Abstract
Bacillus subtilis cytochrome c oxidase caa3 is encoded by the ctaCDEF genes at the ctaABCDEF locus, with the ctaBCDEF genes organized as an operon-like unit. A dyad symmetry sequence and a catabolite response element homolog can be recognized in the 240-bp intercistronic region between ctaB and ctaC. ctaB′-lacZ and ctaBCD′-lacZ transcriptional fusions integrated at the native locus were used to study catabolite effects on transcription of the ctaB and ctaCDEF genes. In Schaeffer’s medium lacking glucose, ctaBCD′-lacZ was expressed at a very low level during the exponential phase, and expression increased about 30-fold 2 h after entry into the stationary phase. In the presence of 0.5% glucose, ctaBCD′-lacZ expression was totally repressed. In contrast to ctaBCD′-lacZ, ctaB′-lacZ was constitutively expressed regardless of carbon source. The ctaCDEF genes were separated from ctaB by insertion of plasmids carrying selectable markers in such a way that the ctaCDEF and ctaB transcription units remained intact. Enzymatic assays of caa3 with these constructs, showed that ctaCDEF was not expressed independently of ctaB. Also, when a ′ctaB-ctaC′-lacZ fusion (containing the ctaB-ctaC intercistronic region) was placed at a remote nonessential locus, β-galactosidase activity could not be detected. The absence of a promoter in the ctaB-ctaC intercistronic space also was indicated by the inability to detect ctaC-specific transcripts with RNase protection assays, primer extension, and rapid amplification of 5′ cDNA ends. Direct mRNA measurements showed that, in the presence of 0.5% glucose, ctaBCDEF transcripts terminated at the 3′ end of the putative stem-loop structure and the distal portion was down-regulated. A possible mechanism for ctaCDEF gene regulation is suggested. Catabolite repression of ctaBCD′-lacZ was partly dependent on CcpA but was independent of HPr. The expression of ctaBCDEF also appears to require the strC, ctaA, and resD-resE gene products.
When a variety of carbon sources are available, Bacillus subtilis preferentially utilizes glucose. While glucose is present, genes encoding enzymes for nonfermentable carbon source utilization are usually down-regulated, a process called catabolite repression (CR) (2). During glycolysis, end products such as acetoin are formed (28), secreted into the medium, and not utilized by the cell until glucose is exhausted. Energy production through consumption of nonfermentable carbon sources can be obtained only via oxidative respiration, for which an active tricarboxylic acid (TCA) cycle and a functional respiratory chain are required. Terminal oxidases are enzymes that catalyze the final step of respiration, reducing one molecule of oxygen to two molecules of water. The free energy available from this reaction is used by the oxidase complexes to pump protons from the cytoplasm to the cell exterior (5, 13, 30, 45). Both biochemical and genetic evidence has shown that there are two heme A-containing terminal oxidases (a-type cytochromes) in B. subtilis (21, 29). One is quinol oxidase aa3, encoded by the qoxABCD operon located at kb 3913 to 3917 on the B. subtilis genetic map (20, 35). The other is cytochrome c oxidase caa3, encoded by the ctaCDEF genes located at kb 1560 to 1563 (20, 37). At the cta locus, there are six genes, ctaABCDEF. ctaA and ctaB encode two enzymes required for the biosynthesis of heme A, the prosthetic group of both aa3 and caa3 (26, 27, 42).
It was first thought that terminal oxidase caa3 was primarily expressed when cells were grown at a low rate (1, 43). In an early spectral analysis, Chaix and Petit (1) reported that the absorption peak at 600 nm (aa3) was shifted to 605 nm (caa3) and the peak at 548 nm corresponding to cytochrome c increased when cultures grown in glucose-containing minimal medium were compared with cultures grown in succinate-containing minimal medium. Biochemical evidence has shown that both a-type terminal oxidases are present in succinate-grown cells, whereas caa3 is undetectable in glucose-grown cells (21). Quantification of aa3 expression was monitored with a qoxA′-lacZ transcriptional fusion, and it was shown that the level of expression of the qoxABCD operon was higher in rich medium (Luria-Bertani [LB] medium) and glucose-containing minimal medium than in succinate-containing minimal medium (35). There have not been molecular studies on how ctaCDEF is regulated in response to carbon sources prior to this report.
Mueller and Taber (26, 27) have studied ctaA extensively and have shown that the expression of ctaA is active in exponential cultures and is elevated postexponentially in a relatively high concentration of glucose (1.0%). The promoter for ctaA (ctaAp) has been identified; transcription of ctaA occurs in the sense opposite that of ctaBCDEF (26, 27). Thus, ctaB must possess a specific promoter (ctaBp) in order to be transcribed. CtaB, as well as CtaA, has an important role in heme A biosynthesis and is required for the formation of both a-type terminal oxidases (42). Because aa3 is constitutively expressed regardless of culture conditions (21, 35) and because of the linked enzymatic function of CtaB and CtaA, ctaB is very likely transcribed with kinetics similar to those of ctaA or at least is not repressed by glucose. Between the ctaB and ctaC open reading frames, there is a 240-bp intercistronic region, sufficient for a specific promoter capable of activating ctaCDEF transcription; however, such a promoter has not been experimentally demonstrated. It is also possible that, in the absence of a ctaCDEF-specific promoter, the ctaBCDEF genes are organized as an operon unit and that ctaCDEF transcription depends on the upstream promoter ctaBp. In this case, a glucose-sensitive regulatory element should occur in the ctaB-ctaC intercistronic region to allow the differential expression of ctaB and ctaCDEF.
Three components involved in CR of gene expression in B. subtilis have been identified (2, 11, 17, 18). A 14-bp palindromic sequence was first identified as necessary for CR of α-amylase (amyE) expression in B. subtilis, and a consensus sequence was subsequently proposed for the catabolite response element (CRE) based on a mutational analysis of this region (46). Sequences similar to the CRE consensus sequence have been demonstrated to mediate CR in a number of other genes originating from B. subtilis, Bacillus megaterium, and Staphylococcus xylosus (11 and references therein). The identity of the critical sequence of the CRE was further strengthened by use of point mutations in various B. subtilis genes, e.g., acsA, acuA, and hutP, leading to increased or decreased repression efficiencies (15, 47). CR of most genes containing the CRE is also affected by the trans-acting factors CcpA and HPr at the level of transcription initiation (11). However, a ptsH1 strain with an alanine substitution at Ser-46 had no effect on the repression of amyE (44) and only partially relieved the repression of iol (6) and levD (23). A ptsH1-crh (crh encodes an HPr-like protein [12]) double mutant almost completely relieved these repressions.
Several genes are known to influence the synthesis or assembly of a-type cytochromes. The most striking are strC and resD-resE; strC mutants were first isolated as spontaneous streptomycin-resistant colonies (38) and contained only 40% of the wild-type complement of a-type cytochromes (24). The two-component signal transduction system resD-resE is a global regulator of B. subtilis respiration; a resD-resE mutant completely lacked a-type terminal oxidases (41). Both strC and resD-resE are required for the postexponential activation of ctaA (27, 41).
Here we report the transcriptional regulation of the ctaBCDEF gene cluster in response to growth phase, carbon sources, the catabolite repression regulators CcpA and HPr, and the ctaA regulator genes strC and resD-resE.
MATERIALS AND METHODS
Bacterial strains and media.
