Graphical abstract
Keywords: Broiler chicken, Meat yield, Domestication, GWAS, Transcriptomic, SOX6
Highlights
-
•
The genomes of modern purebred broilers carried fewer deleterious mutations compared to local chickens and wild ancestors.
-
•
163 protein-coding genes were selected positively in purebred broilers during long term selection.
-
•
The MYH1 gene family harbored the top significant selective sweep windows and displays muscle-specific expression in purebred broilers.
-
•
SOX6 gene provided key regulatory function to breast muscle traits and adaptation to artificial selection.
-
•
A haplotype within SOX6 appeared differential pattern in gene expression, breast muscle traits, and haplotype frequency.
Abstract
Introduction
Investigating the genetic markers and genomic signatures related to chicken meat production by combing multi-omics methods could provide new insights into modern chicken breeding technology systems.
Object
Chicken is one of the most efficient and environmentally friendly livestock, especially the fast-growing white-feathered chicken (broiler), which is well known for high meat yield, but the underlying genetic basis is poorly understood.
Method
We generated whole-genome resequencing of three purebred broilers (n = 748) and six local breeds/lines (n = 114), and sequencing data of twelve chicken breeds (n = 199) were obtained from the NCBI database. Additionally, transcriptome sequencing of six tissues from two chicken breeds (n = 129) at two developmental stages was performed. A genome-wide association study combined with cis-eQTL mapping and the Mendelian randomization was applied.
Result
We identified > 17 million high-quality SNPs, of which 21.74% were newly identified, based on 21 chicken breeds/lines. A total of 163 protein-coding genes underwent positive selection in purebred broilers, and 83 genes were differentially expressed between purebred broilers and local chickens. Notably, muscle development was proven to be the major difference between purebred broilers and local chickens, or ancestors, based on genomic and transcriptomic evidence from multiple tissues and stages. The MYH1 gene family showed the top selection signatures and muscle-specific expression in purebred broilers. Furthermore, we found that the causal gene SOX6 influenced breast muscle yield and also related to myopathy occurrences. A refined haplotype was provided, which had a significant effect on SOX6 expression and phenotypic changes.
Conclusion
Our study provides a comprehensive atlas comprising the typical genomic variants and transcriptional characteristics for muscle development and suggests a new regulatory target (SOX6–MYH1s axis) for breast muscle yield and myopathy, which could aid in the development of genome-scale selective breeding aimed at high meat yield in broiler chickens.
Introduction
Chicken is one of the most efficient animals for protein production, providing>30% of meat products for humans and playing an essential role in global food security [1]. Although there are hundreds of domestic chicken breeds across the world [2], they originally could not supply sufficient meat to satisfy human demand due to low meat productivity. Since the 1950 s, fast-growing white-feathered chickens (broilers) have been bred, and the productivity of chickens has been continually enhanced by intensive selection for production traits [3], [4], [5]. Broilers can now rapidly achieve a body weight of 4–5 kg within 56 days, in contrast to the<1 kg weight of their wild ancestor Gallus gallus spadiceus (GGS) and of most other domestic chickens [6]. Broilers are normally produced by a three-way or four-way cross system in which the purebred paternal line is one of the most important contributors to the high meat yield. Additionally, the feed conversion ratio of white-feathered broilers is ten times that of cattle, but the carbon emission of broilers is only 1/10 of that of cattle [7]. This is closely related to the high meat yield of broilers. However, the genetic basis for meat production of purebred broiler lines developed through intensive selection has not been fully investigated.
Chicken is a typical model for investigating the genetic cause of traits such as the fast growth in purebred broilers that arises from human-driven selection. Rubin et al. reported partial genes related to appetite (PMCH) and growth (e.g., IGF-1, INSR, TBC1D1) by comparing the genomic variants of broilers, layers, and red jungle fowls (RJFs) [4]. Wang et al. indicated that IGF-1 and POU1F1 were selected in dwarf breeds, indicating an important function of these two genes adaptative to growth and development [8]. Similarly, additional genes (e.g., HNF4G, TBXAS1, GJD2) were determined to be significantly different between commercial broilers and RJFs, especially on the end of chicken (Gallus gallus) chromosome 14 (GGA14) [9]. Yang et al. reported that two significant peaks on GGA3 and GGA24 associated with meat production (body weight and thigh-related traits) in broilers and that the candidate gene ADGRG6 showed genomic differentiation between large and small body size chickens [10]. IGF2BP1 was proven to be correlated with body size in multiple animals [11], [12], [13], and it was also related to breast muscle weight in chickens [14]. However, as this research develops, an expansion in breeds and population sizes is needed to enable deeper exploration, and meat traits should be a primary focus.
In this study, a large population (n = 1,061) and many breeds (n = 21) were applied to reduce the risk of bias. We performed a systematic comparison of whole genomic variants, transcriptomes involving multiple tissues and stages, and a genome-wide association study (GWAS) to identify the genes and regions affecting meat production, especially for breast muscle. This study provides new insights into the genetics of modern broiler selection and will facilitate the development of techniques for meat production improvement.
