Abstract
Introduction
Alzheimer’s Disease (AD) is complex and novel approaches are urgently needed to aid in diagnosis. Blood is frequently used as a source for biomarkers; however, its complexity prevents proper detection. The analytical power of metabolomics, coupled with statistical tools, can assist in reducing this complexity.
Objectives
Thus, we sought to validate a previously proposed panel of metabolic blood-based biomarkers for AD and expand our understanding of the pathological mechanisms involved in AD that are reflected in the blood.
Methods
In the validation cohort serum and plasma were collected from 25 AD patients and 25 healthy controls. Serum was analysed for metabolites using nuclear magnetic resonance (NMR) spectroscopy, while plasma was tested for markers of neuronal damage and AD hallmark proteins using single molecule array (SIMOA).
Results
The diagnostic performance of the metabolite biomarker panel was confirmed using sparse-partial least squares discriminant analysis (sPLS-DA) with an area under the curve (AUC) of 0.73 (95% confidence interval: 0.59–0.87). Pyruvic acid and valine were consistently reduced in the discovery and validation cohorts. Pathway analysis of significantly altered metabolites in the validation set revealed that they are involved in branched-chain amino acids (BCAAs) and energy metabolism (glycolysis and gluconeogenesis). Additionally, strong positive correlations were observed for valine and isoleucine between cerebrospinal fluid p-tau and t-tau.
Conclusions
Our proposed panel of metabolites was successfully validated using a combined approach of NMR and sPLS-DA. It was discovered that cognitive-impairment-related metabolites belong to BCAAs and are involved in energy metabolism.
Supplementary information
The online version contains supplementary material available at 10.1007/s11306-023-02078-8.
Keywords: Alzheimer, Metabolites, Biomarker, Serum, Nuclear magnetic resonance, Single molecule array
Background
Neurodegenerative diseases, such as Alzheimer’s Disease (AD), account for a significant proportion of mortality, morbidity, and healthcare cost globally (Mattsson-Carlgren et al. 2020). Clinical examination alone is inadequate for guiding diagnosis, prognosis, and monitoring progress in research, clinical practice, and drug development. Imaging and biomarkers can aid diagnostics by providing an objective indicator of the underlying pathology. In the case of AD, this includes structural, functional, and molecular imaging, as well as measurements of signature proteins in the cerebrospinal fluid (CSF), i.e. amyloid-β (Aβ) and tau isoforms (Livingston et al. 2017; G. M. McKhann et al. 2011). In certain clinical situations, CSF levels of neurofilament light (Nf-L) protein, a marker for neuronal injury, have been used to diagnose neurodegenerative disorders (Khalil et al. 2018). However, with these current diagnostic methods, several drawbacks have to be accounted for, limiting their applicability as first-line screening tools. Although technological advances could increase the precision of these methods, their expense and lack of patient compliance prevent this from occurring. In addition, advanced scanning methods, including positron emission tomography, are expensive and less accessible for general practitioners (O’Brien and Herholz 2015), while CSF collection through a lumbar puncture is invasive (De Almeida et al. 2011). A blood sample may provide a matrix that could outweigh the drawbacks of the currently used biomarkers to diagnose patients with AD. With the benefits of blood being a biofluid in close contact with every organ in the body, its composition could reflect the potential state of the surrounding organ (Jacobs et al. 2005). The blood-brain barrier (BBB) separates the central nervous system (CNS) from the periphery, allowing only gaseous exchange, together with small ions, water- and small liposoluble-molecules to pass (Zlokovic 2011). However, during AD pathogenesis, the BBB becomes permeable (Baird et al. 2015), potentially enabling the identification of neuronal metabolites in blood samples.
Even though blood provides a non-invasive biological matrix for investigating disease pathology, its complexity impedes the findings of potential new biomarkers. The omics-era has aided in the realisation of the need to explore such complex samples, with metabolomics being one of the more recent members of the omics family (Hampel et al. 2016). Metabolomics covers the study of all metabolites in a cell, organ, or organism. Metabolites are small molecules < 1,500 Da and comprise amino acids, lipids, peptides, vitamins, etc. (Lamichhane et al. 2018), and are endpoints of the regulations at the genetic, transcript, and protein levels. Thus, small alterations of upstream molecules could substantially affect the concentration of a metabolite (J. Nielsen and Oliver 2005). Not only can disease progression cause metabolic perturbations, but environmental factors, treatments, and nutrition also play a role (Stringer et al. 2016). As for clinical applications, metabolic pathways have been shown to be evolutionarily conserved across species, thus bridging the gap between animal studies and human clinical trials (Wilkins and Trushina 2018). Nuclear magnetic resonance (NMR) spectroscopy is among the most informative techniques for studying metabolomics (Wishart 2008), and is able to efficiently analyse and detect hundreds of small molecules in a single measurement in human samples, including plasma and serum (Wang et al. 2013). Essentially, all metabolites present their own unique and reproducible NMR signature and thus can be used to explore metabolic processes and screen for the presence of known metabolites (Song et al. 2019). In addition, NMR is also non-destructive and more informative than other techniques, such as mass spectrometry; however, it lacks sensitivity and requires a larger quantity of sample material (Stringer et al. 2016; Wilkins and Trushina 2018).
Although biomarker studies contribute to the global search for a solution to the growing problem of the aging population, the literature demonstrates that replication efforts for many promising biomarker findings have failed (Voyle et al. 2015). Thus, the present study aimed to validate suggested metabolite biomarkers presented in our previous discovery study (J. E. Nielsen et al. 2021) and to provide additional information on metabolic perturbations associated with cognitive impairment. Using NMR-based metabolomics, the serum metabolic signatures from patients with mild to moderate AD were compared to those of cognitively healthy individuals. Furthermore, we validated our initial model using a larger validation cohort by incorporating the commonly identified metabolites and supplementing our findings with additional metabolic perturbations.
