GIST with KIT/PDGFRA nVAF > 50% showed less favorable RFS than patients in the group of nVAF ≤ 50% (2-year RFS, 72.6% vs. 93%, respectively; P = .003). The multivariable Cox regression model confirmed these results. In the homogeneous sub-population of intermediate-risk, patients with KIT-mutated GIST, the presence of nVAF >50% was statistically associated with higher disease recurrence.
Conclusion
In our study, we demonstrated that higher nVAF levels were independent predictors of GIST prognosis and survival in localized GIST patients with tumors harboring KIT or PDGFRA mutations. In the cohort of intermediate-risk patients, nVAF could be helpful to improve prognostication and the use of adjuvant imatinib.
Keywords: KIT mutation, PDGFRA mutation, variant allele frequency, GIST, adjuvant imatinib
Innovative measures, such as KIT/PDGFRA -mutant-specific variant allele frequency levels detection from next-generation sequencing, could act as a surrogate of tumor burden and correlate with prognosis and overall survival of patients with gastrointestinal stromal tumors, helping the choice for adjuvant treatment in this patient population.
Implications for Practice.
In the adjuvant setting, imatinib treatment is recommended for patients with GIST at high risk of recurrence. In the intermediate-risk subgroup, the choice of adjuvant imatinib is challenging. Our data, in a large population of 200 patients with KIT/PDGFRA mutated GIST, showed that the KIT/PDGFRA-nVAF levels detection negatively correlate with prognosis and overall survival of localized GIST patients. KIT-nVAF >50% was associated with poorer RFS compared to KIT-nVAF ≤50%. These findings are particularly relevant in the intermediate-risk patients. In this subpopulation, nVAF levels could be helpful to improve prognostication and to address adjuvant imatinib.
Introduction
Gastrointestinal stromal tumors (GISTs) are the most common malignant mesenchymal tumors in the gastrointestinal tract.1 Historically, GISTs represent the paradigmatic model of oncogene addiction and precision oncology.2 The full understanding of proto-oncogene receptor tyrosine kinase (KIT) and platelet-derived growth factor receptor alpha (PDGFRA) as GIST oncogenic drivers, together with the remarkable success of tyrosine kinase inhibitors (TKIs), significantly improved the overall survival (OS) of these patients to the range of 6-8 years.3 Over the past years, the advances in technology and the up-front use of comprehensive molecular profiling further improved the molecular breadth of information available to clinicians.4,5 In addition to KIT and PDGFRA GIST mutations, account for 70% and 15% of cases, respectively, and the 9% of GIST driven by the succinate dehydrogenase (SDH) loss of function, a variety of distinct oncogenic drivers was described in the sub-population of GIST originally referred to as “wild-type,” including activating mutations of PIK3CA, BRAF, and RAS family members, and translocations of NTRK, FGFR, and ALK.6-11 Although the mutational status has not been incorporated in prognostic risk-stratification, several literature evidence highlights the importance of tumor genotyping in clinical practice for tailoring treatments based on the mutational profile.12,13 Specifically, some typical mutations involving specific exons or codon locations show a prognostic impact on the GIST natural history and tumor behavior, such as the p.D842V mutation in exon 18 of PDGFRA, classically associated with favorable outcome, and KIT exon 11 deletions or deletion/insertions involving codons 557-558, conversely related to poor prognosis and high risk of tumor recurrence.14,15
To date, translation of the insights from molecular testing and profiling to GIST patient care is an ongoing challenge. Sequencing technologies and models continue to evolve quickly, and next-generation sequencing (NGS) has been rapidly integrated into molecular pathology.16 Notably, the information by high-throughput sequencing may have clinical relevance in the localized and metastatic disease. In the adjuvant setting, imatinib treatment is now recommended for patients with localized GIST at high risk of recurrence, however, a more accurate prognostication could improve patients’ selection for adjuvant therapy.17,18 When performing NGS an interesting parameter arising from the analysis is the variant allele frequency (VAF). VAF represents the percentage of sequence reads carrying the mutation with respect to the wild-type fraction.19 Although the exact frequency of allelic fraction is difficult to estimate because is influenced by the proportion of tumor cells in the tumor sample and the presence of copy number variations, KIT/PDGFRA-VAF of tumoral tissue could represent a surrogate measure of the proportion of GIST cells that harbor the specific DNA mutation. We hypothesized that tumor VAF could have a prognostic role in patients with GIST.
Methods
Study Design and Patient Selection
This was a multicenter, hospital-based, retrospective/prospective cohort study to investigate the prognostic role of KIT or PDGFRA-VAF of GIST in patients with localized disease and tumors radically resected. In the current manuscript, we present the results from the retrospective phase of the study.
Patients with metastatic de novo GIST, or lacking information on molecular testing and/or follow-up data, were excluded from the outcome analysis. The pathological information collected on primary GIST included the histological subtype, the primary tumor diameter, and mitotic count, and the site of origin of primary tumors from pathology reports for clinical use. All included patients had a tumor molecular profiling testing result by using a targeted NGS panel for the presence of GIST hot spot mutations. Mutation analysis was locally assessed at each participating center as part of routine clinical care. The clinical data on GIST surgery, the type and duration of adjuvant treatment, and tumor recurrence were abstracted from the clinical records. Recurrence-free survival (RFS) and overall survival (OS) were calculated. The association between VAF (%) and the clinical outcomes was evaluated.
The study was conducted according to good clinical practice (GCP) and has been designed with the ethical principles laid down in the Declaration of Helsinki on human experimentation. The University Hospital AOUP “Paolo Giaccone” (Palermo, Italy) coordinated the study. The study protocol was approved by the ethical committee of the coordinating center (Comitato Etico Palermo 1; Study Protocol “EVA GIST Project – Evaluation of Variant Allele Frequency in GIST” approval number: 04-13.04.22) and by the Institutional Review Boards of all Italian participating centers.
Mutational Analysis
Tissue samples were acquired following procedures of tumor resection surgery. The GIST diagnosis was made based on histopathologic assessment and immunohistochemical staining for CD117 antigen expression from local pathologists, with special expertise in GIST. The pathologists reported the primary tumor diameter and the tumor mitoses according to local protocols. Mitotic counts were expressed as the number of mitoses on 50 high-power fields (HPFs) or number of mitoses on a total area of 5 mm2. According to the ESMO-EURACAN guidelines,17 the mitotic count, expressed as number of mitoses/50 HPFs, was converted into the equivalent value expressed as number of mitoses/mm2. The exact proportion of neoplastic cells was assessed by the histopathologists. Before molecular testing, tissue adequacy and the presence of a suitable percentage of neoplastic cellularity have been evaluated. The proportion of neoplastic cells vs. “contaminant” non-neoplastic cells (neoplastic cellularity) in the area marked on the slide and used for DNA extraction was estimated by pathologists. Variant allele frequencies (VAFs) were reported as percentages and further normalized (nVAF) to the percentage of neoplastic cellularity (NC%) for each patient using the following formula: nVAF = (VAF/NC%) x 100.
