Abstract
The tpl gene of Citrobacter freundii encodes an enzyme that catalyzes the conversion of l-tyrosine to phenol, pyruvate, and ammonia. This gene is known to be positively regulated by TyrR. The amplitude of regulation attributable to this transcription factor is at least 20-fold. Three TyrR binding sites, designated boxes A, B, and C, centered at coordinates −272.5, −158.5, and −49.5, respectively, were identified in the upstream region of the tpl promoter. The results of mutational experiments suggest that TyrR binds in cooperative fashion to these sites. The nonavailability of any TyrR site impairs transcription. Full TyrR-mediated activation of tpl required integration host factor (IHF) and the cAMP receptor protein (CRP). By DNase I footprinting, it was shown that the IHF binding site is centered at coordinate −85 and that there are CRP binding sites centered at coordinates −220 and −250. Mutational alteration of the IHF binding site reduced the efficiency of the tpl promoter by at least eightfold. The proposed roles of CRP and IHF are to introduce bends into tpl promoter DNA between boxes A and B or B and C. Multimeric TyrR dimers were demonstrated by a chemical cross-linking method. The formation of hexameric TyrR increased when tpl DNA was present. The participation of both IHF and CRP in the activation of the tpl promoter suggests that molecular mechanisms quite different from those that affect other TyrR-activated promoters apply to this system. A model wherein TyrR, IHF, and CRP collaborate to regulate the expression of the tpl promoter is presented.
Tyrosine phenol lyase (TPL; EC 4.1.99.2), formerly known as β-tyrosinase, catalyzes a reversible pyridoxal phosphate-dependent α, β-elimination reaction that degrades l-tyrosine to phenol, pyruvate, and ammonia (22, 23, 25). In the reverse direction, TPL can catalyze the synthesis of l-tyrosine or 3,4-dihydroxyphenylalanine from pyruvate, ammonia, and phenol or catechol. Detailed studies, including the cloning and characterization of the structural gene for TPL, have been carried out for Citrobacter freundii (3, 24), Erwinia herbicola (14, 54), and Escherichia intermedia (26).
In E. herbicola, TPL is induced by l-tyrosine and repressed when cells are grown in glucose (13). The regulatory mechanism(s) that controls the expression of the TPL gene (tpl) is not well understood. A previous study on the regulation of transcription from the tpl promoter of C. freundii demonstrated a role for the TyrR protein in regulating the tpl system (50).
The TyrR protein of Escherichia coli regulates the expression of a number of genes involved in the biosynthesis and transport of aromatic amino acids known as the TyrR regulon (for a review, see reference 39). Seven of the genes of the regulon are repressed by the TyrR protein (39), one gene (mtr) is activated (18, 46), and another (tyrP) is regulated either positively or negatively, depending on whether phenylalanine or tyrosine is present (21, 58). In general, tyrosine serves as the effector for repression, while phenylalanine is the cofactor for activation (39). However, in the TyrR-mediated activation of the mtr promoter, either tyrosine or phenylalanine can function as an inducer (18, 46). The detailed mechanisms of TyrR-mediated repression and activation are unknown.
To investigate possible mechanisms for the regulation of TPL, we employed a variety of genetic and biochemical procedures to analyze transcription from the tpl promoter of C. freundii. The present work enlarges our understanding of the TyrR-regulated tpl system. Two proteins, integration host factor (IHF) and cyclic AMP (cAMP) receptor protein (CRP) were shown to participate in the TyrR-mediated activation of the tpl promoter. The most likely role of these two proteins is to bend DNA in the region upstream of the tpl promoter, thereby enhancing the oligomerization of TyrR dimers. We propose that the functional role of the cis-acting sites within the tpl promoter is not merely to tether TyrR at a high local concentration near the tpl promoter, thereby increasing its ability to contact ς70 RNA polymerase, but also to facilitate the formation of a complex containing at least two TyrR dimers that is required for transcriptional activation. The requirement for additional protein factors in the TyrR-mediated activation of the tpl promoter sets this system apart from other TyrR-activated systems.
MATERIALS AND METHODS
Bacterial strains, plasmids, bacteriophages, and oligonucleotides.
The biological materials used in this study are described in Table 1, along with the chemically synthesized oligonucleotides used in site-directed mutagenesis.
TABLE 1.
E. coli strains, phages, plasmids, and oligonucleotides
| Strain, phage, plasmid, or oligonucleotide | Relevant genotype, description, or sequence | Reference or source |
|---|---|---|
| Strains | ||
| SP1312 | F−zah-735::Tn10 Δ(argF-lac)U169 | 18 |
| SP1313 | F−zah-735::Tn10 Δ(argF-lac)U169 ΔtyrR | 18 |
| SP1626 | As SP1312 but with Δcrp-39 rpsL136 | This work |
| SP1627 | As SP1313 but with Δcrp-39 rpsL136 | This work |
| SP1628 | As SP1312 but with himD::Cm | This work |
| SP1629 | As SP1313 but with himD::Cm | This work |
| SP1630 | As SP1626 but with himD::Cm | This work |
| SP1631 | As SP1627 but with himD::Cm | This work |
| NK5031 | Δ(lacIZY)MS265 gyrA supF | 34 |
| CA8439 | λ−relA1 rpsL136 Δcrp-39 spoT1 ΔcyaA854 thi-1 | 44 |
| CA8445 | Δcrp-45 Δcya-854 strA (=rpsL) thi | 44 |
| CSH26(λRZ11) | ara Δ(lac-pro) thi (λplac5 cI857 Sam7) | 63 |
| BW12848 | lac-169 hip(himD)::cat pho-510 thi | B. Wanner |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| BL21(DE3) | F−ompT (lon) hsdSB (rB− mB−) with DE3, a λ prophage carrying the T7 RNA polymerase gene | 52 |
| Phages | ||
| λHQS10 | tpl-lacZ reporter; Ptpl-lacZ+ | 50 |
| λHQS10mut1 | tpl-lacZ reporter with G→A mutation in box A | 50 |
| λHQS10mut2 | tpl-lacZ reporter with G→A mutation in box B | 50 |
| λHQS10mut3 | tpl-lacZ reporter with G→A and C→T mutations in box C | This work |
| λHQS10mut2,3 | tpl-lacZ reporter with G→A mutation in box A and with G→A and C→T mutations in box C | This work |
| λHQS10Δ64 | tpl-lacZ reporter with 64-nucleotide deletion | 50 |
| λHQS10Δ130 | tpl-lacZ reporter with 130-nucleotide deletion | 50 |
| λHQS10Δ64C | tpl-lacZ reporter with 64-nucleotide deletion; box C replaced by box A | This work |
| λHQS10Δ179C | tpl-lacZ reporter with 179-nucleotide deletion; box C replaced by box A | This work |
| λHQS10IHFmut | tpl-lacZ reporter; T→C and A→C mutations in IHF site | This work |
| Plasmids | ||
| pHA5 | Derived from pBR322; insert of 3.6-kb BamHI fragment containing crp gene and promoter | R. Ebright |
| pXZCRP | Derived from pHA5 by inserting an EcoRI fragment of 500 bp containing the f1 origin | R. Ebright |
| pJC100 | pET3a-tyrR+ | 51 |
| pJC136 | pACYC184-tyrR+ | This work |
| pET3a | T7 vector | 43 |
| pHNβα | Derivative of pHX3-8 by conditional digestion of HindIII which yield incomplete digests | 28 |
| pHNβ2α | Derivative of pHNβα, containing two copies of himD gene | 28 |
| pUC19 | Cloning vector | 63 |
| pUC19tpl | Wild-type tpl promoter in pUC19 | 50 |
| pUC19tplmut1 | G→A change in box A of the tpl promoter in pUC19 | 50 |
| pUC19tplΔ64 | 64-nucleotide deletion derivative of pUC19tpl | This work |
| pUC19tplΔ64C | 64-nucleotide deletion; box C is replaced by box A of tpl promoter in pUC19tpl | This work |
| pUC19tplΔ179 | 179-nucleotide deletion derivative of pUC19-tpl | 50 |
| pUC19mut3 | G→A and C→T changes in box C of tpl promoter | This work |
| pUC19mut1,3 | G→A in box A; G→A and C→T changes in box C of tpl promoter | This work |
| pUC19mut2,3 | G→A in box B; G→A and C→T changes in box C of tpl promoter | This work |
| pUC19tplIHFmut | T→C and A→C mutations in IHF site | This work |
| pMLB1034 | Promoterless lacZ | 6 |
| pMLB1034tplmut3 | G→A and C→T changes in box C of tpl promoter in pMLB1034 | This work |
| pMLB1034tplmut2,3 | G→A in box B; G→A and C→T changes in box C of tpl promoter in pMLB1034 | This work |
| pMLB1034tplIHFmut | T→C and A→C mutations in IHF site | This work |
| pMLB1034tplΔ64C | 64-nucleotide deletion; box C is replaced by box A of tpl promoter in pMLB1034 | This work |
| pMLB1034tplΔ179C | 179-nucleotide deletion; box C is replaced by box A of tpl promoter in pMLB1034 | This work |
| Oligonucleotides | ||
| 351K | GCTGTAAATATAGGTGATGCAAATTACACGTC | |
| 352K | GACGTGTAATTTGCATCACCTATATTTACAGC | |
| 488K | CGGGTAAAAAAATGACGTGTGTAAAGCATCATTTACATTTACAGC | |
| 489K | GCTGTAAATGTAAATGATGCTTTACACACGTCATTTTTTTACCCG | |
| IHFc | CACGCAAAAAAAAACTTGTCGACTATGAACGGG | |
| IHFd | CCCGTTCATAGTCGACAAGTTTTTTTTTGCGTG |
Media.
The liquid minimal medium was salt mix E of Vogel and Bonner (56) containing vitamin B1 (1 mg/liter), biotin (0.1 mg/liter), and either glucose (0.2%) or glycerol (0.2%). Lurig broth (LB) medium (30) contained Bacto Yeast Extract (5 g/liter), Bacto Tryptone (10 g/liter), sodium chloride (5 g/liter), and glucose (1 g/liter). Solid medium was Bacto Nutrient Agar (Difco) (31 g/liter). The following compounds were included when appropriate: l-tyrosine (50 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal), 40 μg/ml; ampicillin, 50 μg/ml; chloramphenicol, 25 μg/ml; streptomycin, 30 μg/ml; and kanamycin, 25 μg/ml.
DNA preparation.
Plasmid DNA was isolated with the Wizard plus miniprep DNA purification system (Promega). Cultures for plasmid preparation were grown to saturation in L broth supplemented with appropriate antibiotics. The preparation of competent cells and their transformation were carried out by the procedure of Mandel and Higa (32). When thermosensitive lysogens were being transformed, the heat step was omitted.
Chemicals and reagents.