The B. subtilis strains and plasmids used in this study are listed in Table 1. RB1 (trpC2) is a derivative of wild-type B. subtilis 168. Growth supplements and antibiotics were obtained from Sigma Chemical Company, and media were obtained from Difco Laboratories. B. subtilis strains were maintained on LB medium. B. subtilis strains containing integrative plasmids conferring resistance to chloramphenicol, tetracycline, and spectinomycin were grown on LB agar plates containing 5 μg of chloramphenicol, 5 μg of tetracycline, and 60 μg of spectinomycin per ml, respectively. Escherichia coli strains containing plasmids conferring resistance to ampicillin were grown on MacConkey agar plates or in LB broth containing 50 μg of ampicillin per ml. Promoter activity indicated by lacZ fusions in B. subtilis colonies was detected by spraying plates with 4-methylumbelliferyl-β-d-galactoside to identify fluorescent colonies under UV light.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| RB1 | trpC2 | This laboratory |
| RB95 | strC2 | This laboratory |
| RB996 | trpC2 ctaA | This laboratory |
| RB1263a | trpC2 resD-resE | M. Hulett |
| RB1298 | RB1Ω pDIA5333 | This study |
| RB1301 | RB1263a Ω pDIA5333 | This study |
| RB1303 | RB95 Ω pDIA5333 | This study |
| RB1305 | RB996 Ω pDIA5333 | This study |
| RB1319A | RB1 Ω pAI195 | This study |
| RB1320 | RB1 Ω pAI196 | This study |
| RB1321 | RB1 Ω pAI197 | This study |
| RB1323 | trpC2 ccpA::spc | J. Stülke |
| RB1325 | RB1 Ω pDIA5334 | This study |
| RB1327 | ptsGHI::tet | J. Stülke |
| RB1330 | RB1298 Ω RB1323 | This study |
| RB1331 | RB1298 Ω PB1327 | This study |
| E. coli | ||
| JM107 | Δ(lac-proAB) thi endA1 gyrA96 hsdR17 relA1 λ− supE44 (F′ traD36 proAB lacIqZΔM15) | This laboratory |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) supE44 λ− thi-1 | GIBCO BRL |
| Plasmids | ||
| pDIA5333 | ctaBCD′-lacZ Ampr Cmr | 36 |
| pDIA5334 | ctaB′-lacZ Ampr Cmr | 36 |
| pSGMU38 | lacZ Ampr Cmr 7.8 kb | J. Errington |
| pAI195 | pSGMU38 with a 444-bp ctaB-ctaC fragment fused to lacZ | This study |
| pAI196 | pSGMU38 with a 324-bp ctaB-ctaC fragment fused to lacZ | This study |
| pAI197 | pSGMU38 with a 760-bp ctaB-ctaC fragment fused to lacZ | This study |
For the study of ctaB or ctaCDEF gene expression in response to various carbon sources, B. subtilis strains were grown in Schaeffer’s sporulation medium (9) containing no glucose, 0.1 or 0.5% glucose (Sigma), 20 mM acetoin (Fluka), or 20 mM malate (Sigma). The medium (liquid-to-flask volume ratio of 1:10) was inoculated with 1 ml of an overnight culture to yield an initial optical density at 600 nm of approximately 0.02, as measured with a UV-visible spectrophotometer (Pharmacia Biotech). The flasks were shaken in a 37°C water bath at 250 rpm, and culture samples were taken every 15 min in the exponential phase and every 30 or 60 min in the stationary phase. The time points chosen for the collection of cell samples were relative to T0, the point at which the departure from exponential growth was first observed in the growth curve.
In vitro DNA manipulations and bacterial cell transformation.
Restriction digestion, ligation, small-scale plasmid isolation from E. coli, genomic DNA isolation from B. subtilis strains, and subcloning were performed by either standard protocols (16, 22) or by following the manufacturer’s instructions for Wizard Miniprep kits (Promega) and Puregene (Gentra System, Inc.). Enzymes were obtained from the following sources: United States Biochemical Corp., Sigma, New England Biolabs, and Amersham Life Science. B. subtilis was transformed by the method of Piggot et al. (31); E. coli was transformed by the method of Hanahan (16) or by following the GIBCO BRL protocol for DH5α transformation.
Construction of integration plasmids.
Integration plasmids pAI195, pAI196, and pAI197 were constructed by cloning the B. subtilis ctaB-ctaC fragment generated by PCR into the PstI-HindIII restriction site of pSGMU38 (8). pAI195, pAI196, and pAI197 contain 444-, 324-, and 760-bp PCR products, respectively, generated by use of the primer pairs XL16 (5′-CGTACGAAGCTTCTTCTATTTA-3′) and XL19 (5′-GACTAGCTGCAGACTGCAACAACAACCACC-3′), XL24 (5′-CGTACGAAGCTTCAGGCGGCTTTACTTTTAAC-3′) and XL19, and XL16 and XL26 (5′-GACTAGCTGCAGTGTCGGTACAATCAGCTCC-3′), respectively; the underlined sequences are the engineered PstI (CTGCAG) and HindIII (AAGCTT) restriction enzyme sites. Each reaction contained 100 pmol of each primer and 20 ng of linearized plasmid pAI536 (26). PCR was performed on a Perkin-Elmer Gene Amp PCR System 9600 as follows: cycle 1, 5 min at 94°C, 2 min at 50°C, and 3 min at 72°C; cycles 2 to 29, 1 min at 94°C, 2 min at 50°C, and 3 min at 72°C; and cycle 30, 1 min at 94°C, 2 min at 50°C, and 10 min at 72°C. The ctaBCD′-lacZ and ctaB′-lacZ fusions were constructed by inserting a BglII-EcoRI fragment containing the first 244 codons of ctaA, all of ctaB and ctaC, and the first 51 codons of ctaD and a BglII-NcoI fragment encompassing the first 244 codons of ctaA and the first 299 codons of ctaB, respectively, into the multiple cloning site of pJM783 (36). The fusions were introduced into the B. subtilis chromosome by Campbell-type recombination events, and the integrations were confirmed by Southern blotting (data not shown).
RNase protection assays.
Total cellular RNA from B. subtilis was isolated with an RNaid Plus kit (Bio 101, Inc.) by following the supplier’s protocol. Two DNA fragments were amplified by PCR to serve as templates for in vitro transcription of the cRNA probes. The PCR procedure was performed in the same way as for the amplification of the ctaB-ctaC intergenic region described above, except that different primer pairs were used. The template for the long probe (473 nucleotides), spanning from within ctaB to within the ctaC open reading frame, was amplified with primers XL1 (5′-TCTATTTCGTTGCCATGG-3′) and XL2 (5′-GGATCCTAATACGACTCACTATAGGGAGGACTGCAACAACAACCACCR-3′); the underlined sequence is the T7 promoter. The short probe (364 nucleotides), spanning from the right arm of the stem-loop structure (see Fig. 1) to within the ctaC open reading frame, was amplified with primers XL1 and XL3 (5′-TCTTGTTTAATCAGGCGG-3′). With the purified PCR fragments as templates, [α-32P]UTP (Dupont)-labelled cRNA probes were generated with an Ambion, Inc., MAXIscript T7/T3 in vitro transcription kit by following the supplier’s protocol. The full-length cRNA probes were isolated from polyacrylamide gels and hybridized to B. subtilis or yeast total cellular RNA. RNase protection assays were performed as described by Driscoll and Taber (7) with an RPA kit (Ambion). Autoradiography was carried out on the dried gels with intensifying screens (Kodak) at −70°C overnight.