Methods
Study demographics
In our previous discovery study, 20 participants were enrolled, with 10 healthy controls and 10 patients with mild to modate AD. For this validation study, we increased the number of participants to 50, with 25 in each group either diagnosed with mild to moderate AD or as healthy controls. All subjects were Caucasian. The patients were recruited from the Department of Neurology, Aalborg University Hospital. Recruitment was performed consecutively at the time of diagnosis for the patients and prior to starting their treatment regimen. The diagnosis was based on the following criteria; the International Classification of Diseases and Related Health Problems 10th Edition (ICD10) (WHO n.d.), and the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA) (G. McKhann et al. 1984). Paraclinical measurements comprised of Mini-Mental State Examination (MMSE), Addenbrooke’s Cognitive Examination (ACE), Functional Activities Questionnaire (FAQ), as well as Aβ, phospho-tau (p-tau), and total-tau (t-tau) measured in CSF. The paraclinical measurements were included when deemed necessary due to diagnostic uncertainty. Age- and sex-related donors were recruited from the blood bank at Aalborg University Hospital to serve as a comparison group for AD patients. In Denmark, blood donors are healthy unpaid volunteers without any apparent illnesses. Inclusion criteria for blood donors were an age > 65 years old and completion of a standard blood donor questionnaire describing physical and mental health, such as experiencing memory impairment, fatigue, or chest pain. Prior to inclusion in the study, all participants signed a written consent form. The study was approved by the local North Denmark Region Committee on Health Research Ethics (N-20150010) and conducted according to the Declaration of Helsinki.
Sample collection and processing
Blood samples were drawn from study participants and processed as described in a previous study (Ellegaard Nielsen et al. 2020). Briefly, blood was collected from the median cubital vein using a 21-gauge needle in 10 mL clot activator tubes (BD Vacutainer, UK) and also 4 mL Ethylenediaminetetraacetic acid (EDTA) tubes (Vacuette, Greiner Bio-One, Austria). After sample collection, the blood was centrifuged twice at 2500 × g for 15 min at room temperature to obtain serum and plasma. After each centrifugation step, serum and plasma samples were aspirated to approximately 1 cm above the buffy coat or pellet. Finally, serum and plasma samples were aliquoted, snap-frozen using liquid nitrogen, and stored at − 80 °C until further analyses.
Routine analyses
Organ markers were routinely measured in serum samples to ensure that none of the study participants had co-morbidities. The clinical biochemistry markers measured were alanine transaminase, albumin, carbamide, cholesterol, creatinine, C-reactive protein, glucose, high and low-density lipoprotein (HDL and LDL, respectively), lactate dehydrogenase (LDH), and triglycerides using the Alinity ci-series (Abbott, Chicago, IL, USA) and haemoglobin using either XN-9000 (Sysmex Europe SE, Germany) or Hb 201 DM (Hemocue AB, Sweden).
Single Molecule Array
Aβ40, Aβ42, glial fibrillary acidic protein (GFAP), Nf-L, and p-tau181 were measured in EDTA plasma using the respective commercially available kits; Neurology 4-Plex E and P-Tau181 (Quanterix©, Billerica, MA, USA) by Single Molecule Array (SIMOA®) HD-X Analyzer. The analyses were performed according to the manufacturer’s instructions. In addition, the manufacturer’s commercial controls were applied for quality control.
Nuclear magnetic resonance spectroscopy
Serum samples were initially thawed for one hour and then carefully diluted 1:1 dilution with sodium phosphate buffer (0.075 M, pH 7.4, 20% D2O in H2O, 6 mM NaN3, 4.6 mM 3-(trimethylsilyl)-2,2,3,3-tetradeuteropropanoic acid (TSP-d4)) and aliquoted into 5 mm NMR tubes. NMR spectra were recorded using a Bruker Avance Neo 600 MHz spectrometer attached to a BBI probe (Bruker Biospin GmbH, Rheinstetten, Germany). IconNMR (Topspin 4.1.1, Bruker Biospin GmbH, Rheinstetten, Germany) and Samplejet autosampler (Bruker Biospin GmbH, Rheinstetten, Germany) were used for sample handling and data acquisition. One-dimensional nuclear Overhauser effect (1D-NOESY) spectra, together with Carr-Purcell-Meiboom-Gill (CPMG), were recorded at 310 K using parameters for acquisition from Dona et al. (Dona et al. 2014). For the 1D-NOESY spectra, 96k data points were recorded, with 30 ppm spectral width. In contrast, CPMG spectra were recorded with 72k data points and a spectral width of 20 ppm. For both experiments, 32 scans with water suppression (25 Hz) during relaxation delay (4 s) and mixing time (NOESY, 10 ms) were used for recording. Free induction decays were Fourier transformed after artificial zero fillings up to 128k data points and 0.3 Hz line broadening. The spectra were automatically zero-order phase corrected. In accordance with the manufacturer, B.I.Methods (Bruker Biospin GmbH, Rheinstetten, Germany), reference samples were routinely measured and processed in automation for temperature calibration, water suppression determination, and external quantitative referencing. B.I.Quant-PS™ 2.0 (Bruker Biospin GmbH, Rheinstetten, Germany) was used to automatically quantify metabolites. A labeled spectrum with expanded regions for peak intensity comparison can be found in Supplementary file 1.
Data analysis
For the validation cohort, 40 metabolites were identified using NMR. Information from the discovery cohort can be found in the discovery study (J. E. Nielsen et al. 2021). Metabolites were filtered for ≥ 70% valid values in at least one group before statistical analyses were conducted. Prior to statistical analysis, metabolites from the discovery- and validation-cohorts were adjusted for age and sex using a linear model. Also, prior to validation of the initial metabolic signature using multivariate data analysis, data were auto-scaled and mean-centered for metabolites common in both the discovery and validation cohorts.
Three models; Random Forest, Extreme Gradient Boosting (XGBoost), and sparse-partial least squared discriminant analysis (sPLS-DA), were tested and estimated by their performance using the following parameters; Area under the curve (AUC) and 95% confidence interval (CI) were reported to indicate the sensitivity and specificity of the model, together with the accuracy, positive predictive value (PPV), negative predictive value (NPV), and selected number of important metabolites. The Random Forest model performance was estimated by the out-of-bag error rate and optimal number of features was selected using the randomForest v4.7-1.1 and Boruta v8.0.0 R packages. XGBoosting was performed using the R package xgboost v1.7.5.1, with performance estimated by root mean squared error (RMSE). Optimal number of features were ranked according to importance score, and selected if importance score was > 0.1. As previously described (J. E. Nielsen et al. 2021), the sPLS-DA model was build with a 5-fold cross-validation (CV) repeated 100 times using the mixOmics v6.20.0 R package. The optimal number of selected features was estimated using the classification error rate. For visual purpose scores plot for sample groupings, loadings plot for weighted importance of selected metabolites, and receiver operating characteristic (ROC) curve are presented for the most optimal model.
