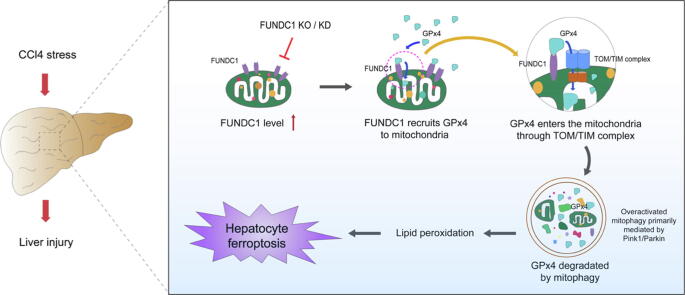
Graphical abstract
FUNDC1 promotes liver fibrosis and hepatic injury through direct binding to GPx4 to facilitate its mitochondrial translocation through TOM/TIM complex, where it is degraded along with the ROS-induced damaged mitochondria by mitophagy, resulting in hepatic ferroptosis.
Keywords: Liver fibrosis, FUNDC1, GPx4, Mitophagy, Ferroptosis, TOM/TIM complex
Highlights
-
•
This work examined the role for FUNDC1, a mitophagy receptor, in carbon tetrachloride (CCl4)-induced liver injury. CCl4 treatment induced excess production of mitochondrial ROS and overactivated mitophagy. We found that FUNDC1 is a culprit in eliciting ferroptosis but not in mitophagy activation, offering new insights for FUNDC1 function beyond mitophagy.
-
•
FUNDC1 interacted directly with glutathione peroxidase (GPx4), a selenoenzyme to neutralize lipid hydroperoxides and ferroptosis, via its 96-133 amino acid domain to facilitate GPx4 recruitment into mitochondria from cytoplasm.
-
•
GPx4 entered mitochondria through mitochondrial protein import system – the translocase of outer membrane/translocase of inner membrane (TOM/TIM) complex, prior to degradation of GPx4 mainly through mitophagy along with ROS-induced damaged mitochondria, resulting in hepatocyte ferroptosis.
Abstract
Introduction
Liver fibrosis is a life-threatening pathological anomaly which usually evolves into advanced liver cirrhosis and hepatocellular carcinoma although limited therapeutic option is readily available. FUN14 domain containing 1 (FUNDC1) is a mitophagy receptor with little information in liver fibrosis.
Objective
This study was designed to examine the role for FUNDC1 in carbon tetrachloride (CCl4)-induced liver injury.
Methods
GEO database analysis and subsequent validation of biological processes including western blot, immunofluorescence, and co-immunoprecipitation were applied to clarify the regulatory role of FUNDC1 on mitophagy and ferroptosis.
Results
Our data revealed elevated FUNDC1 levels in liver tissues of patients with liver fibrotic injury and CCl4-challenged mice. FUNDC1 deletion protected against CCl4-induced hepatic anomalies in mice. Moreover, FUNDC1 deletion ameliorated CCl4-induced ferroptosis in vivo and in vitro. Mechanically, FUNDC1 interacted with glutathione peroxidase (GPx4), a selenoenzyme to neutralize lipid hydroperoxides and ferroptosis, via its 96–133 amino acid domain to facilitate GPx4 recruitment into mitochondria from cytoplasm. GPx4 entered mitochondria through mitochondrial protein import system-the translocase of outer membrane/translocase of inner membrane (TOM/TIM) complex, prior to degradation of GPx4 mainly through mitophagy along with ROS-induced damaged mitochondria, resulting in hepatocyte ferroptosis.
Conclusion
Taken together, our data favored that FUNDC1 promoted hepatocyte injury through GPx4 binding to facilitate its mitochondrial translocation through TOM/TIM complex, where GPx4 was degraded by mitophagy to trigger ferroptosis. Targeting FUNDC1 may be a promising therapeutic approach for liver fibrosis.
Introduction
Liver fibrosis refers to a devastating pathological process characterized by buildup of excess extracellular matrix proteins in a cadre of chronic liver diseases [1]. It is mainly provoked by hepatic stresses including viral hepatitis, nonalcoholic steatohepatitis (NASH), alcoholic liver disease (ALD), iron overload syndrome, hereditary haemochromatosis, and diabetes mellitus [2], [3]. With persistent deterioration of liver fibrosis, chronic liver failure, cirrhosis, and hepatocellular carcinoma ensue, imposing a major threat for human health although little effective treatment remedy is readily available for patients with advanced liver fibrosis beyond liver transplantation due to its poorly defined underlying mechanism(s) [4], [5]. In this context, it is imperative to seek novel antifibrotic therapeutic options to retard and prevent the onset and progression of liver fibrosis.
Mitophagy, a selective form of autophagy, governs mitochondrial turnover and recycling to maintain hepatic homeostasis through removal of long-lived/damaged mitochondria [6]. Dysregulation of mitophagy is implicated in the onset and development of liver diseases including chronic hepatic fibrosis. Mitophagy was reported to promote tissue damage in the face of carbon tetrachloride (CCl4) insult [7], [8], [9]. Among which, PTEN-induced kinase 1 (Pink1)/E3 ubiquitin ligase Parkin is the most well examined component to regulate mitophagy in stress conditions including liver fibrosis [7], [9]. BCL2 interacting protein 3 like (BNIP3) is a mitophagy receptor highly expressed in the liver and directly interacts with microtubule-associated light chain 3 (LC3) to initiate mitophagy [10], [11]. FUN14 domain-containing protein 1 (FUNDC1) is a new mitophagy receptor through direct interaction with LC3 motif in a fashion reminiscent of BNIP3 [12], [13]. Data from our group and others have noted a pivotal role for FUNDC1 in the maintenance of hepatic homeostasis [14], [15]. FUNDC1-dependent mitophagy suppressed hepatocarcinogenesis through amelioration of inflammasome activation [15]. In addition, downregulation of FUNDC1 suppressed mitophagy in alcoholic liver diseases, which activated the DNA-dependent protein kinase catalytic subunit (DNA-PKcs)-p53-dynamin-related protein 1 (Drp1) signaling cascade, leading to unchecked mitochondrial fission and liver injury [14]. These findings depict a crucial role for FUNDC1 and mitophagy in liver diseases. Moreover, FUNDC1 offers a much broader biological action beyond control of mitophagy [16], [17]. For example, FUNDC1 participates in mitochondrial reprogramming and cellular plasticity through interacting with Lon protease 1 (LonP1), a member of protein folding quality control system on the mitochondrial inner membrane to manage proteolytic disposal of misfolded and aggregated proteins [17]. Our study reported an essential role for FUNDC1 in preserving mitochondrial Ca2+ homeostasis and cardiac function in obese hearts through its interaction with the ubiquitin E3 ligase FBXL2 [18]. Nevertheless, a role for FUNDC1 in liver fibrosis remains elusive.
Ferroptosis is a new form of iron-dependent regulated cell death, resulting from accumulation of reactive oxygen species (ROS) and lipid peroxidation [19], [20]. It is unique in morphology, biochemistry, and genetics in comparison with other forms of regulated cell death (RCD) [21]. Previous findings have indicated involvement of ferroptosis in a wide variety of pathological processes including cancers, liver and cardiovascular diseases [22], [23], [24], [25]. Recent data have indicated a close link between mitochondria and ferroptosis, both of which heavily impacting oxidative metabolism [26]. Morphological alterations including mitochondrial fragmentation and cristae enlargement were noted in mitochondria under ferroptosis [26]. Furthermore, ferroptosis inhibitors were reported to exquisitely target mitochondria, consistent with the potential involvement of mitochondria in ferroptosis process [27], [28]. Nonetheless, whether FUNDC1, a vital component of mitophagy, elicits any impact on the occurrence of ferroptosis remains elusive. Glutathione peroxidase 4 (GPx4) is a glutathione peroxidase that utilizes glutathione (GSH) as a cofactor to convert lipid hydroperoxides into non-toxic lipid alcohols to mitigate ferroptosis [29]. Downregulation or depletion of GPx4 evokes accumulation of lipid peroxides and other reactive oxygen species, resulting in cell ferroptosis [23], [30]. Excessive hepatic iron deposition and ferroptosis is speculated to potentiate liver fibrosis in mice, the process of which can be reversed by ferrostatin-1 [31]. Hence, ferroptosis likely promotes the pathogenesis of liver fibrosis. Besides, GPx4 activation was shown to protect against ferroptosis-induced liver injury in mice [32]. GPx4 is widely distributed in cytoplasm, mitochondria, and nuclei, while transportation of GPx4 from cytosol to mitochondria occurs under pathological conditions [33]. Elevated GPx4 levels in mitochondria play an essential regulatory role in mitochondrial apoptosis and mitochondrial ATP production under stress [33], [34], [35]. To-date, possible mechanism(s) governing GPx4 translocation from cytoplasm to mitochondria is largely unknown.
Hence, our aim in this study was to examine the role of FUNDC1 in carbon tetrachloride (CCl4)-induced liver fibrosis and liver injury. Our data revealed upregulated FUNDC1 in livers from mice with hepatic fibrosis and serum and liver tissues from patients afflicted with hepatic fibrosis. FUNDC1 knockout (KO) effectively ameliorated hepatic inflammation, fibrosis and ferroptosis. Mechanistically, FUNDC1 bound and recruited GPx4 to mitochondria, to facilitate its entrance into the mitochondria through mitochondrial protein import system TOM/TIM complex, where GPx4 was degraded by mitophagy (primarily mediated by Pink1/Parkin), leading to hepatocyte ferroptosis. Our data further depicted that FUNDC1 predisposed CCl4-induced hepatic injury through mitophagy-dependent ferroptosis in hepatocytes, denoting promises of targeting FUNDC1 or mitophagy in the therapeutics of liver fibrosis.
Materials and methods
Animal models of hepatic fibrosis and liver function
Global FUNDC1 knockout (FUNDC1-/-) mice were generated as described in our earlier report [18]. Adult (8–10-week-old) male wild-type (WT) and FUNDC1-/- mice were maintained at 24 ± 2 °C and 50 ± 5% humidity. Mice were offered sterile standard normal diet and water ad libitum. Liver fibrosis was established using an intraperitoneal injection of CCl4 (Sigma‐Aldrich, St. Louis, MO, USA; 1:4 v/v in corn oil) at a dosage of 5 mL/kg of body weight (BW) twice per week for 4 weeks, a widely perceived model of liver fibrosis in rodents [36]. An equal amount of vehicle (corn oil) was delivered to non-fibrotic group. A cohort of mice received GPx4 inhibitor RSL3 (Selleck Chemicals, Houston, TX, USA, 2.5 mg/kg., i.p.) once a day for 10 days. Standard necropsy techniques were used and liver injury was evaluated by serum alanine transaminase (ALT) and aspartate transaminase (AST) determined using the VetAce Analyser (Alfa Wassermann, Inc., West Caldwell, NJ, USA).
Human samples
Human liver samples were obtained from healthy adults (n = 7) and patients with liver fibrosis (n = 7) (Table S1).