Restriction endonucleases, T4 DNA ligase, and DNA polymerase I large (Klenow) fragment were purchased from New England Biolabs. [α-35S]dATP and [α-32P]dATP were purchased from Amersham. Special-purpose oligonucleotides for use in PCR and site-directed mutagenesis were synthesized in the Laboratory for Macromolecular Structure, Purdue University. The protein markers and reagents for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting (immunoblotting) were purchased from Bio-Rad. Anti-TyrR antibodies were prepared as previously described (9). Dimethyl pimelimidate (DMP) was purchased from Pierce. o-Nitrophenyl-β-d-galactopyranoside (ONPG) was purchased from Sigma. All other chemicals were of the highest quality that was commercially available.
Strain construction.
The construction of tyrR+/tyrR isogenic strains and λ lysogens containing a lacZ reporter system specific for the tpl promoter was described in reference 50. High-titer lysates of Ptpl-lacZ+ λ phages were used to lysogenize the Lac− strains SP1312 (tyrR+) and SP1313 (tyrR) to generate SP1312 Ptpl-lacZ+ and SP1313 Ptpl-lacZ+. Strains SP1626 (tyrR+) and SP1627 (tyrR) are Δcrp derivatives of the aforementioned lysogens that were constructed by standard P1 transduction methods with CA8439 as the donor (50). These strains were λ resistant, streptomycin resistant, and temperature sensitive. Strains SP1628 (tyrR+) and SP1629 (tyrR) are IHF-negative derivatives of SP1312 Ptpl-lacZ+ and SP1313 Ptpl-lacZ+ that were constructed with P1 grown on BW12848. This P1 lysate was also used to construct ΔhimD mutants of SP1626 (Ptpl-lacZ+) and SP1627 (Ptpl-lacZ+). tpl promoter variants containing mutations in either the IHF site, box C, or boxes B and C (Fig. 1) were generated in derivatives of pUC19 as described below. Segments of tpl promoter DNA were inserted as EcoRI-BamHI fragments into pMLB1034. The resulting pMLB1034 derivatives were transformed into CSH26(λRZ11). Lac+ λ recombinants were isolated as described in reference 50, with NK5031 as the plating indicator.
FIG. 1.
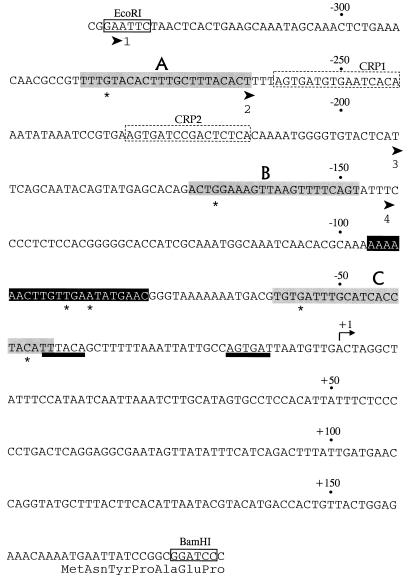
The tpl promoter and associated regulatory elements. Only the nucleotide sequence of the messenger-equivalent strand is shown. The coordinate system has been changed from the system in the original publication (3) by assigning +1 to the start point of transcription. DNA fragments used in the functional analysis of the tpl promoter were synthesized by PCR, with pRVT1 (3) as the template. Cleavage sites for restriction endonucleases EcoRI and BamHI were installed at the indicated locations (coordinates −329 and +180) in order to facilitate the construction of single-copy reporter systems. The EcoRI-BamHI fragment was cloned into pUC19, which contains a PstI site downstream of the BamHI site. The fragment from pUC19-tpl generated by EcoRI-PstI digestion was used for DNase I footprinting experiments. The 5′ end points of three truncated derivatives of the tpl promoter are shown as broad arrowheads labeled 2, 3, and 4 at coordinates −262, −193, and −144. The −10 and −35 recognition elements are underlined with heavy black lines. The target sites for cAMP-CRP (CRP1 and CRP2) are outlined with broken lines. TyrR boxes, named A, B, and C, are shaded. Residues marked by asterisks (−280, −166, −55 and −40) are the locations of TyrR operator mutations or IHF mutations (see text). The IHF binding site is presented in white letters on black.
Enzyme assays.
β-Galactosidase was assayed by the method of Miller (35). Cells were grown to early mid-log phase at 30°C in liquid minimal medium containing l-tyrosine (50 μg/ml). Assays, carried out in triplicate, had standard errors of <10%. The enzyme assay values are reported in Miller units.
Proteins used in footprinting studies.
TyrR protein was purified as previously described (10). CRP was purified by a modification of a method developed by R. Ebright, Rutgers University, using cAMP-Sepharose affinity chromatography. An affinity column was prepared by coupling 8-(6-aminohexyl)amino-adenosine 3′,5′-cyclic monophosphate (AHAcAMP; Sigma) to CNBr-activated Sepharose 4B gel (Pharmacia). E. coli CA8445/pXZCRP, which carries a multicopy plasmid encoding the CRP protein, was grown in L-broth medium at 37°C for 15 h and harvested. Crude extracts were prepared using a French pressure cell (Aminco) operated at 1,000 lbs/in2 and then loaded on a 6-ml cAMP affinity column. The column was washed with 2 bed volumes of wash buffer (20 mM sodium phosphate [pH 7.0], 2 mM EDTA [pH 8.0], 5 mM β-mercaptoethanol, 300 mM NaCl, 6% glycerol) and then with 2 bed volumes of wash buffer plus AMP (5 mM 5′ AMP and 5 mM 3′ AMP). After the washes, CRP was eluted with buffer containing 500 mM NaCl and 5 mM cAMP. About 90% pure CRP was obtained. A further purification step was carried out by fast protein liquid chromatography with a Mono S column (Pharmacia). The purity of CRP, by SDS-PAGE, was at least 99%. The IHF protein was a gift from Steven D. Goodman, University of Southern California. All protein concentrations were determined by Bio-Rad protein assay reagent with bovine serum albumin as a standard.
DNase I footprinting.
A DNA fragment of 527 bp carrying the tpl promoter was released from pUC19-tpl by digestion with EcoRI plus PstI, isolated by electrophoresis on 1% agarose, followed by purification with a QIAEX II gel extraction kit (Qiagen). Elution was carried out with TE buffer (20 mM Tris [pH 8.0], 0.1 mM EDTA). This fragment (0.5 μg) was selectively labeled at the EcoRI site by treatment with DNA polymerase I (Klenow fragment) in the presence of [α-32P]dATP by the method of Brenowitz et al. (7). The unincorporated radiolabel was removed with a G-25 spin column (Boehringer Mannheim), and residual protein was removed with a QIA spin column (Qiagen). The optimal concentration of DNase I in footprinting analyses was established to be 1.5 to 2.0 μg/ml. For CRP footprinting analyses, radiolabeled DNA (1 nmol/tube) was incubated with CRP (10 to 100 nM) at room temperature in assay buffer containing 10 mM Tris-Cl (pH 8.0), 5 mM MgCl2, 1 mM CaCl2, 2 mM dithiothreitol, 100 mM KCl, 50 μg of bovine serum albumin per ml, 2 μg of calf thymus DNA per ml, and 100 μM cAMP. The total volume of each binding reaction mixture was 200 μl. After 30 min, each tube was treated with 5 μl of DNase I (1.5 μg/ml) for 2 min. Each reaction was stopped by the addition of 40 μl of 50 mM EDTA (pH 8.0), followed by a single treatment with 200 μl of phenol-chloroform-isoamyl alcohol (24:25:1). The DNA was precipitated with ethanol containing 5 μg of tRNA per ml and 0.1 volume of 3 M sodium acetate. The DNA was resuspended in 5 μl of loading dye, heated for 5 min at 90°C, and loaded onto an 8% acrylamide–6 M urea gel. The products of the A+G cleavage reactions (33) were coelectrophoresed with sample to identify protected nucleotides. After electrophoresis, the gel was exposed overnight at −70°C to Kodak XAR-5 film. For the TyrR and IHF footprinting reactions, the procedure was the same as described as above. Unless otherwise stated, the assay buffer for TyrR also contained 0.2 mM l-tyrosine and 0.2 mM ATP. DNA fragments containing G-to-A changes in box A were prepared by EcoRI-PstI digestion of pUC19-tplmut1. DNA fragments from the deletion mutant Δ64 or Δ179 were generated by EcoRI-PstI cleavage of pUC19-tplΔ64 or pUC19-tplΔ179, respectively.
Mutagenesis of TyrR and IHF binding sites.
The introduction of the G-to-A changes in box A or box B and the deletion of segments of the tpl promoter is described in reference 50. Mutations in box C (G-to-A and C-to-T) were introduced by using a Quick-change site-directed mutagenesis kit (Stratagene) with the oligonucleotide pair 351K and 352K (Table 1). The template DNA was either pUC19-tpl or pUC19-tplmut2. The consensus sequence of the TyrR target is TGTAAAN6TTTACA (N is any nucleotide). Box C DNA(TGATTTGCATCACCTACA) was replaced by the box A sequence (TGTACATTTGCTTTACA), except for the six nonconserved central nucleotides. These mutational changes were introduced into either pUC19-tplΔ64 or pUC19-tplΔ179 using the oligonucleotide pair 488K and 489K. Mutations in the IHF site (A6CTTGTTGAATATGAAC→A6CTTGTCGACTATGAAC) were introduced in similar fashion with the oligonucleotide pair IHFc and IHFd (Table 1). The template DNA was pUC19 tpl. Each mutational change was verified by DNA sequencing.
Chemical cross-linking.
Reactions were allowed to proceed at room temperature (25°C) in 0.2 M triethanolamine buffer at pH 8.0 containing 1 mg of TyrR protein and 2 mg of DMP per ml. Samples were removed at different times. Each reaction was stopped by the addition of an equal volume of 2× loading buffer (10) and then frozen at −20°C. Circular DNA from pUC19-tpl was prepared with a mega kit from Qiagen. In the presence of DNA, cross-linking was carried out with TyrR and CRP in a buffer containing 200 μM cAMP, 200 μM l-tyrosine, 5 mM MgCl2, 1 mM CaCl2, and 200 μM ATP. The molar ratio of DNA to TyrR to CRP was 1:3:1. After incubation for 20 min, 2 mg of DMP per ml was added. Successive steps were performed as described above. Cross-linking mixtures were analyzed either by 0.7% SDS-PAGE or Western blotting. The primary polyclonal anti-TyrR antibody was a 1:3,000 dilution of high-titer antiserum (9). Peroxidase-conjugated goat anti-rabbit immunoglobulin G (Bio-Rad) was employed as the second antibody (1:3,000 dilution). Staining was carried out with H2O2 and 4-chloronaphthol by standard procedures (17).
RESULTS
Regulation of the tpl promoter by TyrR and cAMP-CRP.
SP1312 (Ptpl-lacZ+) and SP1313 (Ptpl-lacZ+) are isogenic tyrR+/tyrR strains that carry single-copy lacZ reporter genes driven by the tpl promoter of C. freundii (50). When these strains were grown in the presence of l-tyrosine, the β-galactosidase levels in the tyrR+ strain were at least 20-fold higher than in the Δ(tyrR)strain (Table 2, lines 1 and 2). When SP1313 (Ptpl-lacZ+) was transformed with a tyrR+ plasmid (pJC100 [51]) that led to haploid level expression of a functional TyrR protein, the reporter enzyme levels were restored to control values. These results confirm that the tpl promoter, like the mtr and tyrP promoters, is subject to positive control by the TyrR protein (Table 2, lines 1 to 3).