Materials and methods
Ethics statement
All experimental procedures associated with the chickens used in this study were conducted following guidelines established by the Ministry of Science and Technology (Beijing, China). Ethical approval was granted by the Animal Welfare and Ethics Committee of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (IAS-CAAS, Beijing, China), with the reference number IAS2019-44.
Birds and sample collection
A total of 691 fast-growing white-feathered chickens (Line B) from two generations were used. The paternal line was generated by Foshan Gaoming Xinguang Agriculture and Animal Husbandry Co., Ltd. (Foshan, Guangdong, China) [15]. After 12 h of fasting, the chickens were slaughtered on day 42 (D42), and breast muscle was stripped and weighed. The breast muscle weight (BrW) was recorded, and the BrW percentage (BrP) was calculated as previously reported [15]. A total of 120 individuals were used for myopathy evaluation, including wooden breast (WB) and white striping (WS). WB and WS were scored and classified by subjective assessment as described in previous studies (normal breast scored 0, mild WB/WS scored 1, moderate WB/WS scored 2) [16], [17]. Drip loss (DL) and compression force (CF) were measured as previous reports [18], [19]. All the phenotypic data were distributed within the range of the mean ± 3 standard deviations and passed quality control for subsequent GWAS analysis.
Additionally, Line B (n = 60) and Beijing You (BJY) chickens (n = 60) were reared from day 1 (D1) with the recommended conditions [20]. Briefly, a common corn-soybean diet containing 2,900 kcal/kg metabolic energy and 183 g/kg crude protein was provided, and the fresh feed and water were available ad libitum. Six individuals, randomly selected from each of the two breeds at D1 and D42, were slaughtered, and six tissues, including heart, liver, lung, breast muscle, thigh muscle, and abdominal fat (only at D42), were collected and stored at −80 °C for RNA isolation.
Genetic materials, DNA extraction and sequencing
Nine chicken breeds (n = 862), including three purebred broiler breeds (Line B: paternal line B; Cornish; and BRp, broiler paternal line) and six local chicken breeds (BJY, Beijing You chicken; HX, Huxu chicken; PC, Piao chicken; TBC, Tibetan chicken; WD, Wuding chicken; WC, Wenchang chicken), were sequenced (supplementary table S1). For each individual, blood samples were collected from the wing vein, and genomic DNA was extracted from blood samples using the phenol–chloroform method. The quality of DNA was assessed based on agarose gel electrophoresis. A DNA library (paired-end, 2 × 150 bp) for each DNA sample was constructed, and all libraries were sequenced using the Illumina Nova 6000 sequencing platform. In total, approximately 7.7 Tb previously unpublished data was generated.
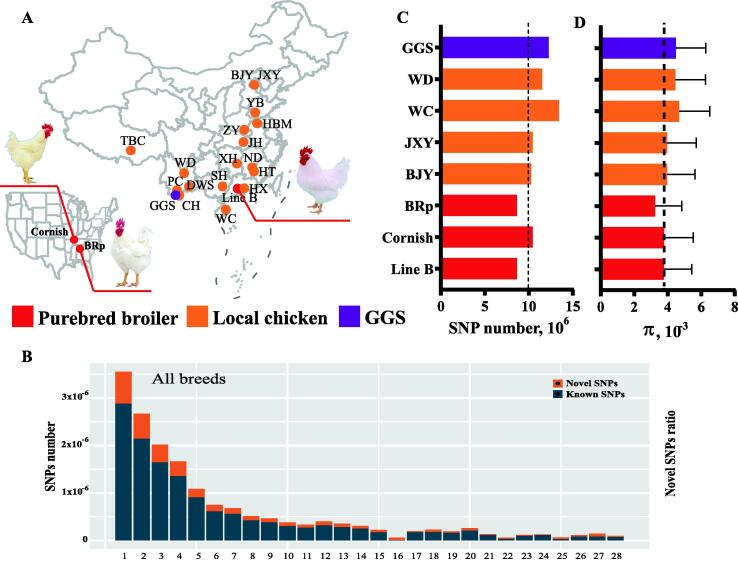
In addition, we downloaded a list of sequencing data for 12 chicken breeds (n = 199), including for the local chicken breeds (CH, Chahua chicken; JXY, Jingxing Yellow chicken, YB, Yuanbao chicken, HBM, Huaibei Ma chicken; HT, Hetian chicken; JH, Jianghan chicken; DWS, Daweishan mini chicken; ND, Ningdu chicken; SH, Sanhuang chicken; WC; XH, Xianghuang chicken; ZY, Zhengyang chicken) and the wild ancestor (GGS) with accession numbers: CRA004023 and CRA002643 from GSA database, SRP155577 from the SRA database, and the ChickenSD database [9], [10], [14], [21], [22], [23], [24]. In summary, a total of 21 chicken breeds/lines (n = 1,061) belonging to three types—purebred broilers, local chickens, and wild ancestors—were used in this study (Fig. 1A).