Ethics statement
The animal experimental procedures were approved by the Institutional Animal Use and Care Committee at the Zhongshan Hospital Fudan University (Shanghai, China), and were in compliance with the NIH Guide for the Care and Use of Laboratory Animals. The experimental protocol involving human subjects received approval from the Institutional IRB committee of Xijing Hospital Air Force Military Medical University (Xi’an, China), the approval number is KY202113504-1.
Gene ontology (GO) and pathway enrichment analysis
mRNA profiles of mouse fibrotic liver tissues and gene annotation platforms of GPL6885 were downloaded from Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/; GSE55747). The Linear Models for Microarray Data (LIMMA) algorithm was applied to detect the differential expressed genes (DEGs) based on the fragments per kilobase of exon model per million mapped fragments (FPKM) data. Genes with Benjamini-Hochberg method adjusted p-value<0.05 and |log2 fold-change (FC)| greater than 1.0 in CCl4-treated mice compared with corn oil-treated mice were selected as bicalutamide response-related genes.
For pathway enrichment analysis, the “clusterProfiler” and “AnnotationHub” packages in R were performed to identify hub maps. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms with the Benjamini-Hochberg method adjusted p-value<0.05 and gene counts of set arranged from 10 to 500 were selected. Furthermore, the Gene Set Variation Analysis (GSVA) algorithm also applied to evaluate involvement of ferroptosis, mitochondrial assembly and function based on the gene expression profiles. In addition, the interaction network was constructed via partial correlation analysis based on normalized GSVA pathway score.
Measurements of cytochrome P450 reductase activity, serum iron, non-heme iron and lipid peroxidation
Cytochrome P450 reductase activity was measured using a cytochrome P450 reductase activity kit (Biovision, San Francisco, CA, USA, #K700-100). Levels of plasmic iron and hepatic non-heme iron were measured using an iron assay kit (Abcam, #ab83366) in accordance with the manufacturer’s instruction. Levels of carbonyls were quantified using a protein carbonyl content assay kit (Abcam, #ab126287). Levels of GSH, GSSG and MDA were quantified using glutathione and MDA assay kits from Beyotime (Nanjing, China).
Histological examination
Following anesthesia, livers were exercised and placed in 10% neutral-buffered formalin for 24 hrs at room temperature. Specimens were embedded in paraffin, cut into 5-μm sections and subjected to Masson trichrome or Sirius red staining. Percentage of fibrosis was calculated using a digital microscope (×400) and Image J (version1.34S) software.
Immunohistochemical staining of neutrophils
Deparaffinized liver sections (5-μm thick) were immunohistochemically stained for myeloperoxidase (MPO, primary antibody from Biocare Medical, Concord, CA), a hallmark of neutrophils, using the DAB kit (Gene Tech, Shanghai, China) per the manufacturer’s instruction. The MPO-positive cells were randomly quantified from 5 fields per mouse (at 100 × magnification) using the Image J software.
Realtime quantitative PCR
Total RNA was extracted using a Trizol Reagent (Invitrogen, NY, Empire State, USA). The purity and concentration of RNA were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, ME, USA). Reverse-transcription was conducted using PrimeScript TM RT Master Mix (Takara, Shiga, Japan) for synthesis of cDNA. Realtime quantitative PCR was performed using FastStart Essential DNA Green Master (Roche, Shanghai, China). β-actin was employed as the reference gene. Relative gene expression was calculated using 2-ddCt method. Sequences of PCR primers were listed in Table S3 (Dongxuan Gene, Kunshan, China).
Transmission electron microscopy (TEM)
Cubic liver pieces were dissected and fixed with 2.5% glutaraldehyde in 0.1 M sodium phosphate (pH 7.4) for 24 hrs at 4 °C. Samples were dehydrated through graded alcohols and were embedded in Epon Araldite following fixation in 1% OsO4 for 1 hr. Ultrathin sections (50-nm) were produced using an ultramicrotome (Leica, Wetzlar, Germany), and were stained with uranyl acetate and lead citrate. Specimens were visualized under an Electron Microscope. Images were captured using the Ditital Micrograph software.
Cell culture and treatment
Human hepatoma HepG2 cell line was obtained from Genscript (Nanjing, China). HepG2 cells were transfected with CYP2E1 plasmid construct a CYP2E1 stable overexpressed cell line (also named E47 cell line) [37]. Given that CCl4-induced hepatotoxicity is predominantly mediated through Cyp2E1 metabolism to yield CCl3 radical, HepG2 cell lines overexpressing CYP2E1 (E47 line) are deemed a useful model to discern the biochemical properties of CYP2E1 [37]. Cells were cultured in a Dulbecco’s Modified Eagle Medium (DMEM, Gibco), containing 10% fetal bovine serum albumin (FBS, Gibco) at 37 °C, 5% CO2. CCl4 was dissolved in dimethyl sulfoxide (DMSO, Sigma‐Aldrich, St. Louis, MO, USA) in serum-free DMEM. To determine involvement of ferroptosis, mitophagy and lysosomal involvement, Erastin (Selleck Chemicals, Houston, TX, USA, 10 μM for 24 hrs), oroxylin A (OA, Med Chem Express, NJ, USA, 150 μM for 48 hrs), RSL3 (Selleck Chemicals, Houston, TX, USA, 1 μM for 24 hrs), MG132 (Med Chem Express, NJ, USA, 10 μM for 6 hrs) or Chloroquine (Sigma‐Aldrich, MO, USA, 20 μM for 24 hrs) was added to E47 cells when cells grow to a proper density for further experimentation.
Plasmid constructs
Full-length, domain truncated (FUNDC1-Delta-2-47aa, FUNDC1-Delta-69-74aa, and FUNDC1-Delta-96–133) FUNDC1 plasmid tagged with Flag were cloned in pcDNA3.1+ (Dongxuan Genes, Kunshan, China). CYP2E1 and GPx4 tagged with His were cloned in pcDNA3.1+ (Dongxuan Genes).
Cell viability assay
Cell viability was evaluated using a MTT kit (Beyotime, Nanjing, China) per the manufacturer’s protocol. Briefly, E47 cells were seeded in 96-well plates at 5000 cells per well. Cells were incubated with fresh media containing MTT solution for 4 hrs prior to addition of dimethyl sulfoxide (DMSO) to dissolve formazan. Samples were assessed using a microplate reader (Thermo Fisher, Waltham, ME, USA), and absorbance was read at 490 nm.
Measurement of mitochondrial ROS (mtROS)
E47 cells were incubated with the MitoSox Red fluorescent dye (5 μM, Thermo Fisher) at 37 °C for 30 min. Cells were rinsed with a warm phosphate buffered saline (PBS) buffer prior to observation under a Leica confocal microscopy (Wetzlar, Germany).
Measurement of mitochondrial morphology and mitochondrial membrane potential (MMP)
Mitochondrial morphology and mitochondrial membrane potential (MMP) were assessed using MitoTracker and tetramethylrhodamine methyl Ester (TMRM) staining, respectively. Cells were stained with MitoTracker Green or Red solution (20 nM, Molecular Probes, Invitrogen, Carlsbad, CA, USA) or TMRM (20 nM, Molecular Probes) at 37 °C for 30 min, and were imaged through a fluorescence microscope. The Image J software was used to evaluate red fluorescence intensity.
Lipid ROS assay
Lipid ROS was measured using the fluorescent Probes BODIPY 581/591 C11 (D3861, Invitrogen, Carlsbad, CA, USA). E47 cells were seeded in 3.5 cm plates and pretreated with inhibitors and CCl4, and incubated with the kit reagent following the instruction at a concentration of 5 μM for 30 min in the dark. Cells were rinsed three times with PBS prior to image acquisition using a fluorescence microscope (Leica, Wetzlar, Germany).
Immunofluorescence analysis
Cell samples were fixed by 4% paraformaldehyde for 15 min. E47 cells were permeabilized and blocked for 1 hr then incubated with specified antibodies (GPx4, Abcam, #ab125066, Tom20, Abcam, #ab283317, and HSP60, Abcam, #ab46798) overnight at 4 °C, and followed by the incubation of the corresponding Alexa Fluor conjugated secondary antibodies (Cell Signaling Technology, Boston, MA, USA) for 1 hr in the dark. After staining, the immunofluorescence was examined using a laser confocal microscope with a 630 × oil immersion objective with 488 and 561 nm laser excitation (Leica, Wetzlar, Germany). Results were analyzed by a coloc2 plug-in (Fiji, version 2.0, Rawak Software Inc., Stuttgart, Germany) of the Image J software.
Structure-based protein interaction interface analysis between FUNDC1 and GPx4
Protein structure of FUNDC1 was predicted by a template-based homology structure modeling tool SWISS-MODEL (https://www.swissmodel.expasy.org), using PDB structure 2IP6, chain A (covering residues 82–131, sequence identity = 10.00%) and 3BK6, chain A (covering residues 74–256, sequence identity = 21.64%) as the template, respectively. Structure of GPx4 was downloaded from the PDB database (PDB ID:5L71). Prediction of potential interaction interface between GPx4 and FUNDC1 was obtained from PRISM tool (https://cosbi.ku.edu.tr/prism). Prediction results were visualized using the PyMol tool (https://pymol.org).
Isolation of mitochondria and nuclei
Preparation of mitochondria and nuclei was conducted using the Mitochondria/Cytosol Fractionation Kit (Abcam, #ab65320) and Nuclear Extraction Kit (Abcam, #ab219177). Isolation of mitochondria and nuclei was performed on ice to prepare mitochondrial, cytosolic and nuclear fractions for Western blot analysis.
Co-Immunoprecipitation (Co-IP)
Co-immunoprecipitation (Co-IP) assay was conducted using a Pierce® Co-IP kit (Pierce, IL) following the manufacturer’s manual. One hundred μg purified anti-flag antibodies were first coupled with resin. Protein samples (1000 μg) were exposed to the antibody-coupled resin for 2 hrs. Following mixing and washing, 50 μL elution buffer was added into the protein-antibody complexes. Protein samples were subjected to immunoblotting using anti-GPx4, anti-FUNDC1 and anti-LC3B antibodies (1:1000, Cell Signaling Technology) [33].
Immunoprecipitation assay and mass spectrometry
E47 cells were transfected with FUNDC1-Flag plasmid. Forty-eight hrs later, cell lysates were immunoprecipitated with the anti-Flag magnetic beads (Cell Signaling Technology). Precipitates were separated using SDS-PAGE and were then stained with coomassie blue. Protein bands were cut into small pieces and subjected in-gel digestion. The extracted peptides from the gel pieces were prepared for proteomic data analysis by liquid chromatography tandem mass spectrometry (LC-MS/MS).
Western blot analysis
Liver tissues and cells were homogenized and sonicated in a lysis buffer containing 20 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton, 1 mM EGTA, 1 mM EDTA, 0.1% sodium dodecyl sulfate, and a protease inhibitor cocktail. Protein samples were incubated with anti-tumor necrosis factor-α (TNF-α), anti-interleukin-6 (IL-6), anti-CYP2E1, anti-HSP60, anti-Histone H3, anti-FUNDC1, anti-BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3), anti-Parkin, anti-translocase of outer and inner membrane/translocase (TIM23), anti-microtubule-associated protein light chain 3 (LC3B), anti-p62, anti-solute carrier family 7 member 11 (SLC7A11), anti-glutathione peroxidase (GPx4), anti-Vinculin, and anti-β-actin antibodies. All antibodies were obtained from Cell Signaling Technology (Danvers, MA, USA), Abcam (Cambs, UK), Santa Cruz Biotech (Santa Cruz, CA, USA) or Proteintech (Wuhan, China). Membranes were incubated with secondary antibodies (Proteintech, Wuhan, China) following primary antibodies incubation. Films were scanned and detected with a Bio-Rad calibrated densitometer [38].