Data were assessed for normality by Shapiro-Wilk test and histograms, and compared between the groups using a Student’s t-test, presented as mean ± standard deviations (SD). Correction for multiple comparison was also provided using the Benjamini-Hochberg false-discovery rate (FDR) corrected p-value. Nf-L and GFAP were corrected for age using a linear model (Vågberg et al. 2015). A significance level of p < 0.05 was chosen. Fold changes (FC) between groups were also calculated for the metabolites, using FC = (MetaboliteAD – MetaboliteCon) / MetaboliteCon. Correlations between important metabolites, selected by multivariate data analysis, and clinical data were investigated using Pearson’s ρ, with only the significant correlations presented. Data analysis and graphical representations were conducted using R version 4.2.2.
Network analysis was performed using the MetScape (version 3.1.3) App in CytoScape (version 3.9.1). The network was based on KEGG IDs from significantly altered metabolites between AD patients and healthy individuals. Raw NMR data for the validation cohort, clinical data, and input data for the network analysis can be found in Supplementary files 2, 3, and 4, respectively.
Results
Characteristics of study participants
The biochemical parameters, clinical test results for cognitive performances, corresponding clinical parameters, and SIMOA measurements for both study groups have been summarised in Table 1. Briefly, the majority of the biochemical measurements were within the standard reference intervals. A few, but significant differences were also observed between the AD patients and cognitively healthy individuals, including a slightly higher age (p = 0.00001), higher LDH levels (p = 0.03), and lower glucose levels in the AD patient group (p = 0.01). Patients who required additional cognitive testing and paraclinical measurements were identified, where AD patients presented with low MMSE (20.0 ± 4.5) and ACE (58.0 ± 16.5) scores and a high FAQ (11.8 ± 6.2) score, whereas paraclinical tests demonstrated elevated levels of CSF tau, p-tau (81.7 ± 25.0 ng/L) and t-tau (520.4 ± 102.4 ng/L), and decreased levels of CSF Aβ (682.8 ± 216.3 ng/L) for some of the patients, indicating extracellular tau accumulation and intracellular Aβ build-up. Plasma measurements of markers for neuronal injury and AD hallmark proteins were included as additional clinical information. Generally, AD patients had significantly higher plasma levels of Aβ40 (p = 0.002), GFAP (p = 0.01), Nf–L (p = 0.04), and p-tau181 (p = 0.00005) than healthy individuals; however, Aβ42 and Aβ42/Aβ40 ratio did not differ between the two groups (p = 0.5) and (p = 0.06), respectively. For GFAP and Nf-L unadjusted values for mean and SD are shown in Table 1.
Table 1.
Characteristics of study participants
| Units | Con (n = 25) | AD (n = 25) | p-value | Reference interval | |
|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | ||||
| Demographics | |||||
| Age | Years | 66.6 ± 1.3 | 75.7 ± 8.2 | 0.00001 | – |
| Male/female | n | 16 / 9 | 15 / 10 | – | – |
| Ethnicity | – | Caucasian | Caucasian | – | – |
| Biochemical measurements | |||||
| ALAT | U/L | 26.3 ± 8.6 | 22.3 ± 11.6 | 0.17 | 10.0–50.0 |
| Albumin | g/L | 41.0 ± 1.9 | 41.5 ± 1.9 | 0.37 | 34–45 |
| Carbamide | mmol/L | 5.8 ± 1.3 | 5.7 ± 1.5 | 0.77 | 3.1–8.1 |
| Cholesterol | mmol/L | 5.4 ± 0.9 | 5.5 ± 1.1 | 0.88 | 4.2–8.5 |
| Creatinine | µmol/L | 79.0 ± 10.2 | 83.4 ± 14.5 | 0.22 | 45–105 |
| CRP | mg/L | 1.9 ± 1.4 | 2.2 ± 2.9 | 0.57 | < 8 |
| Glucose | mmol/L | 6.4 ± 1.7 | 5.4 ± 0.9 | 0.01 | 4.2–7.8 |
| Haemoglobin | mmol/L | 8.8 ± 0.7 | 8.5 ± 1.0 (n = 15) | 0.45 | 7.3–10.5 |
| HDL | mmol/L | 1.5 ± 0.3 | 1.6 ± 0.4 | 0.35 | 0.7–1.9 |
| LDL | mmol/L | 3.2 ± 0.8 | 3.3 ± 0.9 | 0.71 | 2.2–5.7 |
| LDH | U/L | 170.2 ± 31.2 | 192.1 ± 38.7 | 0.03 | 105–255 |
| Triglycerides | mmol/L | 1.5 ± 0.8 | 1.3 ± 0.8 | 0.34 | 0.6–3.9 |
| Clinical parameters | |||||
| MMSE | – | – | 20.0 ± 4.5 | – | – |
| ACE | – | – | 58.0 ± 16.5 (n = 21) | – | – |
| FAQ | – | – | 11.8 ± 6.2 (n = 21) | – | – |
| CSF Aβ | ng/L | – | 682.8 ± 216.3 (n = 9) | – | > 500 |
| CSF p-tau | ng/L | - | 81.7 ± 25.0 (n = 9) | – | < 61 |
| CSF t-tau | ng/L | – | 520.4 ± 102.4 (n = 9) | – | < 450 |
| SIMOA | |||||
| Aβ40 | pg/mL | 95.1 ± 10.2 | 108.7 ± 17.4 | 0.002 | – |
| Aβ42 | pg/mL | 5.3 ± 1.0 | 5.6 ± 1.3 | 0.5 | – |
| Aβ42/Aβ40 | – | 0.06 ± 0.009 | 0.05 ± 0.009 | 0.06 | – |
| GFAP | pg/mL | 88.6 ± 32.8 | 247.1 ± 277.9 | 0.01 | – |
| Nf-L | pg/mL | 12.5 ± 4.4 | 36.9 ± 24.5 | 0.04 | – |
| p-tau181 | pg/mL | 1.8 ± 0.8 | 3.1 ± 1.3 | 0.00005 | – |
Demographics data of study participants together with biochemical measurements, cognitive test results, paraclinical measurements, and SIMOA measurements. Abbreviations; Aβ – Amyloid-β, ACE – Addenbrooke’s Cognitive Examination, AD – Alzheimer’s Disease, ALAT – Alanine transaminase, p-tau – Phosphorylated tau, CRP – C-reactive protein, CSF – Cerebrospinal fluid, FAQ – Functional Activities Questionnaire, GFAP – Glial fibrillary acidic protein, HDL – High-density lipoprotein, LDH – Lactate dehydrogenase, LDL – Low-density protein, MMSE – Mini-Mental State Examination, Nf-L – Neurofilament light, SD – Standard deviation, SIMOA – Single molecule array, t-tau – Total tau.