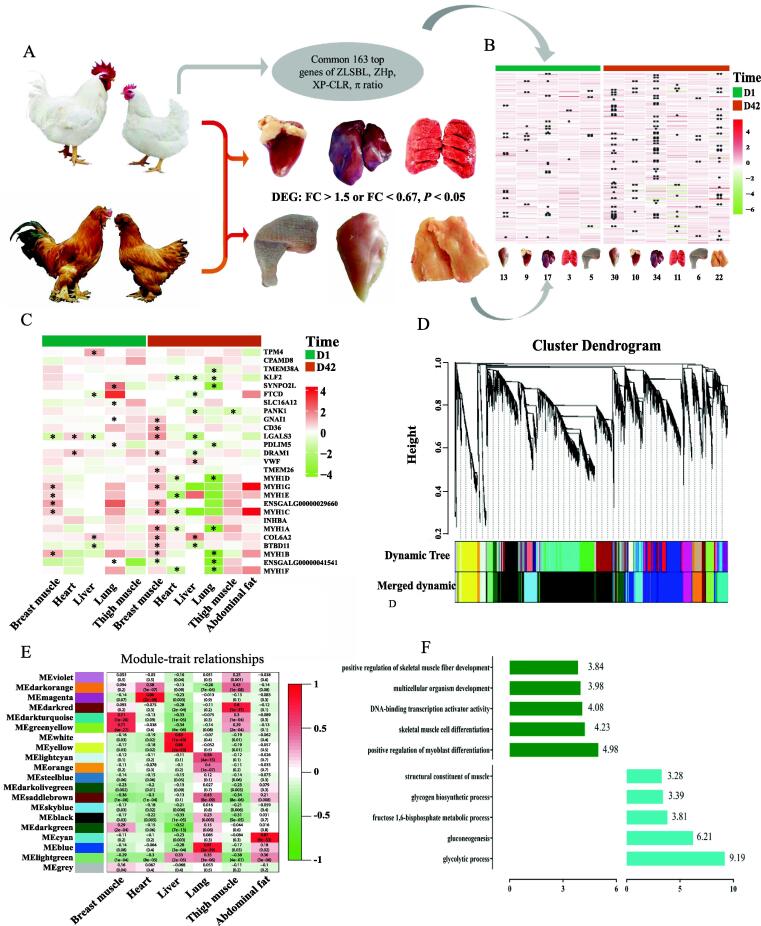
Fig. 1.
Statistics for genomic information from 21 chicken breeds/lines. (A) The geographic distribution of 21 chicken breeds/lines. (B) The number and ratio of known and novel SNPs in each chromosome of purebred broiler Line B. (C) SNP number of global genome in representative chickens of purebred broiler, local chicken and GGS. The result for other breeds was presented in Figure S1. The black horizontal line indicates the ten million SNPs. (D) Genomic nucleotide diversity in representative breeds of purebred broiler, local chicken and GGS. The result for other breeds was presented in Figure S2. The black horizontal line indicates π = 3.8 × 10-3, the highest level for the breeds in purebred broilers cluster. BRp: broiler paternal line, BJY: Beijing You chicken, CH: Chahua chicken, DWS: Daweishan mini chicken, GGS: Gallus gallus spadiceus, HBM: Huaibei Ma chicken, HT: Hetian chicken, HX: Huxu chicken, JH: Jianghan chicken, JXY: Jingxing Yellow chicken, ND: Ningdu chicken, PC: Piao chicken, SH: Sanhuang chicken, TBC: Tibetan chicken, WC: Wenchang chicken, WD: Wuding chicken, XH: Xianghuang chicken, YB: Yuanbao chicken, ZY: Zhengyang chicken. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Variant calling, quality control, and annotation
The raw reads were first trimmed in Trimmomatic v0.36 with default parameters [25], and only paired quality reads were preserved for the following analysis. Next, the filtered reads for all individuals were aligned with the chicken reference genome (GRCg6a) using Burrows-Wheeler Aligner (BWA) v0.7.17 [26]. The obtained Sequence Alignment Map (SAM) files were converted to Binary Alignment Map (BAM) format and then sorted by coordinates and indexed using Samtools v1.12 [27]. Polymerase chain reaction (PCR) duplicates were removed using PICARD v2.26. Single-nucleotide polymorphisms (SNPs) were called first using the HaplotypeCaller function in Genome Analysis Toolkit (GATK) v4.2.2 [28], and the genomic variant call format (gVCF) file for each chicken was acquired. Then, the CombineGVCFs and GenotypeGVCFs functions were used to combine all the gVCF files and to jointly call SNPs. In addition, we removed SNPs using GATK with specific standards: Quality score < 30.0, QualByDepth < 2.0, FisherStrand > 60.0, RMSMappingQuality < 40.0, StrandOddsRatio > 3.0, MappingQualityRankSumTest < -12.5, and ReadPosRankSum < -8. A total of 17,622,141 biallelic SNPs were retained for the following analysis.
After filtering, the reserved SNPs were annotated based on location categorization using SnpEff v5.0 [29], for which the corresponding annotation file was obtained from the Ensembl database. Here, the SNPs were clustered into ten classifications based on their genomic positions, including 3′ and 5′ UTRs, up- and downstream regions, exonic and intronic regions, splicing sites, and intergenic regions. Additionally, the ENSEMBL Variation Database for Single Nucleotide Polymorphisms (https://ftp.ensembl.org/pub/release-106/variation/gvf/gallus_gallus/) was used to detect novel mutations.