TABLE 2.
Regulation of the tpl promoter by TyrR, cAMP-CRP, and IHF
| Strain | Relevant genotype | β-Galactosidase activitya
|
|
|---|---|---|---|
| Glucose | Glycerol | ||
| SP1312 (Ptpl-lacZ+) | tyrR+ | 783 | 2,540 |
| SP1313 (Ptpl-lacZ+) | Δ(tyrR) | 35.5 | 68 |
| SP1313 (Ptpl-lacZ+)/pJC100 | Δ(tyrR) tyrR+ | 1,530 | 2,180 |
| SP1626 (Ptpl-lacZ+) | tyrR+ Δ(crp) | 828 | 812 |
| SP1627 (Ptpl-lacZ+) | Δ(tyrR) Δ(crp) | 61 | 73 |
| SP1626 (Ptpl-lacZ+)/pXZCRP | tyrR+ Δ(crp) crp+ | 446 | 780 |
| SP1627 (Ptpl-lacZ+)/pXZCRP | Δ(tyrR) Δ(crp) crp+ | 53 | 158 |
| SP1628 (Ptpl-lacZ+) | tyrR+ Δ(himD) | 19.5 | ND |
| SP1629 (Ptpl-lacZ+) | Δ(tyrR) Δ(himD) | 16.3 | ND |
| SP1312 (Ptpl-lacZ+ IHF-mut) | tyrR+; altered IHF site | 89.2 | ND |
| SP1313 (Ptpl-lacZ+ IHF-mut) | Δ(tyrR); altered IHF site | 39.0 | ND |
| SP1630 (Ptpl-lacZ+) | tyrR+ Δ(crp) Δ(himD) | ND | 5.6 |
| SP1631 (Ptpl-lacZ+) | Δ(tyrR) Δ(crp) Δ(himD) | ND | 5.3 |
| SP1628/pHNβ2α | tyrR+ Δ(himD) himD+ | 708.5 | ND |
β-Galactosidase activity is expressed in Miller units. Cultures were grown in liquid minimal medium with l-tyrosine and with either glucose or glycerol as a carbon source. The values shown are the averages of triplicate assays. The variation was never more than ±10%.
Previous studies in E. herbicola showed that the formation of tpl mRNA was drastically curtailed when cells were grown in the presence of glucose (53). It is also known that a null mutation in cya causes about a 50-fold drop in tpl promoter activity (50). Inspection of the tpl promoter of C. freundii suggested that one or more potential CRP binding sites was present. To extend our understanding of catabolite repression via the CRP system as it applies to the tpl promoter of C. freundii, reporter enzyme levels were measured on cells cultivated in glycerol-containing liquid minimal medium. With glycerol as a carbon source, there is little or no catabolite repression and the intracellular levels of cAMP are elevated. In the presence of l-tyrosine, the tpl promoter was slightly more active (twofold) when Δ(tyrR) cells were grown in glycerol-based medium than in glucose-containing medium (Table 2, line 2). Also, significantly more TyrR-specific activation, approximately 37-fold, was observed in glycerol-grown tyrR+ cells compared to a 22-fold effect of TyrR when cells were grown on glucose (Table 2, lines 1 and 2). This supports previous studies (50, 53) which suggested that the cAMP-CRP system can modulate the tpl promoter.
The crp gene was inactivated in the tyrR+ and Δ(tyrR) strains used in the previous experiment. The resulting Δ(crp) derivatives [SP1626 (Ptpl-lacZ+) and SP1627 (Ptpl-lacZ+)] were grown in liquid minimal medium containing either glycerol or glucose and l-tyrosine. In the Δ(crp) background, the differential activation of the tpl promoter that was seen when glycerol-grown cells were compared to glucose-grown cells was no longer evident (Table 2, lines 4 and 5). Introduction of a Crp+ plasmid (pXZCRP) into the Δ(crp) strains partially restored the glycerol-specific induction of the tpl promoter (Table 2, lines 6 and 7). In the absence of the TyrR protein, cAMP-CRP had no effect on reporter enzyme levels (Table 2, lines 2 and 5). These data suggest that the role of cAMP-CRP is to potentiate the TyrR protein-mediated stimulation of transcription from the tpl promoter and that in tyrR+ cells, the contribution of each transcription factor to promoter strength is independent and additive.
Chemical identification of TyrR and CRP binding sites in the tpl promoter.
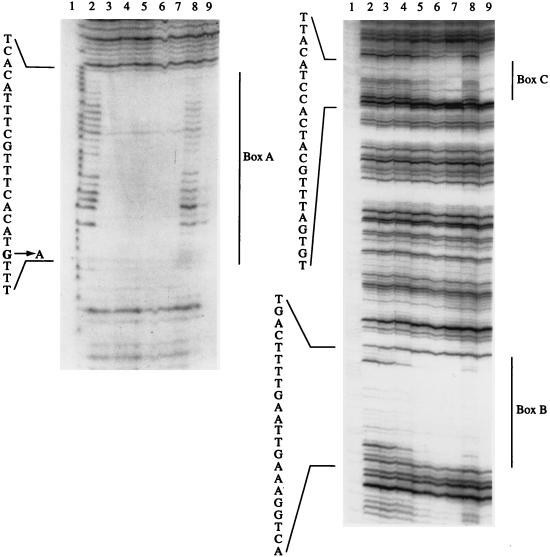
DNase I footprinting of tpl promoter DNA was carried out to define with precision the locations of the TyrR binding sites. A fragment of DNA identical to the one that had been used to construct the tpl reporter systems was isolated from pUC19-tpl and 32P labeled at one end as described in Materials and Methods. In DNase I footprinting studies, three TyrR binding sites, designated A, B, and C were identified (Fig. 2). Boxes A and B, centered at coordinates −272.5 and −158.5 respectively, are very similar in sequence to the consensus TyrR operator sequence (TGTAAAN6TTTACA) (39). Box C, centered at coordinate −49.5, bore little resemblance to the consensus TyrR targets other than the two invariant G and C residues (GN14C). According to the classification scheme of Pittard (39), boxes A and B are likely to be strong TyrR boxes, while box C would appear to be a weak TyrR box. The approximate affinity of TyrR for each operator target was estimated. Box A was fully protected by 1 to 5 nM TyrR (Fig. 2, lane 3 of left gel). Under the same conditions, 20 nM (lane 5 of right gel) or 80 nM (lane 7 of right gel) TyrR were required to fully protect box B or C. The apparent affinity of the TyrR protein for each target site decreased in relation to the proximity of the site to the transcription start point of the tpl promoter.
FIG. 2.
DNase I footprints of TyrR bound to the promoter regions of tpl or tpl-mut1. The DNA fragments were generated by EcoRI-PstI digestion of pUC19-tpl (lanes 2 to 7, both panels), or pUC19-tplmut1 (lanes 8 and 9, both panels). The latter construct contains a G-to-A change in box A. Both fragments were labeled with 32P at the 5′ ends as described in Materials and Methods. Treatment with DNase I was carried out in the presence (lanes 3 to 9, both gels) or absence (lanes 2, both gels) of TyrR. All reaction mixtures contained 0.2 mM l-tyrosine, 0.2 mM ATP, and 0.7 nM DNA. The concentrations of TyrR used were as follows: lanes 3, 5 nM; lanes 4, 10 nM; lanes 5 and 8, 20 nM; lanes 6, 40 nM; lanes 7 and 9, 80 nM. Lanes 1 contain the A+G sequence of tpl DNA. The regions protected by TyrR are indicated. Box A is shown in the left gel, and boxes B and C are shown in the right gel. Each of the DNA sequences shown reads from the 5′ end (bottom) toward the 3′ end (top) as shown in Fig. 1.
To explore whether TyrR binds cooperatively to the three sites, the effect of a disabling G-to-A change in box A on the binding ability of TyrR was examined. As expected, this mutation decreased the affinity of TyrR for box A by at least 20-fold (Fig. 2, lane 8 of left gel). A slight effect of this box A mutation on the binding of TyrR to boxes B and C (approximately twofold reduction) was detected (Fig. 2, lane 8 of right gel). These results suggest that the binding of TyrR to the operator targets upstream of tpl is slightly cooperative, in the sense that the binding of protein to box B would be favored when box A was occupied. However, these results must be interpreted with caution, given the fact that this experiment used linear DNA as the target molecule, whereas cellular DNA is negatively supercoiled.
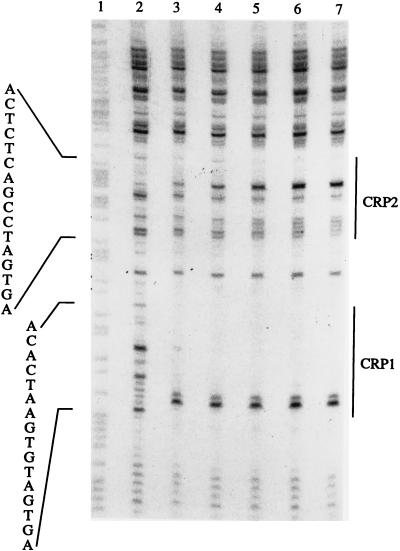
Based on a computer search, which suggested two putative CRP boxes upstream of the tpl promoter, as well as the in vivo results, CRP is predicted to bind to tpl promoter DNA. This hypothesis was investigated by DNase I footprinting. A 500-bp EcoRI-PstI fragment, containing the tpl promoter region, was labeled at one end with [α-32P]dATP (Materials and Methods). The DNase I footprinting results identified two regions upstream of the tpl promoter that were protected by CRP in the presence of cAMP (Fig. 3 and 4). The target designated CRP1 (Fig. 1), centered at coordinate −250 of the tpl promoter, was well protected by cAMP-CRP. Complete protection was observed at a CRP concentration of approximately 10 nM. A second target, CRP2, situated 30 bp downstream of the CRP1 site, was incompletely protected by CRP (Fig. 1 and 3). In the CRP2 region, the cleavage of certain phosphodiester bonds by DNase I was enhanced as the concentration of cAMP-CRP was increased. The reason for this is not clear. The sequences of the two CRP binding sites are quite similar to each other and to the other known CRP consensus sequences (31).
FIG. 3.
DNase I footprints of CRP bound to the tpl promoter region. The DNA fragment used was generated by EcoRI-PstI digestion of pUC19-tpl and labeled with 32P at the 5′ end. The DNA was treated with DNase I in the presence (lanes 3 to 7) or absence (lane 2) of CRP and cAMP. The concentrations of DNA and cAMP were 1 nM and 100 μM, respectively. The concentrations of CRP were as follows: lane 3, 10 nM; lane 4, 25 nM; lane 5, 50 nM; lane 6, 75 nM; lane 7, 100 nM. Lane 1 contains the A+G sequence of the DNA. Segments protected by CRP are indicated to the right. The sequences read from the 5′ end (bottom) toward the 3′ end (top) as shown in Fig. 1.