All tumors were locally examined for somatic mutations in GIST actionable genes. Genomic DNA extraction was performed according to local protocols after manual dissection under microscopic guidance from FFPE (formalin-fixed and paraffin-embedded) sections. Next-generation sequencing multigene panel analysis was performed according to local platforms and protocols. For all detected pathogenic variants (PVs), a gene name, a nucleotide change (c.notation), and an amino acid substitution (p.notation) were typed. The classification of the variants was performed by consulting the databases “Catalogue of Somatic Mutations in Cancer” (COSMIC), Varsome tool, and ClinVar. According to the aim of the study, only KIT (exons 9, 11, 13, and 17) and PDGFRA (exons 12, 14, and 18) PVs were included in the analyses. Patients with tumor harboring PVs in other genes, or gene translocations, were excluded from this analysis because the very small numbers precluded a statistically significant conclusion.
Statistical Considerations
Our primary outcome measure was to assess the impact of nVAF on RFS and OS. Secondary objectives included the association of nVAF and the site of origin of primary tumors, the baseline diameter and mitosis, and the mutational status. The comparison between subgroups was performed with Fisher exact test, Pearson’s chi-square test, and ANOVA test. RFS was measured between the date of surgery and the date of first documentation of GIST recurrence or death, censoring patients who are alive without recurrence on the date of the last follow-up. OS was calculated from GIST diagnosis to death by any cause or last follow-up (censored patients). The receiver operating characteristic (ROC) curves analysis was used to determine the optimal cut-off for VAF and nVAF, to classify “low” vs. “high” values. The optimal cut-off of KIT/PDGFRA-VAF was 45% (AUC = 0.86; 95% CI, 0.72-0.99; P-value < .01). The optimal cut-off of nVAF, normalized to the percentage of neoplastic cellularity, was 50% (AUC = 0.84; 95% CI, 0.68-0.99; P-value < .02). The analysis of RFS and OS between groups was compared using the Kaplan-Meier method and log-rank test. To identify independent prognostic factors for RFS and OS, univariate and multivariate Cox proportional hazard regression models were performed. All tests were performed with a significance level of P < .05. Statistical analyses were conducted using IBM SPSS Statistics for Windows Version 27.0 (IBM Corporation).
Results
Study Population
Two-hundred (200) patients with localized GIST between 2015 and 2022 at 6 Italian Oncologic Centers in the European Reference Network on Rare Adult Cancers (EURACAN) Network, were included in the study.
We used the optimal KIT/PDGFRA nVAF threshold of 50%, determined by ROC curves analysis. To assess whether tumor nVAF can affect clinical-pathological features, the nVAF ≤ 50% (named “low nVAF”) vs nVAF >50% (named “high nVAF”) of KIT and PDGFRA mutations were correlated with the features of GIST patients (Table 1).
Table 1.
Patient and disease characteristics of patients with GIST localized.
| Characteristic | No. of patients (%) | nVAF ≤ 50% | nVAF > 50% | P-value |
|---|---|---|---|---|
| No of patients | 200 | 79 | 121 | — |
| Gender | .08 | |||
| Male | 111 (55.5) | 50 (63.3) | 61 (50.4) | |
| Female | 89 (44.5) | 29 (36.7) | 60 (49.6) | |
| Age groups (years) | .03 | |||
| ≤50 | 45 (22.5) | 24 (30.4) | 21 (17.4) | |
| >50 | 153 (76.5) | 54 (68.3) | 99 (81.8) | |
| Missing | 2 (1) | 1 (1.3) | 1 (0.8) | |
| Site of origin | .04 | |||
| Gastric | 104 (52) | 48 (60.7) | 56 (46.3) | |
| Others | 95 (47.5) | 30 (38) | 65 (53.7) | |
| Missing | 1 (0.5) | 1 (1.3) | 0 | |
| Baseline diameter | .02 | |||
| ≤5 cm | 89 (44.5) | 44 (55.7) | 45 (37.2) | |
| >5 cm | 102 (51) | 34 (43) | 68 (56.2) | |
| Missing | 9 (4.5) | 1 (1.3) | 8 (6.6) | |
| Baseline mitosis | .4 | |||
| ≤5/mmq | 111 (55.5) | 42 (53.2) | 69 (57) | |
| >5/mmq | 73 (36.5) | 32 (40.5) | 41 (33.9) | |
| Missing | 16 (8) | 5 (6.3) | 11 (9.1) | |
| Histology | 1 | |||
| Spindle cell | 111 (55.5) | 43 (54.5) | 68 (56.2) | |
| Epithelioid/mixed | 71 (35.5) | 28 (35.4) | 43 (35.5) | |
| Missing | 18 (9) | 8 (10.1) | 10 (8.3) | |
| Mutated KIT gene | .04 | |||
| Yes | 159 (79.5) | 57 (72.2) | 102 (84.3) | |
| No | 41 (20.5) | 22 (27.8) | 19 (15.7) | |
| KIT exons | .5 | |||
| Exon 11 | 130 (81.8) | 45 (78.9) | 85 (83.4) | |
| Other exons | 28 (17.6) | 12 (21.1) | 16 (15.7) | |
| Missing | 1 (0.6) | 0 | 1 (0.9) | |
| KIT exon 11 | .2 | |||
| Del-557/8* | 41 (31.5) | 11 (24.4) | 30 (35.3) | |
| Others** | 89 (68.5) | 34 (75.6) | 55 (64.7) |
*Del-557/8: deletions (del) or deletion/insertion (delins) involving 557 and/or 558 codons of KIT exon 11.
**Others: del or delins in other codons than KIT exon 11 557 and/or 558 codons, or KIT exon 11 duplications, insertions or SNVs.
Abbreviations: IQR, interquartile range; nVAF, normalized variant allele frequency.
One hundred and eleven patients were males (55.5%) and 89 were females (44.5%). The median age at study entry was 58 years [interquartile range (IQR), 21-87 years] in the nVAF ≤ 50% group, and 62 years [IQR, 30-81 years] in the nVAF >50% group. Differences were detected in terms of median nVAF in the age groups. In the group of 153 patients (76.5%) aged >50 years, the nVAFs were more frequently >50%, compared to the 45 patients (22.5%) in the age group ≤ 50 years (P = .03). Interestingly, the nVAF was significantly higher in patients with no-gastric site of origin than those with gastric GIST (P = .04), diameter of primary tumor >5 cm than baseline diameter ≤5 cm (P = .02), while there were no statistically significant differences in the low vs. high nVAF with respect to primary mitotic count (baseline mitosis ≤5/mmq vs. >5/mmq, P = .4), and histology (spindle cell vs. epithelioid/mixed, P = 1).