Population genetics analysis
First, all SNPs were pruned using PLINK v1.9 [30] with a window size of 50 bp, a window step of 5 bp, and a pairwise r2 threshold of 0.2 (--indep-pairwise 50 5 0.2). This led to the retention of a total of 2,328,925 independent SNPs for population genetics analysis. Principal component analysis (PCA) was performed with Genome-wide Complex Trait Analysis (GCTA) v1.93.2 software [31], with the first three principal components cumulatively explaining 10.93% of the total variance. The population structure of different admixture proportions was evaluated using the program ADMIXTURE v1.3 [32]. Nine solutions (2 ≤ k ≤ 10) were selected for genetic clustering. To infer the genetic structure, we first calculated the identical by state (IBS) distance between chickens using PLINK [30]. Then, we constructed a neighbour-joining tree based on the 1-IBS distance matrix in Molecular Evolutionary Genetics Analysis (MEGA) v7 [33], and FigTree v1.4.0 software (tree.bio.ed.ac.uk/software/figtree/) was used to visualize the phylogenetic trees.
Analysis of nucleotide diversity, linkage disequilibrium (LD) decay, and runs of homozygosity (ROH)
To better evaluate the genetic characteristics among different species, we calculated the genetic diversity (π) in VCFtools v0.1.13 [34]. The LD decay level was quantified and plotted using Poplddecay software [35], with a maximum distance of 500 kb. ROH for each population were estimated using PLINK software with the parameters --homozyg-density 50, --homozyg-gap 1000, --homozyg-kb 300, --homozyg-window-het 3, --homozyg-window-snp 50, --homozyg-window-missing 5 [30]. These results were analysed using all SNPs.
Demographic history inferences
We restructured the chicken demographic history during the past 1,000 to 1,000,000 years using SMC++ software [36]. Four to ten genomes of each population were used to maximize the number of populations to be included. Referring to the study by Nam et al., the model was scaled by a mutation rate (μ) of 1.91e-9 substitutions per genomic site per year and a generation time of 1 year [37]. The result was plotted using an R script.
Identification of deleterious SNPs (dSNPs) and estimation of genetic loads
To assess the evolution of dSNPs in chickens in the wild, domestication, and breeding stages, we estimated the deleterious score of missense mutations annotated from 1,061 genomes according to the reported pipeline [9]. First, the missense SNPs and the synonymous SNPs were retrieved based on the annotation result by using SnpEff v5.0 [29]. Next, the online tool Ensembl Variant Effect Predictor was applied to predict Sorting Intolerant From Tolerant (SIFT) scores for missense SNPs [38], and SIFT scores < 0.05 were defined as evolutionarily intolerant, namely, dSNPs. Finally, to infer the landscape of genetic loads in various stages, the number and frequency of dSNPs per individual were calculated, as well as the ratio of dSNPs to synonymous SNPs.
Detection of selective sweeps
We scanned the purebred broiler genomes for positive selection signatures using the Z-transformed locus-specific branch length (ZLSBL), the nucleotide diversity (π) ratio (πbroiler/πGGS or local), the cross-population composite likelihood ratio (XP–CLR), and the Z-transformed pooled heterozygosity (ZHp). The ZLSBL and π ratio were calculated using a 40–kb sliding window with 20–kb stepwise increments in VCFTools [34], estimates of the cross-population composite likelihood ratio (XP–CLR) were obtained in xpclr software [39] to compare the broilers and local chicken breeds or GGS, and ZHp was calculated using an in-house script as described previously with the same window size and step [4]. The ZLSBL approach was applied as follows: ZLSBL = (LSBL – μLSBL)/σLSBL, where μ is the overall average LSBL score and σ is the standard deviation of all windows within each cluster. We used an empirical 99th percentile as the cut-off to retrieve the putative selective sweeps in purebred broilers. To exclude the selected regions in local chicken breeds and GGS that were the same as those in the broilers, we also performed selective sweep analysis between local chicken breeds and GGS using the π ratio, XP–CLR, and ZHp methods. For the π ratio approach, the windows that reached the threshold of -Logπ (broiler/local) < -1.64 or -Logπ (broiler/local) < -1.72 were retained, among which the windows over the threshold of 0.5 < -Logπ(local/GGS) < 2 were excluded. For the XP–CLR method, windows with XP–CLR (broiler vs local) > 300.29 or XP–CLR (broiler vs GGS) > 268.72 were retained, and windows over the threshold of XP–CLR (local vs GGS) > 173.71 (1% threshold) were removed. For the ZHp index, the windows with ZHpbroiler < -3.04 were kept, while the overlapping windows with ZHplocal < -3.87 (1% threshold) or ZHpGGS < -4.16 (1%) were eliminated. Collectively, the putative sweeps, only selected in purebred broilers but not in local chicken breeds and GGS, were defined with overlapping windows identified by at least two calculation methods. The putative selected genes (PSGs) were annotated based on the reference genome (GRCg6a).