FIG. 4.
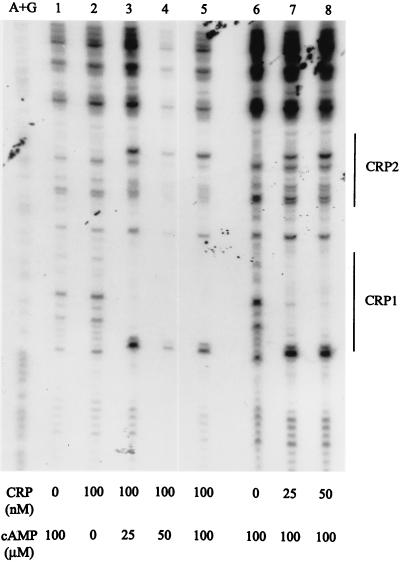
Requirement for cAMP in DNase I footprinting of tpl promoter DNA by CRP. The DNA fragment used in lanes 1 to 5 was an EcoRI-PstI fragment from pUC19-tpl. The fragment used in lanes 6 to 8 was an EcoRI-PstI fragment from pUC19-tplmut1 which contains a critical G-to-A change in box A. Each DNA fragment was labeled with 32P at the 5′ end. The concentrations of CRP and cAMP are shown at the bottom. The segments protected by CRP are indicated to the right. A+G standards are in the leftmost lane. TyrR (50 nM), l-tyrosine (0.2 mM), and ATP (0.2 mM) were present in each tube.
Because CRP site 1 and TyrR box A are separated by only 3 nucleotide pairs, it seemed possible that TyrR and cAMP-CRP, both bound at adjacent sites in the tpl promoter, might interact in a cooperative fashion. When a DNA fragment containing a G-to-A change in box A that drastically reduced the affinity for TyrR was used as a target in DNase I footprinting studies in the presence of TyrR, DNase I protection by cAMP-CRP at the CRP1 and CRP2 sites was unaffected (Fig. 4, compare lanes 6 to 8 with lanes 2 to 5). This suggests that cAMP-CRP and TyrR bind independently to their respective target sites within the tpl promoter which is consistent with the reporter enzyme experiments (Table 2).
To confirm that cAMP plays a role in the regulation of the tpl promoter, DNase I footprinting was carried out in the absence or presence of cAMP (Fig. 4, lanes 1 to 5). There was no protection by CRP (100 nM) alone (Fig. 4, lane 2). Increasing the concentration of cAMP from 25 to 100 μM (Fig. 4, lanes 3 to 5) afforded complete protection by CRP.
Effects of ATP and tyrosine on DNase I protection.
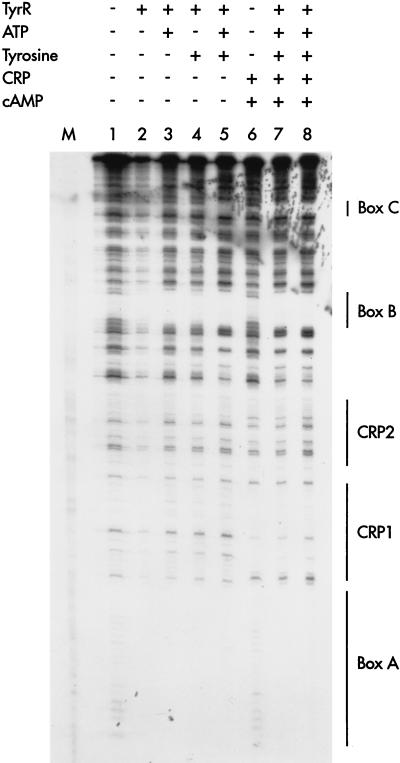
In order to assess the roles of ligands in the binding of TyrR to its respective target sites, a series of DNase I footprinting assays were carried out under a variety of different conditions (Fig. 5). Box A was protected by unliganded TyrR. The pattern of protection was unaltered by the addition of ATP and tyrosine, alone or in combination. Box B was partially protected by unliganded TyrR; full protection was observed when ATP or ATP plus tyrosine were present. Box C was protected only when TyrR, ATP, and tyrosine were present. When cAMP-CRP was present, there were no changes in the protection of the TyrR boxes.
FIG. 5.
Effects of ATP and tyrosine on DNase I footprinting by TyrR and cAMP-CRP. The DNA used in each case was 1 nmol of a 32P-labeled EcoRI-PstI fragment from pUC19-tpl. The label was at the 5′ end (see Materials and Methods). The presence (+) or absence (−) of other compounds added to each tube are shown at the top of the figure. When present, TyrR was included at a final concentration of 100 nM, except in lane 7, when the TyrR concentration was 50 nM. The concentrations of other additives were as follows: ATP, 0.2 mM; l-tyrosine, 0.2 mM; CRP, 50 nM; cAMP, 100 μM. The segments protected by TyrR (boxes A, B and C) and cAMP-CRP are indicated to the right. Lane M, A+G standards.
Regulation of the tpl promoter by IHF.
The center-to-center distance between boxes A and B is almost the same as the distance between boxes B and C (Fig. 1). Two CRP binding sites are located between boxes A and B. The binding of cAMP-CRP to its target site may serve to bend DNA in this region, thereby mediating interactions between TyrR dimers bound to boxes A and B. This raised the possibility that a binding site for an unknown factor lay between boxes B and C and that its role was to facilitate interactions between TyrR dimers bound to boxes A, B, and C. A putative IHF binding site was detected by a computer search of the region upstream of the tpl promoter. An approach similar to that used in the investigation of the role of cAMP-CRP was chosen to explore a possible role for IHF. The himD gene, encoding the β subunit of IHF, was knocked out in a pair of tyrR+ and Δ(tyrR) strains [SP1628 (Ptpl-lacZ+) and SP1629 (Ptpl-lacZ+)]. The ability of TyrR to activate the tpl promoter was severely affected in IHF-negative strains. An IHF-specific reduction of about 35-fold in reporter enzyme levels was detected (Table 2, compare lines 1 and 9). The utilization of the tpl promoter was also studied in strains with knockouts of both the himD and crp genes [SP1630 (Ptpl-lacZ+) and SP1631 (Ptpl-lacZ+)]. In glycerol-grown cultures, severe reductions in enzyme level were observed. Reporter enzyme levels were approximately 500-fold lower than in the tyrR+ reference strain SP1312 (Ptpl-lacZ+) grown in the same medium (Table 2, compare lines 1 and 12). On the other hand, about a 10-fold reduction below that of the Δ(tyrR) host SP1313(Ptpl-lacZ+) was observed in glycerol-grown cells (compare Table 2, lines 2 and 13), indicating that IHF can slightly activate the tpl promoter in the absence of TyrR. This activation was strongly enhanced to about 150-fold when TyrR was present (Table 2, lines 4 and 13). Introduction of pHNβ2α, which encodes two copies of the β subunit of IHF and one copy of the α subunit, fully restored the activation function of the TyrR protein (Table 2, lines 1, 8, and 14). The results indicated that IHF is not only involved in the regulation of tpl but also plays a critical supporting role in enabling TyrR to function as an activator.
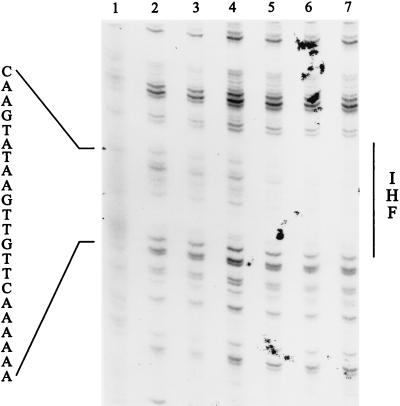
A 22-bp binding site for IHF, centered at coordinate −85 of the tpl promoter, was located by DNase I footprinting (Fig. 6). The importance of this segment in transcription from the tpl promoter was confirmed by site-directed mutagenesis. As described in Materials and Methods, the IHF binding site was changed from A6CTTGTTGAATATGAAC to A6CTTGTCGACTATGAAC. Reporter enzyme measurements from single-copy versions of this altered tpl promoter in tyrR+ cells showed that there had been an 8.8-fold reduction in promoter strength (compare lines 1 and 10 of Table 2). Basal expression from the tpl promoter, measured in Δ(tyrR) cells, was essentially unchanged when the IHF binding site was mutated (compare lines 2 and 11 of Table 2). This set of results strongly supports a role for IHF in the activation of the tpl promoter.
FIG. 6.
DNase I footprint of the IHF binding site within the tpl promoter region. The EcoRI-PstI fragment from pUC19-tpl was labeled with 32P at the 5′ end as described in Materials and Methods. The DNA was treated with DNase I in the presence (lanes 3 to 7) or absence (lane 2) of IHF. The concentration of DNA was 0.7 nM in each reaction mixture. The concentrations of IHF were as follows: lane 3, 9 nM; lane 4, 18 nM; lane 5, 36 nM; lane 6, 72 nM; lane 7, 144 nM. Lane 1 contains the A+G sequence. The region protected by IHF is indicated to the right. The protected sequence reads from the 5′ end (bottom) toward the 3′ end (top) as shown in Fig. 1.
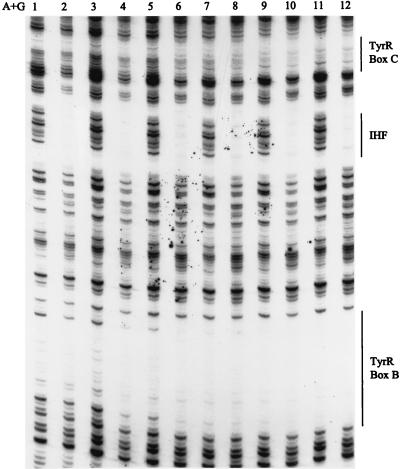
To address the possibility that IHF alters the affinity of the tpl promoter for TyrR, a DNase I footprinting experiment was performed in the presence of various combinations of cAMP-CRP, IHF, and TyrR. Since the protection by TyrR of boxes B and C is the most sensitive indicator of possible cooperativity in the occupancy of these target sites by the three transcription factors, attention was focused on this segment of the tpl promoter. To a first approximation, neither IHF nor cAMP-CRP altered the affinity of TyrR for either box B or box C (Fig. 7). The only IHF-specific effect that was noted was a slight reduction in the cleavability of phosphodiester bonds on either side of the IHF target site (compare lanes 2 and 3, 4 and 5, 6 and 7, etc.). However, this effect was not correlated with TyrR concentration. In agreement with a previous result (Fig. 4), cAMP-CRP had no effect on the binding of TyrR to its target sites.
FIG. 7.