Validation of metabolic signatures for Alzheimer’s Disease diagnostics
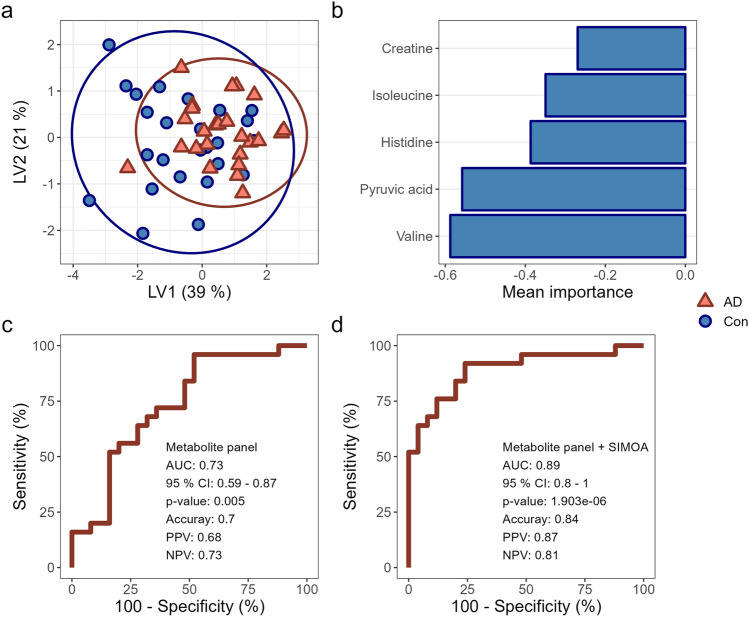
To validate the metabolic signature identified in our discovery study, NMR spectroscopy was applied to measure the concentration of serum metabolites in our validation study cohort. Three prediction models were tested for their performance based on AUC, accuracy, PPV, and NPV. These models included sPLS-DA, random forest, and XGBoost (Supplementary file 5). Based on these criteria, sPLS-DA showed the highest performance with five selected metabolites building the model (pyruvic acid, valine, histidine, isoleucine, and creatine), while random forest performed the second best with four selected metabolites (histidine, valine, pyruvic acid, and creatine), and lastly the XGBoost with four metabolites (histidine, pyruvic acid, valine, and tyrosine). Thus, sPLS-DA was selected as our data's most optimal validation model.
The validation model showed a small overlap between the patient and control groups, as seen in the scores plot of the measured serum samples (Fig. 1a). Based on the validated model, five metabolites significantly contribute to sample grouping, accounting for 39% of the group variation (Fig. 1b). Consequently, the model had an AUC performance of 0.73 (95% CI = 0.59–0.87) for discriminating AD patients from cognitively healthy individuals (Fig. 1c). Furthermore, the model had an accuracy = 0.70, PPV = 0.68, and NPV = 0.73, indicating its diagnostic value. Interestingly, when adding the significantly altered proteins (Aβ40, GFAP, Nf-L, and p-tau181) to the validation model, improved its diagnostic performance, resulting in an AUC of 0.89 (95% CI = 0.80–1.00) with an accuracy of 0.84, PPV of 0.87, and NPV of 0.81 (Fig. 1d).
Fig. 1.
Validation of metabolic signature for cognitive impairment. a Scores plot for sparse-partial least squared discriminant analysis (sPLS-DA), with each score representing a sample. b Loadings plot for selected metabolites representing their mean importance for sample grouping reflected in the scores plot. The colour-coding of the bars indicates their importance for the corresponding group. c Receiver operating characteristics (ROC) curve indicates the ability of the model to distinguish the groups. Together with the ROC curve is the area under the curve (AUC) with the presented 95% CI, accuracy, PPV, and NPV. d ROC curve of selected metabolites combined with significantly altered markers of neurodegeneration (Aβ40, GFAP, Nf-L, p-tau181) showing an improved diagnostic efficacy, also presented with AUC and the 95% CI, accuracy, PPV, and NPV. Abbreviations; AD – Alzheimer’s Disease, AUC – Area under the curve, CI – Confidence interval, Con – Healthy controls, LV – Latent variable, NPV – Negative predictive value, PPV – Positive predictive value.
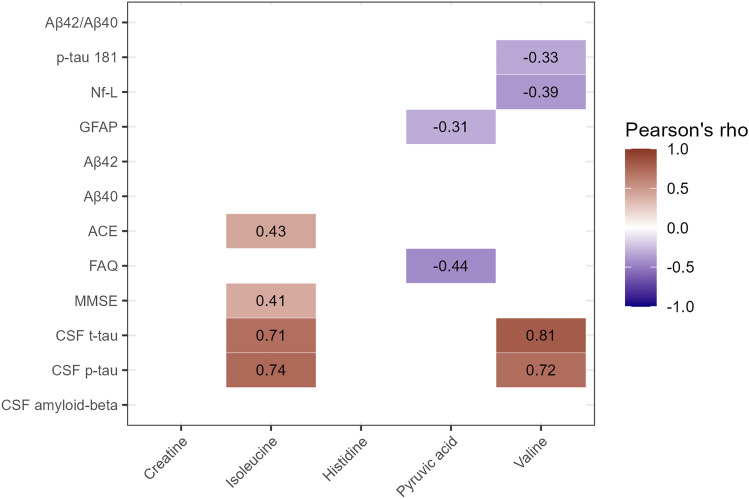
Furthermore, the selected panel of five metabolites was correlated against the clinical data and markers of neuronal damage to determine their possible association with neurodegenerative diseases (Fig. 2). Especially isoleucine and valine exhibited a strong significant positive correlation with CSF levels of p-tau and t-tau with a Pearson’s correlation of 0.74 and 0.71 for isoleucine and 0.72 and 0.81 for valine, respectively. Pyruvic acid showed a moderate negative correlation with cognitive scoring test FAQ (ρ = − 0.44), while isoleucine showed moderate positive correlations with cognitive scoring tests MMSE (ρ = 0.41) and ACE (ρ = 0.43).