Statistical analysis
Data were presented as mean ± standard error of mean (SEM). Results were analyzed using a Prism 8.0 software (GraphPad, San Diego, CA). Comparation between two groups were conducted using the non-parametric t-test (two-tailed). Comparation among multiple groups were conducted using one-way ANOVA followed Tukey's test for post hoc analysis. Statistical significance was set at p < 0.05.
Results
Elevated serum and tissue levels of FUNDC1 in patients and mouse models of liver fibrosis
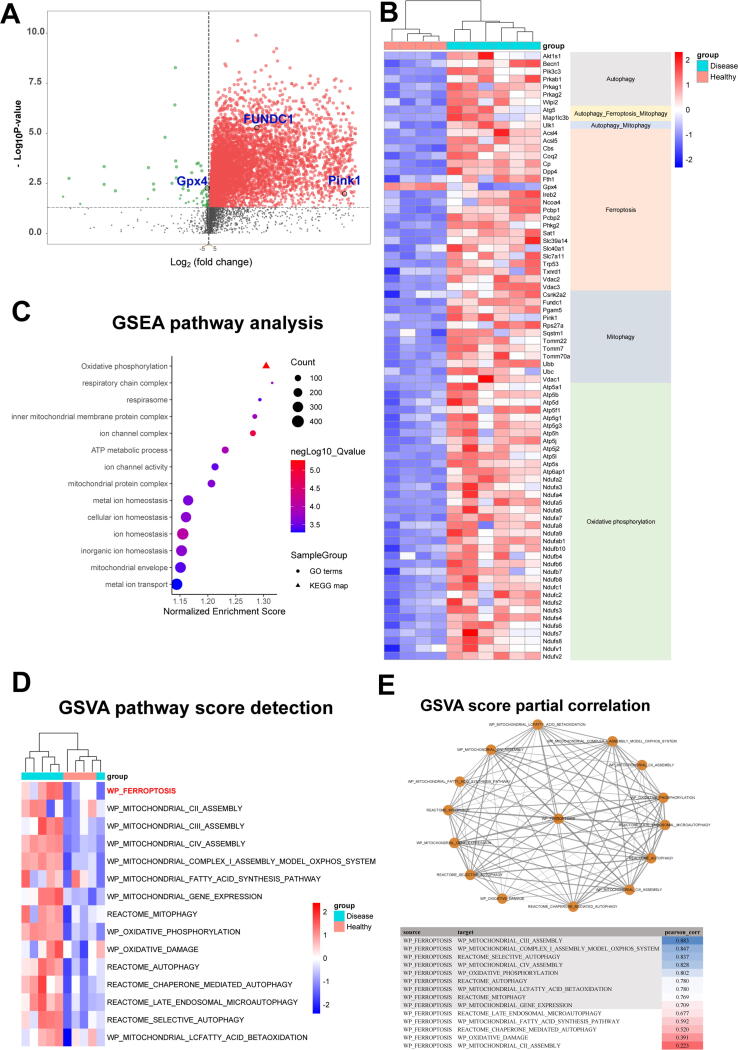
To discern possible biological processes and cell signaling pathways involved in liver fibrotic injury, data from GEO database (GSE55747) were analyzed. The volcano plot displayed upregulated FUNDC1 and Pink1, as well as downregulated GPx4 in CCl4-induced liver fibrosis in mice (Fig. 1a). A hierarchical clustering of the most highly regulated genes revealed “autophagy”, “ferroptosis”, “mitophagy”, and “oxidative phosphorylation” with distinct patterns of presentation between liver fibrotic and control groups (Fig. 1b). GSEA results showed that multi-biological processes, involving mitochondrial function, oxidative phosphorylation and metal ion homeostasis, may play an important regulatory role in CCl4-induced liver fibrosis (Fig. 1c). Similarly, these terms involving ferroptosis, mitochondrial assembly and function also showed significant differences after quantifying molecular pathways (Fig. 1d). Moreover, results from GSVA score partial correlation denoted that ferroptosis was closely associated with mitochondria, mitochondrial complex, as well as mitophagy (Fig. 1e). These analyses strongly promoted ferroptosis and mitochondria or mitophagy may be involved in the progression of liver fibrosis.
Fig. 1.
Bioinformatics analysis of hepatic pathways and genes in liver fibrosis. (a) Volcano plot of microarray data, generated by clustering based on probes enriched or depleted (fold change greater than 2; adjusted-P < 0.05) in CCl4-treated mice compared to vehicle mice; (b) Heatmap displaying relative expression of genes related to autophagy, ferroptosis, mitophagy and oxidative phosphorylation in the liver (n = 4–6); (c, d) GSEA analysis and GSVA score of distinct biological processes in healthy and disease groups. (n = 5); and (e) GSVA score partial correlation.
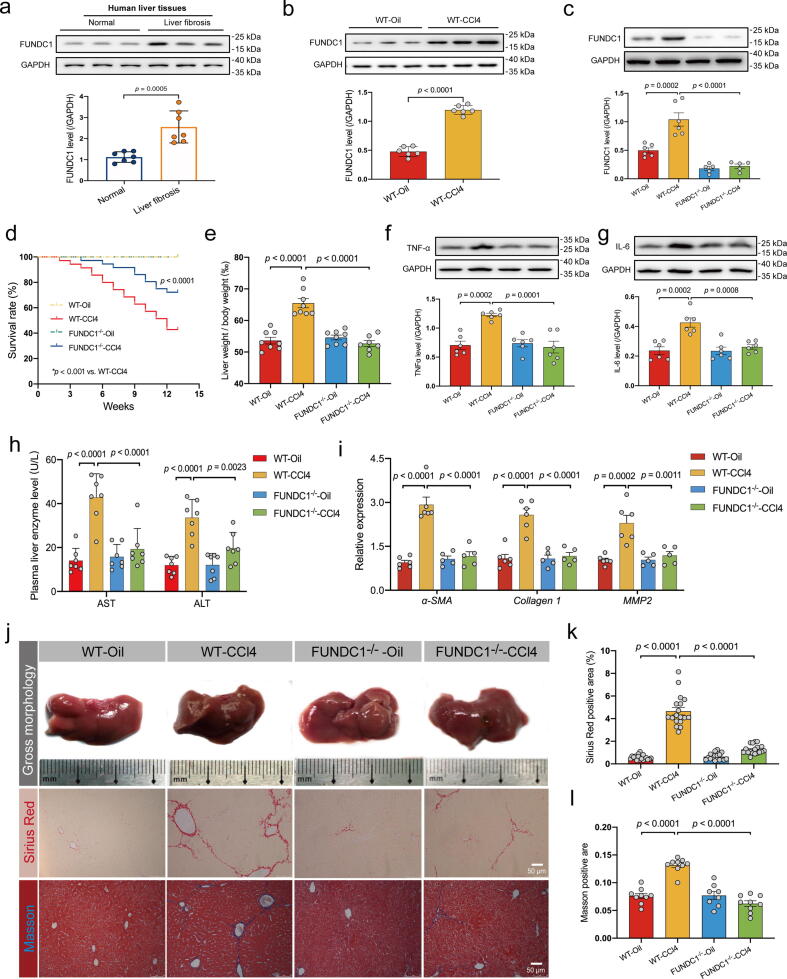
To examine the possible role of FUNDC1 in liver fibrosis, levels of FUNDC1 were determined in human liver tissues and serum. Our results noted overtly elevated FUNDC1 levels in liver tissues from patients with liver fibrosis (Fig. 2a), in line with the volcano plot analysis (Fig. 1a). Next, intraperitoneal injection of CCl4 was used to establish a murine model of liver fibrosis. Our results suggested elevated FUNDC1 levels in liver tissues from CCl4-challenged mice (Fig. 2b). These findings favored a role for FUNDC1 in liver tissues and serum from both human and rodent models of liver fibrosis.
Fig. 2.
Levels of FUNDC1 in fibrotic livers and impact of FUNDC1 ablation on CCl4 (4 weeks)-induced hepatic injury and liver fibrosis. (a) Representative immunoblots and quantitative histogram of FUNDC1 in liver tissues from normal individuals or patients with liver fibrosis (GAPDH as the loading control) (n = 7/group); (b) Representative immunoblots and quantitative histogram of FUNDC1 in mice with or without CCl4 treatment (n = 5–6/group); (c) Representative immunoblots and quantitative histogram of FUNDC1 in mice (n = 5–6/group); (d) Survival rate (n = 13/group); (e) Ratio of liver-to-body weight (n = 8/group); (f-g) Representative immunoblots and quantitative histogram of TNF-α and IL-6 (GAPDH as the loading control) (n = 5–6/group); (h) Level of plasmic ALS and AST (n = 7–8/group). (i) Quantitative mRNA expression of a-SMA, Collagen 1 and MMP2 in liver tissues (n = 5–6/group). (j) Representative images of gross liver morphology (upper panel), Sirius Red staining (middle panel) and Masson staining (bottom panel). (k) Quantified Fibrotic area of Sirius Red staining (black) and (l) Masson staining (blue) (n = 6–9 mice per group). Mean ± SEM (detailed statistical results shown in Table S4); Statistical significance was set at p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
FUNDC1 ablation protects against CCl4-induced hepatic injury and liver fibrosis
To discern a possible role of FUNDC1 in liver fibrosis, FUNDC1-/- and WT mice were challenged with CCl4 or an equal volume of corn oil. Levels of FUNDC1 were significantly upregulated by CCl4 challenge, which was vanished in liver tissues from FUNDC1-/- mice (Fig. 2c). Survival rate was diminished in response to CCl4 challenge in WT mice, the effect of which was overtly attenuated by FUNDC1 ablation with little effect by FUNDC1 knockout itself (Fig. 2d). Meanwhile, CCl4 challenge overtly increased liver-to-body weight ratio in WT mice, the effect of which was negated by FUNDC1 ablation with little effect from FUNDC1 deficiency itself (Fig. 2e). In addition, CCl4 challenge overtly upregulated levels of proinflammatory markers including tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), which were alleviated by FUNDC1 deletion, with little discernible effect from FUNDC1 knockout itself (Fig. 2f, 2 g). Analysis of serum samples revealed significantly elevated levels of hepatocellular injury markers ALT and AST in CCl4-challenged mice, the effect of which was obliterated by FUNDC1 knockout with little effect from FUNDC1 ablation itself (Fig. 2h). To evaluate the effect of FUNDC1 knockout on CCl4-induced liver fibrosis, mRNA levels of fibrotic markers including alpha-smooth muscle actin (a-SMA), Collagen 1, and matrix metalloproteinase 2 (MMP2) were examined in liver tissues. Our data revealed that CCl4 challenge upregulated fibrotic markers, the effects of which were negated by FUNDC1 ablation with little effect from FUNDC1 ablation itself (Fig. 2i). In addition, CCl4-challenged mouse livers were much darker in color with poorer contour and more grainy texture compared with corn oil-treated mice, the effects were less pronounced in FUNDC1-/- mice challenged with CCl4 (Fig. 2j, Table. S2). Histological staining using Sirius red and Masson Trichrome staining revealed extensive destructions in structure, along with abnormal collagen deposition in CCl4-challenged mice. FUNDC1 ablation itself had little effects in hepatic morphology although it ameliorated CCl4-induced unfavorable morphological changes (Fig. 2j-l). These results suggested that CCl4 evoked liver injury, the effect of which was rescued by FUNDC1 deletion. These results favored an important role of FUNDC1 in the onset and development of liver fibrosis and liver injury. Given the perceived prerequisite role for reductive metabolism of CCl4 to reactive intermediates using cytochrome P450 enzymes for CCl4 toxicity [39], cytochrome P450 enzymatic activity was determined. Our data revealed overtly decreased cytochrome P450 activity upon CCl4 challenge, the effect of which was spared by FUNDC1 knockout with little influence from FUNDC1 ablation itself (Fig. S1a).