Effects of IHF and cAMP-CRP on DNase I footprinting of boxes B and C. The DNA used was 0.7 nmol of an EcoRI-PstI fragment from pUC19-tpl that had been 32P labeled at the 5′ end (see Materials and Methods). The tubes alone were used (lane 1), or CRP (20 nM) and IHF (145 nM) (lanes 2, 4, 6, 8, 10, and 12) were added to the tubes. The TyrR concentrations were as follows: lanes 2 and 3, 10 nM; lanes 4 and 5, 20 nM; lanes 6 and 7, 40 nM; lanes 8 and 9, 60 nM; lanes 10 and 11, 80 nM; lane 12, 100 nM. Each reaction mixture contained ATP and tyrosine (0.2 mM each). Tubes containing CRP also contained cAMP (100 μM).
Effects of mutations within the TyrR boxes of the tpl promoter.
Previous comparisons (39) of TyrR binding sites have shown that there are only two absolutely conserved nucleotides (GN14C) within this class of operators. Alteration of either residue severely reduced the TyrR-mediated regulation of promoter activity. To examine the roles of boxes A, B, and C in the regulation of the tpl promoter, G-to-A changes were introduced at the first invariant position in each of the TyrR boxes. When box A of the tpl promoter was mutated, the β-galactosidase activity of glucose-grown cells fell to a level characteristic of Δ(tyrR) host strains, i.e., there was a reduction of approximately 13-fold in reporter enzyme level from that in tpl promoter systems where box A was intact (compare line 1 of Table 3 to line 1 of Table 2). The effect of a G-to-A change in box B was not as severe as that in box A. In glucose-grown cells, a sevenfold reduction in reporter enzyme levels was observed (compare line 2 of Table 3 to line 1 of Table 2). Considering only glycerol-grown cells, where the cAMP-CRP effect is maximal, the β-galactosidase activities were reduced about 4.6-fold when either box A or box B was mutated (compare lines 1 and 2 of Table 3 to line 1 of Table 2). These data are consistent with the results of Table 2 (lines 1 and 4), where the contribution of cAMP-CRP to tpl promoter activity was found to be about threefold. There was no detectable TyrR or cAMP-CRP-mediated activation when box C was mutationally inactivated. Even the stimulatory effect of cAMP-CRP was lacking (Table 3, lines 3 and 4). This was unexpected, since box C showed little sequence homology to other TyrR boxes and fits the criteria for a weak box. Evidently cAMP-CRP can stimulate transcription from the tpl promoter when either box A or box B is disabled, but not when box C is incapable of engaging the TyrR protein.
TABLE 3.
Mutational analysis of transcriptional activation at the tpl promoter
| Strain | Relevant genotype | β-Galactosidase activitya
|
|
|---|---|---|---|
| Glucose | Glycerol | ||
| SP1312 (Ptpl-lacZ+)mut1 | tyrR+, G→A in box A | 57.9 | 555 |
| SP1312 (Ptpl-lacZ+)mut2 | tyrR+, G→A in box B | 111.0 | 549 |
| SP1312 (Ptpl-lacZ+)mut3 | tyrR+, G→A and C→T in box C | 47.8 | 85.3 |
| SP1313 (Ptpl-lacZ+)mut3 | Δ(tyrR), G→A and C→T in box C | 22.0 | 22.6 |
| SP1312 (Ptpl-lacZ+)Δ64 | tyrR+, 64-nucleotide deletion | 41.2 | 278 |
| SP1313 (Ptpl-lacZ+)Δ64 | Δ(tyrR), 64-nucleotide deletion | 13.1 | 30.0 |
| SP1312 (Ptpl-lacZ+)Δ130 | tyrR+, 130-nucleotide deletion | 34.1 | 88.9 |
| SP1313 (Ptpl-lacZ+)Δ130 | Δ(tyrR), 130-nucleotide deletion | 17.1 | 55.2 |
| SP1312 (Ptpl-lacZ+)mut2,3 | tyrR+, G→A and C→T in box C, G→A in box B | 42.0 | 72.7 |
| SP1313 (Ptpl-lacZ+)mut2,3 | Δ(tyrR), G→A and C→T in box C, G→A in box B | 28.3 | 38.2 |
| SP1312 (Ptpl-lacZ+)Δ64C | tyrR+, 64-nucleotide deletion, box C is replaced by box A | 93.6 | 170 |
| SP1313 (Ptpl-lacZ+)Δ64C | Δ(tyrR), 64-nucleotide deletion, box C is replaced by box A | 12.1 | 28.7 |
| SP1312 (Ptpl-lacZ+)Δ179C | tyrR+, 179-nucleotide deletion, box C is replaced by box A | 65.9 | 72.9 |
| SP1313 (Ptpl-lacZ+)Δ179C | Δ(tyrR), 179-nucleotide deletion, box C is replaced by box A | 14.3 | 22.0 |
| SP1313 (Ptpl-lacZ+)Δ64C/pJC100 | Δ(tyrR) ptyrR+ Δ64C | 316 | 527 |
| SP1313 (Ptpl-lacZ+)Δ179C/pJC100 | Δ(tyrR) ptyrR+ Δ179C | 410 | ND |
| SP1313 (Ptpl-lacZ+)Δ179C/pJC100/pJC136 | Δ(tyrR) (ptyrR+) Δ179C | 614 | ND |
β-Galactosidase activity is expressed in Miller units. Cultures were grown in glucose or glycerol as indicated. ND, not determined.
In previous work, it was shown that the tpl promoter became nonfunctional when upstream segments of DNA containing boxes A or A and B were deleted (50). The deletions in question shortened the tpl promoter by 64, 130, and 179 nucleotides from the 5′ upstream region (Fig. 1). This result was confirmed and extended in the present study. In no case was there significant β-galactosidase activity. In glycerol-grown cells, the Δ130 mutant with both CRP binding sites deleted showed much lower reporter enzyme levels (threefold drop) than the Δ64 mutant with intact CRP sites (Table 3, lines 5 to 8). This result is consistent with the general notion that cAMP-CRP stimulation functions largely in conjunction with the occupancy by TyrR of box C. If either target site is removed from the tpl promoter, reporter enzyme levels fall to basal values.
The DNase I footprinting result (Fig. 2) showed that the 3′ end of box C either overlapped or was immediately adjacent to the −35 region of the tpl promoter. This suggested that box C plays a critical role in TyrR-mediated transcriptional activation. In order to address the functional importance of box C, we constructed a multiple mutant with a G-to-A change in box B in combination with two inactivating mutations in box C. The promoter activity of the resulting construct was totally abolished in both glucose- and glycerol-based media (Table 3, lines 9 and 10). Recall that it had previously been shown that a single mutational change in box B had only a slight effect. The possibility of cooperativity in the binding of TyrR to boxes A, B, and C was raised in the experiment of Fig. 2. If the role of cooperativity in the binding of TyrR to the tpl promoter were only to provide a local high concentration of activator near the RNA polymerase binding site, then converting box C to a strong box identical to box A (a high-affinity target for the TyrR protein) in promoters lacking box A or boxes A and B would be predicted to generate a TyrR-responsive tpl promoter. Surprisingly, only a fourfold TyrR-mediated stimulation of reporter enzyme was detected in a promoter containing a strong box at location C in combination with a deletion of boxes A and B [SP1312 (Ptpl-lacZ+)Δ179C] (Table 3, lines 13 and 14). A TyrR-mediated stimulation of about sixfold was detected in a promoter containing a strong box C together with a deletion of box A [SP1312 (Ptpl-lacZ+)Δ64C] (Table 3, lines 11 and 12). Introduction of pJC100, which enables cells to make TyrR protein at slightly higher than haploid levels, into SP1313 (Ptpl-lacZ+)Δ64C significantly increased the level of reporter enzyme. A TyrR-specific induction of 26-fold was observed (Table 3, compare lines 12 and 15). A similar amplitude of change was obtained when pJC100 was introduced into SP1313 (Ptpl-lacZ+)Δ179C (Table 3, compare lines 14 and 16). Further elevations in reporter enzyme levels were observed when TyrR levels were boosted via the introduction of a second compatible plasmid (pJC136) encoding TyrR (Table 3, line 17). These data suggest that elevated levels of TyrR facilitate transcriptional activation of the tpl promoter via box C. It was noted earlier that DNase I footprinting of box C also required high levels of TyrR (Fig. 2). We hypothesize that the TyrR operators in the tpl promoter facilitate the formation of a higher-order protein complex necessary for the productive interaction between TyrR and RNA polymerase.
Interaction between TyrR dimers at the tpl promoter.
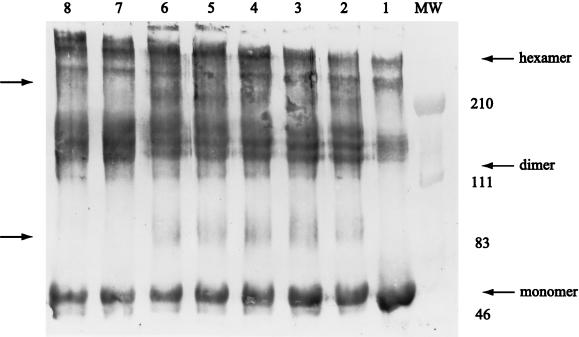
Mutational studies on DNA carrying TyrR boxes suggested that the DNA-mediated association of TyrR dimers plays a role in activating transcription at the tpl promoter. Earlier studies demonstrated that TyrR exists as a dimer in solution (9) and that dimers could interact to give rise to hexamers (59). To further investigate the range of aggregation states available to TyrR, a series of chemical cross-linking studies were conducted. Upon incubation with DMP, the subunits of TyrR protein were readily converted, in a time-dependent manner, to species with increasing multiples of the molecular weight of the monomer (Fig. 8). DMP mediates cross-linking via the ɛ amino groups of lysine residues, provided that these functional groups are separated by no more than 9.2 Å. In the absence of DNA, complexes corresponding to the dimeric, tetrameric, hexameric, and octameric forms of TyrR were detected both on stained SDS-polyacrylamide gels or on Western blots developed with anti-TyrR antibodies (Fig. 8, lanes 7 and 8). When tpl DNA, bearing operator targets for TyrR, was present, the percentage of hexamer was dramatically increased, while the octamer form of TyrR became almost undetectable (data not shown). When cAMP-CRP was included in the cross-linking reaction mixtures with TyrR and DNA, two major complexes likely to contain TyrR-CRP cross-linked species were demonstrated by immunoblotting (Fig. 8). The molecular mass of each complex was estimated to be 83 to 95 kDa (broad) and 200 kDa. The 83- to 95-kDa complexes consist of two species. One is likely to contain one subunit of TyrR and one subunit of CRP; the other is likely to contain one subunit of TyrR and two subunits of CRP. The 200-kDa protein complex is likely to have originated from cross-linking between one TyrR dimer and two CRP dimers. This is reasonable because there are two CRP binding sites near TyrR boxA (Fig. 1).
FIG. 8.