Fig. 2.
Correlogram of metabolites of interest and clinical parameters. Only correlations shown to be significant are included. The colour of the square indicates if the correlation is positive (red) or negative (blue), and the intensity of the colour corresponds to the level of the correlation. Abbreviations; ACE – Addenbrooke’s cognitive examination, CSF – Cerebrospinal fluid, FAQ – Functional activities questionnaire, GFAP – Glial fibrillary acidic protein, MMSE – Mini-mental state examination, Nf-L – Neurofilament light, p-tau – Phospho-tau, t-tau – Total-tau.
Two of the metabolites measured in both the discovery and validation data sets were significantly altered when comparing healthy and diseased individuals (Table 2). These two metabolites exhibited similar changes in both the discovery and validation studies. Furthermore, tyrosine and leucine also presented as significantly changed in the validation cohort, however, not in the discovery cohort, while histidine presented as significantly changed only in the discovery cohort. The unadjusted mean ± SD values are shown in Table 2.
Table 2.
Common significantly altered metabolites
| Metabolite [mmol/L] |
Con | AD | FC | p-value | FDR | ||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||||
| Discovery study | |||||||
| Valine | 0.118 | 0.019 | 0.092 | 0.011 | − 0.2 | 0.007 | 0.07 |
| Histidine | 0.037 | 0.002 | 0.032 | 0.002 | − 0.1 | 0.009 | 0.07 |
| Pyruvic acid | 0.032 | 0.007 | 0.026 | 0.004 | − 0.2 | 0.03 | 0.19 |
| Validation study | |||||||
| Tyrosine | 0.071 | 0.016 | 0.051 | 0.011 | − 0.3 | 0.006 | 0.09 |
| Valine | 0.275 | 0.052 | 0.214 | 0.040 | − 0.2 | 0.01 | 0.09 |
| Pyruvic acid | 0.118 | 0.031 | 0.079 | 0.027 | − 0.3 | 0.02 | 0.09 |
| Leucine | 0.111 | 0.035 | 0.078 | 0.017 | − 0.3 | 0.02 | 0.09 |
Two metabolites were dysregulated in serum samples between cognitively affected and healthy individuals in both the discovery and validation studies, sorted according to p-value. Abbreviations; AD – Alzheimer’s Disease, Con – Healthy controls, FC – Fold change, SD – Standard deviation
Metabolic alterations in the validation cohort
To extrapolate novel metabolic information, serum samples from the validation study were examined for significantly altered metabolites between the groups. This brought the total number of significantly different metabolites between the groups to five. Four of these metabolites were previously identified as significant in the discovery cohort, only valine was previously reported as significantly altered (Table 3). The unadjusted mean ± SD values are shown in Table 3. Significance testing of unadjusted metabolites can be found in Supplementary file 6. Potential significant differences due to age could be removed by adjustment, however, this difference could also be due to the AD patients on average being older or having different metabolite concentrations due to the disease state, thereby eliminating this relevant difference.
Table 3.
Significantly altered metabolites in the validation cohort
| Metabolite (ppm, multiplicity) [mmol/L] |
Con | AD | FC | p-value | FDR | ||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||||
| Tyrosine (7.19, app. d) | 0.071 | 0.016 | 0.051 | 0.011 | − 0.3 | 0.006 | 0.13 |
| Valine (1.02, d) | 0.275 | 0.052 | 0.214 | 0.040 | − 0.2 | 0.01 | 0.13 |
| Lysine (3.03, t) | 0.220 | 0.043 | 0.175 | 0.036 | − 0.2 | 0.02 | 0.13 |
| Pyruvic acid (2.36 s) | 0.118 | 0.031 | 0.079 | 0.027 | − 0.3 | 0.02 | 0.13 |
| Leucine (0.94(s) & 0.96(d)) | 0.111 | 0.035 | 0.078 | 0.017 | − 0.3 | 0.02 | 0.14 |
Significantly altered metabolites measured in serum samples comparing cognitively affected with healthy individuals, sorted according to the p-value. Abbreviations; AD – Alzheimer’s Disease, Con – Healthy controls, FC – Fold change, FDR – False-discovery rate, SD – Standard deviation.
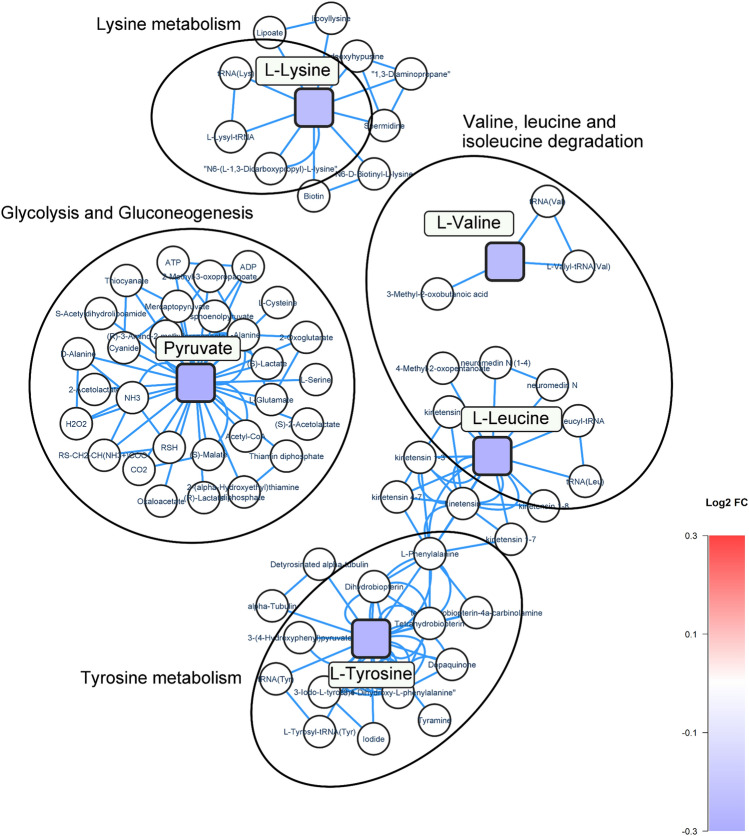
A network analysis was performed to investigate biological pathways for the significantly altered metabolites related to cognitive impairment (Fig. 3). Pyruvate involved in glycolysis and gluconeogenesis (log2 FC = − 0.33, p-value = 0.02) was the most reduced metabolite in relation to AD. In addition, the metabolic pathways of Lysine, Tyrosine, and branch-chained amino acids (BCAAs); valine and leucine, were also modified. This validation study identified and confirmed changes to BCAA metabolism previously found in the discovery study; leucine (log2 FC = 0.3, p-value = 0.02) and valine (log2 FC = − 0.2, p-value = 0.01).