Identification of differentially expressed genes (DEGs) by the transcriptomic atlas in multiple tissues and stages
A total of 129 samples of six tissues from purebred broiler Line B and local breed BJY were collected at D1 and D42, including breast muscle, thigh muscle, heart, lung, liver, and abdominal fat. The RNA of these samples was extracted using TRIzol reagent (TAKARA, Beijing, China). After quality control, the qualified RNA samples were used to perform reverse transcription and cDNA library construction as previously reported. Each qualified library was sequenced on the Illumina PE150 platform, and>10 G raw reads per sample were generated. The bioinformatic analysis pipeline was implemented according to the methods of a previous report [14]. Briefly, the raw reads were trimmed using Trimmomatic [25] with the default parameters, and clean reads were produced. Then, the clean reads were aligned to the chicken reference genome GRCg6a (https://ftp.ensembl.org/pub/release-104/fasta/gallus_gallus/dna/Gallus_gallus.GRCg6a.dna_sm.toplevel.fa.gz) using Hierarchical Indexing for Spliced Alignment of Transcripts 2 (HISAT2) v2.2.1 [40] and assembled using StringTie v2.1.6 [41]. Next, raw gene counts were acquired according to the Python script provided by StringTie (l = 150) [41]. The gene expression level was normalized by using the DESeq2 package in the R environment [42], and differentially expressed genes (DEGs) were identified as having a fold change > 1.5 or < 0.67 and P value < 0.05.
Weighted gene co-expression network analysis (WGCNA)
To explore the expression differences between the two types of chickens in the global gene atlas, especially for PSGs, we constructed a co-expression network using the weighted gene co-expression network analysis package based on the transcriptomic profile from the 129 tissues described above. Briefly, we constructed an expression matrix of 15,620 genes (average count > 10) across six tissues. The soft threshold (β = 6) was determined according to the scale-free distribution (R2 > 0.85). Then, we chose step-by-step and dynamic cutting methods to construct the gene network and to detect modules (minModuleSize = 50, mergeCutHeight = 0.25). The module with a correlation coefficient > 0.5 was regarded as a tissue-specific module. Within the module, the hub genes were defined as having gene significance > 0.2 and module membership > 0.8. Furthermore, Gene Ontology enrichment analysis was performed based on the genes of each tissue-specific module. In addition, we identified the tissue-specific expression genes based on PSGs as previously described [43]. Briefly, the tissue-specific genes (TSGs) were defined as follows: (1) the average count value of the candidate gene in one tissue was more than three times that of other tissues, (2) the average count value of the candidate gene target tissue was > 50% of the average expression level in all other tissues, and (3) the expression level of the candidate gene was in the top 25% for all gene expression levels in corresponding tissues.
GWAS for BrW and BrP in Line B
Line B was selected based on meat production traits over multiple generations, and SNP information and phenotypic records were comprehensively retained. To explore the genetic basis of breast muscle growth, association analysis of BrW and BrP was performed using the linear mixed model in Genome-wide Efficient Mixed Model Association (GEMMA) v0.98.4 [44] based on chickens genotyped by whole genome sequencing. After quality control (--mind 0.1, --maf 0.05) using PLINK v1.9 software [30], a total of 8,663,580 SNPs were retained, and GWAS was performed for BrW and BrP as follows:
where indicates the vector of the BrW record; W indicates the covariates matrix, including a column of 1 s, as well as generation and sex effects; is the vector of the corresponding coefficient (including the intercept); is the effect size of each marker; , where indicates the random polygenic effect, denotes the n-dimensional multivariate normal distribution, is the ratio of two variance components, is the variance of the residual errors, and is the kinship matrix; and , where is the residual error and is the identity matrix. The proportion of variance explained by the markers was calculated using the equation [45], where and represent allele frequency for minor and major alleles, respectively. represents the allele effect size, and represents the genetic variance, which was calculated by ASReml 4.1 [46]. Similarly, the whole-genome and suggestive significance thresholds were corrected by the Bonferroni test (0.05/8,663,580 and 1/8,663,580, respectively). Additionally, Manhattan and Q-Q plots were visualized via the qqman package in the R environment [47].
Cis-expression QTL (cis-eQTL) mapping and Mendelian randomization in Line B
To verify the influence of SOX6 on BrW and BrP, we performed a novel Mendelian randomization analysis, namely, summary-data-based Mendelian randomization (SMR), integrating cis-eQTLs and summary data from GWAS to estimate the effect [48]. First, a total of 160 individuals in Line B were subjected to RNA isolation, reverse transcription and reverse transcription-polymerase chain reaction (RT-PCR) assays, and cis-eQTL analysis. These were performed in Matrix eQTL software [49] based on the SOX6 expression level and surrounding SNPs located 500 kb upstream and downstream of the SOX6 region. A total of 2143 significant SNPs (FDR < 0.05) were identified. Next, we compared the eQTL and GWAS results and selected 14 SNPs after clumping that were significantly correlated with SOX6 expression but not to breast muscle traits as the instrument variable. The cis-eQTL and GWAS results for BrW and BrP were set as exposure and outcome variable, respectively. The SMR was conducted using the TwoSampleMR software [50].