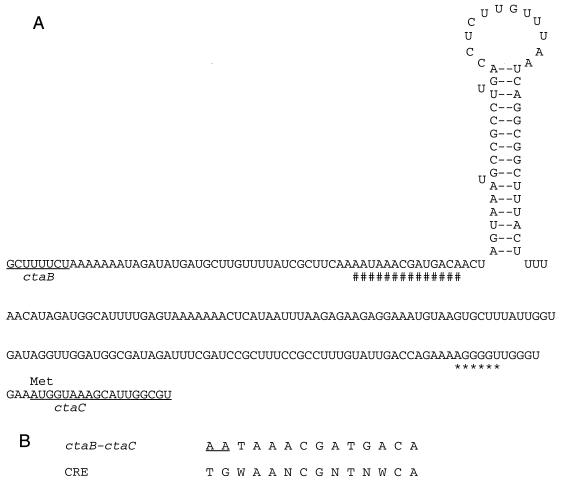
FIG. 1.
Sequence of the ctaB-ctaC intergenic region showing the CRE and the presumed rho-independent terminator structure. (A) The CRE homologue (#) and the putative ribosome binding site (∗) are marked, the ctaB and ctaC open reading frames are underlined, and the CtaC translational start site is labeled at AUG (methionine). Secondary structure prediction was performed by use of the Squiggles option of the Plotfold program of Genetics Computer Group sequence analysis software. (B) In the CRE consensus sequence, N represents any nucleotide, and W represents either A or T. The underlined sequence in the ctaB-ctaC CRE homologue represents the mismatch compared with the consensus sequence.
RT-PCR assay.
Total cellular RNA from B. subtilis grown with or without glucose was isolated with an RNeasy Total RNA kit (Qiagen). Extracts were treated with RNase-free DNase I from the MAXIscript kit at 37°C for 1 h to destroy any possible DNA contamination. Samples were then precipitated with 0.5 M ammonium acetate and 2.5 volumes of ethanol, washed with 70% ethanol, and resuspended in 50 μl of nuclease-free water. RNA concentrations were read as the absorbance at 260 nm in a Genequant II UV spectrometer (Pharmacia Biotech). The reverse transcription (RT) reactions were performed by use of tubes containing You-Prime First-Strand Beads (Pharmacia Biotech), 4.2 μg of B. subtilis total cellular RNA, and 15 pmol of synthetic oligonucleotide ctaCB1 (5′-ACAGTTCTTCACCCTGCTTAGCC-3′) by following the bead supplier’s protocol. H2O was used as one negative control. The other negative control was 4.2 μg of total cellular RNA aliquots without the You-Prime First-Strand Beads. The positive control was 27 ng of plasmid pAI536 containing the ctaABCD sequence (26). Five-microliter aliquots of cDNA or controls were then amplified with Ready To Go PCR Beads (Pharmacia Biotech) by following the supplier’s protocol. Primers ctaCB1 and ctaCF1 (5′-CAAGCCAGGAGCTGATTGTACC-3′), each at 20 pmol, were included in all reactions and controls. Following RT-PCR, 10-μl aliquots were electrophoresed in a 1% agarose gel (FMC BioProducts) containing 0.5 μg of ethidium bromide (Life Technologies) per ml. The relative intensities of DNA bands were photographed with an AlphaImager (Alpha Innotech Corporation).
Enzymatic activity assays.
The determination of β-galactosidase activities in cultures of B. subtilis was performed by the method of Zuber and Losick (48). One-milliliter aliquots of cells were removed from cultures, flash frozen in liquid nitrogen, and stored at −70°C overnight for enzymatic activity assays. β-Galactosidase activity was expressed in Miller units as described previously (25).
TMPD (N,N,N′,N′-tetramethyl-p-phenylenediamine) plate assays were performed as described previously (26). TMPD oxidation-positive colonies became blue within less than 5 min, whereas TMPD oxidation-negative colonies remained white.
RESULTS
Genetic and transcriptional organization of the ctaBCDEF locus.
ctaBCDEF is organized as an operon-like unit in which only two regions have sufficient sequence to accommodate promoters: 5′ to ctaB and the 240-bp ctaB-ctaC intercistronic region. In the latter region (3′ to ctaB), a dyad symmetry sequence could be detected, and mRNA could form a stem-loop secondary structure with a ΔG of −16 kcal/mol (Fig. 1). This potential RNA structure might serve as a rho-independent terminator. Three nucleotides upstream of the dyad symmetry sequence, a potential CRE homologue (with the first 2 nucleotides mismatched to the consensus sequence) could be detected (Fig. 1). The second of the two mismatches occurred at one of the five critical bases of the optimal CRE sequence deduced from the amyE mutational analysis (46). However, CREs with weak similarity to the optimal sequence, especially at the first two bases, are still able to function in CR of acuABC (15) and hut, gnt, and xyl (47 and references therein). The observation that the first two base mismatches are always accompanied by less similarity to the remaining optimal CRE makes it difficult to exclude the potential role of the CRE identified in the ctaB-ctaC intercistronic region. Combined with the adjacent CRE, the putative stem-loop structure might play a role in the regulation of the ctaBCDEF operon.
Effects of glucose, glycerol, and secondary carbon sources on ctaBCDEF expression.
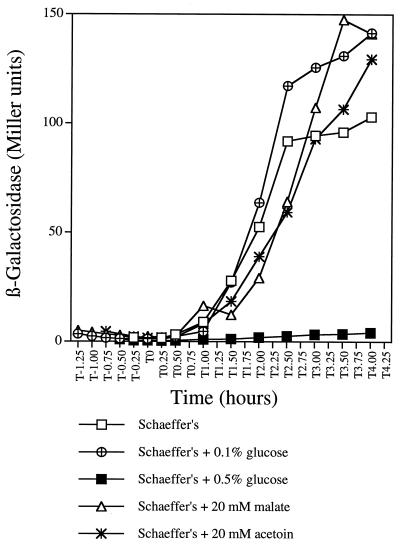
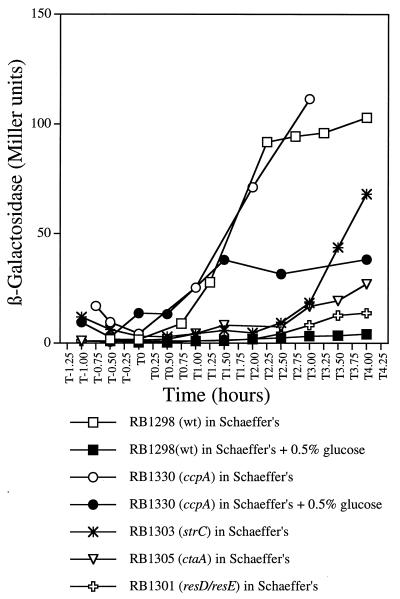
Evidence for the regulation of ctaCDEF expression has not been reported, except for limited biochemical and spectroscopic evidence, which suggests that B. subtilis contains caa3 only during growth on nonfermentable carbon sources (1, 21, 43). In order to further study the expression of ctaCDEF and its relationship to the expression of ctaB, a plasmid (pDIA5333) containing a ctaBCD′-lacZ transcriptional fusion (36) was integrated as a single copy at the native locus of wild-type strain RB1, and strain RB1298 was obtained. The expression of the ctaBCDEF genes was tested by growing strain RB1298 in Schaeffer’s medium alone or supplemented with different concentrations of glucose or with 20 mM acetoin or malate. In Schaeffer’s medium without additional carbon sources, ctaBCD′-directed β-galactosidase expression was less than 4 Miller units during the exponential phase, began to increase at the initiation of the stationary phase (T0), and was maintained at about 100 Miller units from T2 to T4 (Fig. 2). In contrast, when a high concentration of glucose (0.5%) was present in the medium, β-galactosidase activity was totally lacking throughout both exponential and postexponential phases. These results demonstrate that the expression of ctaBCDEF is growth phase dependent and is subject to glucose repression. However, at a lower concentration of glucose (0.1%), which would be exhausted and converted into glycolytic end products by T0, glucose repression was not present at T1, and the ctaBCD′-directed expression level was eventually substantially higher (about 150 Miller units) than that in glucose-free medium (Fig. 2). This result was presumably due to the stimulatory effects of glycolytic end-product reutilization in the postexponential period. In order to test this conclusion, 20 mM acetoin (a glycolytic end product) or 20 mM malate (a TCA cycle intermediate) was added directly to the medium to mimic their production via glycolysis. Results similar to those obtained with 0.1% glucose were observed (Fig. 2). These data suggest that the transcription of ctaBCDEF can also be postexponentially stimulated by secondary carbon sources.