Fig. 3.
Network analysis of dysregulated pathways related to cognitive impairment. Square nodes represent altered metabolites identified in the validation study, and circular nodes represent metabolites as part of the pathways not identified in the study. Colour of the square nodes represents the log2 FC value of the corresponding metabolite, according to alterations between the groups, with blue indicating a downregulation, and red indicating an upregulation. Metabolites are mapped according to their KEGG IDs
Discussion
In this study, we examined serum-derived metabolites associated with cognitive impairment in patients with mild to moderate AD compared to healthy individuals. The primary objective was to validate the significance of a panel of metabolites previously identified in the discovery study (J. E. Nielsen et al. 2021). The secondary objective was to add novel information not previously identified.
As previously mentioned, reproducibility is one of the more significant obstacles in biomarker studies (Mattsson-Carlgren et al. 2020). The authors of the referred study raised several crucial points to improve the reproducibility of future biomarker studies for neurodegenerative disease. These aspects range from cohort-related factors to independent validation. In the presented study, we have sought to comply with these recommendations, including; 1) consecutively recruitment of study participants to enroll more heterogenous groups, such that multiple factors attenuating the effect of the biomarker to avoid overestimation, 2) avoiding confounding factors, such as the presence of co-morbidities affecting measured biomarker levels, 3) performing validation in a separate validation cohort, as internal CV, i.e. when splitting a single cohort, has shown to fail when replicating the models in independent cohorts (Burnham et al. 2014; Voyle et al. 2015), possibly due to systematic bias between the groups, 4) reporting not only the overall measure of performance, such as the AUC but also including the parameters PPV, NPV, etc., and lastly 5) metabolomics was performed in a different lab in the validation study compared to the discovery study, thereby also accounting for between-lab variability. (Mattsson-Carlgren et al. 2020)
We examined the serum metabolome of our study participants through NMR spectroscopy in combination with multivariate data analysis. Three models were tested using the discovery and validation data sets. Overall, these models primarily selected the same metabolites, indicating the relevance of these metabolites for distinguishing cognitively impaired persons from healthy individuals. The commonly selected metabolites included pyruvic acid, valine, and histidine. Pyruvic acid, or pyruvate, is the end-product of glycolysis and the substrate for mitochondrial adenosine triphosphate (ATP) synthesis. The nervous system is vulnerable to alterations in pyruvate metabolism due to the high ATP demand (Gray et al. 2014), which is used to maintain neuronal activity and homeostasis of the extracellular space and to defend against oxidative stress (Bak et al. 2006; García-Nogales et al. 2003). In contrast, to our findings, increased levels of pyruvate have been observed in CSF of AD patients (Parnetti et al. 1995), but similar alterations were identified in blood samples from patients with Parkinson’s disease (Ahmed et al. 2009). As stated in the discovery study, valine and histidine are well-studied amino acids with respect to AD pathology (J. E. Nielsen et al. 2021). Consequently, the results of the present study further validate their importance related to AD. In accordance with previous findings identifying decreased levels of valine in serum (Xiong et al. 2022) and CSF and positive correlations between CSF-valine and MMSE score, researchers have continued investigating this particular amino acid in relation to AD (Vignoli et al. 2020). Valine was identified as a potential marker for predicting the transition from mild cognitive impairment to AD (Xiong et al. 2022). Histidine is an essential amino acid (Kim and Kim 2020), and a known scavenger of hydroxyl radicals, part of the reactive oxygen species (Cai et al. 1995). Using a cell model for anti-aging effects, increased proliferation and neurogenesis, as well as up-regulation of anti-oxidant enzymes, have been demonstrated as positive effects of histidine (Kim and Kim 2020). As mentioned earlier, BBB breakdown occurs during AD pathogenesis with the potential presence of neuronal metabolites in the circulatory system. Thus, the metabolic shift between healthy and diseased individuals observed in this study could also be due to the presence of neuronal-derived metabolites entering the bloodstream in AD subjects, while not being present in healthy individuals due to an intact BBB.
As an additional clinical characteristic parameter, we included measurements of established non-disease specific markers of neurological damage (GFAP and Nf-L), as well as hallmark targets of AD (Aβ40, Aβ42, Aβ42/ Aβ40, and p-tau181) measured in blood, as the literature strongly implicates their diagnostic performance and significance in relation to neurological disease. (Chatterjee et al. 2022; Smirnov et al. 2022). Overall, the present study confirms previous findings; however, we found significantly elevated levels of Aβ40 and no difference in Aβ42 concentrations and Aβ42/Aβ40 ratios. This may be due to underlying cardiovascular conditions, such as hypertension, ischemic heart disease, and cardio-protective medications, which have been shown to influence plasma concentrations of Aβ. (Janelidze et al. 2016). However, this indifference could also be associated to the cohort size of the study needing more power to confirm a significant difference. In the biochemical measurements, glucose and LDH levels were significantly different between the control group and the patients, but both groups had levels within the normal range. Interestingly, reduced glucose utilization has been shown in AD brains (Kumar et al. 2022), and this could also explain the observed significantly lower pyruvate concentration. Although, blood donors are encouraged to have eaten prior to blood donation, which also could be the reason for the significance in blood glucose levels.
Even though our results confirmed important findings and contributed to the search for valid blood-based biomarkers to aid in diagnosing AD, it is essential to note the limitations of our study. First, although the patient group was clinically confirmed to have AD, not all patients underwent neuropsychological testing or had CSF proteins measured because their physician deemed it unnecessary for the patient's diagnosis. Secondly, to thoroughly verify our metabolite panel as a diagnostic tool for AD, it would be necessary to test its accuracy against other neurodegenerative diseases and different stages of AD. Thirdly, we discovered a significant age difference between our study groups, with AD patients being, on average, older than the control group. Unfortunately, it is not possible to recruit older blood donors, however, adjustment for age was performed on metabolite values prior to statistical analsysis. Fourthly, adding CSF samples to a study of the metabolome in relation to cognitive impairment can strengthen biomarker panels identified in the peripheral system. Lastly, both p-value and FDR corrected values were reported to minimize type II errors, however, multivariate statistics were also applied to encompass the overall information in the data, including covariance and correlation between metabolites.