We then assessed the relevance of the mutational status on tumor nVAF low vs. high. Out of all 200 patients, 159 (79.5%) had a GIST harboring a KIT PV, and 41 patients (20.5%) showed a PDGFRA PV. Notably, the nVAF was significantly higher in patients harboring KIT PVs than in those with PDGFRA-mutated tumors (median KIT vs. PDGFRA nVAF, 55% vs. 46%; P = .04). However, in the group of patients with KIT-mutated GIST, there were no statistically significant differences in the nVAF low or high with respect to KIT Exons (Exon 11 or other exons; P = .5). To investigate to impact of KIT exon 11 PV type, the patients were classified according to the presence of KIT exon 11 deletions (del) or deletion/insertion (delins) involving 557 and/or 558 codons, versus other mutations, where the other mutations were deletion or delins in other codons than 557 and/or 558, or KIT exon 11 duplications, insertions, or single nucleotide variants (SNVs). No statistically significant differences in the nVAF were with respect to KIT exon 11 mutation type (KIT exon 11 codon 557 and/or 558 deletion or deletion/insertion vs. others; P = .2).
Outcome Analysis
We evaluated the survival outcomes of patients with localized GIST with regard to nVAF. The outcomes investigated were RFS and OS. Outcome data were available for n.178 localized GIST patients. The median follow-up time was 24 months (range, 6-116 months). The RFS rate at 2-years was 80.9% (median RFS 62 months; 95% CI, 38.5-85.5). During the follow-up, a total of 34 RFS events (recurrence or death) were observed (19.1%). Five events occurred in the group of 72 patients with tumors showing nVAF ≤ 50% (6.9%), and 29 events occurred in the 106 GIST patients with tumor nVAF > 50% (27.3%). When RFS between the 2 groups was compared, GIST patients with tumor nVAF > 50% showed less favorable RFS than patients in the group of nVAF ≤ 50% (2-year RFS, 72.6% vs. 93%, respectively; P = .003; Fig. 1F).
Figure 1.
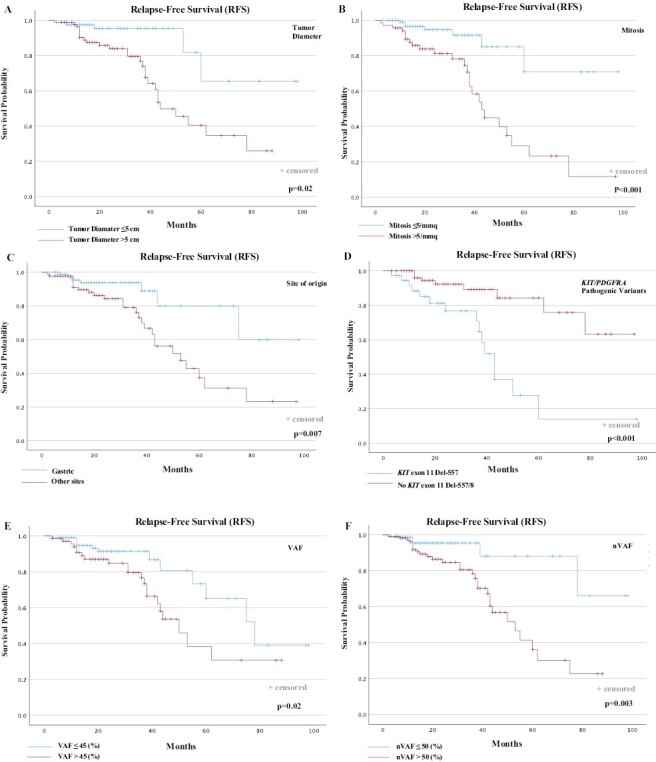
RFS according to prognostic factors. RFS according to (A) diameter of the primary tumor; (B) baseline mitosis; (C) site of origin; (D) KIT or PDGFRA PVs; (E) VAF; (F) nVAF. RFS, relapse-free survival; VAF, variant allele frequency; nVAF, normalized VAF.
When we examined the main prognostic factors in localized GISTs (primary mitotic count, tumor size, and tumor site), all 3 known factors were significantly associated with RFS (tumor mitosis >5/mmq vs. ≤ 5/mmq: 2-year RFS, 63.2% vs. 93.1%, P < .001; primary tumor diameter >5 cm vs. ≤ 5 cm: 2-year RFS, 70.7% vs. 93.8%, P = .002; no-gastric vs. gastric site of origin: 2-year RFS, 70.5% vs. 91.1%, P = .007; Fig. 1A–1C). Although mutational status has not been incorporated in any established prognostic systems at present, mutational analysis has a prognostic relevance beyond a predictive value for sensitivity to molecular-targeted therapy. Therefore, the impact of KIT Exon 11 PVs on RFS was evaluated. RFS in the subsets of patients who had KIT exon 11 codons 557 and/or 558 deletion or deletion/insertion (delins), had unfavorable RFS than the rest of the patients with GIST (2-year RFS, 59.5% vs. 89.5%, P < .001; Fig. 1D).
Overall median OS was not reached. During the follow-up, 8 total events (deaths) were observed (4.5%). All events occurred in the group of patients with GIST with nVAF >50%; conversely, no event was observed in the group with nVAF ≤50%. Therefore, 100% of patients with nVAF ≤50% were alive at 2 years, compared to 92.5% for the patients with nVAF >50% (P = .04; Fig. 2F).
Figure 2.
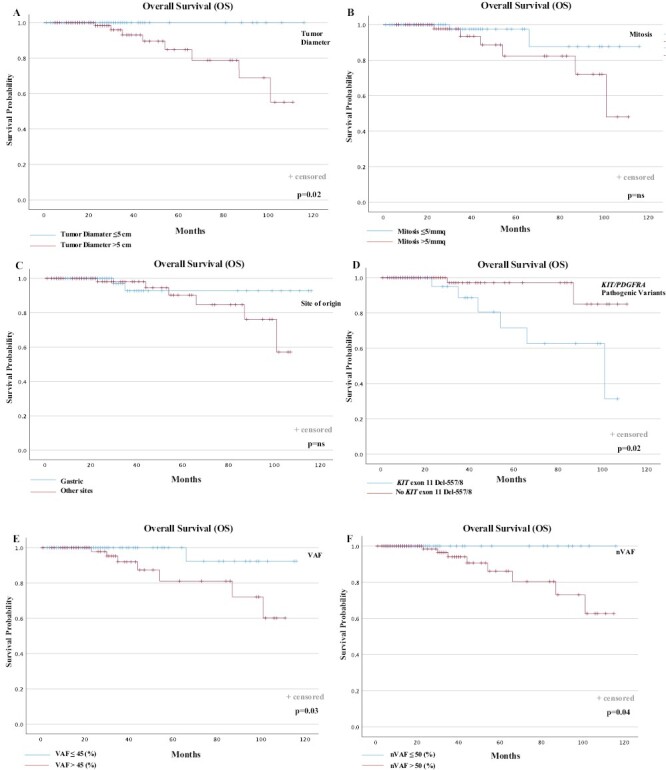
OS according to prognostic factors. OS according to (A) diameter of primary tumor; (B) baseline mitosis; (C) site of origin; (D) KIT or PDGFRA PVs; (E) VAF; (F) nVAF. OS, overall survival; VAF, variant allele frequency; nVAF, normalized VAF.