FIG. 2.
Expression of ctaBCD′-lacZ in the presence of different carbon sources. β-Galactosidase activity in strain RB1298 (containing the ctaBCD′-lacZ fusion) grown in Schaeffer’s medium supplemented with different carbon sources is indicated. β-Galactosidase activity was plotted against the time at which the cell samples were collected. T0 indicates the end of exponential growth.
In gram-positive bacteria, the glycerol effect on catabolite-repressible genes is not well understood, but it seems to involve a mechanism different from that for glucose repression (2). Experiments similar to those shown in Fig. 2 were carried out with strain RB1298, except that the effect of growth in glycerol-supplemented Schaeffer’s medium was measured. As with glucose, high concentrations of glycerol abolished postexponential-phase expression of the ctaBCD′-lacZ fusion, and low concentrations of glycerol activated expression to the same extent as low concentrations of glucose. The kinetics of expression in the presence of glycerol were similar to those in the presence of glucose in terms of timing. However, the response to glycerol was more sensitive than that to glucose, since the glycerol repressive effect was still present at 0.05% (5.4 mM) glycerol, while 0.1% (5.5 mM) glucose was no longer repressive (Fig. 2). It is not known whether this result is due to the inherent differences in transcriptional responses to the two substrates, to differences in uptake efficiency, or to growth phase-dependent differences in rates of substrate utilization.
Glucose-independent expression of ctaB.
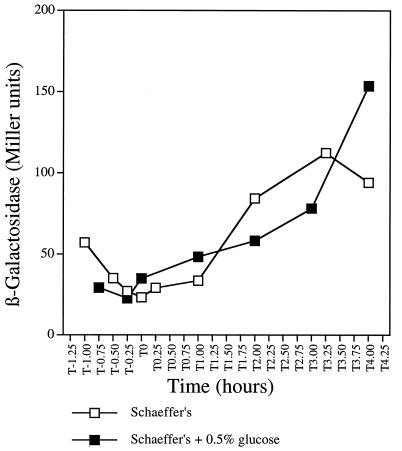
The heme A-containing cytochrome aa3 is present when cells are grown in glucose-containing media (21). Because heme A synthesis requires CtaB, the ctaB gene is therefore expected not to be subject to glucose repression. In order to monitor ctaB expression, the β-galactosidase activity of a ctaB′-lacZ transcriptional fusion in strain RB1325 was measured. In Schaeffer’s medium alone, ctaB′-lacZ was actively expressed in growing cells (Fig. 3), and expression increased postexponentially to a level comparable to that of the ctaBCD′-lacZ fusion (Fig. 2). In contrast to that of ctaBCD′-lacZ, ctaB′-lacZ expression was not repressed during growth in 0.5% glucose but rather reached a level (approximately 150 Miller units) similar to that of ctaBCD′-lacZ expression in 0.1% glucose or in the presence of the secondary carbon source acetoin or malate. This result may have been due in part to the stimulatory effect of glycolytic end products and TCA cycle intermediates, but the more important results is that, unlike transcription in the ctaCDEF gene cluster (as measured with the ctaBCD′-lacZ fusion), the expression of ctaB is glucose insensitive. Direct measurement of ctaB mRNA in RNase protection assays with a ′ctaB-ctaC′ probe (see the long probe in Fig. 5) transcribed from a PCR fragment showed accumulated ctaB transcripts when cultures were grown in the presence or absence of 0.5% glucose (data not shown).
FIG. 3.
Glucose effect on the expression of ctaB′-lacZ. ctaB′-directed β-galactosidase synthesis of strain RB1325 grown in Schaeffer’s medium alone or supplemented with 0.5% glucose was measured and plotted as described in the legend to Fig. 2.
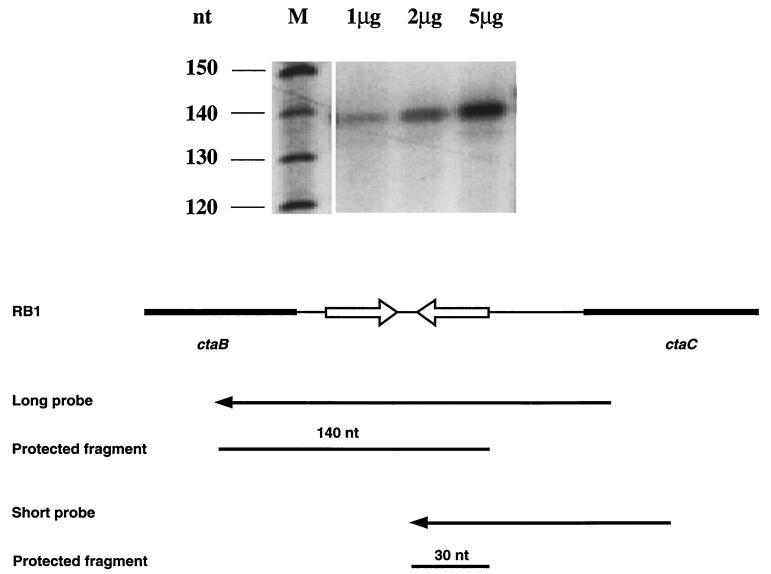
FIG. 5.
Low-resolution mapping of the ctaB transcript 3′ terminus by RNase protection assays. In lane M, the size marker SequaMark (Research Genetics) was used by following the supplier’s protocol. Sizes in nucleotides (nt) are indicated on the left. The other lanes contained 1, 2, and 5 μg of total cellular RNA isolated from cells at T0, hybridized to the long probe, and treated with 0.025 U of RNase A and 1 U of RNase T1. Protected hybrids were separated by electrophoresis on a 5% polyacrylamide gel containing 8 M urea. An autoradiograph of the dried gel is shown. The structures of the long probe and the short probe are shown.
ctaCDEF is not expressed when separated from ctaB.