Effect of FUNDC1 deletion and overexpression on CCl4-induced changes in hepatocyte ferroptosis in vivo and in vitro
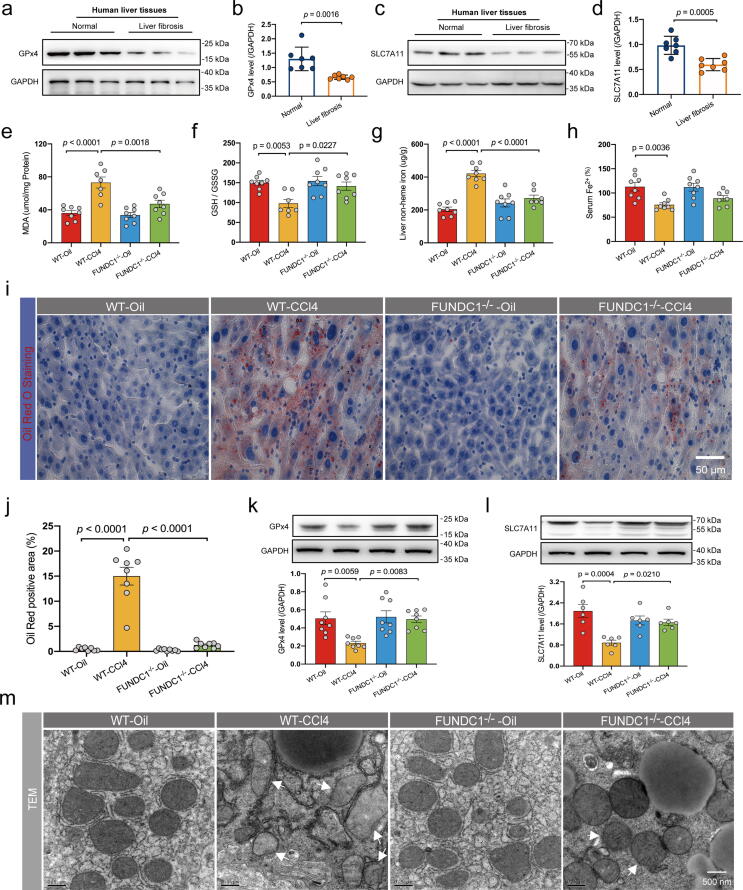
Ferroptosis was believed to participate in liver fibrosis and hepatocyte injury. We analyzed levels of GPx4 and solute carrier family 7 member 11 (SLC7A11) in human liver tissues and noted downregulation of GPx4 and SLC7A11 in patients with liver fibrosis (Fig. 3a-d). To examine possible involvement of ferroptosis in FUNDC1 ablation-offered protection against CCl4-induced hepatocyte injury, liver glutathione (GSH) and glutathione disulfide (GSSG), malondialdehyde (MDA), oil Red O staining, iron in serum and liver tissues, and ferroptosis protein markers GPx4 and SLC7A11 were examined in liver tissues. Our results indicated that CCl4 challenge evoked overt lipid peroxidation and lipid deposition as evidenced by GSH/GSSG ratio, MDA and oil Red O staining, all characteristic of ferroptosis (Fig. 3e-f, 3i-j). FUNDC1 deletion alleviated CCl4-evoked lipid peroxidation and lipid deposition without any effect itself (Fig. 3e-f, 3i-j). Next, levels of hepatic non-heme iron were elevated while serum Fe2+ levels were decreased in face of CCl4 treatment, indicating overload of free iron in the liver, a vital instigator for ferroptosis (Fig. 3g-h). FUNDC1 deletion mitigated CCl4-induced iron overload, although FUNDC1-/- and WT mice displayed little difference in iron content in the absence of CCl4 challenge (Fig. 3g-h). Levels of GPx4 and SLC7A11, two important ferroptosis biomarkers resilient to lipid peroxidation, were downregulated by CCl4, suggesting a role for ferroptosis in CCl4-induce hepatic damage (Fig. 3k-l). Ablation of FUNDC1 upregulated levels of GPx4 and SLC7A11 with little effect itself (Fig. 3k-l). Transmission electron microscopy (TEM) examination noted that CCl4-treated liver tissues exhibited pronounced mitochondrial damage including overt shrinkage or inflation of mitochondria, loss of mitochondrial cristae and mitochondrial membrane integrity (hallmarks of ferroptosis), the effect of which was attenuated by FUNDC1 deletion (Fig. 3m). To discern the effect of CCl4 challenge on mitochondria, mitochondrial morphology and mitochondrial membrane potential (MMP) were examined using the fluorescent dyes MitoTracker and TMRM, respectively, following CCl4 challenge in vitro (0.1% v/v for 12 hrs) in E47 cells (a CYP2E1 stable overexpressed HepG2 cell line, a valuable model for biochemical and toxicological assessment of the P450 enzyme CYP2E1 – which participates in CCl4 toxicity) [40], [41], [42]. Transfection efficiency of CYP2E1 was shown in Fig. S1b. CCl4 treatment evoked elevated ROS in mouse livers as evidenced by upregulated levels of protein carbonyl, the effect of which was alleviated by FUNDC1 deletion (Fig. 1c). MitoTracker and TMRM staining revealed that CCl4 evoked fractionated mitochondria and collapsed MMP, the effect of which was reversed by FUNDC1 knockdown (FUNDC1 KD) using FUNDC1 siRNA (siFUNDC1), without any effect from siFUNDC1 itself (Fig. S2a-c). Evaluation of mtROS using MitoSOX in E47 cells revealed that CCl4 promoted mtROS production, the effect of which was resisted by FUNDC1 deletion (Fig. S2d, S2e). Moreover, examination of C11-BODIPY staining, a fluorophore sensitive to lipid peroxidation, suggested that CCl4 challenge evoked lipid peroxidation in E47 cells, the effect of which was nullified by FUNDC1 ablation (Fig. S2f, S2g). Not surprisingly, FUNDC1 overexpression aggravated hepatocyte damage with severe ferroptosis (downregulated GPx4 and SLC7A11) (Fig. S3a-d), lipid peroxidation (GSH/GSSG ratio, MDA production and C11-BODIPY staining) (Fig. S3e-f, S3h-i), and MTT cell survival (Fig. S3g), and mitochondrial injury (MitoTracker and TMRM staining) (Fig. S3j-l).
Fig. 3.
Effect of FUNDC1 deficiency on CCl4 (4 weeks)-induced changes in hepatocyte ferroptosis in vivo. (a-d) Representative immunoblots and quantitative histogram of GPx4 and SLC7A11 in liver tissues of normal individuals or patients with liver fibrosis (GAPDH as the loading control) (n = 7/group); (e) Level of MDA (n = 7–8/group); (f) Ratio of GSH to GSSG (n = 8/group); (g-h) Level of liver non-heme iron and serum Fe2+ tested using an iron assay kit (n = 7–8/group); (i) Representative images of Oil Red O staining; (j) Quantified Lipid droplet area (n = 8 mice per group); (k-l) Representative immunoblots and quantitative histogram of GPx4, SCL7A11 (GAPDH as the loading control), (n = 6–8/group); and (s) Representative TEM images. White arrowheads denote damaged mitochondria (shrunk or inflated mitochondria, reduced number of mitochondrial cristae and destroyed mitochondrial membrane). Mean ± SEM (detailed statistical results shown in Table S4); Statistical significance was set at p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To further decipher if the beneficial action of FUNDC1 deficiency against CCl4-induced liver injury involved ferroptosis, the ferroptosis inducer erastin was employed (10 μM for 24 hrs) [43], [44] in E47 cells. As shown in Fig. S4a-d, erastin turned on ferroptosis as manifested by downregulation of SLC7A11 and GPx4. Lipid peroxidation was assessed using MDA, GSH/GSSG and C11-BODIPY staining. Our data revealed that erastin cancelled off FUNDC1 knockdown-offered protection against CCl4-induced changes in MDA production, C11-BODIPY fluorescence and GSH/GSSG ratio in E47 cells (Fig. S4e-f, S4h-i). Besides, MTT cell survival results revealed that erastin abolished FUNDC1 knockdown-induced protection against CCl4, with little effect by erastin itself (Fig. S4g). In addition, MitoTracker and TMRM staining noted that erastin reversed FUNDC1 knockdown-evoked protection against CCl4-induced mitochondrial dysmorphology and dysfunction (Fig. S4j-l). Taken together, these results supported a protective role for FUNDC deficiency against CCl4-induced hepatocyte ferroptosis.
Fig. 4.