DNA dependence of chemical cross-linking of TyrR demonstrated by immunoblotting. The cross-linking reagent was DMP. The reaction was carried out with 3.6 μM TyrR, 1.2 μM CRP, 100 μM ATP, 50 μM cAMP, and 200 μM l-tyrosine in the presence (lanes 1 to 6) or absence (lanes 7 and 8) of pUC19-tpl DNA. Lane 1 is a control (no DMP). The concentration of DMP was 4 mg/ml. The incubation times for each reaction mixture were 10 min (lane 2), 20 min (lane 3), 40 min (lanes 4 and 7), 80 min (lanes 5 and 8), and 160 min (lane 6). The locations of molecular weight markers (in thousands) are indicated to the right. The positions of TyrR monomer, dimer, and hexamer are indicated with arrows to the right. The positions of presumptive TyrR-CRP complexes are indicated with arrows to the left. For details, see Materials and Methods.
Attempts to verify the proposed composition of these species through the use of polyclonal goat anti-CRP immunoglobulin G (a gift from Sankar Adhya) were unsuccessful, presumably because the epitopes of CRP recognized by this reagent were unavailable by Western blotting. The inclusion of IHF in the cross-linking reactions produced no detectable changes from the patterns shown in Fig. 8 (data not shown). The apparent heterogeneity in molecular mass of the observed TyrR dimers, tetramers, and hexamers probably reflects cross-linking between different lysine residues of TyrR located at widely different positions within the TyrR monomers. These results suggest that a higher-order nucleoprotein complex containing TyrR, such as a hexamer, forms at the tpl promoter. This could be the species that interacts with RNA polymerase to activate tpl expression.
DISCUSSION
The TyrR protein of E. coli is known to regulate the expression of a number of genes involved in the biosynthesis and transport of aromatic amino acids (39). The present results, together with previous data (50), enlarge our understanding of TyrR-regulated systems. TPL, an enzyme of C. freundii that degrades tyrosine to phenol, pyruvate, and ammonia is clearly a member of the TyrR regulon. The regulation of tpl requires l-tyrosine as a cofactor. Like mtr and tyrP (18, 21, 46, 58), tpl is positively regulated by TyrR. In tester strains bearing a deletion of the tyrR gene, there was a reduction of at least 20-fold in the production of β-galactosidase from a single-copy tpl-lacZ reporter system from that of tyrR+ controls. Promoter function was completely restored when a plasmid expressing the tyrR+ gene was introduced into the Δ(tyrR) strain. As revealed by DNase I footprinting experiments, TyrR binds to three operator targets, named A, B, and C, within the tpl promoter region. The binding sites are centered at positions −49.5, −158.5, and −272.5 relative to the transcriptional start point. Mutational alteration of any of the three operators abolished or severely reduced transcription.
Inspection of the sequence of each TyrR binding site (Fig. 1) reveals that boxes A and B are quite symmetrical and closely resemble the TyrR consensus sequence (TGTAAAN6TTTACA). This was not the case for box C. Unliganded TyrR readily bound to the box A region of tpl. The DNase I footprint of Tyr bound to box A was unaffected by l-tyrosine and ATP. Box B was also protected by unliganded TyrR, but binding was enhanced when ATP was present. Under the same conditions, the box C region was protected by TyrR only when ATP and l-tyrosine were present (Fig. 5). Thus, box C conforms to the general criteria for a weak TyrR box first enunciated by Pittard and coworkers (39). The inability of TyrR to bind to box C in the absence of tyrosine and ATP probably reflects a poor fit between the operator recognition site of TyrR and the DNA of box C. TyrR binds to the three targets within the tpl promoter with progressively diminishing affinity, in the order box A > box B > box C. We assume that these different affinities for TyrR are functionally related to the regulatory response that is observed during the expression of tpl. It appears that transcription from the tpl promoter occurs only when TyrR occupies the lower-affinity site. Our data are consistent with the notion that TyrR binds cooperatively to the three boxes. When box A or boxes A and B were deleted, the utilization of the tpl promoter fell to a very low level. The interdependence of TyrR targets observed in vivo was supported by DNase I footprinting. The protection of box C was significantly diminished when box A or boxes A and B were deleted. In the Δ64 version of the tpl promoter (box A deleted), the DNase I protection of box C required >80 nM TyrR compared to a requirement of 80 nM TyrR for wild-type tpl DNA. With the Δ179 version of the tpl promoter (boxes A and B deleted), the protection of box C required >120 nM TyrR compared to a requirement of 80 nM for wild-type tpl DNA (data not shown). This apparently cooperative binding of the TyrR protein to its multiple binding sites is likely to offer a physiological advantage by increasing the overall effectiveness and specificity of occupancy by TyrR of binding sites within the tpl promoter. Similar situations exist for other systems, including the Lrp-regulated ilvIH promoter (57), the simian virus 40 early promoter (5), Drosophila heat shock promoters (62), and yeast promoters that are subject to general amino acid control (4, 20).
In contrast to the situation that prevails in several other TyrR-regulated promoters, two of the operator targets (boxes A and B) of the tpl promoter lie far upstream of the transcriptional start point, while box C is immediately adjacent to the RNA polymerase-binding site (Fig. 1). The three targets are separated from each other by 10 helical turns of B-form DNA. In the mtr and tyrP promoters, there are only two TyrR binding sites within each promoter whose centers are separated by no more than 30 bp. Neither mtr nor tyrP has TyrR target sites further upstream than coordinate −66 (21). The importance to transcriptional activation of the spacing between TyrR boxes has been studied in the tyrP system. Activation of tyrP can be detected if the boxes are separated by one turn of the helix but not if the separation involves three turns of the helix (2). In contrast, we found that even TyrR binding sites situated far upstream from the promoter were essential for the regulation of the tpl promoter. It is uncommon to find such remote regulatory elements in association with ς70 promoters, where activators tend to bind predominantly to targets between coordinates −80 and −30 (16). The ς70 tpl promoter is an exception (50). The regulatory function of boxes A and B appears to be obligatorily linked with box C, located near the −35 recognition element of the tpl promoter. Inactivation of box C essentially eliminates the function of the other two sites, even when boxes A and B are fully intact. Since boxes A and B are far from the tpl promoter region, only looping of DNA would allow interactions between TyrR proteins, thus generating higher-order multimers of TyrR. Our cross-linking study of TyrR in the presence of tpl promoter DNA (Fig. 6) provides supporting evidence for multimerization of the TyrR protein.
Effects of global transcriptional regulatory proteins.
Two DNA-binding proteins, IHF and CRP, were shown to participate in the regulation of tpl expression. Although IHF and CRP are generally considered to be global regulators, the involvement of both factors in the activation of a single promoter has been reported only for the tdc promoter (60). Here we explored a transcriptional regulatory system in which IHF and CRP are both required for full TyrR-mediated activation of tpl. Initially, this was studied with appropriately constructed background strains carrying a tpl reporter system. When a Δ(himD) mutation was introduced into host strains that contained a tpl-lacZ reporter system, the transcriptional activity of the tpl promoter became virtually undetectable, even when TyrR was available. On the other hand, a Δ(crp) mutation lowered the expression of tpl only about threefold. Although both IHF and CRP were shown to interact directly with the tpl promoter region, the IHF appears to be more critical in tpl regulation than CRP. In the absence of TyrR, IHF showed about a 10-fold effect on induction of the tpl promoter. This induction was dramatically increased when TyrR was present, indicating that IHF acts as a coactivator of the tpl system. Given what is known about the interaction of IHF with DNA (1), a plausible role for IHF is to bend tpl DNA in the region between boxes B and C. The bend angle, predicted to be 140° or greater, would alter the shape of DNA from approximately a straight line to something resembling a hairpin (42). IHF is involved in numerous processes in E. coli and some of its bacteriophages and plasmids, including site-specific recombination, DNA replication, and gene expression (12, 15). Most IHF-specific transcriptional regulatory events involve ς54-dependent promoters, which become fully functional only when activator proteins bind to remote upstream sites. In NtrC-responsive ς54 promoters such as nifA, IHF binds to a site midway between an upstream activation site and the promoter, where it mediates the formation of a DNA loop that brings these elements into close proximity (19, 27). In contrast, ς70-dependent promoters are rarely regulated via remote upstream activator binding sites.
How IHF is involved in many regulatory systems is a question that has been pursued for many years. The focus of interest has been on systems where removal of IHF causes qualitative changes. However, many of the reported effects on transcription of mutations in IHF are modest (two- to fivefold). Large effects attributable to IHF have been reported in only three cases: (i) induction by the NifA regulator of the nifHDK operon (19, 36); (ii) induction by the NarL regulator of the gene encoding nitrate reductase (41, 47); (iii) induction by the TdcR regulator of the gene encoding threonine dehydratase (60). We have demonstrated a large effect of IHF on the TyrR-mediated regulation of the tpl promoter. It has been reported that the capacities of NifA, NarL, and TdcR to produce a marked increase in enzyme levels are virtually abolished by mutations in the genes encoding IHF subunits and/or the target sites for IHF that lie upstream of the relevant promoters. Remarkably, all of these cases, including the TyrR-regulated tpl promoter, lead to the formation of ammonia from alternate sources (nitrogen, nitrate, threonine, and tyrosine). It has been suggested (37) that IHF may be essential for a range of cellular responses to nitrogen limitation, especially during simultaneous oxygen deprivation. Our data support a role for IHF as an enhancer of the tpl regulatory system and also provide evidence for a larger role for IHF in gene regulation.
It is not surprising that the cAMP-CRP complex is a transcriptional activator of the tpl promoter, given the fact that CRP is a generally recognized global regulator of gene expression (7a). In many cases, CRP acts by binding to a single site, located slightly upstream of the RNA polymerase binding site. In the absence of CRP, the promoter often displays a low affinity for ς70 RNA polymerase (Eς70), while CRP and Eς70 bind cooperatively to the promoter in the presence of CRP (11). In a few other cases, several adjacent CRP binding sites have been observed, but the nature of their involvement in promoter activation is not well understood (8, 29, 38, 49, 55). A recent study of an artificial promoter having two CRP sites whose positions were systematically varied has clarified the functions of dual CRP targets (5a).
Our data suggest that CRP binds to two adjacent sites located far upstream of the RNA polymerase binding site of the tpl promoter and that this binding leads to full TyrR-mediated activation. In the presence of the TyrR protein, tpl promoter activity was increased by a factor of three by cAMP-CRP (Table 2). Two cAMP-CRP binding sites, situated between TyrR boxes A and B, were identified by DNase I footprinting (Fig. 3). The possibility of direct interaction between cAMP-CRP and the TyrR protein was addressed in chemical cross-linking experiments. Although this approach gave evidence consistent with TyrR-CRP proximity, DNase I footprinting analysis was inconclusive in showing any effects of TyrR on the binding of cAMP-CRP or vice versa.
What might be the role of CRP in the tpl system? Two properties of CRP that were observed in studies of other promoters may be relevant to this question. First, the results of biochemical and genetic experiments have suggested that other proteins are capable of binding to CRP (11). Second, the binding of CRP to its target induces a 90° bend in the DNA (48). Thus, the role of CRP in the tpl regulatory system may be to contribute to the assembly of a TyrR-containing nucleoprotein complex. This could be accomplished either by specific interactions of CRP with TyrR or via cAMP-CRP-mediated bending of the DNA between boxes A and B that might favor interactions between TyrR dimers bound at these sites (1).