We next investigated the survival outcomes with regard to unadjusted VAF, to evaluate the possibility of avoiding further adjustment of the VAF value to the percentage of neoplastic cellularity for each patient in clinical practice. The optimal cut-off for the KIT/PDGFRA-VAF, determined through the ROC curves analysis, was 45% (AUC = 0.86; 95% CI, 0.72-0.99; P-value < .01). Five13 events occurred in the group of 108 patients with tumors showing VAF ≤45% (12.0%), and 21 events occurred in the 69 patients with GIST with tumor VAF >45% (30.4%). Despite VAF was also statistically associated with RFS (VAF >45% vs. VAF ≤45%: 2-year RFS, 69.6% vs. 88.0%, P = .02; (Fig. 1E), and OS (VAF >45% vs. VAF ≤45%: 2-year OS, 90.2% vs. 99.1%, P = .03; Fig. 2E), the statistical significance of the unadjusted VAF as a prognostic factor of tumor recurrence and patient survival was lower compared to “normalized” VAF on individual neoplastic cellularity shown above.
Univariable and Multivariable Analysis
To assess if the prognostic value of nVAF for RFS and OS was independent of other known clinicopathological factors, we performed univariable and multivariable Cox proportional hazard regression models. The following factors were found to be statistically significantly associated with RFS in univariable analyses: diameter of primary tumor >5 cm (HR: 3.94; 95% CI, 1.58-10.26; P = .005), mitosis >5/mmq (HR: 4.59; 95% CI, 1.98-10.63; P < .001), no-gastric site of origin (HR: 2.83; 95% CI, 1.28-6.27; P = .01), KIT Exon 11 deletions or delins involving codons 557 and/or 558 (HR: 0.21; 95% CI, 0.09-0.49; P < .001), and nVAF > 50% (HR: 3.83; 95% CI, 1.47-9.93; P = .006). In the final multivariable Cox regression model, mitosis (HR: 4.26; 95% CI, 1.36-13.38; P = .01), KIT exon 11 PV type (HR: 0.22; 95% CI, 0.08-0.63; P = .005), and nVAF (HR: 4.97; 95% CI, 1.29-19.11; P = .01), remain statistically significant.
Regarding OS, only the presence of KIT Exon 11 deletions or delins involving codons 557 and/or 558 were statistically significantly associated with OS in univariable analyses (HR: 0.18; 95% CI, 0.03-0.91; P = .03) (Table 2).
Table 2.
Univariable and multivariable analysis of prognostic factorsfor RFS and OS in localized patients with GIST.
| RFS | Univariable Cox regression | Multivariable Cox regression | ||
|---|---|---|---|---|
| HR (95%CI) | P-value | HR (95%CI) | P-value | |
| Gender (F vs. M) |
0.58 (0.29-1.19) | Ns | ||
| Primitive tumor diameter (≤5 cm vs. >5 cm) |
3.94 (1.58-10.26) | .005 | 1.08 (0.31-3.77) | ns |
| Mitosis (≤ 5/mmq vs. >5/mmq) |
4.59 (1.98-10.63) | <.001 | 4.26 (1.36-13.38) | .01 |
| Gastric site of origin (No vs. yes) |
2.83 (1.28-6.27) | .01 | 1.50 (0.52-4.31) | ns |
| Exon 11 Del or Delins 557 and/or 558 (No vs. yes) |
0.21 (0.09-0.49) | <.001 | 0.22 (0.08-0.63) | .005 |
| nVAF (≤ 50% vs. > 50%) |
3.83 (1.47-9.93) | .006 | 4.97 (1.29-19.11) | .01 |
| OS | Univariable Cox regression | Multivariable Cox regression | ||
|---|---|---|---|---|
| HR (95%CI) | P-value | HR (95%CI) | P-value | |
| Gender (F vs. M) |
0.84 (0.19-3.61) | ns | ||
| Primitive tumor diameter (≤ 5 cm vs. >5 cm) |
1.19 (0.48-2.98) | ns | ||
| Mitosis (≤ 5/mmq vs. >5/mmq) |
3.37 (0.68-16.79) | ns | ||
| Gastric site of origin (No vs. yes) |
2.15 (0.43-10.85) | ns | ||
| Exon 11 Del or Delins 557 and/or 558 (No vs. yes) |
0.18 (0.03-0.91) | .03 | ||
| nVAF (≤ 50% vs. > 50%) |
6.92 (0.85-56.62) | ns | ||
Diameter and site of primitive tumor, the mitosis number, the presence of KIT exon 11 codons 557 and/or 558 deletion or deletion/insertion, and nVAF on tumor tissue, were evaluated in the Cox regression model.
Abbreviations: Del, deletions; Delins, deletions/insertions; HR, hazard ratio; OS, overall survival; RFS, relapse-free survival; VAF, variant allele frequency; nVAF, normalized VAF.
Therefore, these results showed that, in the population of GIST patients with localized disease, the nVAF ≤50% on tumor tissue, along with the primitive tumor diameter ≤5 cm, the mitosis number ≤5/mmq, the gastric site of origin, and the absence of KIT exon 11 deletions or delins 557/558, were significant independent prognostic factors for longer RFS. RFS and OS curves were plotted according to each independent prognostic factor (Figs. 1A–F and 2A–F).
The Impact of nVAF in the Intermediate-Risk, KIT Mutated Patients
We included a further analysis of the subpopulation of patients with intermediate-risk GIST. To achieve a homogeneous cohort of patients, we excluded patient with GIST with tumors harboring other mutations than KIT, and patients treated with adjuvant imatinib. The aim of current analysis was to investigate the ability of nVAF to identify a subpopulation of intermediate-risk patients with higher risk of recurrence disease, who may benefit from adjuvant treatment.
Sixty-six (66) patients were included in the analysis. During the follow-up, a total of 10 RFS events (recurrence or death) were observed (15.1%). RFS rate was 74.4% for the high-nVAF group and 100% for the low-risk group. When RFS between the 2 groups was compared, patients with GIST with intermediate risk and high-nVAF showed a poorer RFS than the low-nVAF group (P = .01). RFS curves in the intermediate-risk patient cohort were plotted according to VAF and nVAF (Fig. 3A, B).
Figure 3.