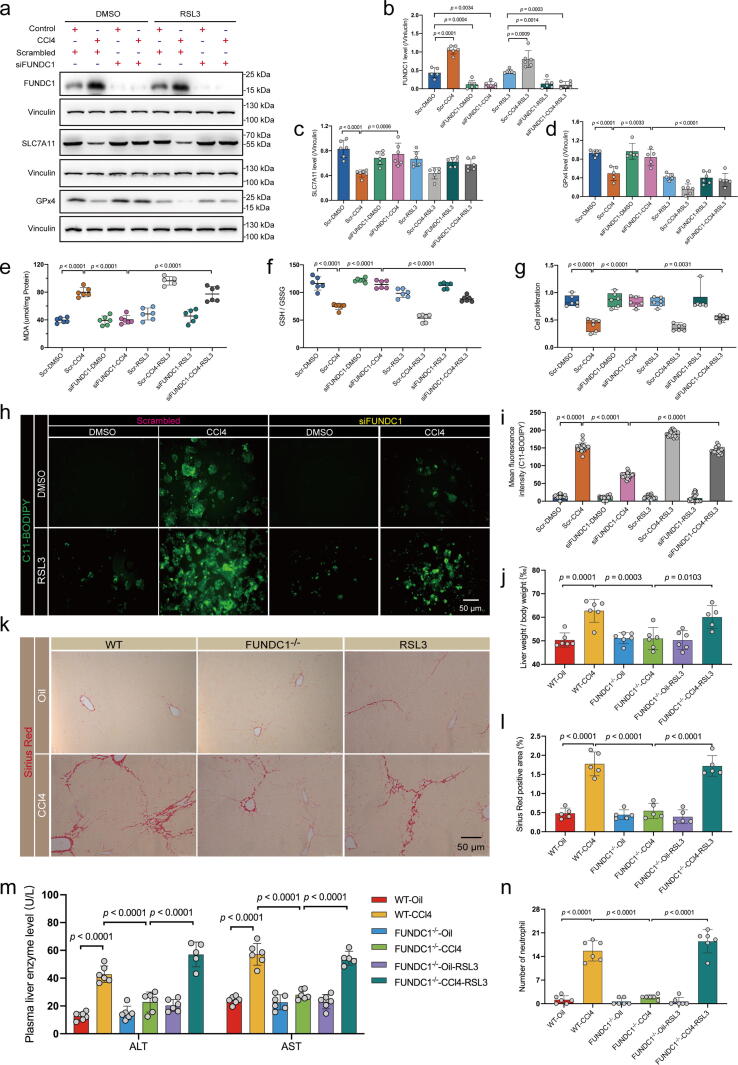
Effect of the GPx4 inhibitor RSL3 in CCl4-treated mice or E47 cells with FUNDC1 deficiency. WT or FUNDC1-/- mice were intraperitoneally injected with RSL3 (2.5 mg/kg) once a day for 10 days. E47 cells were transfected with scrambled or FUNDC1 siRNA for 48 hrs and were then treated DMSO or RSL3 (1 μM for 24 hrs) prior to exposure to CCl4 for 12 hrs. (a) Representative immunoblots of FUNDC1, SLC7A11 and GPx4 in E47 cells (Vinculin as the loading control); (b) Quantitated FUNDC1 level; (c) Quantitated SLC7A11 level; (d) Quantitated GPx4 level (n = 5–6/group); (e) Level of MDA (n = 5–6/group); (f) Ratio of GSH to GSSG (n = 6/group); (g) MTT assay of cell survival (n = 6/group); (h) Representative images of C11-BODIPY staining; (i) Quantified mean fluorescence intensity of C11-BODIPY images (n = 20/group); (j) Ratio of liver-to-body weight (n = 5–6/group); (k) Representative images of Sirius Red staining; (l) Quantified Fibrotic area of Sirus Red staining (n = 5–6 mice per group); (m) Level of plasma ALS and AST (n = 5–6/group); and (n) Quantified neutrophil number. Mean ± SEM (detailed statistical results shown in Table S4); Statistical significance was set at p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Activation of mitophagy evokes ferroptosis and reverses the protective effect of FUNDC1 deficiency.
Selective autophagy including mitophagy was recently suggested to interplay with ferroptosis in a ROS-dependent way [45]. Given the nature of FUNDC1 as a mitophagy receptor, mitophagy was monitored in CCl4-treated mice and E47 cells. Western blot analysis revealed upregulated mitophagy/autophagy markers including FUNDC1, BNIP3, and Parkin along with downregulated TIM23 (Fig. 2c, S5a-c), and more mitophagosomes were observed using TEM following CCl4 challenge in liver tissues (Fig. S5d-e), denoting hyperactivated mitophagy. Not surprisingly, FUNDC1 deletion partially dampened CCl4-induced rises in mitophagy with little discernible effect itself (Fig. S5a-e). To evaluate the main mitophagy components involved in response to CCl4 exposure, FUNDC1, Parkin and BNIP3 were individually knocked down, using siRNA transfection in vitro. Results from Western blot and Mito-Keima fluorescence showed that knockdown of Parkin mitigated CCl4-induced downregulation of TIM23 and upregulation of mito-LC3II, mito-p62 as well as Mito-Keima index (ratio of 561 nm to 457 nm fluorescence intensity of Mito-Keima) to a much greater extent compared with FUNDC1 or BNIP3 knockdown (Fig. S5f-m), denoting a rather prominent role for Pink1/Parkin as opposed to FUNDC1 or BNIP3 in mediating mitophagy in liver fibrosis. To explore possible contributing factors for downregulated GPx4, GPx4 levels were re-examined in the face of CCl4 challenge and/or FUNDC1 overexpression, in the presence or absence of the lysosomal inhibitor chloroquine (20 μM for 24 hrs) or the proteasomal inhibitor MG132 (10 μM for 6 hrs). Our result showed that FUNDC1 overexpression accentuated CCl4-induced GPx4 degradation, the effect of which was mitigated (restored to near control level) by inhibition of lysosomes but not proteasomes (Fig. S6a-b). To further examine the role of mitophagy in ferroptosis, oroxylin A (OA, 150 μM for 48 hrs), a mitophagy inhibitor targeting Pink1/Parkin- and BNIP3- mediated mitophagy [10], was added prior to CCl4 challenge in E47 cells. Our data revealed that oroxylin A inhibited mitophagy in CCl4-treated E47 cells with FUNDC1-OE as evidenced by downregulation of Parkin and BNIP3 as well as upregulation of TIM23, with subtle change in FUNDC1 (Fig. S7a-e). Interestingly, oroxylin A reversed FUNDC1 OE-induced deterioration in CCl4-evoked loss of GPx4 (compared with partial attenuation in SLC7A11) in E47 cells (Fig. S7f-g). The effects of Parkin knockdown on mitophagy- and ferroptosis-related proteins including TIM23, SLC7A11 and GPx4 were consistent with finding from oroxylin A treatment (Fig. S8a-g), although BNIP3 knockdown displayed minimal effect on levels of TIM23 and GPx4 (Fig. S8h-n). Oroxylin A also nullified FUNDC1 OE-induced exacerbation in CCl4-evoked MDA production and C11-BODIPY fluorescence and drop in GSH-GSSG in E47 cells (Fig. S7h-I, S7k-l). This is further supported by MTT cell survival assay (Fig. S7j), favoring a detrimental role for mitophagy in CCl4-evoked cell damage. MitoTracker and TMRM staining revealed that oroxylin A cancelled off the beneficial effect of FUNDC1 knockdown against CCl4-induced mitochondrial dysmorphology and MMP collapse (Fig. S7m-n, Fig. S6c). Together, these findings suggested that oroxylin A cancelled off FUNDC1 overexpression-elicited aggravating effect on CCl4-induced ferroptosis, indicating an obligatory regulatory role of mitophagy in ferroptosis.
Fig. 5.
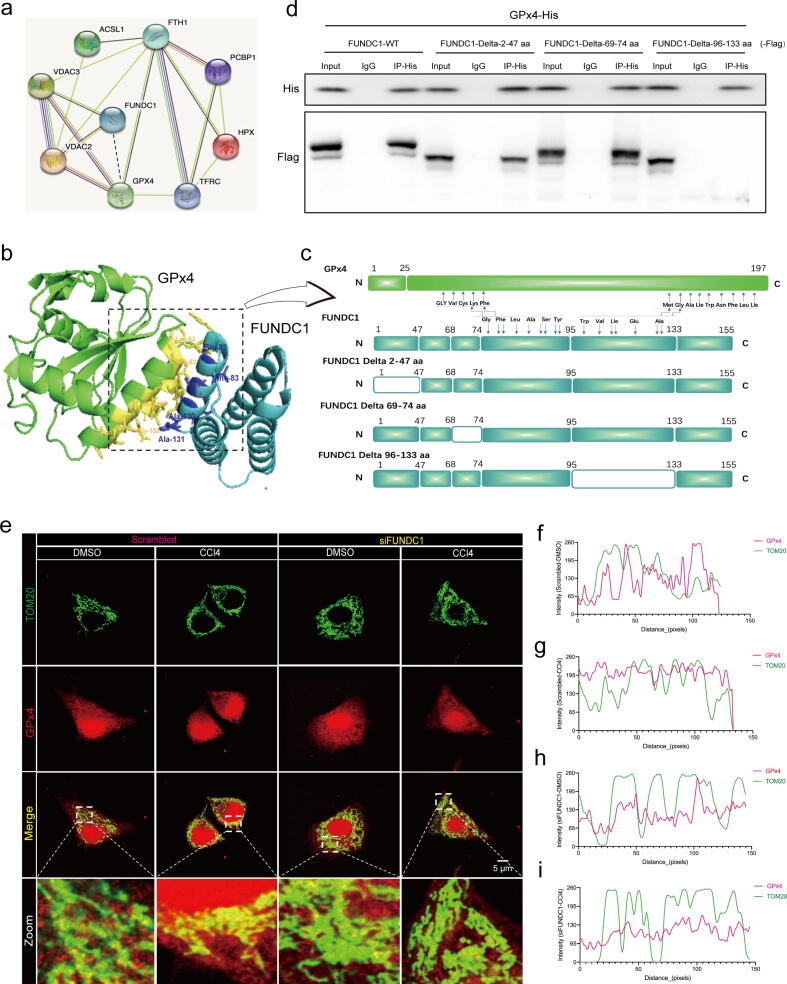
FUNDC1 directly interacts with GPx4 through its 96–133 amino acid domain and recruits GPx4 to mitochondria. (a) Co-IP mass spectrometry noted GPx4 as a potential interacting protein of FUNDC1; (b) Structure-based protein interaction interface analysis between FUNDC1 and GPx4. Cartoon represents the predicted FUNDC1-GPx4 complex structure, where the interaction hotspot residues are labeled; (c) Schematic diagram of domain deletion of FUNDC1 for Co-IP experiments, and predicted interaction sites on FUNDC1 with GPx4; (d) Co-IP analysis of GPx4 and FUNDC1 with or without different amino acid fragment deletion (FUNDC1 WT, FUNDC1 Delta-2–47 aa, FUNDC1 Delta-69–74 aa or FUNDC1 Delta-96–133 aa) in E47 cells; (e) Representative fluorescent images of co-localization of GPx4 (red) with TOM20 (green) in E47 cells transfected with scrambled or FUNDC1 siRNA for 48 hrs prior to exposure to DMSO or CCl4 for 12 hrs; and (f-i) Fluorescence intensity curve of GPx4 and TOM20 in co-localization images of Scramble-DMSO, Scramble-CCl4, siFUNDC1-DMSO and siFUNDC1-CCl4 groups. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
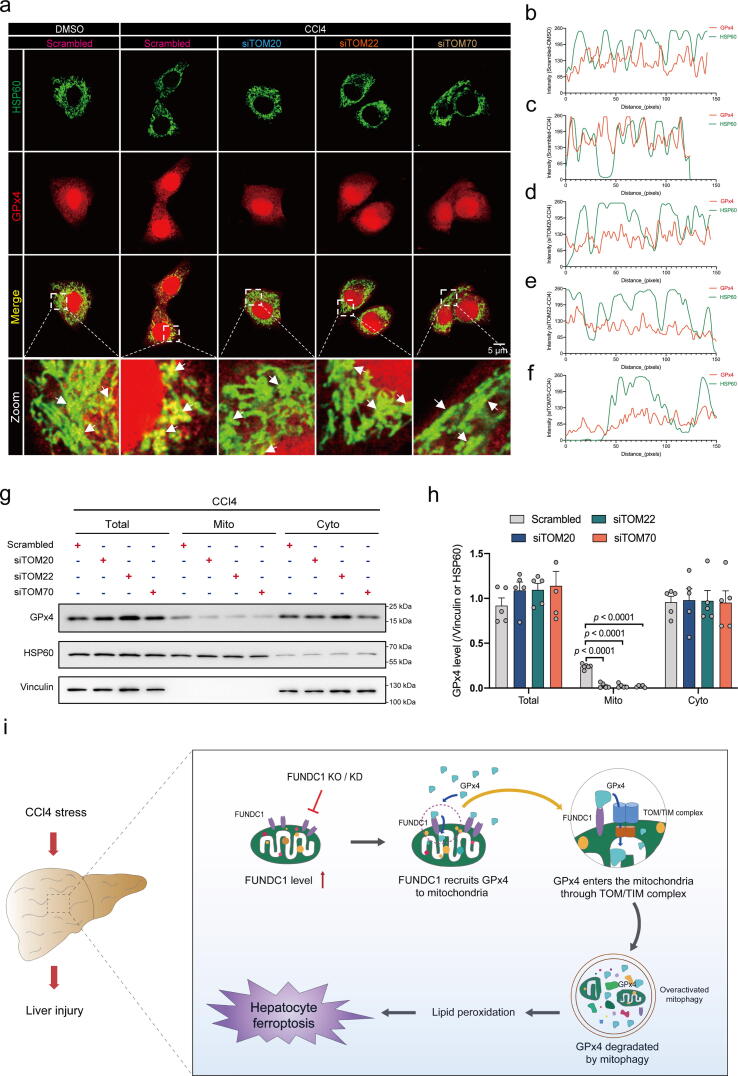
GPx4 enters the mitochondria through TOM/TIM complex. E47 cells were transferred with TOM20, TOM22 or TOM70 siRNA for 48 hrs prior to CCl4 exposure for 12 hrs. (a) Representative fluorescent images of co-localization of GPx4 (red) with HSP60 (green). White arrowheads point to the co-localization of GPx4 with HSP60 (yellow surrounded by green); (b-f) Fluorescence intensity curve of GPx4 and HSP60 in co-localization images of Scramble-DMSO, Scramble-CCl4, siTOM20-CCl4, siTOM22-CCl4 and siTOM70-CCl4 groups; (g) Representative immunoblots of GPx4 level in total (Vinculin as the loading control), mitochondrial (HSP60 as the loading control) and cytoplasmic (Vinculin as the loading control) compartments; (h) Quantitated GPx4 level in total, mitochondria and cytoplasm. (n = 5–6/group); and (i) Schematic diagram depicting proposed mechanism where FUNDC1 promoted hepatocyte injury in liver fibrosis via direct binding of GPx4 to facilitate its mitochondrial translocation through TOM/TIM complex, where GPx4 was degraded by mitophagy, leading to hepatic ferroptosis. Mean ± SEM (detailed statistical results shown in Table S4); Statistical significance was set at p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
FUNDC1 deficiency inhibits ferroptosis through a GPx4-dependent mechanism