Gene expression and western blot analysis of SOX6
We examined the gene expression level of SOX6 and flanking genes in breast muscle tissue sampled from purebred broiler Line B (D42), local breeds JXY (D98), and Jinling chickens (JL, D56) at their market age with RT–PCR. Total RNA was isolated using TRIzol reagent (Takara, Beijing, China), and the concentration and integrity of the RNA were measured with an Agilent 2100 Nano and gel electrophoresis. Then, first-strand cDNA was synthesized according to the manufacturer’s instructions for the PrimeScriptTM RT Reagent Kit with gDNA Eraser (Takara, Beijing, China). Here, 1 μg of RNA was reverse transcribed as the template for RT–PCR in a 20 μl volume, and the residual DNA was eliminated in this step. The obtained reverse transcription products were subsequently mixed with SYBR Green Mix (2 × ), ROX dye II (50 × ), primers, and double-distilled water (ddH2O) to produce a 10 μl RT–PCR system. The primers were designed using Oligo 6.0 based on the coding sequence of SOX6 (F: 5′–TCCCTGACATGCACAACTCC–3′, R: 5′–TTGAGGCTGTTGTCCTACGG–3′) and flanking genes (supplementary table S2) in the 5′ to 3′ direction. Each sample included three technical replicates. The RT–PCR conditions were as follows: 94 °C for 5 min, followed by 35 cycles of 94 °C for 3 s and an annealing temperature of 32 s. RPL32 and HSPA2 were selected as the internal controls (supplementary table S2). According to the RT–PCR results, the expression level was calculated using the 2-△△CT method [51].
Protein was extracted from breast muscle tissue and lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.25% deoxycholic acid, 1% NP-40 and 0.5% sodium dodecyl sulfate (SDS)) supplemented with protease inhibitor (Roche Applied Science, Mannheim, Germany) and centrifuged at 12,000 g at 4 °C for 10 min. For immunoblot analyses, the protein lysates or immunoprecipitated samples were separated by electrophoresis on sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) gels and then transferred onto polyvinylidene fluoride membranes (Millipore). The membranes were first blocked with 5% (w/v) fat-free milk in Tris-Buffered Saline Tween-20 (TBST) and then incubated with the corresponding primary antibodies (SOX6, sc-393314, Santa Cruz, 1:500; and GAPDH, A01020S, Abbkine, 1:10000) diluted in 5% fat-free milk in TBST. After washing with TBST, the membranes were incubated with the appropriate secondary antibodies diluted in 5% fat-free milk in TBST. Protein bands were visualized using Immobilon Western Chemiluminescent Horseradish Peroxidase (HRP) Substrate (Millipore) according to the manufacturer’s instructions.
Statistical analysis
Statistical analyses were conducted using SPSS 25.0 software or a custom script in the R environment. The Wilcoxon signed-rank test was used to determine the differences in dSNPs among the three types of chickens. A general linear model was used to compare the differences in breast muscle phenotypes and expression of SOX6 and other genes. P < 0.05 was set as the significance threshold. Pearson correlation analysis was used to estimate the relationship between SOX6 and breast muscle traits (WB, WS, DL, and CF).
Results
Whole-genome sequencing and variation
We generated whole-genome sequences of 862 chickens, totalling 7.7 Tb raw reads with an average depth of 9.2× (supplementary table S1, other sequences of 199 chickens were obtained from the SRA database). After using stringent criteria for variant filtration, a total of 17,622,141 high-quality SNPs were detected in the 21 chicken breeds/lines. The retained SNPs were distributed across the genome with an average density of 1 SNP every 60 bases. After annotation, the highest ratio of novel to known SNPs was calculated on GGA16, and 21.74% of these SNPs had not been included in dbSNP (Fig. 1B, supplementary table S3). Line B is a fast-growing purebred broiler with white feathers, carrying 8,663,580 SNPs of all variants, with an average density of 1 SNP every 114 SNPs. Only 4.7% of SNPs were identified as novel variants (supplementary table S3). Most SNPs were intronic and intergenic for all breeds (supplementary table S4). Intensive selection for specific traits has been carried out on purebred broilers in recent decades, resulting in the loss of genetic diversity, including reduced SNP number and nucleotide diversity (π) in purebred broilers (Fig. 1C-D, supplementary Fig. S1-2). Here, we only show the results for purebred broilers, wild ancestors, and some typical slow-growing local chicken breeds (BJY, HX, WC, and WD). The complete results are shown in supplementary Fig. S1-2. Both sets of results are consistent with the rules of intensive human-driven selection in fast-growing purebred broilers.