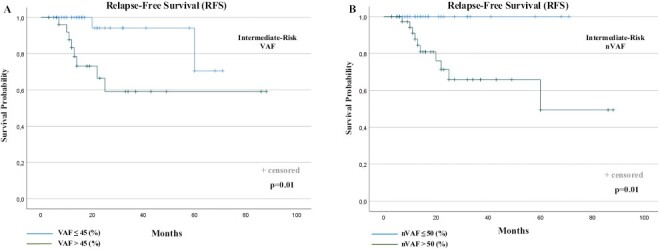
RFS according to VAF (A) and nVAF (B) in the sub-population of intermediate-risk, KIT mutated, GIST patients. RFS, relapse-free survival; VAF, variant allele frequency; nVAF, normalized VAF.
Discussion
In the past 20 years, considerable progress has been made in the molecular characterization of GIST and, many years later imatinib introduction, GISTs remain a perfect model for the development of precision medicine in cancer.20 Mutation analysis is now the standard diagnostic procedure at diagnosis as an essential predictive tool for treatment decision-making.21 Several reports have shown that the type and gene location of mutation strongly affects the activity of TKIs and, consequently, influence the decision on imatinib adjuvant and neoadjuvant treatment, and the management of locally advanced and metastatic disease.22-25 Although mutational status is not currently used to risk assessment in localized GIST, some mutations involving specific exons or codons showed a prognostic influence on tumor relapse.26-28 To date, the patient selection for optimal adjuvant therapy is provided by classifications and nomograms based on tumor size, mitotic index, tumor location, and tumor rupture.29 Novel risk-stratification methods were developed, such as the prognostic contour maps, where tumor size and mitosis count are treated as continuous non-linear variables.30 However, prognostication in routine cancer care is still a challenge for optimal patient selection. Current NGS technology allows us to assess the unique and complex set of clonal and subclonal mutations that represent the drivers of cancer evolution in GIST.5,31 Thus, we hypothesized that innovative measures, such as KIT/PDGFRA-mutant-specific nVAF levels detection, may act as a surrogate of tumor burden, and negatively correlate with prognosis and overall survival of patients with localized GIST.
In our cohort of patients with KIT or PDGFRA mutated GIST, we demonstrate that higher levels of nVAF were associated with worse RFS and OS in patients with radically resected GIST patients. This appears to be an independent predictive factor for RFS in our multivariable model.
Specifically, GIST patients with tumor nVAF > 50% showed less favorable RFS than patients in the group of nVAF ≤ 50%. According to literature data on established prognostic factors, primary mitotic count, tumor size, and tumor site, nVAF were significantly associated with RFS. When we explored the prognostic impact of KIT exon 11 codon 557 and/or 558 deletion or deletion/insertion, repeatedly associated with poor prognosis, the patients with tumors harboring these pathogenic variants showed unfavorable RFS compared to patients with any other KIT mutations, such as duplication, insertion, and single nucleotide variant (SNV), or deletion and deletion/insertion in other codons than 557 and/or 558. These data are consistent with the results of previous studies and confirm that in patients with localized GIST completely resected, KIT exon 11 deletions affecting codons 557 and/or 558 are associated with malignant tumor behavior and poor clinical outcome with an increased risk of relapse after surgery.14,26 The reasons for this more aggressive biology could be explained by the critical autoinhibitory role on the process of tyrosine kinase activation exert by 557 and 558 codon regions as a part of the code of the juxtamembrane (JM) domain that contacts the activation loop. Therefore, when these codons are deleted, results in a considerably increased spontaneous receptor phosphorylation and activation of the downstream pathway.32,33
We also found a correlation between high allelic frequency and specific clinical and pathological characteristics of patients. High KIT/PDGFRA-mutant-nVAF levels on tumoral tissue were associated with the age of GIST onset >50 years, no-gastric site of primary tumors, and diameter of primary tumor >5 cm. Furthermore, median nVAF was statistically higher in KIT-mutated than PDGFRA-mutated tumors, the last following a more indolent clinical course and often of prognostically favorable gastric origin compared to patients with tumor harboring KIT exon 11 mutations.15 Overall, these data confirm the prognostic significance of nVAF levels and their potential role in further improving prognostication in GIST patients.
Other authors exploited the VAF as a tool to assess the impact of allelic frequency on the survival of patients with cancer.34-38 Most studies were based on metastatic patients receiving TKI, suggesting a predictive role of this variable in other tumor types. No study analyzed the allelic frequency among patients with GIST. The recent experience of Berrino et al. group in 2021,34 interestingly supported the feasibility of BRAF-VAF for its undoubtedly prognostic and predictive value in a cohort of melanoma patients at different disease stages. In particular, the authors demonstrated a significant correlation between higher BRAF-VAF levels and the clinical outcome along with several pathological features such as melanoma thickness according to Clark level classification, lymphocyte infiltration, and lymph node metastases vs. visceral or cutaneous metastases. Notably, older patients showed increased VAF levels, similar to our population of GIST patients with KIT/PDGFRA-mutated. The biological reason why VAF levels could be associated with patient age remains speculative. Moreover, BRAF-VAF levels greater than 25% positively correlated with a favorable PFS and OS in patients undergoing combinatorial treatment with anti-BRAF/anti-MEK targeted therapies.34 In 2021, the other 2 groups, have further supported and highlighted the clinical impact of EGFR-VAF in lung adenocarcinoma patients treated with EGFR TKIs. In this setting, a significant correlation has been demonstrated between a high EGFR exon 19 deletion VAF and PFS and OS if compared to EGFR exon-21 point mutations VAF. Overall, an EGFR-VAF greater than 70% could be positively correlated with both PFS and OS,35 and a low VAF could be considered as a surrogate of less responsiveness to targeted treatment options.36 Interestingly, in liquid malignancies, a high TP53-VAF value has been described as a negative prognostic factor in patients with myelodysplastic syndromes in terms of overall survival37 as well as an independent predictor of leukemic transformation.38
With respect to VAF range, the data presented in this study highlight interesting greater VAF levels in GIST specimens if compared to VAF values of other solid tumors (ie, melanoma and NSCLC), where clinically relevant mutations are present at lower VAFs than GIST.34-37,39 The biological reason needs to be further investigated. A possible explanation is that GIST tumor evolution is closely guided by the onset of driver mutations of known oncogenes involved in neoplastic transformation.21,40,41 In cancer evolution, driver mutations are usually clonal mutations that occur early and have a higher allelic frequency than late, subclonal mutations.42 Consequently, the GIST dependence from KIT/PDGFRA mutations could strongly support the identification of high allelic frequencies at the molecular analysis. In some patients, the high VAF value might be due to germline KIT variants. However, it should be noted that: (1) such events of KIT germline are extremely rare43; (2) patients with germline variants of KIT have characteristic phenotypic manifestations43,44 and should have a family history of GIST or other diseases (such as cutaneous mastocytosis),45 both of which were not found in our recruited cases. Although it was not possible to perform a germline assessment of the identified variants in our cohort, due to the aforementioned reasons, we have excluded familial cancer syndrome caused by germline mutations in KIT/PDGFRA genes. Moreover, even though recent studies showed that personal or family syndromic features may be absent in a large proportion of patients with hereditary cancer predisposition, the germline alterations in GIST-associated gene are largely prevalent in patients with KIT/PDGFRA-wild-type GISTs (SDHA, SDHB, SDHC, NF1); conversely, constitutional variants in genes such as KIT are primarily somatic.46To our knowledge, our research is the first investigating the prognostic relevance of KIT/PDGFRA-nVAF in GIST patients. These current findings, on a large patient cohort, suggest that the high KIT/PDGFRA mutation allelic frequency of GIST should be one of the criteria to decide whether to use adjuvant treatment. Notably, in our sub-population of patients with intermediate-risk, KIT mutated, GIST, the presence of high variant allele frequency was statistically more associated with disease recurrence than low nVAF. In this cohort of patients, the use of adjuvant imatinib could be considered to reduce the risk of tumor relapse.