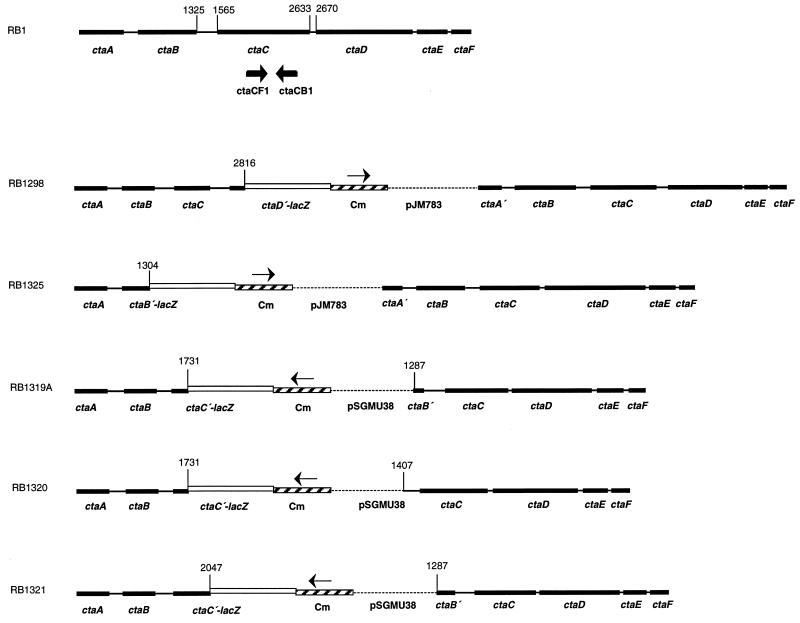
Based on sequence analysis, two possible regulatory mechanisms could be responsible for the differential expression of ctaCDEF in response to various carbon sources: (i) ctaCDEF has a specific promoter that regulates its expression or (ii) ctaCDEF transcription relies on the ctaB promoter, and catabolite-sensitive regulation occurs in the ctaB-ctaC intercistronic region to reduce ctaCDEF transcription in the presence of glucose. In order to test for the existence of a ctaCDEF-specific promoter, plasmids pAI195, pAI196, and pAI197 carrying chloramphenicol markers were integrated into the chromosome of strain RB1 by a single-crossover recombination event. By selection for Cmr colonies, strains RB1319A, RB1320, and RB1321 were obtained. In these integrants (Fig. 4), the continuity of ctaBCDEF transcription through the ctaB-ctaC intercistronic region is disrupted by the chloramphenicol resistance cassette. This cassette is transcribed in an opposite sense compared to the transcription of ctaBCDEF, but transcription within both ctaB and ctaCDEF would not be interrupted. Gene sequences downstream from the inserted plasmid pSGMU38 were initiated from different positions in the different integrants. In strains RB1319A and RB1321, ′ctaBCDEF was initiated from the last eight codons of ctaB. In strain RB1330, ′ctaCDEF was initiated from the second half of the dyad symmetry sequence (Fig. 1 and 4). The formation of the caa3 oxidase in the integrants would be completely dependent on a ctaCDEF-specific promoter within the ctaB-ctaC intercistronic region if such a promoter existed. Measurement of caa3 activity in intact cells was performed with TMPD, which can serve as an artificial electron donor for cytochrome c oxidase caa3 but not for quinone oxidase aa3 (21, 29).
FIG. 4.
Gene organization of the cta locus in the wild-type strain and strains carrying integrated plasmids. Genes ctaA, ctaB, and ctaCDEF are indicated as black boxes. Nucleotide numbering is based on the sequence determined by Saraste et al. (37). The approximate position of primer pair ctaCB1-ctaCF1 is indicated by arrows in the RB1 gene locus map. lacZ, chloramphenicol resistance gene (Cm), and plasmid sequences are indicated by open boxes, hatched boxes, and broken lines, respectively. The transcription orientations of Cm are represented by arrows. In strains RB1319A, RB1320, and RB1321, the initiation codon and the ribosome binding site (RBS) of the lacZ gene originate from the B. subtilis spoIIA gene (8). In strains RB1298 and RB1325, the initiation codon and the RBS of the lacZ gene originate from the B. subtilis spoVG gene (36). The maps are not drawn to scale.
Table 2 shows that caa3 was no longer present under either growth condition after ctaCDEF was separated from ctaB by the integration events. Also, upstream of the chloramphenicol selectable marker in these constructs, ctaC′-lacZ fusions occurred at different sites within ctaC open reading frames. RB1319A, RB1320, and RB1321 contain the putative first 55, 55, and 160 amino acids of CtaC, respectively, according to the sequence published by Saraste et al. (37). β-Galactosidase activity fold increases (that in Schaeffer’s medium divided by that in Schaeffer’s medium plus 0.5% glucose) measured with these integrants were comparable to that of RB1298 (data not shown).
TABLE 2.
Phenotypic properties of ctaBCDEF transcriptional continuity disruption mutants
| Strain and relevant genotype |
caa3 contenta in the following medium:
|
|
|---|---|---|
| Schaeffer’s | Schaeffer’s + glucose | |
| RB1 (wild type) | Blue | White |
| RB996 ΔctaA | White | White |
| RB1298 ctaBCD′-lacZ CmrbctaBCDEFc | Blue | White |
| RB1319A ctaBC′-lacZ CmrdctaCDEFc | White | White |
| RB1320 ctaBC′-lacZ CmrdctaCDEFc | White | White |
| RB1321 ctaBC′-lacZ CmrdctaCDEFc | White | White |
caa3 activity measured with TMPD as a substrate. Blue, caa3 positive; white, caa3 not detectable.
The chloramphenicol resistance cassette is transcribed in the same direction as ctaBCDEF.
Genotypes of integrants show the positions of chloramphenicol resistance cassette and lacZ fusion plasmid integration.
The chloramphenicol resistance cassette is transcribed in the direction opposite that of ctaBCDEF.
The lack of a ctaCDEF-specific promoter was also supported by the following two experiments. First, when the intact ctaB-ctaC intercistronic sequence was cloned in front of promoterless lacZ at the amyE locus, as either an in-frame translational or a transcriptional fusion, no β-galactosidase activity could be detected (data not shown). However, the same translational construct has been successfully used as an expression indicator for menp1, the major promoter in menaquinone biosynthesis (32). Second, when primer extension and rapid amplification of 5′ cDNA ends were carried out with primers complementary to the 5′ region of the ctaC open reading frame, no ctaC′ transcriptional start sites were found. The absence of ctaC′-initiated transcripts was also indicated by the direct measurement of transcripts in RNase protection assays (see below).
Low-resolution mapping of the ctaB transcript 3′ terminus.
The absence of a ctaCDEF-specific promoter pointed to the likelihood of catabolite-sensitive transcription termination in the ctaB-ctaC intercistonic region. To map the approximate 3′ ends of ctaB transcripts, independent RNase protection assays were performed with two cRNA probes initiating at the same nucleotide in the ctaC open reading frame (nucleotide 1731, according to reference 37) (Fig. 5). If a promoter exists in the ctaB-ctaC intercistronic region, both probes should protect a common fragment of the 5′ end of ctaC mRNA. However, the 3′ terminus of ctaB mRNA would be protected uniquely by each of the probes. Total cellular RNA was isolated from RB1 cells grown in LB medium to growth stage T0. [32P]UTP-labelled probes were hybridized to increasing amounts of B. subtilis or yeast RNA, and unprotected RNA was digested with RNase A and RNase T1. Protected RNA species were only observed when the long probe was tested (Fig. 5; the 30-nucleotide protected fragment of the short probe was not visualized due to its small size). This approximately 140-nucleotide long fragment maps to the 3′ end of the stem-loop structure, indicating that ctaB transcripts terminate at the stem-loop structure. Neither the long probe nor the short probe was able to detect any ctaC′ mRNA. This result confirms that no transcripts were initiated in the ctaB-ctaC intercistronic space. As a control, neither probe protected any fragment in yeast total cellular RNA (data not shown). Cells grown in Schaeffer’s medium with or without glucose to T0 were also tested, and ctaB mRNA was observed to terminate at the same site (data not shown).