Our findings validated the significance of the identified metabolite biomarker panel from the discovery study in distinguishing cognitively healthy individuals from patients with cognitive impairment. In addition, we evaluated various models to validate the performance of our panel, finding sPLS-DA to be the best fit. Lastly, new insights into disruptions in energy metabolism were uncovered.
Conclusions
In the current study, we validated our blood-based biomarker model derived from a discovery study consisting of five metabolites; pyruvic acid, valine, histidine, isoleucine, and creatine. The novel information provided by the validation cohort confirmed the involvement of significantly altered metabolites in the BCAAs metabolism. Moreover, metabolic changes in the energy metabolism were observed. Combining the proposed metabolite biomarker panel with neurodegenerative markers in plasma increased the diagnostic value. Although our validation yielded very intriguing results, the diagnostic performance of this panel of metabolic markers must also be assessed against other types of neurodegenerative diseases and various stages of AD progression.
Supplementary information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge the assistance provided by Mette Ullits Thomsen, Helle Dalsgaard Holst, Helle Hylander, and Mette Jespersgaard for their invaluable help in enrollment and blood sample collection from AD patients and blood donors. The NMR Center at DTU and the Villum Foundation are also acknowledged for providing access to the 800 MHz NMR spectrometer used for the discovery cohort. The NMR experiments on the validation cohort were performed at the MR Core Facility, Norwegian University of Science and Technology (NTNU). MR Core facility is funded by the Faculty of Medicine and Health sciences at NTNU and Central Norway Regional Health Authority. Sebastian Krossa at MR Core Facility is acknowledged for preparing supplementary figure.
Author contributions
JEN processed and prepared the samples, analysed and interpreted the data, performed the statistical analysis, and wrote the original draft for the manuscript. TA performed the NMR spectroscopy on the validation cohort. CHG performed the NMR spectroscopy on the discovery cohort. DAO performed the SIMOA analysis on the validation cohort. KV enrolled patients in the study and provided clinical data. JSM provided review and editing of the manuscript. SRK conceptualized and supervised the study, and was a major contributor in writing the manuscript. SP conceptualized and supervised the study, and was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Funding
Open Access funding provided by the Qatar National Library. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data Availability
All data generated or analysed during this study are included in this published article (and its supplementary information files).
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
The study was approved by the local North Denmark Region Committee on Health Research Ethics (N-20150010) and conducted according to the Declaration of Helsinki.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmed SS, Santosh W, Kumar S, Christlet HTT. Metabolic profiling of Parkinson’s disease: Evidence of biomarker from gene expression analysis and rapid neural network detection. Journal of Biomedical Science. 2009;16(1):63. doi: 10.1186/1423-0127-16-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird AL, Westwood S, Lovestone S. Blood-based proteomic biomarkers of Alzheimer’s disease pathology. Frontiers in Neurology. 2015 doi: 10.3389/fneur.2015.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak LK, Schousboe A, Waagepetersen HS. The glutamate/GABA-glutamine cycle: Aspects of transport, neurotransmitter homeostasis and ammonia transfer. Journal of Neurochemistry. 2006;98(3):641–653. doi: 10.1111/j.1471-4159.2006.03913.x. [DOI] [PubMed] [Google Scholar]
- Burnham SC, Faux NG, Wilson W, Laws SM, Ames D, Bedo J, et al. A blood-based predictor for neocortical Aβ burden in Alzheimer’s disease: results from the AIBL study. Molecular Psychiatry. 2014;19(4):519–526. doi: 10.1038/mp.2013.40. [DOI] [PubMed] [Google Scholar]
- Cai Q, Takemura G, Ashraf M. Antioxidative properties of histidine and its effect on myocardial injury during ischemia/reperfusion in isolated rat heart. Journal of Cardiovascular Pharmacology. 1995;25(1):147–155. doi: 10.1097/00005344-199501000-00023. [DOI] [PubMed] [Google Scholar]
- Chatterjee P, Pedrini S, Ashton NJ, Tegg M, Goozee K, Singh AK, et al. Diagnostic and prognostic plasma biomarkers for preclinical Alzheimer’s disease. Alzheimer’s & Dementia. 2022;18(6):1141–1154. doi: 10.1002/alz.12447. [DOI] [PubMed] [Google Scholar]
- De Almeida SM, Shumaker SD, LeBlanc SK, Delaney P, Marquie-Beck J, Ueland S, et al. Incidence of post-dural puncture headache in research volunteers. Headache. 2011;51(10):1503–1510. doi: 10.1111/j.1526-4610.2011.01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dona AC, Jiménez B, Schäfer H, Humpfer E, Spraul M, Lewis MR, et al. Precision high-throughput proton NMR spectroscopy of human urine, serum, and plasma for large-scale metabolic phenotyping. Analytical Chemistry. 2014;86(19):9887–9894. doi: 10.1021/ac5025039. [DOI] [PubMed] [Google Scholar]
- Ellegaard Nielsen J, Sofie Pedersen K, Vestergård K, Maltesen G, Christiansen R, Lundbye-Christensen GS, et al. Novel blood-derived extracellular vesicle-based biomarkers in Alzheimer’s disease identified by proximity extension assay. Biomedicines. 2020 doi: 10.3390/biomedicines8070199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Nogales P, Almeida A, Bolaños JP. Peroxynitrite Protects Neurons against nitric oxide-mediated apoptosis. Journal of Biological Chemistry. 2003;278(2):864–874. doi: 10.1074/jbc.M206835200. [DOI] [PubMed] [Google Scholar]
- Gray LR, Tompkins SC, Taylor EB. Regulation of pyruvate metabolism and human disease. Cellular and Molecular Life Sciences. 2014;71(14):2577–2604. doi: 10.1007/s00018-013-1539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel H, O’Bryant SE, Castrillo JI, Ritchie C, Rojkova K, Broich K, et al. Precision medicine - The Golden gate for detection, treatment and prevention of Alzheimer’s disease. The Journal of Prevention of Alzheimer’s Disease. 2016;3(4):243–259. doi: 10.14283/jpad.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG, Smith RD. Utilizing human blood plasma for proteomic biomarker discovery. Journal of Proteome Research. 2005;4(4):1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- Janelidze S, Stomrud E, Palmqvist S, Zetterberg H, van Westen D, Jeromin A, et al. Plasma β-amyloid in Alzheimer’s disease and vascular disease. Scientific Reports. 2016;6(1):26801. doi: 10.1038/srep26801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil M, Teunissen CE, Otto M, Piehl F, Sormani MP, Gattringer T, et al. Neurofilaments as biomarkers in neurological disorders. Nature Reviews Neurology. 2018;14(10):577–589. doi: 10.1038/s41582-018-0058-z. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kim Y. L-histidine and L-carnosine exert anti-brain aging effects in D-galactose-induced aged neuronal cells. Nutrition Research and Practice. 2020;14(3):188. doi: 10.4162/nrp.2020.14.3.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Kim SH, Bishayee K. Dysfunctional glucose metabolism in Alzheimer’s disease onset and potential pharmacological interventions. International Journal of Molecular Sciences. 2022;23(17):9540. doi: 10.3390/ijms23179540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane, S., Sen, P., Dickens, A. M., Hyötyläinen, T., & Orešič, M. (2018). Chapter Fourteen - An Overview of Metabolomics Data Analysis: Current Tools and Future Perspectives. In J. Jaumot, C. Bedia, & R. Tauler (Eds.), Data Analysis for Omic Sciences: Methods and Applications (Vol. 82, pp. 387–413). Elsevier. 10.1016/bs.coac.2018.07.001 .