Phylogenetic and demographic analyses
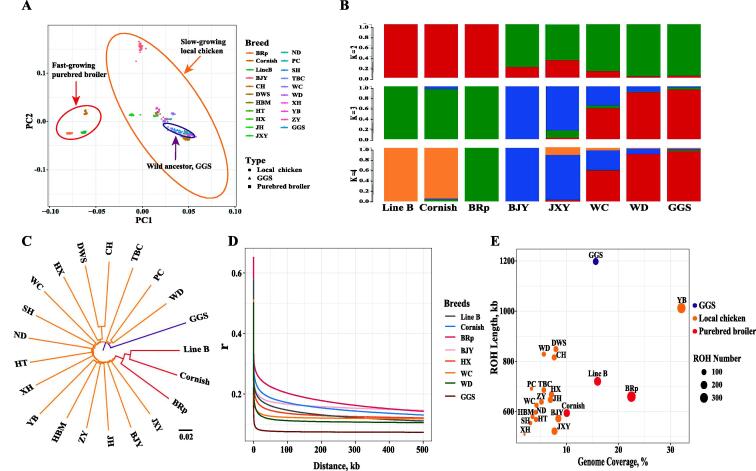
A comprehensive analysis of genetic relationships among these breeds was conducted. First, we performed a PCA of 21 chicken breeds/lines based on the pruned SNPs (Fig. 2A). Despite the division among purebred broilers, local chicken breeds and wild ancestors, a sizeable genetic difference between BJY and other local chicken breeds was also found. We further performed genetic coancestry analyses to partition all chickens into groups by varying the number of presumed ancestral populations (K = 2–10, Fig. 2B, supplementary Fig. S3). We found that Line B was genetically close to the Cornish breed and that both were separated from the BRp breed, local chicken breeds, and GGS. A neighbour-joining tree was constructed based on pairwise genetic distances with GGS as the outgroup (Fig. 2C). As expected, the fast-growing purebred broilers were genetically distant from GGS. The results showed clusters based on geographical distribution. For instance, GGS and the species in the southwestern region of China (WD, TBC, PC, DWS, and CH) were defined as belonging to the same clade. This result was consistent with the coancestry analysis result. We used SMC++ to infer the dynamic changes in effective population size (Ne) for all breeds (supplementary Fig. S4). A decreasing tendency in Ne in the three broiler populations revealed strong evidence of domestication bottlenecks for fast-growing broilers, as well as WC and HX, compared with GGS. In addition, this analysis also demonstrated that broilers have a stronger bottleneck and much smaller recent Ne than most local chicken breeds (supplementary Fig. S4). This proved a stronger selection pressure on fast-growing purebred broilers and was consistent with the result of a higher LD decay distance for purebred broilers (Fig. 2D, supplementary Fig. S5). However, the ROH length of purebred broilers (594–720 kb) was slightly lower than that of local (509–1012 kb) and wild ancestors (1198 kb) (Fig. 2E). The ROH number and average genome coverage were higher in purebred broilers (257, 16.2%) than in local chickens (103, 7.0%) and GGS (137, 15.6%) (Fig. 2E, supplementary Fig. S6).
Fig. 2.
Population genetic diversity and demographic history inference. (A) PCA plot with 21 chicken breeds/lines. Different shapes indicate purebred broiler, local chicken, and GGS, respectively. (B) Population structure analysis of three types of chickens, where number of ancestral clusters were set from K = 2–4, the comprehensive result was shown in Figure S3. (C) Neighbor-joining tree constructed by genetic distance (1-IBS method) among 21 chicken breeds/lines. (D) LD decay in purebred broilers, typical local chickens, and GGS. (E) ROH length and genome coverage for 21 chicken breeds/lines. The size of each dot indicated the ROH number of corresponding chicken breed/line. BRp: broiler paternal line, BJY: Beijing You chicken, CH: Chahua chicken, DWS: Daweishan mini chicken, GGS: Gallus gallus spadiceus, HBM: Huaibei Ma chicken, HT: Hetian chicken, HX: Huxu chicken, JH: Jianghan chicken, JXY: Jingxing Yellow chicken, ND: Ningdu chicken, PC: Piao chicken, SH: Sanhuang chicken, TBC: Tibetan chicken, WC: Wenchang chicken, WD: Wuding chicken, XH: Xianghuang chicken, YB: Yuanbao chicken, ZY: Zhengyang chicken. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Genetic load after domestication and artificial selection
Domestication has caused deleterious mutations to accumulate in chickens and other domesticated animals [52], [53]. Here, we calculated the numbers and frequency of deleterious and synonymous SNPs in each genome (Table 1). For 82,784 missense SNPs, 16.75% (13,869) were predicted to be evolutionarily intolerant (score < 0.05, supplementary table S5). As shown in Table 1, the results showed that purebred broilers and local chicken breeds harboured approximately 15.63% and 24.05% more deleterious SNPs (dSNPs) than GGS across their genomes (supplementary Fig. S7A). Although the number of synonymous SNPs was significantly different among the three groups (supplementary Fig. S7B), the ratio of dSNPs to synonymous SNPs and frequency of dSNPs were higher in broilers and local chicken breeds than in GGS (supplementary Fig. S7C-D). Collectively, a rapid accumulation of dSNPs was found in local chicken breeds and broilers due to domestication and directional breeding. Next, we compared the levels of both heterozygous and homozygous dSNPs and observed that 56.75% and 69.85% of dSNPs in broilers and local chicken breeds were preserved in heterozygous status, respectively, compared to dSNPs in GGS (50.90%, supplementary Fig. S7E-F). Compared to local chicken breeds, purebred broilers carried more homozygotes (broiler compared with local chicken: 544 compared with 406) and fewer heterozygotes (broiler compared with local chicken: 1,427 compared with 1,885), and the ratio of homozygous and heterozygous dSNPs exhibited a similar trend between broilers and local chicken breeds (Table 1). These results suggested that dSNPs mainly accumulated under heterozygous status, and partial deleterious SNPs were eliminated due to intensive breeding.