Glucose effect on the transcription of ctaCDEF.
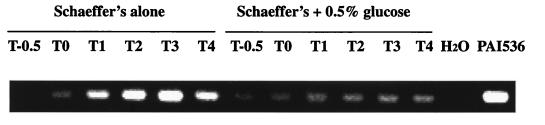
The results described above suggested that the absence of the terminal oxidase caa3 in exponential-phase cultures and postexponential-phase cultures in the presence of high glucose concentrations was due to down-regulation of the distal portion of ctaBCDEF transcripts. In order to directly measure ctaC transcripts, RT-PCR assays were carried out. Cellular RNA was extracted from cells grown in Schaeffer’s medium with or without glucose at growth stages T−0.5 to T4. First-strand cDNA was reverse transcribed with primer ctaCB1 hybridized to ctaC mRNA (beginning at nucleotide 2360, according to reference 34), and a 338-bp ctaC open reading frame fragment was subsequently amplified with primer pair ctaCB1-ctaCF1 (starting t nucleotide 2022, according to reference 37) (Fig. 4). As shown in Fig. 6, in the absence of glucose, ctaC mRNA abundance was undetectable during the exponential phase, began to increase at T0, and reached a maximum after T2. When cells were grown with 0.5% glucose added to the culture, ctaC mRNA levels were barely detectable. DNA contamination was monitored by PCR amplification of RNA aliquots without reverse transcriptase treatment, and no contamination was observed (data not shown). This result indicates that the absence of transcription through ctaC was associated with the down-regulation of caa3 synthesis when the cells were grown with high concentrations of glucose. However, in the absence of glucose, ctaBCDEF transcripts proceeded across the ctaB-ctaC intercistronic region and into the ctaCDEF coding region.
FIG. 6.
Direct measurement of ctaC mRNA. A culture of wild-type strain RB1 was grown in Schaeffer’s medium with or without glucose, and total cellular RNA was isolated at T−0.5, T0, T1, T2, T3, and T4. The reverse-transcribed first-strand cDNA from primer ctaCB1 was amplified by PCR with primer pair ctaCB1-ctaCF1. The location of the primers is shown in Fig. 4. The 338-bp amplified ctaC fragments were separated by electrophoresis on a 1% agarose gel containing 0.5 μg of ethidium bromide per ml. As controls, plasmid pAI536 and H2O were treated as total cellular RNA was. The growth conditions and timing are indicated.
Expression of the ctaBCDEF operon in ccpA and ptsH mutants.
When either the ccpA or the ptsH gene (encoding HPr) is inactivated by integration of antibiotic resistance cassettes, the integrants lose the glucose repression of many catabolic genes (11, 18). To investigate the role of CcpA and HPr in CR of the ctaBCDEF operon, either a ptsHI (ptsHI::tet) or a ccpA (ccpA::spc) gene disruption (40) was introduced into strain RB1298 carrying the ctaBCD′-lacZ transcriptional fusion (Table 1). CcpA effects were assessed by measuring the β-galactosidase activity of strain RB1330 (ccpA::spc ctaBCD′-lacZ) in the presence of high concentrations of glucose. In the ccpA mutant background, glucose repression was partially lifted, such that ctaBCD′-lacZ activity was about one-third that observed in the wild type (Fig. 7). Partial relief of glucose repression in the ccpA strain could occur if CcpA were not the only effector of glucose repression (see Discussion). The strain containing the ptsHI::tet gene disruption remained completely repressible by glucose and showed normal growth phase-dependent regulation (data not shown), suggesting that HPr is not required for specific regulation of the ctaBCDEF operon.
FIG. 7.
Effect of ccpA, strC, resD-resE, and ctaA mutations on the expression of ctaBCD′-lacZ. β-Galactosidase activities of the ctaBCD′-lacZ fusion in strains RB1298 and RB1330 grown in Schaeffer’s medium alone or supplemented with 0.5% glucose or in strains RB1301, RB1303, and RB1305 grown in Schaeffer’s medium alone were measured and plotted as described in the legend to Fig. 2. wt, wild type.
Effects of strC, resD-resE, and ctaA mutations on ctaBCD′-lacZ expression.
To examine the possible effects of strC and resD-resE on ctaBCDEF expression, the ctaBCD′-lacZ transcriptional fusion was integrated into strains RB95 and RB1263a, creating strains RB1303 and RB1301, respectively. β-Galactosidase activity was measured with Schaeffer’s medium. Strain RB1305 (ΔctaA ctaBCD′-lacZ) was also tested, since this ctaA deletion mutant is a-type cytochrome deficient (26). The expression of ctaBCD′-lacZ in the resD-resE and ctaA strains was reduced to about 10 to 25% that in the wild type (Fig. 7). An additional effect was observed: strC caused a delay of about 1.5 h in the postexponential-phase increase in ctaBCD′-lacZ expression. Evidently, the products of strC and resD-resE are necessary (directly or indirectly) for optimal postexponential-phase transcriptional activation of ctaBCDEF. The effect of strC or resD-resE mutations on the synthesis or assembly of the terminal oxidase caa3 complex may be mediated through their negative effect on ctaA expression.
DISCUSSION
Under the culture conditions used in this study, it was found that genes encoding the terminal oxidase caa3 are expressed by B. subtilis principally during the postexponential growth period and are subject to glucose repression. The rate of growth during the postexponential period in Schaeffer’s medium is low, and we suggest that this low growth rate provides one type of signal for the expression of ctaCDEF. The nature of this signal is not known, but the effect has been observed during exponential growth in defined medium with succinate as a carbon source by Lauraeus et al. (21). Before the modern distinction between terminal oxidases aa3 and caa3 was recognized, Chaix and Petit (1) systematically studied by spectral means the influence of carbon sources on the formation of the 600-nm (aa3) and 605-nm (caa3) components in intact cells. The latter was present in the exponential phase only when the growth rates in defined media were low. In the current studies, exponential growth rates did not vary widely when Schaeffer’s medium, which is an undefined, broth-based medium, was augmented with a variety of carbon sources. Thus, for example, the addition of malate did not result in the transcription of ctaCDEF during the exponential phase, because the cells were not dependent on this nonfermentable carbon source for growth.
Earlier studies from our laboratory addressed the growth-dependent regulation of respiratory chain components when B. subtilis cultures were grown in Schaeffer’s medium. Expression of the ctaA gene was shown to be maximal 2 hours after the onset of the stationary phase (T2) (27), while menp1-initiated transcript formation was maximal from the exponential phase (T−1) to the early stationary phase (T1) and then declined rapidly (32). In contrast to the data for ctaA and menp1, the data presented here show that both the terminal oxidase caa3 genes ctaCDEF and the heme A biosynthesis gene ctaB are activated and maintained at maximal transcription levels after T2. Overall, the transcription of ctaA, ctaB, and menp1 is maintained at substantial levels after T2, even though not all of these genes are expressed at maximal levels. A sufficient supply of the heme A prosthetic group and the reducing substrate (menaquinone) may allow the cells to switch on the caa3 respiration branch in order to cope with limited nutrient availability during the late stationary phase.