Moreover, in the future, nVAF could offer a promising approach to detecting the treatment-induced secondary mutations in metastatic patients. In this setting, emerging data in the literature increasingly demonstrate the clinical utility of liquid biopsies for a non-invasive and serial characterization of GIST mutations.47-50
Even though our findings would add important information on current prognostic factors, we acknowledge some limitations. First, the short follow-up of radically resected GIST, is related to the limited period of time from the use of NGS in the clinical practice. Another limitation of our study is the inclusion of all risk categories and KIT or PDGFRA mutated tumors, which show different clinical outcomes. Although this limitation is partially overcome by the statistically significant results in the homogeneous subgroup of patients with intermediate-risk, KIT mutations, without adjuvant treatment, further prospective validation of these results on a larger population is required to better define the impact of KIT/PDGFRA allele frequencies on patients’ with GIST clinical outcomes.
Conclusion
Deciphering the molecular architecture of GISTs has greatly improved our understanding of these tumors and is now increasingly implemented by innovative sequencing and genomic technologies.
Although a variety of clinical and pathological factors have been defined as predictors of tumor recurrence and patient survival, further refinements could be useful to further refine prognostication, especially when discussing prognosis and adjuvant treatment with patients classified as intermediate-risk and for better-tailored adjuvant treatment duration. The variant allelic frequency is readily available to clinicians and could be a useful surrogate of the clonal burden of KIT or PDGFRA mutations. In our study, we demonstrated that higher nVAF levels were independent predictors of GIST prognosis and survival in patients with localized GIST with tumors harboring KIT or PDGFRA mutations. This finding is particularly relevant in the intermediate-risk population, where higher nVAF is a prognostic factor of tumor recurrence, and the use of adjuvant imatinib could be considered to improve the clinical outcome. Further prospective validations of these findings will be able to develop more accurate prognostic tools in the clinical decision-making process.
Contributor Information
Lorena Incorvaia, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Dario De Biase, Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy; Solid Tumor Molecular Pathology Laboratory, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Bologna, Italy.
Margherita Nannini, Department of Experimental, Diagnostic and Specialized Medicine, S. Orsola-Malpighi Hospital, University of Bologna, Bologna, Italy.
Elena Fumagalli, Department of Medical Oncology, Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy.
Bruno Vincenzi, Department of Medical Oncology, Campus Biomedico University of Rome, Rome, Italy.
Ida De Luca, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Chiara Brando, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Alessandro Perez, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Maria A Pantaleo, Department of Experimental, Diagnostic and Specialized Medicine, S. Orsola-Malpighi Hospital, University of Bologna, Bologna, Italy.
Silvia Gasperoni, Department of Oncology and Robotic Surgery, Translational Oncology Unit, University Hospital Careggi, Firenze, Italy.
Lorenzo D’Ambrosio, Division of Medical Oncology, Candiolo Cancer Institute, FPO - IRCCS, Candiolo, TO, Italy.
Giovanni Grignani, Division of Medical Oncology, Candiolo Cancer Institute, FPO - IRCCS, Candiolo, TO, Italy.
Thais Maloberti, Solid Tumor Molecular Pathology Laboratory, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Bologna, Italy; Department of Medical and Surgical Sciences (DIMEC), University of Bologna, Bologna, Italy.
Erika Pedone, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Tancredi Didier Bazan Russo, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Alessandro Mazzocca, Department of Medical Oncology, Campus Biomedico University of Rome, Rome, Italy.
Laura Algeri, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Alessandra Dimino, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Nadia Barraco, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Roberta Serino, Department of Medical Oncology, Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy.
Valerio Gristina, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Antonio Galvano, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Viviana Bazan, Department of Biomedicine, Neuroscience and Advanced Diagnostics (BIND), Section of Medical Oncology, University of Palermo, Palermo, Italy.
Antonio Russo, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Giuseppe Badalamenti, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Funding
This research received no external funding.
Conflict of Interest
Lorenzo D’Ambrosio serves on the advisory board for PSI CRO Italy, GSK, AstraZeneca, Eisai, and Boehringer Ingelheim, and received support for meeting participation from PharmaMar and AstraZeneca. Giovanni Grignani serves on the advisory board for Bayer, Eisai, GSK, Lilly, and Merck, has been an invited speaker for Novartis and PharmMar, and received support in the form of an institutional grant from PharmaMar. The other authors indicated no financial relationships.