Given that our aforementioned results noted a predominant response in GPx4 over SLC7A11 in oroxylin A-evoked action in E47 cells with FUNDC1 OE (Fig. S4a), we speculated that FUNDC1 may impact GPx4. A GPx4 inhibitor Ras-selective lethal small molecule 3 (RSL3, 1 uM for 24 hrs) was thus added to CCl4 treated- E47 cells with FUNDC1 knockdown [46]. Western blot data confirmed inhibition of RSL3 on GPx4 (Fig. 4a-d). Then, lipid content examination showed that RSL3 abrogated FUNDC1 knockdown-induced suppression of MDA production and C11-BODIPY fluorescence, and GSH depletion (Fig. 4e-f, h-i). RSL3 did not affect lipid deposition and peroxidation by itself (Fig. 4e-f, h-i). MTT assay indicated that RSL3 ablated FUNDC1 deficiency-evoked cell survival response in the face of CCl4 challenge (Fig. 4g). MitoTracker and TMRM fluorescence results showed that RSL3 pretreatment nullified FUNDC1 deficiency-offered beneficial effect against CCl4-induced mitochondrial damage (e.g., fractionated mitochondria and MMP collapse) (Fig. S9a-c). In addition, FUNDC1-/- mice were intraperitoneally injected with RSL3 (2.5 mg/kg, once a day for 10 days) [47]. RSL3 downregulated expression of GPx4 and SLC7A11 (with a more pronounced effect on GPx4 expression) (Fig. S9d, S9e). RSL3 negated the protective effect of FUNDC1 ablation on liver fibrosis as revealed by liver-to-body weight ratio as well as Sirius red staining (Fig. 4j-l), liver injury shown by levels of ALT and AST in serum (Fig. 4m), and liver inflammation evidenced by neutrophil infiltration staining (Fig. 4n). These results revealed that inhibition of GPx4 abolished the beneficial response offered by FUNDC1 deficiency in CCl4-induced cell damage.
FUNDC1 interacts with GPx4 and recruits GPx4 to mitochondria.
To discern how FUNDC1 interfered ferroptosis, possible interacting partners of FUNDC1 were examined using Mass spectrometry following immunoprecipitation (IP). FUNDC1 with Flag tag (FUNDC1-Flag) was expressed prior to immunoprecipitation with an anti-flag antibody in E47 cells. LC-MS analysis recovered GPx4 and several ferroptosis proteins using specific Co-IP with FUNDC1-Flag (Fig. 5a). In conjunction with changes of GPx4 response to the mitophagy inhibitor oroxylin A (Fig. S7a, S7g), it is possible that FUNDC1 plays a regulatory role in ferroptosis through interacting with GPx4. PRISM tool analysis revealed that FUNDC1 and GPx4 shared structural motif for direct binding. Prediction results from PRISM revealed that FUNDC1 and GPx4 may form a protein interaction interface (Fig. 5b). To confirm and further explore specific interaction between FUNDC1 and GPx4, truncated mutants of FUNDC1 were constructed with deletion of 2–47 amino acid fragment (Delta-2–47 aa), 69–74 amino acid fragment (Delta-69–74 aa) or 96–133 amino acid fragment (Delta-96–133 aa) (Fig. 5c). WT and truncated variants of flag-tagged FUNDC1 plasmids were transfected with His-tagged GPx4 plasmid in E47 cells. Co-IP assays showed that WT and truncated variants of FUNDC1 interacted with GPx4 with the exception of mutation of Delta-96–133 aa, suggesting an obligatory role for the 96–133 aa domain of FUNDC1 in the direct interaction with GPx4 (Fig. 5d). To explore if CCl4 challenge affected mitochondrial localization of GPx4 under intact or deficient FUNDC1, co-localization of GPx4 with mitochondria was determined using immunofluorescence staining. Figures manifested that CCl4 facilitated co-localization of GPx4 with mitochondria outer membrane protein TOM20, which was annulled by FUNDC1 deficiency (Fig. 5e, 5i), indicating a facilitating effect of CCl4 on mitochondrial localization of GPx4, possibly with the assistance of mitochondrial membrane protein FUNDC1. To discern the roles of FUNDC1 in mitophagy and GPx4 recruitment, we detected the influence of GPx4 on the binding affinity of FUNDC1 with LC3B motif (FUNDC1 regulates mitophagy through interacting with LC3B). As shown in Fig. S10, upregulation of GPx4 weakened the binding affinity of FUNDC1 for LC3B (Fig. S10a). This finding denotes that recruitment of GPx4 by FUNDC1 may possibly lessen FUNDC1-mediated mitophagy.
GPx4 enters the mitochondria through TOM/TIM complex.
What happened for GPx4 following its recruitment to mitochondria by FUNDC1? FUNDC1 was reported to interact with certain cytoplastic proteins, to foster their mitochondrial translocation through the TOM/TIM complex for further degradation using the degradation system within mitochondrial matrix, uncovering a new functional category for FUNDC1 [16]. Given that GPx4 was downregulated in response to CCl4 challenge, whether FUNDC1 contributed to GPx4 mitochondrial translocation through TOM/TIM complex for degradation was explored. The main members of TOM/TIM complex TOM20, TOM22, and TOM70 were knocked down using siRNA in E47 cells (Fig. S10b-d, Table. S3). Effect of knockdown of TOM20-, TOM22-, or TOM70 on GPx4 co-localization with mitochondrial matrix protein heat shock protein 60 (HSP60) was also examined using immunofluorescence. Our data suggested that knockdown of TOM20-, TOM22-, or TOM70 independently impeded GPx4-HSP60 co-localization (Fig. 6a-f). Western blot analysis of mitochondria from E47 cells with or without TOM20-, TOM22-, or TOM70-knockdown revealed that deficiency in any of the three proteins resulted in loss of GPx4 content in mitochondria under CCl4 insult (Fig. 6g, h) but not DMSO treatment (Fig. S10e, S10f). Also, GPx4 content in nuclei was not affected (Fig. S10g, S10h). These findings indicate that GPx4 may enter mitochondria through the TOM/TIM complex following its recruitment to mitochondrial surface by FUNDC1 upon CCl4 challenge, where unchecked mitophagy (mainly Pink1/Parkin-mediated) may contribute to GPx4 degradation in the face of CCl4 insult (Fig. 6i).
Discussion
Major findings from this study revealed elevated levels of mitophagy receptor FUNDC1 in liver tissues from liver fibrotic rodents and patients. In line with the GEO database analysis and subsequent validation of biological processes and cell signaling pathways involved in liver fibrosis, removal of FUNDC1 using an engineered model protected against CCl4-induced liver damage and ferroptosis, without obvious effect on cytochrome P450 enzyme activities – which mediates CCl4 metabolism and participates in CCl4 toxicity [40], [41], [42]. This is in line with the notion that induction of ferroptosis using erastin ablated FUNDC1 deficiency-induced hepatic protection. Elevated mitophagy was also noted in response to CCl4 challenge in vivo and in vitro. Interestingly, Pink1/Parkin but not FUNDC1 or BNIP3 was likely the primary mitophagic degradation machinery in our current experimental setting. Along the same line, inhibition of Pink1/Parkin-mediated mitophagy using oroxylin A or siRNA abolished the aggravating effect of FUNDC1 OE in CCl4 insult. These findings suggested an essential role for FUNDC1 in the manipulation of ferroptosis in CCl4-evoked cell damage, expanding its biological action beyond mitophagy (although such action is also mitophagy-depended). Furthermore, GPx4 inhibition using RSL3 revealed that inhibition of GPx4 countered FUNDC1 deficiency-offered benefit, demonstrating a seemingly obligatory role for GPx4 in FUNDC1-regulated ferroptosis. FUNDC1 overexpression accentuated CCl4-induced GPx4 degradation, the effect of which was mitigated by inhibition of lysosomes but not proteasomes, further consolidating mitophagy regulated ferroptosis in this circumstance. Results from Mass spectrometry and PRISM analysis tool verified a direct interaction between FUNDC1 and GPx4. Co-IP results further revealed an interaction between these two proteins, and noted a role for 96–133 amino acid domain of FUNDC1 for interaction with GPx4. Outcomes of immunofluorescence, and subcellular distribution detection using Western blot noted that GPx4 enters mitochondria through TOM/TIM complex. Downregulation of GPx4 and subsequently ferroptosis induction are believed to be evoked by a sequential event initiated from GPx4 recruitment to mitochondria by FUNDC1, entrance of GPx4 into mitochondria through the TOM/TIM complex, CCl4-induced unchecked mitophagy and Parkin mitophagy-induced GPx4 degradation. FUNDC1 appears to be the primary culprit for this cascade of pathological processes, favoring its unique role as a therapeutic target in ferroptosis and liver fibrosis.