When grown with low concentrations of glucose, B. subtilis reutilizes accumulated glycolytic end products after glucose is exhausted. As shown here, the level of ctaBCDEF transcription in the late postexponential phase was higher in the presence of a nonfermentable carbon supplement or a low concentration (0.1%) of glucose than in glucose-free Schaeffer’s medium. A similar stimulatory effect was also observed for menp1-initiated transcripts in the stationary phase (32). This effect on menp1 was abolished in mutants blocked in secondary carbon reutilization: acuA (acetoin utilization) and acsA (acetyl coenzyme A synthesis; acetate utilization). Therefore, nonfermentable carbon sources may serve as or induce a positive signal for B. subtilis to coordinately regulate genes involved in energy production. A TGAAA sequence motif previously described for menaquinone biosynthesis genes (menB and menE) and for the heme A biosynthesis gene ctaA (7) also appears in the ctaB-ctaC intercistronic region. This sequence may serve as a cis-acting element for such regulation.
From sequence analysis alone, it appears that ctaCDEF may possess its own specific promoter in the ctaB-ctaC intercistronic region (37). However, genetic disruption experiments described in this work excluded the possibility of ctaCDEF being transcribed independently of ctaB. The downstream portion of the polycistronic transcript (ctaBCDEF) initiated from ctaBp provides information for the translation of the caa3 subunit peptides. Another prokaryotic ctaBCDEF homologue is the alkaliphilic Bacillus firmus OF4 cta operon, in which ctaCDEF encodes pH-regulated cytochrome caa3 (33). The cta gene organizations in B. firmus OF4 and B. subtilis are identical and exhibit 54% overall amino acid sequence identity. Northern blot analysis revealed a 5-kb (ctaBCDEF) message when the B. firmus OF4 total cellular RNA was probed with a ctaB probe, with no sign of any shorter transcripts (ctaB) (33). We suggest that, as with B. firmus OF4, B. subtilis initiates a polycistronic mRNA spanning ctaB to ctaF.
Many B. subtilis genes that respond to CR are regulated by CcpA, which binds to the CRE located in the respective promoter regions or in the 5′-terminal regions of their open reading frames (11, 18, 19). Thus, the frequency of transcription initiation is regulated. Our present data do not easily fit into such a regulator-operator model because of the lack of a ctaCDEF-independent promoter. However, a less common transcriptional termination-antitermination mechanism has been identified for a number of catabolic genes in B. subtilis (34, 39). One of the best-studied models is the B. subtilis glpD gene, encoding glycerol-3-phosphate (G3P) dehydrogenase (10, 14). The operon in which glpD is located belongs to the glp regulon, which is involved in the uptake and metabolism of glycerol and G3P. The expression of glpD is induced by G3P and repressed by glucose. An inverted repeat has been identified as a transcription terminator in the leader region of the glpD gene, and spontaneous mutants with deletions or insertions in the inverted repeat produce G3P dehydrogenase constitutively in the presence of glucose. Transcriptional antitermination is effected by the upstream gene product, GlpP, in conjunction with G3P. It has been suggested (14) that the GlpP protein interacts directly with the terminator to control glpD mRNA stability. Consequently, glpP mutants fail to grow on glycerol as a sole carbon and energy source (14).
A similar antitermination mechanism could also occur in ctaCDEF gene regulation, with the stem-loop structure in the ctaB-ctaC region acting as a terminator. This hypothesis leads to the speculation that antitermination also involves an interaction between a regulatory protein and the growing transcript. In the presence of glucose, the formation of this terminator could be enhanced by regulators such as CcpA. After glucose is exhausted, CcpA might not be able to stabilize the terminator, allowing the ctaBp-initiated transcription machinery to proceed through the dyad symmetry region into the ctaCDEF genes. The proximally located CRE might be recognized as an mRNA binding signal for the CcpA regulator. This model differs from the previously studied models in which the CRE is recognized as a DNA binding sequence for CcpA (18, 19). Recently, additional CcpA-like catabolite repressor proteins were reported (3, 4) and might be implicated in the residual CR of ctaCDEF transcription in the ccpA::spc background. Measuring ctaCDEF expression in ccpB and ccpA-ccpB strains (4) will assist in resolving this question.
Additional HPr-like proteins also appear to be involved in B. subtilis catabolite control. Recently, it was shown that, in a B. subtilis HPr-deficient strain, the synthesis of inositol dehydrogenase and β-xylosidase was not relieved from CR (12), as we have observed with the genes encoding terminal oxidase caa3. In that study (12), an additional disruption in the crh gene, encoding an HPr-like protein, caused an almost complete loss of glucose repression. These results indicate that, in addition to CcpA and CcpB, both HPr and Crh participate in CR of certain genes in B. subtilis. It would be interesting to test whether caa3 synthesis is also derepressed in a crh or a crh-ptsH double-mutation background, as is the expression of inositol dehydrogenase and β-xylosidase.
ACKNOWLEDGMENTS
We thank Margarida Santana, Philippe Glaser, Jörg Stülke, and Marion Hulett for B. subtilis strains; Xuan Qin, Belinda Rowland, and Linda Parsons for suggestions on the manuscript; Jeffrey Driscoll for assistance with graphics and for suggestions on the manuscript; and the Wadsworth Center Molecular Genetics Core Facility for oligonucleotide synthesis and sequencing.
Partial support was provided by Public Health Service grant GM-44547 from the National Institutes of Health.
REFERENCES
- 1.Chaix P, Petit J. Influence du taux de croissance sur la constitution du spectre hematinique de Bacillus subtilis. Biochim Biophys Acta. 1967;25:481–486. doi: 10.1016/0006-3002(57)90517-6. [DOI] [PubMed] [Google Scholar]
- 2.Chambliss H G. Carbon source-mediated catabolite repression. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 213–218. [Google Scholar]
- 3.Chauvaux S. CcpA and HPr(Ser-P): mediators of catabolite repression in Bacillus subtilis. Forum Microbiol. 1996;14:518–522. doi: 10.1016/0923-2508(96)84006-x. [DOI] [PubMed] [Google Scholar]
- 4.Chauvaux S, Paulsen I T, Saier M H., Jr CcpB, a novel transcription factor implicated in catabolite repression in Bacillus subtilis. J Bacteriol. 1998;180:491–497. doi: 10.1128/jb.180.3.491-497.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collman J P, Fu L, Herrmann P C, Zhang X. A functional model related to cytochrome c oxidase and its electrocatalytic four-electron reduction of O2. Science. 1997;275:949–951. doi: 10.1126/science.275.5302.949. [DOI] [PubMed] [Google Scholar]
- 6.Deutscher J, Reizer J, Fischer C, Galinier A, Saier M H, Jr, Steinmetz M. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J Bacteriol. 1994;176:3336–3344. doi: 10.1128/jb.176.11.3336-3344.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Driscoll J, Taber H W. Sequence organization and regulation of the Bacillus subtilis menBE operon. J Bacteriol. 1992;174:5063–5071. doi: 10.1128/jb.174.15.5063-5071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Errington J. A general method for fusion of the Escherichia coli lacZ gene to chromosomal genes in Bacillus subtilis. J Gen Microbiol. 1986;132:2953–2966. doi: 10.1099/00221287-132-11-2953. [DOI] [PubMed] [Google Scholar]