Author Contributions
Conception/design: L.I., D.d.B., G.B. Provision of study material or patients: D.d.B., M.N., E.F., B.V., I.D.L., C.B., A.P., M.A.P., S.G., L.d.A., G.G., T.M., E.P., T.D.B.R., A.M., L.A., A.D., N.B., R.S. Collection and/or assembly of data: L.I., C.B., E.P., A.P., V.B., A.R. Data analysis and interpretation: L.I., D.d.B., V.G., A.G. Manuscript writing: L.I., D.d.B., G.B. Final approval of manuscript: All authors.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Corless CL, Barnett CM, Heinrich MC.. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11(12):865-878. 10.1038/nrc3143 [DOI] [PubMed] [Google Scholar]
- 2. Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279(5350):577-580. 10.1126/science.279.5350.577 [DOI] [PubMed] [Google Scholar]
- 3. Khosroyani HM, Klug LR, Heinrich MC.. TKI treatment sequencing in advanced gastrointestinal stromal tumors. Drugs. 2023;83(1):55-73. 10.1007/s40265-022-01820-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cannella R, Tabone E, Porrello G, et al. Assessment of morphological CT imaging features for the prediction of risk stratification, mutations, and prognosis of gastrointestinal stromal tumors. Eur Radiol. 2021;31(11):8554-8564. 10.1007/s00330-021-07961-3 [DOI] [PubMed] [Google Scholar]
- 5. Vanden Bempt I, Vander Borght S, Sciot R, et al. Comprehensive targeted next-generation sequencing approach in the molecular diagnosis of gastrointestinal stromal tumor. Genes Chromosomes Cancer. 2020;60(4):239-249. [DOI] [PubMed] [Google Scholar]
- 6. Wu CE, Tzen CY, Wang SY, Yeh CN.. Clinical diagnosis of gastrointestinal stromal tumor (GIST): from the molecular genetic point of view. Cancers (Basel). 2019;11(5):679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Corless CL, Schroeder A, Griffith D, et al. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol. 2005;23(23):5357-5364. 10.1200/JCO.2005.14.068 [DOI] [PubMed] [Google Scholar]
- 8. Boikos SA, Pappo AS, Killian JK, et al. Molecular subtypes of KIT/PDGFRA wild-type gastrointestinal stromal tumors: a report from the National Institutes of Health Gastrointestinal Stromal Tumor Clinic. JAMA Oncol. 2016;2(7):922-928. 10.1001/jamaoncol.2016.0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miettinen M, Killian JK, Wang ZF, et al. Immunohistochemical loss of succinate dehydrogenase subunit A (SDHA) in gastrointestinal stromal tumors (GISTs) signals SDHA germline mutation. Am J Surg Pathol. 2013;37(2):234-240. 10.1097/PAS.0b013e3182671178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cocco E, Scaltriti M, Drilon A.. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15(12):731-747. 10.1038/s41571-018-0113-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li F, Huynh H, Li X, et al. FGFR-mediated reactivation of MAPK signaling attenuates antitumor effects of imatinib in gastrointestinal stromal tumors. Cancer Discov. 2015;5(4):438-451. 10.1158/2159-8290.CD-14-0763 [DOI] [PubMed] [Google Scholar]
- 12. Incorvaia L, Fanale D, Vincenzi B, et al. Type and gene location of KIT mutations predict progression-free survival to first-line imatinib in gastrointestinal stromal tumors: a look into the exon. Cancers (Basel). 2021 27;13(5):993. 10.3390/cancers13050993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rossi S, Gasparotto D, Miceli R, et al. KIT, PDGFRA, and BRAF mutational spectrum impacts on the natural history of imatinib-naive localized GIST: a population-based study. Am J Surg Pathol. 2015;39(7):922-930. 10.1097/PAS.0000000000000418 [DOI] [PubMed] [Google Scholar]
- 14. Incorvaia L, Badalamenti G, Fanale D, et al. Not all KIT 557/558 codons mutations have the same prognostic influence on recurrence-free survival: breaking the exon 11 mutations in gastrointestinal stromal tumors (GISTs). Ther Adv Med Oncol. 2021 30;13:17588359211049779. 10.1177/17588359211049779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wozniak A, Rutkowski P, Schöffski P, et al. Tumor genotype is an independent prognostic factor in primary gastrointestinal stromal tumors of gastric origin: a European multicenter analysis based on ConticaGIST. Clin Cancer Res. 2014 Dec 1;20(23):6105-6116. 10.1158/1078-0432.CCR-14-1677 [DOI] [PubMed] [Google Scholar]
- 16. Russo A, Incorvaia L, Capoluongo E, et al. The challenge of the Molecular Tumor Board empowerment in clinical oncology practice: A Position Paper on behalf of the AIOM- SIAPEC/IAP-SIBioC-SIC-SIF-SIGU-SIRM Italian Scientific Societies. Crit Rev Oncol Hematol. 2022;169:103567. 10.1016/j.critrevonc.2021.103567 [DOI] [PubMed] [Google Scholar]
- 17. Casali PG, Blay JY, Abecassis N, et al. ; ESMO Guidelines Committee, EURACAN and GENTURIS. Electronic address: clinicalguidelines@esmo.org. Gastrointestinal stromal tumours: ESMO-EURACAN-GENTURIS Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2022;33(1):20-33. 10.1016/j.annonc.2021.09.005 [DOI] [PubMed] [Google Scholar]
- 18. Pantuso G, Macaione I, Taverna A, et al. Surgical treatment of primary gastrointestinal stromal tumors (GISTs): management and prognostic role of R1 resections. Am J Surg. 2020;220(2):359-364. 10.1016/j.amjsurg.2019.12.006 [DOI] [PubMed] [Google Scholar]
- 19. Strom SP. Current practices and guidelines for clinical next-generation sequencing oncology testing. Cancer Biol Med. 2016;13(1):3-11. 10.28092/j.issn.2095-3941.2016.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Serrano C, George S.. Gastrointestinal stromal tumor: challenges and opportunities for a new decade. Clin Cancer Res. 2020;26(19):5078-5085. 10.1158/1078-0432.CCR-20-1706 [DOI] [PubMed] [Google Scholar]
- 21. Debiec-Rychter M, Sciot R, Le Cesne A, et al. ; EORTC Soft Tissue and Bone Sarcoma Group. KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer. 2006;42(8):1093-1103. 10.1016/j.ejca.2006.01.030 [DOI] [PubMed] [Google Scholar]
- 22. Joensuu H, Wardelmann E, Sihto H, et al. Effect of KIT and PDGFRA mutations on survival in patients with gastrointestinal stromal tumors treated with adjuvant imatinib: an exploratory analysis of a randomized clinical trial. JAMA Oncol. 2017;3(5):602-609. 10.1001/jamaoncol.2016.5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vincenzi B, Nannini M, Badalamenti G, et al. Imatinib rechallenge in patients with advanced gastrointestinal stromal tumors following progression with imatinib, sunitinib and regorafenib. Ther Adv Med Oncol. 2018;10:1758835918794623. 10.1177/1758835918794623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nannini M, Nigro MC, Bruno V, et al. Personalization of regorafenib treatment in metastatic gastrointestinal stromal tumours in real-life clinical practice. Ther Adv Med Oncol. 2017;9(12):731-739. 10.1177/1758834017742627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nannini M, Rizzo A, Nigro MC, et al. Standard versus personalized schedule of regorafenib in metastatic gastrointestinal stromal tumors: a retrospective, multicenter, real-world study. ESMO Open. 2021;6(4):100222. 10.1016/j.esmoop.2021.100222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin-Broto J, Gutierrez A, Garcia-Del-Muro X, et al. Prognostic time dependence of deletions affecting codons 557 and/or 558 of KIT gene for relapse-free survival (RFS) in localized GIST: a Spanish Group for Sarcoma Research (GEIS) Study. Ann Oncol. 2010;21(7):1552-1557. 10.1093/annonc/mdq047 [DOI] [PubMed] [Google Scholar]