FUNDC1 is a ubiquitin-independent mitophagy receptor regulating multiple chronic pathological processes including liver diseases [14], [18], [48], [49]. Here we reported that FUNDC1 deficiency protected hepatocytes against CCl4 toxicity in vivo and in vitro, as evidenced by mortality of mice, severity of liver fibrosis, and cell inflammation. Our study also found that CCl4 challenge led to hepatocyte ferroptosis, in line with an earlier report [24]. Besides, changes in FUNDC1 levels had a vital influence on hepatocyte ferroptosis in CCl4-evoked liver damage. We went on to reveal that FUNDC1 directly interacted with GPx4, the important negative regulatory molecule for ferroptosis, via 96–133 aa domain on FUNDC1, leading to downregulation of GPx4. The interaction of FUNDC1 with GPx4 suppressed the binding affinity of FUNDC1 with LC3B, revealing a complex interplay between FUNDC1-mediated recruitment of GPx4 and FUNDC1-mediated mitophagy. These results not only unveiled a likely role for FUNDC1 in CCl4-induced liver toxicity, but also identified a role for ferroptosis in FUNDC1-mediated hepatic regulation, thus offering new insights for FUNDC1 function beyond mitophagy.
Although it is beyond the scope of the current study, possible contribution from disturbed redox balance may contrite to CCl4 challenge-evoked ferroptosis. It was reported that CCl4 insult provoked overwhelmed product of polyunsaturated fatty acid peroxidation hydroxynonenals (HNE), cholesterol oxidation and decomposition, en route to disrupted redox homeostasis to prompt ferroptosis (50). Several essential cellular defense pathways including sirtuin, nuclear factor erythroid-derived 2 (Nrf2) and related cell signaling pathways appear to integrate adaptive stress responses in the combat against proinflammatory response to offset ferroptosis [51], [52]. GPx4 is an essential antioxidant enzyme to resist ferroptosis through reduction of hydroperoxide-associated membranes [33], [53]. GPx4 is found in cytosol, mitochondria, and nuclei of all tissues. Data from our work revealed that CCl4 challenge fostered mitochondrial recruitment of mito-GPx4 evidenced by elevated co-localization of GPx4 with the mitochondrial matrix protein HSP60, and up-regulated mito-GPx4 content. Other than retardation against lipid peroxidation, GPx4 exerts a mitochondrial protective role by regulating mitochondrial apoptosis and mitochondrial energy metabolism [33], [54]. Therefore, in pathological conditions with evident mitochondrial damage including mtROS production and increased mitochondrial permeability, GPx4 is prone to recruitment and translocation into mitochondria though the subsequent fate remains uncertain [33], [35]. We uncovered for the first time that GPx4 mitochondrial recruitment could be undertaken by mitochondrial membrane protein FUNDC1. More intriguingly, our work exhibited that disruption of the TOM/TIM complex by TOM20-, TOM22- or TOM70-knowdown [16] nullified co-localization of GPx4 with HSP60. Thus, GPx4 enters mitochondria through TOM/TIM complex, a common protein-translocating machinery in mitochondrial outer membranes (MOM) and mitochondrial inner membranes (MIM). Last but not least, GPx4 is believed to be degraded by unchecked mitophagy in liver fibrosis, resulting in ferroptosis. Our data revealed that knockdown of Parkin exerted a much greater effect against CCl4-evoked mitophagy compared with FUNDC1 or BNIP3. This is further supported by the mitophagy inhibitor oroxylin A (mainly targeting Parkin and BNIP3)-induced rescue of unchecked mitophagy, GPx4 degradation and ferroptosis. These findings revealed that FUNDC1-mediated translocation of GPx4 into mitochondria followed by mitophagic degradation may determine the ferroptotic fate in the face of liver fibrosis, suggesting a role for GPx4 as a unique target in CCl4-induced liver injury. However, GEO analysis revealed a decrease in mRNA level of GPx4, indicating contribution from transcriptional factors beyond GPx4 degradation in GPx4 transcription during liver fibrosis. Further study is warranted to elucidate transcriptional regulation of GPx4.
Ferroptosis interplays with autophagy, with crosstalk between mitophagy and ferroptosis drawing much recent attention 50(55). Although mitophagy promotes cell survival in general through removing damaged or long-lived mitochondria and proteins, it leads to cell death under certain circumstances. Our study indicated that unchecked mitophagy (detected by mitophagy related proteins in vivo and in vitro, number of mitophagsome observed using TEM in vivo, and MitoKeima in vitro) may contribute to hepatocyte ferroptosis in the face of CCl4 challenge, consistent with earlier findings that mitophagy promotes lipid peroxidation-induced ferroptosis through degradation of cargo contents within the cell [50], [51], [55], [56]. In our hands, it may be speculated that mitophagy-mediated degradation of GPx4 (as evidenced by our in vitro data of chloroquine) may be one possible underlying reason for mitophagy-driven regulation of ferroptosis. Hence, we suspect that clinical benefits brought by inhibition of mitophagy might involve, at least in part, mitophagy-mediated regulation in ferroptosis.
Conclusions
In conclusion, findings from our current study have provided new insights that FUNDC1 exerts a deleterious effect towards liver fibrosis evoked by CCl4. Mechanically, FUNDC1 directly binds with and recruits GPx4 to mitochondria to foster the GPx4 mitochondrial entrance through the TOM/TIM complex for degradation by unchecked mitophagy (Fig. 6i). To this end, specific targeting of FUNDC1, the FUNDC1-GPx4 axis, or mitophagy should be considered as a promising strategy in the therapeutics of hepatic ferroptosis and liver fibrosis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by grants from the Program of Shanghai Academic Research Leader No.20XD1420900, the National Natural Science Foundation of China No.82200419 and No.82130011, and Shanghai Municipal Key Clinical Specialty No.shslczdzk01701.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.02.012.
Contributor Information
Jun Ren, Email: jren_aldh2@outlook.com, jren@uw.edu.
Yingmei Zhang, Email: zhang.yingmei@zs-hospital.sh.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Mercado-Gómez M., Lopitz-Otsoa F., Azkargorta M., Serrano-Maciá M., Lachiondo-Ortega S., Goikoetxea-Usandizaga N., et al. Multi-omics integration highlights the role of ubiquitination in CCl4-induced liver fibrosis. Int J Mol Sci. 2020;21(23) doi: 10.3390/ijms21239043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyoğlu D., Idle J.R. The metabolomic window into hepatobiliary disease. J Hepatol. 2013;59(4):842–858. doi: 10.1016/j.jhep.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg D, Ditah IC, Saeian K, Lalehzari M, Aronsohn A, Gorospe EC, et al. Changes in the prevalence of hepatitis c virus infection, nonalcoholic steatohepatitis, and alcoholic liver disease among patients with cirrhosis or liver failure on the waitlist for liver transplantation. Gastroenterology. 2017;152(5):1090-9.e1. [DOI] [PMC free article] [PubMed]
- 4.Lemmer A., VanWagner L.B., Ganger D. Assessment of advanced liver fibrosis and the risk for hepatic decompensation in patients with congestive hepatopathy. Hepatology. 2018;68(4):1633–1641. doi: 10.1002/hep.30048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J.H., Ku S.K., Cho I.L.J., Lee J.H., Na C.S., Ki S.H. Neoagarooligosaccharide protects against hepatic fibrosis via inhibition of TGF-β/Smad signaling pathway. Int J Mol Sci. 2021;22(4) doi: 10.3390/ijms22042041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ajoolabady A., Aslkhodapasandhokmabad H., Aghanejad A., Zhang Y., Ren J. Mitophagy receptors and mediators: therapeutic targets in the management of cardiovascular ageing. Ageing Res Rev. 2020;62 doi: 10.1016/j.arr.2020.101129. [DOI] [PubMed] [Google Scholar]
- 7.Kang J.W., Hong J.M., Lee S.M. Melatonin enhances mitophagy and mitochondrial biogenesis in rats with carbon tetrachloride-induced liver fibrosis. J Pineal Res. 2016;60(4):383–393. doi: 10.1111/jpi.12319. [DOI] [PubMed] [Google Scholar]
- 8.Ke P.Y. Mitophagy in the Pathogenesis of Liver Diseases. Cells. 2020;9(4):831. doi: 10.3390/cells9040831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma X., Luo Q., Zhu H., Liu X., Dong Z., Zhang K., et al. Aldehyde dehydrogenase 2 activation ameliorates CCl4 -induced chronic liver fibrosis in mice by up-regulating Nrf2/HO-1 antioxidant pathway. J Cell Mol Med. 2018;22:3965–3978. doi: 10.1111/jcmm.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao J., Wang J., Xu Y., Guo Q., Sun Y., Liu J., et al. CDK9 inhibition blocks the initiation of PINK1-PRKN-mediated mitophagy by regulating the SIRT1-FOXO3-BNIP3 axis and enhances the therapeutic effects involving mitochondrial dysfunction in hepatocellular carcinoma. Autophagy. 2021;1–19 doi: 10.1080/15548627.2021.2007027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li R., Xin T., Li D., Wang C., Zhu H., Zhou H. Therapeutic effect of Sirtuin 3 on ameliorating nonalcoholic fatty liver disease: The role of the ERK-CREB pathway and Bnip3-mediated mitophagy. Redox Biol. 2018;18:229–243. doi: 10.1016/j.redox.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuang Y., Ma K., Zhou C., Ding P., Zhu Y., Chen Q., et al. Structural basis for the phosphorylation of FUNDC1 LIR as a molecular switch of mitophagy. Autophagy. 2016;12(12):2363–2373. doi: 10.1080/15548627.2016.1238552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., Zhu P., Li R., Ren J., Zhou H. Fundc1-dependent mitophagy is obligatory to ischemic preconditioning-conferred renoprotection in ischemic AKI via suppression of Drp1-mediated mitochondrial fission. Redox Biol. 2020;30 doi: 10.1016/j.redox.2019.101415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou H., Zhu P., Wang J., Toan S., Ren J. DNA-PKcs promotes alcohol-related liver disease by activating Drp1-related mitochondrial fission and repressing FUNDC1-required mitophagy. Signal Transduct Target Ther. 2019;4:56. doi: 10.1038/s41392-019-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W., Li Y., Siraj S., Jin H., Fan Y., Yang X., et al. FUN14 domain-containing 1-mediated mitophagy suppresses hepatocarcinogenesis by inhibition of inflammasome activation in mice. Hepatology. 2019;69(2):604–621. doi: 10.1002/hep.30191. [DOI] [PubMed] [Google Scholar]