Model for activation of transcription from the tpl promoter.
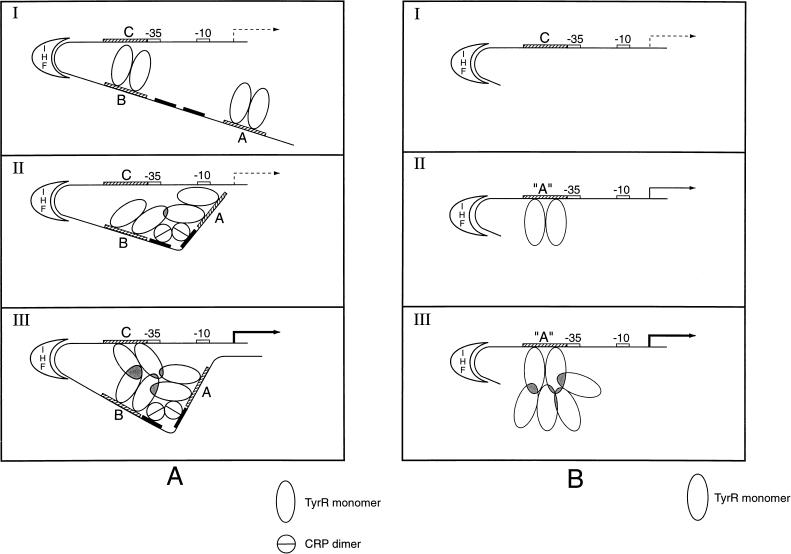
The in vivo and in vitro results presented herein, viewed within the framework of the established properties of the IHF and CRP proteins, suggest a model for the regulation of tpl expression (Fig. 9). This model is based on the hypothesis that the occupancy of the promoter-proximal TyrR binding site (box C) is a prerequisite for transcription. Box C binds TyrR weakly. The cooperative binding of TyrR to the tpl promoter via boxes A and B, with the assistance of IHF and cAMP-CRP, is proposed to facilitate the binding of TyrR to box C. At low tyrosine concentrations, it is proposed that TyrR, bound to box A, can become positioned near box B of the tpl promoter region, provided that there is sufficient cAMP-CRP available to bend tpl DNA between boxes A and B. This would happen readily in glycerol-grown cells but would not occur if the production of cAMP or CRP were blocked by mutation. A TyrR dimer bound at box B is presumed to interact with TyrR at box A, forming a stable complex. When the concentration of tyrosine increases, TyrR acquires the ability to bind to box C; meanwhile, the tetrameric TyrR-box A-box B complex could approach the tpl promoter near box C by virtue of the DNA bending activity of IHF. It is proposed that this gives rise to a very stable TyrR hexamer-DNA structure that can interact with RNA polymerase to initiate transcription. The nonavailability of any of the three proteins or relevant target sites would impair transcription.
FIG. 9.
Model for involvement of IHF and CRP in TyrR-mediated activation of tpl. TyrR boxes A, B, and C are shown as hatched bars. Two CRP binding sites are shown as black bars. The ς70 promoter region is indicated by white bars. See Fig. 1 for the sequences. (A) Activation of the wild-type tpl promoter. (I) Resting state. At low tyrosine concentrations, TyrR dimers occupy strong boxes A and B, while weak box C is unoccupied. The DNA between boxes B and C is bent by IHF. (II) Catabolite repression lifted. cAMP-CRP binds to target sites, introducing a bend between boxes A and B. Interaction between TyrR dimers bound to boxes A and B, induced by DNA bending, stabilizes DNA-TyrR interaction. In states I and II, the transcriptional activity, induced by IHF, is at a very low level (indicated by a broken arrow). The mechanism of activation by IHF is not clear. (III) Tyrosine induction. At increased concentrations of tyrosine, TyrR dimers are able to occupy weak box C. Interactions between TyrR dimers bound to boxes A, B, and C become possible. The interactions between TyrR dimers are indicated by shaded regions. This gives rise to a very stable TyrR-hexamer DNA structure. This higher-order nucleoprotein complex is proposed to fully activate the tpl promoter either by inducing a conformational change within promoter DNA or by interacting directly with ς70 RNA polymerase. Activation of the tpl promoter is indicated by a heavy black arrow. (B) Model for activation of Δ179 mutant tpl promoter. A DNA segment of 179 nucleotides that included boxes A and B was deleted from the region upstream of the tpl promoter region. In a separate operation, weak box C, centered at coordinate −49.5, was replaced by a strong box essentially identical to box A (see Materials and Methods). This construct is designated box A. (I) Promoter inactivity. The Δ179 tpl promoter cannot be utilized when wild-type box C is the only TyrR target, probably because the affinity of TyrR for box C is too low to enable higher-order structures required for promoter activity to form. (II) Weak promoter activity. When box C is replaced by strong box A, TyrR dimers can attach to this region of the promoter. Occasional formation of higher-order structures would allow ς70 RNA polymerase to launch transcription. A low level of promoter activity is observed (indicated by a solid arrow). (III) Promoter activation. When the cellular content of TyrR dimers is elevated, there is an increased chance for the formation of complexes between TyrR dimers bound at box A and free TyrR dimers in solution. The formation of such higher-order complexes between TyrR dimers permits launching of ς70 RNA polymerase. Activation of the tpl promoter is indicated by a heavy black arrow.
It is not clear whether TyrR, IHF, and CRP must physically interact to achieve optimal promoter expression or if simultaneous occupancy of binding sites in DNA is the critical feature. In DNase I footprinting experiments in the presence of all three proteins (TyrR, IHF, and CRP), there were no indications of binding cooperativity (Fig. 7). For none of the other genes regulated by IHF has physical contact between IHF and an upstream regulator or RNA polymerase been demonstrated. Thus, it is unlikely that IHF interacts directly with TyrR and/or RNA polymerase. The presumed role of IHF in tpl expression is to bend tpl promoter DNA to enhance the interaction between TyrR dimers.
Our model is consistent with the work of Wilson et al. (59), who demonstrated that TyrR undergoes ligand-induced hexamerization. By sedimentation equilibrium methods, these workers found that TyrR dimers undergo reversible association to form hexamers in the presence of tyrosine and ATP. In the present study (Fig. 8), we used chemical cross-linking to demonstrate the oligomerization of TyrR in the presence of tpl promoter DNA. Mutational alterations within tpl-specific TyrR targets proved that three sites must be functional in order for full activation of the tpl promoter to proceed.
The available information is also compatible with an alternative model where the formation of a higher-order complex containing three or more dimers of TyrR is the key feature in the transcriptional activation of tpl. Such a model has been proposed by Porter et al. (40) and Wyman et al. (61), who reported that NtrC-P from Salmonella typhimurium must self-assemble into oligomers in order to activate transcription. It has long been known that the central region of TyrR bears significant homology to the central region of NtrC. The central domains of proteins in the NtrC superfamily are thought to interact directly with ς54 RNA polymerase and to mediate protein dimerization. It is reasonable to hypothesize that TyrR and NtrC utilize similar mechanisms of gene regulation. The physiological advantage of oligomerization was suggested (40, 61) to be as follows: DNA target sites, by virtue of the inability to bind dimeric transcription factors, facilitate interactions between regulatory proteins. Activation therefore occurs only at the correct locations on the chromosome. This is important because other activators that are homologs of TyrR or NtrC could activate transcription in response to a variety of physiological signals that are unrelated to the specific regulons to which they belong (40).
Distinctions in mode of action of TyrR at tpl, mtr, and tyrP.
It is likely that the mechanism of TyrR-regulated tpl expression is quite different from the role of TyrR as a positive regulator of mtr and tyrP. In the case of tyrP, there is dual regulation, namely, tyrosine-mediated repression or phenylalanine-mediated activation, at the same promoter. Mutational studies (2) of two tyrP targets (boxes 1 and 2) showed that both TyrR boxes were required for repression but that only the upstream box (box 2) was required for activation. The degree of activation of the tyrP promoter was critically related to the location of box 2. Maximal activation was observed when box 2 was moved 3 or 12 to 14 residues upstream, while no activation was seen at intermediate positions such as +7 and −4 (2). This positional restriction of box 2 in tyrP was attributed to the dual role of box 2 in both repression and activation. We predict that the relative positions of the TyrR targets (boxes A and B) in the tpl promoter will be less critical owing to their remote distance from the promoter. In mtr, mutational experiments were used to demonstrate that the strong box plays an important role in activation by phenylalanine and tyrosine. Mutations in the weak box had no effect on activation by phenylalanine but decreased activation by tyrosine (45). Comparing these results with our data obtained from mutational studies of the tpl promoter, one is forced to conclude that phenylalanine- and tyrosine-mediated TyrR activation involves distinct regulatory mechanisms. Whether a multimeric TyrR complex is involved in tyrosine-mediated regulation of mtr remains to be determined. Further studies will be necessary to establish the precise mechanistic roles of IHF and CRP in tpl expression and how the formation of hexameric TyrR is regulated by tyrosine.
ACKNOWLEDGMENTS
For their generous donations of biological materials and their advice on various procedures used during the course of this work, we thank Hong Qiu Smith, Barry Wanner, Howard Nash, Steven Goodman, Richard Ebright, Wei Niu, and Sankar Adhya.
Financial support for this work was provided in the form of grants from the U.S. Public Health Service (GM 22131) and the U.S. Army Research Office (DAAH 04-95-1-01 38).