- 27. Andersson J, Bümming P, Meis-Kindblom JM, et al. Gastrointestinal stromal tumors with KIT exon 11 deletions are associated with poor prognosis. Gastroenterology. 2006;130(6):1573-1581. 10.1053/j.gastro.2006.01.043 [DOI] [PubMed] [Google Scholar]
- 28. Cho S, Kitadai Y, Yoshida S, et al. Deletion of the KIT gene is associated with liver metastasis and poor prognosis in patients with gastrointestinal stromal tumor in the stomach. Int J Oncol. 2006;28(6):1361-1367. [PubMed] [Google Scholar]
- 29. Gold JS, Gönen M, Gutiérrez A, et al. Development and validation of a prognostic nomogram for recurrence-free survival after complete surgical resection of localised primary gastrointestinal stromal tumour: a retrospective analysis. Lancet Oncol. 2009;10(11):1045-1052. 10.1016/S1470-2045(09)70242-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Joensuu H, Vehtari A, Riihimäki J, et al. Risk of recurrence of gastrointestinal stromal tumour after surgery: an analysis of pooled population-based cohorts. Lancet Oncol. 2012;13(3):265-274. 10.1016/S1470-2045(11)70299-6 [DOI] [PubMed] [Google Scholar]
- 31. Russo A, Incorvaia L, Malapelle U, et al. The tumor-agnostic treatment for patients with solid tumors: a position paper on behalf of the AIOM- SIAPEC/IAP-SIBioC-SIF Italian Scientific Societies. Crit Rev Oncol Hematol. 2021;165:103436. 10.1016/j.critrevonc.2021.103436 [DOI] [PubMed] [Google Scholar]
- 32. Ma Y, Cunningham ME, Wang X, et al. Inhibition of spontaneous receptor phosphorylation by residues in a putative alpha-helix in the KIT intracellular juxtamembrane region. J Biol Chem. 1999;274(19):13399-13402. 10.1074/jbc.274.19.13399 [DOI] [PubMed] [Google Scholar]
- 33. Bauer S, Duensing A, Demetri GD, Fletcher JA.. KIT oncogenic signaling mechanisms in imatinib-resistant gastrointestinal stromal tumor: PI3-kinase/AKT is a crucial survival pathway. Oncogene. 2007 29;26(54):7560-7568. 10.1038/sj.onc.1210558 [DOI] [PubMed] [Google Scholar]
- 34. Berrino E, Balsamo A, Pisacane A, et al. High BRAF variant allele frequencies are associated with distinct pathological features and responsiveness to target therapy in melanoma patients. ESMO Open. 2021;6(3):100133. 10.1016/j.esmoop.2021.100133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gieszer B, Megyesfalvi Z, Dulai V, et al. EGFR variant allele frequency predicts EGFR-TKI efficacy in lung adenocarcinoma: a multicenter study. Transl Lung Cancer Res. 2021;10(2):662-674. 10.21037/tlcr-20-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Friedlaender A, Tsantoulis P, Chevallier M, De Vito C, Addeo A.. The impact of variant allele frequency in EGFR mutated NSCLC patients on targeted therapy. Front Oncol. 2021;11:644472. 10.3389/fonc.2021.644472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deng J, Wu X, Ling Y, et al. The prognostic impact of variant allele frequency (VAF) in TP53 mutant patients with MDS: a systematic review and meta-analysis. Eur J Haematol. 2020;105(5):524-539. 10.1111/ejh.13483 [DOI] [PubMed] [Google Scholar]
- 38. Jiang L, Ye L, Ma L, et al. Predictive values of mutational variant allele frequency in overall survival and leukemic progression of myelodysplastic syndromes. J Cancer Res Clin Oncol. 2022;148(4):845-856. 10.1007/s00432-021-03905-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Biase D, Genestreti G, Visani M, et al. The percentage of Epidermal Growth Factor Receptor (EGFR)-mutated neoplastic cells correlates to response to tyrosine kinase inhibitors in lung adenocarcinoma. PLoS One. 2017;12(5):e0177822. 10.1371/journal.pone.0177822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heinrich MC, Owzar K, Corless CL, et al. Correlation of kinase genotype and clinical outcome in the North American Intergroup Phase III Trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 Study by Cancer and Leukemia Group B and Southwest Oncology Group. J Clin Oncol. 2008;26(33):5360-5367. 10.1200/JCO.2008.17.4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21(23):4342-4349. 10.1200/JCO.2003.04.190 [DOI] [PubMed] [Google Scholar]
- 42. Gomez K, Miura S, Huuki LA, et al. Somatic evolutionary timings of driver mutations. BMC Cancer. 2018;18(1):85. 10.1186/s12885-017-3977-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sekido Y, Ohigashi S, Takahashi T, et al. Familial gastrointestinal stromal tumor with germline KIT mutations accompanying hereditary breast and ovarian cancer syndrome. Anticancer Res. 2017;37(3):1425-1431. 10.21873/anticanres.11466 [DOI] [PubMed] [Google Scholar]
- 44. Ke H, Kazi JU, Zhao H, Sun J.. Germline mutations of KIT in gastrointestinal stromal tumor (GIST) and mastocytosis. Cell Biosci. 2016;6:55. 10.1186/s13578-016-0120-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nishida T, Hirota S, Taniguchi M, et al. Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat Genet. 1998;19(4):323-324. 10.1038/1209 [DOI] [PubMed] [Google Scholar]
- 46. Mandelker D, Marra A, Mehta N, et al. Expanded genetic testing of GIST patients identifies high proportion of non-syndromic patients with germline alterations. NPJ Precis Oncol. 2023;7(1):1. 10.1038/s41698-022-00342-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dimino A, Brando C, Algeri L, et al. Exploring the dynamic crosstalk between the immune system and genetics in gastrointestinal stromal tumors. Cancers (Basel). 2022;15(1):216. 10.3390/cancers15010216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fanale D, Incorvaia L, Badalamenti G, et al. Prognostic role of plasma PD-1, PD-L1, pan-BTN3As and BTN3A1 in patients affected by metastatic gastrointestinal stromal tumors: can immune checkpoints act as a sentinel for short-term survival? Cancers (Basel). 2021;13(9):2118. 10.3390/cancers13092118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Badalamenti G, Fanale D, Incorvaia L, et al. Role of tumor-infiltrating lymphocytes in patients with solid tumors: can a drop dig a stone? Cell Immunol. 2019;343:103753. 10.1016/j.cellimm.2018.01.013 [DOI] [PubMed] [Google Scholar]
- 50. Russo A, Incorvaia L, Del Re M, et al. The molecular profiling of solid tumors by liquid biopsy: a position paper of the AIOM-SIAPEC-IAP-SIBioC-SIC-SIF Italian Scientific Societies. ESMO Open. 2021;6(3):100164. 10.1016/j.esmoop.2021.100164 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.