- 16.Li Y., Xue Y., Xu X., Wang G., Liu Y., Wu H., et al. A mitochondrial FUNDC1/HSC70 interaction organizes the proteostatic stress response at the risk of cell morbidity. Embo j. 2019;38(3) doi: 10.15252/embj.201798786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bento G., Shafigullina A.K., Rizvanov A.A., Sardão V.A., Macedo M.P., Oliveira P.J. Urine-derived stem cells: applications in regenerative and predictive medicine. Cells. 2020;9(3) doi: 10.3390/cells9030573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren J., Sun M., Zhou H., Ajoolabady A., Zhou Y., Tao J., et al. FUNDC1 interacts with FBXL2 to govern mitochondrial integrity and cardiac function through an IP3R3-dependent manner in obesity. Sci Adv. 2020;6(38) doi: 10.1126/sciadv.abc8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W.H., Lin C.C., Wu J., Chao P.Y., Chen K., Chen P.H., et al. The Hippo Pathway Effector YAP Promotes Ferroptosis via the E3 Ligase SKP2. Mol Cancer Res. 2021 doi: 10.1158/1541-7786.MCR-20-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Swanda R.V., Nie L., Liu X., Wang C., Lee H., et al. mTORC1 couples cyst(e)ine availability with GPX4 protein synthesis and ferroptosis regulation. Nat Commun. 2021;12(1):1589. doi: 10.1038/s41467-021-21841-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang L., Kon N., Li T., Wang S.J., Su T., Hibshoosh H., et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520(7545):57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park T.J., Park J.H., Lee G.S., Lee J.Y., Shin J.H., Kim M.W., et al. Quantitative proteomic analyses reveal that GPX4 downregulation during myocardial infarction contributes to ferroptosis in cardiomyocytes. Cell Death Dis. 2019;10(11):835. doi: 10.1038/s41419-019-2061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y., Jiang L., Wang H., Shen Z., Cheng Q., Zhang P., et al. Hepatic transferrin plays a role in systemic iron homeostasis and liver ferroptosis. Blood. 2020;136(6):726–739. doi: 10.1182/blood.2019002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ajoolabady A., Aslkhodapasandhokmabad H., Libby P., Tuomilehto J., Lip G.Y.H., Penninger J.M., et al. Ferritinophagy and ferroptosis in the management of metabolic diseases. Trends Endocrinol Metab. 2021;32(7):444–462. doi: 10.1016/j.tem.2021.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Gao M, Yi J, Zhu J, Minikes AM, Monian P, Thompson CB, et al. Role of mitochondria in ferroptosis. Mol Cell 2019;73(2):354-63.e3. [DOI] [PMC free article] [PubMed]
- 27.Nežić L., Škrbić R., Amidžić L., Gajanin R., Milovanović Z., Nepovimova E., et al. Protective effects of simvastatin on endotoxin-induced acute kidney injury through activation of tubular epithelial cells' survival and hindering cytochrome C-mediated apoptosis. Int J Mol Sci. 2020;21(19) doi: 10.3390/ijms21197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei S., Qiu T., Yao X., Wang N., Jiang L., Jia X., et al. Arsenic induces pancreatic dysfunction and ferroptosis via mitochondrial ROS-autophagy-lysosomal pathway. J Hazard Mater. 2020;384 doi: 10.1016/j.jhazmat.2019.121390. [DOI] [PubMed] [Google Scholar]
- 29.Bersuker K., Hendricks J.M., Li Z., Magtanong L., Ford B., Tang P.H., et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seiler A., Schneider M., Förster H., Roth S., Wirth E.K., Culmsee C., et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. CellMetab. 2008;8(3):237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Aldrovandi M., Conrad M. Ferroptosis: the Good, the Bad and the Ugly. Cell Res. 2020;30(12):1061–1062. doi: 10.1038/s41422-020-00434-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W., Wang Y., Zhang C., Huang Y., Yu J., Shi L., et al. Maresin1 Protect Against Ferroptosis-Induced Liver Injury Through ROS Inhibition and Nrf2/HO-1/GPX4 Activation. Front Pharmacol. 2022;13 doi: 10.3389/fphar.2022.865689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang H., Yoo S.E., Na R., Walter C.A., Richardson A., Ran Q. Short form glutathione peroxidase 4 is the essential isoform required for survival and somatic mitochondrial functions. J Biol Chem. 2009;284(45):30836–30844. doi: 10.1074/jbc.M109.032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cole-Ezea P., Swan D., Shanley D., Hesketh J. Glutathione peroxidase 4 has a major role in protecting mitochondria from oxidative damage and maintaining oxidative phosphorylation complexes in gut epithelial cells. Free Radic Biol Med. 2012;53(3):488–497. doi: 10.1016/j.freeradbiomed.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 35.To TL, Cuadros AM, Shah H, Hung WHW, Li Y, Kim SH, et al. A Compendium of Genetic Modifiers of Mitochondrial Dysfunction Reveals Intra-organelle Buffering. Cell. 2019;179(5):1222-38.e17. [DOI] [PMC free article] [PubMed]
- 36.González-Fernández B., Sánchez D.I., Crespo I., San-Miguel B., de Urbina J.O., González-Gallego J., et al. Melatonin Attenuates Dysregulation of the Circadian Clock Pathway in Mice With CCl4-Induced Fibrosis and Human Hepatic Stellate Cells. Front Pharmacol. 2018;9:556. doi: 10.3389/fphar.2018.00556. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Chen Q., Cederbaum A.I. Cytotoxicity and apoptosis produced by cytochrome P450 2E1 in Hep G2 cells. Mol Pharmacol. 1998;53(4):638–648. doi: 10.1124/mol.53.4.638. [DOI] [PubMed] [Google Scholar]
- 38.Wang S., Tao J., Chen H., Kandadi M.R., Sun M., Xu H., et al. Ablation of Akt2 and AMPKalpha2 rescues high fat diet-induced obesity and hepatic steatosis through Parkin-mediated mitophagy. Acta Pharm Sin B. 2021;11(11):3508–3526. doi: 10.1016/j.apsb.2021.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X.X., Zheng Q.C., Wang Y., Zhang H.X. Theoretical insights into the reductive metabolism of CCl4 by cytochrome P450 enzymes and the CCl4-dependent suicidal inactivation of P450. Dalton Trans. 2014;43(39):14833–14840. doi: 10.1039/c4dt02065k. [DOI] [PubMed] [Google Scholar]
- 40.Luo J., Zhang P., Zhao T., Jia M., Yin P., Li W., et al. Golgi Apparatus-Targeted Chondroitin-Modified Nanomicelles Suppress Hepatic Stellate Cell Activation for the Management of Liver Fibrosis. ACS Nano. 2019;13(4):3910–3923. doi: 10.1021/acsnano.8b06924. [DOI] [PubMed] [Google Scholar]
- 41.Han J., He Y., Zhao H., Xu X. Hypoxia inducible factor-1 promotes liver fibrosis in nonalcoholic fatty liver disease by activating PTEN/p65 signaling pathway. J Cell Biochem. 2019;120(9):14735–14744. doi: 10.1002/jcb.28734. [DOI] [PubMed] [Google Scholar]
- 42.Jia D., Wang Y.Y., Wang P., Huang Y., Liang D.Y., Wang D., et al. SVIP alleviates CCl4-induced liver fibrosis via activating autophagy and protecting hepatocytes. Cell Death Dis. 2019;10(2):71. doi: 10.1038/s41419-019-1311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai T., Liang R., Zhu R., Wang W., Zhou L., Sun Y. MicroRNA-214-3p enhances erastin-induced ferroptosis by targeting ATF4 in hepatoma cells. J Cell Physiol. 2020;235(7–8):5637–5648. doi: 10.1002/jcp.29496. [DOI] [PubMed] [Google Scholar]
- 44.Qi W., Li Z., Xia L., Dai J., Zhang Q., Wu C., et al. LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress during erastin-induced ferroptosis in HepG2 hepatocellular carcinoma cells. Sci Rep. 2019;9(1):16185. doi: 10.1038/s41598-019-52837-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu M., Fan Y., Li D., Han B., Meng Y., Chen F., et al. Ferroptosis inducer erastin sensitizes NSCLC cells to celastrol through activation of the ROS-mitochondrial fission-mitophagy axis. Mol Oncol. 2021 doi: 10.1002/1878-0261.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sui X., Zhang R., Liu S., Duan T., Zhai L., Zhang M., et al. RSL3 Drives Ferroptosis Through GPX4 Inactivation and ROS Production in Colorectal Cancer. Front Pharmacol. 2018;9:1371. doi: 10.3389/fphar.2018.01371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qi J., Kim J.W., Zhou Z., Lim C.W., Kim B. Ferroptosis Affects the Progression of Nonalcoholic Steatohepatitis via the Modulation of Lipid Peroxidation-Mediated Cell Death in Mice. Am J Pathol. 2020;190(1):68–81. doi: 10.1016/j.ajpath.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 48.Wu W., Tian W., Hu Z., Chen G., Huang L., Li W., et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15(5):566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu W., Lin C., Wu K., Jiang L., Wang X., Li W., et al. FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. Embo j. 2016;35(13):1368–1384. doi: 10.15252/embj.201593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gárate-Rascón M., Recalde M., Jimenez M., Elizalde M., Azkona M., Uriarte I., et al. Splicing Factor SLU7 Prevents Oxidative Stress-Mediated Hepatocyte Nuclear Factor 4α Degradation, Preserving Hepatic Differentiation and Protecting From Liver Damage. Hepatology. 2021;74(5):2791–2807. doi: 10.1002/hep.32029. [DOI] [PubMed] [Google Scholar]
- 51.Calabrese V., Guagliano E., Sapienza M., Mancuso C., Butterfield D.A., Stella A.M. Redox regulation of cellular stress response in neurodegenerative disorders. Ital J Biochem. 2006;55(3–4):263–282. [PubMed] [Google Scholar]
- 52.Calabrese V., Cornelius C., Dinkova-Kostova A.T., Calabrese E.J., Mattson M.P. Cellular stress responses, the hormesis paradigm, and vitagenes: novel targets for therapeutic intervention in neurodegenerative disorders. Antioxid Redox Signal. 2010;13(11):1763–1811. doi: 10.1089/ars.2009.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dabkowski E.R., Williamson C.L., Hollander J.M. Mitochondria-specific transgenic overexpression of phospholipid hydroperoxide glutathione peroxidase (GPx4) attenuates ischemia/reperfusion-associated cardiac dysfunction. Free Radic Biol Med. 2008;45(6):855–865. doi: 10.1016/j.freeradbiomed.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 55.Xie Y., Li J., Kang R., Tang D. Interplay between lipid metabolism and autophagy. Front Cell Dev Biol. 2020;8:431. doi: 10.3389/fcell.2020.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Basit F., van Oppen L.M., Schöckel L., Bossenbroek H.M., van Emst-de Vries S.E., Hermeling J.C., et al. Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells. Cell Death Dis. 2017;8(3):e2716. doi: 10.1038/cddis.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.