REFERENCES
- 1.Allemann R K, Egli M. DNA recognition and bending. Chem Biol. 1997;4:643–650. doi: 10.1016/s1074-5521(97)90218-0. [DOI] [PubMed] [Google Scholar]
- 2.Andrews A E, Dickson B, Lawley B, Cobbett C, Pittard A J. Importance of the position of TyrR boxes for repression and activation of the tyrP and aroF genes in Escherichia coli. J Bacteriol. 1991;173:5079–5085. doi: 10.1128/jb.173.16.5079-5085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antson A A, Demidkina T V, Gollnick P, Dauter Z, VonTersch R L, Long J, Berezhnoy S N, Phillips R S, Harutyunyan E H, Wilson K S. Three-dimensional structure of tyrosine phenol-lyase. Biochemistry. 1993;32:4195–4206. doi: 10.1021/bi00067a006. [DOI] [PubMed] [Google Scholar]
- 4.Arndt K, Fink G R. GCN-4 protein, a positive transcription factor in yeast, binds general control promoters at all 5′TGACTC3′ sequences. Proc Natl Acad Sci USA. 1986;83:8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrera-Saldana H, Takahashi K, Vigneron M, Wildeman A, Davidson I, Chambon P. All six GC-motifs of the SV40 early upstream element contribute to promoter activity in vivo and in vitro. EMBO J. 1985;4:3839–3849. doi: 10.1002/j.1460-2075.1985.tb04156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Belyaeva T A, Rhodius V A, Webster C L, Busby S J W. Transcription activation at promoters carrying tandem DNA sites for the Escherichia coli cyclic AMP receptor protein: organization of the RNA polymerase α subunits. J Mol Biol. 1998;277:789–804. doi: 10.1006/jmbi.1998.1666. [DOI] [PubMed] [Google Scholar]
- 6.Berman M L, Jackson D T. Selection of lac gene fusions in vivo: ompR-lacZ fusions that define a functional domain of the ompR gene product. J Bacteriol. 1984;159:750–756. doi: 10.1128/jb.159.2.750-756.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenowitz M, Senear D, Jamison E, Dalma-Weiszhausz D. Quantitative DNase I footprinting. In: Revzin A, editor. Footprinting of nucleic acid-protein complexes. San Diego, Calif: Academic Press; 1993. pp. 1–43. [Google Scholar]
- 7a.Busby S, Kolb A. The CAP modulon. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes; 1996. pp. 255–279. [Google Scholar]
- 8.Chen Y-M, Lu Z, Lin E C C. Constitutive activation of the fucAO operon and silencing of the divergently transcribed fucPIK operon by an IS5 element in Escherichia coli mutants selected for growth on l-1,2-propanediol. J Bacteriol. 1989;171:6097–6105. doi: 10.1128/jb.171.11.6097-6105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui J, Somerville R L. A mutational analysis of the structural basis for transcriptional activation and monomer-monomer interaction in the TyrR system of Escherichia coli K-12. J Bacteriol. 1993;175:1777–1784. doi: 10.1128/jb.175.6.1777-1784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J, Somerville R L. The TyrR protein of Escherichia coli, analysis by limited proteolysis of domain structure and ligand-mediated conformational changes. J Biol Chem. 1993;268:5040–5047. [PubMed] [Google Scholar]
- 11.de Crombrugghe B, Busby S, Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984;224:831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- 12.Drlica K, Rouviere-Yaniv J. Histone-like proteins of bacteria. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enei H, Matsui H, Nakazawa H, Okumura S, Yamada H. Culture conditions for the preparation of cells containing high tyrosine phenol lyase activity. Agric Biol Chem. 1973;37:485–492. [Google Scholar]
- 14.Foor F, Morin N, Bostian K A. Production of l-dihydroxyphenylalanine in Escherichia coli with the tyrosine phenol-lyase gene cloned from Erwinia herbicola. Appl Environ Microbiol. 1993;59:3070–3075. doi: 10.1128/aem.59.9.3070-3075.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman D I. Integration host factor: a protein for all reasons. Cell. 1988;55:545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- 16.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 17.Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 18.Heatwole V M, Somerville R L. The tryptophan-specific permease gene, mtr, is differentially regulated by the tryptophan and tyrosine repressors in Escherichia coli K-12. J Bacteriol. 1991;173:3601–3604. doi: 10.1128/jb.173.11.3601-3604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoover T R, Santero E, Porter S, et al. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990;63:11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- 20.Hope I A, Struhl K. GCN-4 protein, synthesized in vitro, binds His3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985;43:177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- 21.Kasian P A, Davidson B E, Pittard A J. Molecular analysis of the promoter operator region of the Escherichia coli K-12 tyrP gene. J Bacteriol. 1986;167:556–561. doi: 10.1128/jb.167.2.556-561.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumagai H, Kashima N, Yamada H. Racemization of D- or L-alanine by crystalline tyrosine phenol-lyase. Biochem Biophys Res Commun. 1970;39:796–801. doi: 10.1016/0006-291x(70)90393-1. [DOI] [PubMed] [Google Scholar]
- 23.Kumagai H, Matsui H, Ohgishi H, Ogata K, Yamada H, Ueno T, Fukami H. Synthesis of 3,4-dihydroxphenyl-L-alanine from L-tyrosine and pyrocatechol by crystalline β-tyrosinase. Biochem Biophys Res Commun. 1969;34:266–270. doi: 10.1016/0006-291x(69)90826-2. [DOI] [PubMed] [Google Scholar]
- 24.Kumagai H, Matsui H, Yamada H. Formation of tyrosine phenol-lyase by bacteria. Agric Biol Chem. 1970;34:1259–1261. [Google Scholar]
- 25.Kumagai H, Yamada H, Matsui H, Ohkishi H, Ogata K. Tyrosine phenol lyase. J Biol Chem. 1970;245:1773–1777. [PubMed] [Google Scholar]
- 26.Kurusu Y, Fukushima M, Kohama K, Kobayashi M, Terasawa M, Kumagai H, Yukawa H. Cloning and nucleotide sequencing of the tyrosine phenol lyase gene from Escherichia intermedia. Biotechnol Lett. 1991;13:769–772. [Google Scholar]
- 27.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of ς54(ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee E C, MacWilliams M P, Gumport R I, Gardner J F. Genetic analysis of Escherichia coli integration host factor interactions with its bacteriophage λ H′ recognition site. J Bacteriol. 1991;173:609–617. doi: 10.1128/jb.173.2.609-617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Grice S, Matzura H, Marcoli R, Iida S, Bickle T. The catabolite-sensitive promoter for the chloramphenicol acetyltransferase gene is preceded by two binding sites for the catabolite gene activator protein. J Bacteriol. 1982;150:312–318. doi: 10.1128/jb.150.1.312-318.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lennox E S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 31.Lin, E. C. C., and A. S. Lynch. Regulation of gene expression in Escherichia coli, p. 255–280. R. G. Landes, Austin, Tex.
- 32.Mandel M, Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970;53:154–159. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- 33.Maxam A M, Gilbert W. Sequencing end labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 34.Meyer B, Maurer R, Ptashne M. Gene regulation at the right operator (Or) of bacteriophage λ. II. Or1, Or2, and Or3: their roles in mediating the effects of repressor and cro. J Mol Biol. 1980;139:163–194. doi: 10.1016/0022-2836(80)90303-4. [DOI] [PubMed] [Google Scholar]
- 35.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 36.Molina-Lopez J, Govantes F, Santero E J. Geometry of the process of transcription activation at the ς54-dependent nifH promoter of Klebsiella pneumoniae. J Biol Chem. 1994;269:25419–25425. [PubMed] [Google Scholar]
- 37.Nash H A. The HU and IHF proteins: accessory factors for complex protein-DNA assemblies. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes; 1996. pp. 150–179. [Google Scholar]
- 38.Ogden S, Haggerty D, Stoner C M, Kolodrubetz D, Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci USA. 1980;77:3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pittard A J. Biosynthesis of the amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 458–484. [Google Scholar]
- 40.Porter S C, North A K, Wedel A B, Kustu S. Oligomerization of NtrC at the glnA enhancer is required for transcriptional activation. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 41.Rabin R S, Collins L A, Stewart V. In vivo requirement of integration host factor for nar (nitrate reductase) operon expression in Escherichia coli K-12. Proc Natl Acad Sci USA. 1992;89:8701–8705. doi: 10.1073/pnas.89.18.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice P A, Yang S-W, Mizuuchi K, Nash H A. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 43.Rosenberg A H, Lade B N, Chui D-S, Lin S-W, Dunn J J, Studier F W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 44.Sabourin D, Beckwith J. Deletion of the Escherichia coli crp gene. J Bacteriol. 1975;122:338–340. doi: 10.1128/jb.122.1.338-340.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarsero J P, Pittard A J. Molecular analysis of the TyrR protein-mediated activation of mtr gene expression in Escherichia coli K-12. J Bacteriol. 1991;173:7701–7704. doi: 10.1128/jb.173.23.7701-7704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarsero J P, Wookey P J, Pittard A J. Regulation of expression of the Escherichia coli K-12 mtr gene by TyrR protein and Trp repressor. J Bacteriol. 1991;173:4133–4143. doi: 10.1128/jb.173.13.4133-4143.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schröder I, Darie S, Gunsalus R P. Activation of the Escherichia coli nitrate reductase (narGHJI) operon by NarL and Fnr requires integration host factor. J Biol Chem. 1993;268:771–774. [PubMed] [Google Scholar]
- 48.Schultz S C, Shields G C, Steitz T A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90°C. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 49.Shirabe K, Ebina Y, Miki T, Nakazawa T, Nakazawa A. Positive regulation of the colicin E1 gene by cyclic cAMP and cyclic AMP receptor protein. Nucleic Acids Res. 1985;13:4687–4698. doi: 10.1093/nar/13.13.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith H Q, Somerville R L. The tpl promoter of Citrobacter freundii is activated by the TyrR protein. J Bacteriol. 1997;179:5914–5921. doi: 10.1128/jb.179.18.5914-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Somerville R L, Shieh T-L N, Hagewood B, Cui J. Gene expression from multicopy T7 promoter vectors proceeds at single copy rates in the absence of T7 RNA polymerase. Biochem Biophys Res Commun. 1991;181:1056–1062. doi: 10.1016/0006-291x(91)92044-k. [DOI] [PubMed] [Google Scholar]
- 52.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki H, Katoyama T, Yamamoto K, Kumagai H. Transcriptional regulation of tyrosine phenol-lyase gene of Erwinia herbicola AJ2985. Biochem J. 1995;59:2339–2341. doi: 10.1271/bbb.59.2339. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki H, Nishihara K, Osui N, Matsui H, Kamagai H. Cloning and nucleotide sequence for Erwinia herbicola AJ2982 tyrosine phenol-lyase gene. J Ferment Bioeng. 1993;75:145–148. [Google Scholar]
- 55.Vidal-Ingigliardi D, Raibaud O. Three adjacent binding sites for cAmp receptor protein are involved in the activation of the divergent malEp-malKp promoters. Proc Natl Acad Sci USA. 1991;88:229–233. doi: 10.1073/pnas.88.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli. Methods Enzymol. 1956;153:3–11. [PubMed] [Google Scholar]
- 57.Wang Q, Calvo J M. Lrp, a global regulatory protein of Escherichia coli, binds cooperatively to multiple sites and activates transcription of ilvIH. J Mol Biol. 1993;229:306–318. doi: 10.1006/jmbi.1993.1036. [DOI] [PubMed] [Google Scholar]
- 58.Whipp M J, Pittard A J. Regulation of aromatic amino acid transport systems in Escherichia coli K-12. J Bacteriol. 1977;132:453–461. doi: 10.1128/jb.132.2.453-461.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson T J, Maroudas P, Howlett G J, Davidson B E. Ligandinduced self-association of the Escherichia coli regulatory protein TyrR. J Mol Biol. 1994;238:309–318. doi: 10.1006/jmbi.1994.1294. [DOI] [PubMed] [Google Scholar]
- 60.Wu Y, Datta P J. Integration host factor is required for positive regulation of the tdc operon of Escherichia coli. J Bacteriol. 1992;174:233–240. doi: 10.1128/jb.174.1.233-240.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 62.Xiao H, Perisic O, Lis J T. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5bp unit. Cell. 1991;64:585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]
- 63.Yu X M, Reznikoff W S. Deletion analysis of the CAP-cAMP binding site of the Escherichia coli lactose promoter. Nucleic Acids Res. 1984;12:5449–5464. doi: 10.1093/nar/12.13.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]