- 9.Harwood C R, Cutting S M. A1.3 special-purpose media. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. New York, N.Y: John Wiley & Sons, Inc.; 1990. pp. 549–550. [Google Scholar]
- 10.Holmberg C, Rutberg B. Expression of the gene encoding glyerol-3-phosphate dehydrogenase (glpD) in Bacillus subtilis is controlled by antitermination. Mol Microbiol. 1991;5:2891–2900. doi: 10.1111/j.1365-2958.1991.tb01849.x. [DOI] [PubMed] [Google Scholar]
- 11.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the Gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 12.Galinier A, Haiech J, Kilhoffer M, Jaquinod M, Stülke J, Deutscher J, Martin-Verstraete I. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Horsman J A, Barquera B, Rumbley J, Ma J, Gennis R B. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glatz E, Nisson R, Rutberg L, Rutberg B. A dual role for the Bacillus subtilis glpD leader and the GlpP protein in the regulated expression of glpD: antitermination and control of mRNA stability. Mol Microbiol. 1996;19:319–328. doi: 10.1046/j.1365-2958.1996.376903.x. [DOI] [PubMed] [Google Scholar]
- 15.Grundy F J, Turinsky A J, Henkin T M. Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J Bacteriol. 1994;176:4527–4533. doi: 10.1128/jb.176.15.4527-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning. I. A practical approach. Oxford, England: IRL Press; 1985. pp. 1–17. [Google Scholar]
- 17.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 18.Henkin T M. The role of the CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol Lett. 1996;135:9–15. doi: 10.1111/j.1574-6968.1996.tb07959.x. [DOI] [PubMed] [Google Scholar]
- 19.Kim J H, Guvener Z T, Cho J Y, Chung K-C, Chambliss G H. Specificity of DNA binding activity of the Bacillus subtilis catabolite control protein CcpA. J Bacteriol. 1995;177:5129–5134. doi: 10.1128/jb.177.17.5129-5134.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunst F, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 21.Lauraeus M, Haltia T, Saraste M, Wikström M. Bacillus subtilis expresses two kinds of haem-A-containing terminal oxidases. Eur J Biochem. 1991;197:699–705. doi: 10.1111/j.1432-1033.1991.tb15961.x. [DOI] [PubMed] [Google Scholar]
- 22.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 23.Martin-Verstraete I, Stülke J, Klier A, Rapoport G. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J Bacteriol. 1995;177:6919–6927. doi: 10.1128/jb.177.23.6919-6927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McEnroe A S, Taber H W. Correlation between cytochrome aa3 concentrations and streptomycin accumulation in Bacillus subtilis. Antimicrob Agents Chemother. 1984;26:507–512. doi: 10.1128/aac.26.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Mueller J P, Taber H W. Isolation and sequence of ctaA, a gene required for cytochrome aa3 biosynthesis and sporulation in Bacillus subtilis. J Bacteriol. 1989;171:4967–4978. doi: 10.1128/jb.171.9.4967-4978.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller J P, Taber H W. Structure and expression of the cytochrome aa3 regulatory gene ctaA of Bacillus subtilis. J Bacteriol. 1989;171:4979–4986. doi: 10.1128/jb.171.9.4979-4986.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano M M, Dailly Y P, Zuber P, Clark D P. Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. J Bacteriol. 1997;179:6749–6755. doi: 10.1128/jb.179.21.6749-6755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oost J V D, Wachenfeldt C V, Hederstedt L, Saraste M. Bacillus subtilis cytochrome oxidase mutants: biochemical analysis and genetic evidence for two a-type oxidases. Mol Microbiol. 1991;5:2063–2072. doi: 10.1111/j.1365-2958.1991.tb00829.x. [DOI] [PubMed] [Google Scholar]
- 30.Ostermerier C, Iwata S, Michel H. Cytochrome c oxidase. Curr Opin Struct Biol. 1996;6:460–466. doi: 10.1016/s0959-440x(96)80110-2. [DOI] [PubMed] [Google Scholar]
- 31.Piggot P J, Curtis C A M, deLancastre H. Demonstration of a polycistronic transcription unit required for sporulation of Bacillus subtilis by use of integrational plasmid vectors. J Gen Microbiol. 1984;130:2123–2136. doi: 10.1099/00221287-130-8-2123. [DOI] [PubMed] [Google Scholar]
- 32.Qin X, Taber H W. Transcriptional regulation of the Bacillus subtilis menp1 promoter. J Bacteriol. 1996;178:705–713. doi: 10.1128/jb.178.3.705-713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quirk P G, Hicks D B, Krulwich T A. Cloning of the cta operon from alkaliphilic Bacillus firmus OF4 and characterization of the pH-regulated cytochrome caa3 oxidase it encodes. J Biol Chem. 1993;268:678–685. [PubMed] [Google Scholar]
- 34.Rutberg B. Antitermination of transcription of catabolic operons. Mol Microbiol. 1997;23:413–421. doi: 10.1046/j.1365-2958.1997.d01-1867.x. [DOI] [PubMed] [Google Scholar]
- 35.Santana M, Kunst F, Hullo M F, Rapoport G, Danchin A, Glaser P. Molecular cloning, sequencing, and physiological characterization of the qox operon from Bacillus subtilis encoding the aa3-600 quinol oxidase. J Biol Chem. 1992;267:10225–10231. [PubMed] [Google Scholar]
- 36.Santana M, Ionescu M, Vertes A, Longin R, Kunst F, Danchin A, Glaser P. Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants. J Bacteriol. 1994;176:6802–6811. doi: 10.1128/jb.176.22.6802-6811.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saraste M, Metso T, Nakari T, Jalli T, Lauraeus M, Oost J V D. The Bacillus subtilis cytochrome c oxidase. Variations on a conserved protein theme. Eur J Biochem. 1991;195:517–525. doi: 10.1111/j.1432-1033.1991.tb15732.x. [DOI] [PubMed] [Google Scholar]
- 38.Staal S P, Hoch J A. Conditional dihydrostreptomycin resistance in Bacillus subtilis. J Bacteriol. 1972;110:202–207. doi: 10.1128/jb.110.1.202-207.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinmetz M. Carbohydrate catabolism: pathways, enzymes, genetic regulation, and evolution. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 157–170. [Google Scholar]
- 40.Stülke, J. Personal communication.
- 41.Sun G, Chesnut R, Sharkova E, Birkey S, Duggan M F, Sorokin A, Pujic P, Ehrlich S D, Hulett M F. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J Bacteriol. 1996;178:1374–1385. doi: 10.1128/jb.178.5.1374-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svensson B. Ph.D. thesis. Lund, Sweden: Lund University; 1995. [Google Scholar]
- 43.Tochikubo K. Changes in terminal respiratory pathways of Bacillus subtilis during germination, outgrowth, and vegetative growth. J Bacteriol. 1971;108:652–661. doi: 10.1128/jb.108.2.652-661.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voskuil M I, Chambliss G H. Significance of HPr in catabolite repression of α-amylase. J Bacteriol. 1996;178:7014–7015. doi: 10.1128/jb.178.23.7014-7015.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wachtenfeldt C V, Hederstedt L. Molecular biology of Bacillus subtilis cytochromes. FEMS Microbiol Lett. 1992;100:91–100. doi: 10.1111/j.1574-6968.1992.tb14025.x. [DOI] [PubMed] [Google Scholar]
- 46.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wray L V, Jr, Pettengill F K, Fisher S H. Catabolite repression of the Bacillus subtilis hut operon requires a cis-acting site located downstream of the transcription initiation site. J Bacteriol. 1994;176:1894–1902. doi: 10.1128/jb.176.7.1894-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuber P, Losick R. Use of a lacZ fusion to study the role of the spoO genes of Bacillus subtilis in developmental regulation. Cell. 1993;35:275–283. doi: 10.1016/0092-8674(83)90230-1. [DOI] [PubMed] [Google Scholar]