- Livingston G, Sommerlad A, Orgeta V, Costafreda SG, Huntley J, Ames D, et al. Dementia prevention, intervention, and care. Lancet. 2017;390(10113):2673–2734. doi: 10.1016/S0140-6736(17)31363-6. [DOI] [PubMed] [Google Scholar]
- Mattsson-Carlgren N, Palmqvist S, Blennow K, Hansson O. Increasing the reproducibility of fluid biomarker studies in neurodegenerative studies. Nature Communications. 2020;11(1):6252. doi: 10.1038/s41467-020-19957-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA work group* under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34(7):939–939. doi: 10.1212/WNL.34.7.939. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s & Dementia: The Journal of the Alzheimer’s Association. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JE, Maltesen RG, Havelund JF, Færgeman NJ, Gotfredsen CH, Vestergård K, et al. Characterising Alzheimer’s disease through integrative NMR- and LC-MS-based metabolomics. Metabolism Open. 2021;12:100125. doi: 10.1016/j.metop.2021.100125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Oliver S. The next wave in metabolome analysis. Trends in Biotechnology. 2005;23(11):544–546. doi: 10.1016/j.tibtech.2005.08.005. [DOI] [PubMed] [Google Scholar]
- O’Brien JT, Herholz K. Amyloid imaging for dementia in clinical practice. BMC medicine. 2015;13:163. doi: 10.1186/s12916-015-0404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnetti L, Gaiti A, Polidori MC, Brunetti M, Palumbo B, Chionne F, et al. Increased cerebrospinal fluid pyruvate levels in Alzheimer’s disease. Neuroscience Letters. 1995;199(3):231–233. doi: 10.1016/0304-3940(95)12058-c. [DOI] [PubMed] [Google Scholar]
- Smirnov DS, Ashton NJ, Blennow K, Zetterberg H, Simrén J, Lantero-Rodriguez J, et al. Plasma biomarkers for Alzheimer’s disease in relation to neuropathology and cognitive change. Acta Neuropathologica. 2022;143(4):487–503. doi: 10.1007/s00401-022-02408-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Wang H, Yin X, Deng P, Jiang W. Application of NMR metabolomics to search for human disease biomarkers in blood. Clinical Chemistry and Laboratory Medicine (CCLM) 2019;57(4):417–441. doi: 10.1515/cclm-2018-0380. [DOI] [PubMed] [Google Scholar]
- Stringer KA, McKay RT, Karnovsky A, Quémerais B, Lacy P. Metabolomics and Its Application to Acute Lung Diseases. Frontiers in Immunology. 2016 doi: 10.3389/fimmu.2016.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vågberg M, Norgren N, Dring A, Lindqvist T, Birgander R, Zetterberg H, Svenningsson A. Levels and age dependency of neurofilament light and glial fibrillary acidic protein in healthy individuals and their relation to the brain parenchymal fraction. PLoS ONE. 2015;10(8):e0135886. doi: 10.1371/journal.pone.0135886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignoli A, Paciotti S, Tenori L, Eusebi P, Biscetti L, Chiasserini D, et al. Fingerprinting Alzheimer’s Disease by 1 H nuclear magnetic resonance spectroscopy of cerebrospinal fluid. Journal of Proteome Research. 2020;19(4):1696–1705. doi: 10.1021/acs.jproteome.9b00850. [DOI] [PubMed] [Google Scholar]
- Voyle N, Baker D, Burnham SC, Covin A, Zhang Z, Sangurdekar DP, et al. Blood protein markers of neocortical amyloid-β burden: A candidate study using SOMAscan technology. Journal of Alzheimer’s disease: JAD. 2015;46(4):947–961. doi: 10.3233/JAD-150020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tang Y, Liu S, Mao S, Ling Y, Liu D, et al. Metabonomic profiling of serum and urine by (1)H NMR-based spectroscopy discriminates patients with chronic obstructive pulmonary disease and healthy individuals. PloS One. 2013;8(6):e65675. doi: 10.1371/journal.pone.0065675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (March 2020). (n.d.). International Classification of Diseases (ICD). https://icd.who.int/browse10/2019/en. Accessed 22.
- Wilkins JM, Trushina E. Application of metabolomics in Alzheimer’s disease. Frontiers in Neurology. 2018;8(JAN):1–20. doi: 10.3389/fneur.2017.00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS. Applications of metabolomics in drug discovery and development. Drugs in R&D. 2008;9(5):307–322. doi: 10.2165/00126839-200809050-00002. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Therriault J, Ren S, Jing X, Zhang H. The associations of serum valine with mild cognitive impairment and Alzheimer’s disease. Aging Clinical and Experimental Research. 2022;34(8):1807–1817. doi: 10.1007/s40520-022-02120-0. [DOI] [PubMed] [Google Scholar]
- Zlokovic BV. Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nature Reviews Neuroscience. 2011;12(12):723–738. doi: 10.1038/nrn3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its supplementary information files).