Table 1.
Summary statistics for dSNPs in three chickens clusters.
| dSNPs number | Het-dSNPs number | Hom-dSNPs number | Syn-SNPs number | Syn-hom-SNPs number | Syn-het-SNPs number | Ratio 1 | Frequency 2 | |
|---|---|---|---|---|---|---|---|---|
| Purebred broilers | 2515.92 | 1427.81 | 544.05 | 97778.23 | 34326.91 | 29133.06 | 0.0257 | 0.0907 |
| Local chickens | 2698.77 | 1885.01 | 406.88 | 99636.42 | 30213.20 | 39218.89 | 0.0271 | 0.0973 |
| GGS | 2175.59 | 1107.34 | 534.13 | 91231.97 | 33200.56 | 24841.81 | 0.0238 | 0.0784 |
Indicating the ratio of dSNPs to syn-SNPs. dSNPs: deleterious SNPs; syn-SNPs: synonymous SNPs.
Indicating the frequency of dSNPs.
Genomic signatures in purebred broiler chickens
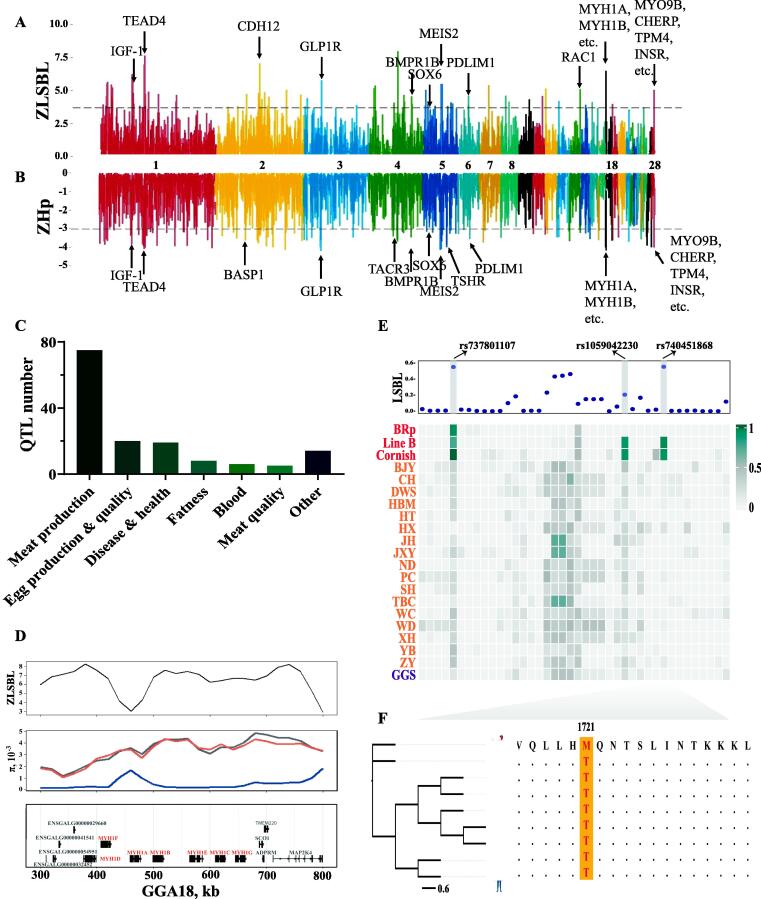
To infer the genomic loci of selective sweeps in modern broilers, we used XP–CLR and ZLSBL test-based allele frequency differentiation (supplementary table S6–7). As genomic regions targeted by human intervention may be expected to have reduced genetic diversity, nucleotide diversity (π) and ZHp were included in this analysis (supplementary table S8–9). Hence, we combined these four methods to define the candidate divergent regions (requiring at least two signals among the four methods) with a window size of 40 kb and step size of 20 kb (Fig. 3A-B, supplementary Fig. S8-10). By overlapping the results of different methods, a list of 391 windows harbouring 163 protein-coding genes and 58 long noncoding RNAs (lncRNAs) were identified in the purebred broiler population (Fig. 3A-B, supplementary table S10–11), and all 58 lncRNAs were annotated as novel genes without any functional description. A comparison of breeding-associated selective sweeps and known quantitative trait loci (QTLs) revealed that the selected regions with higher ZLSBL scores and XP–CLR values but reduced diversity spanned meat production-, immune-, and meat quality-related QTLs (Fig. 3C, supplementary table S12), reflecting the human demands on meat production and the changes in disease resistance for chickens under the breeding scheme.
Fig. 3.
Detection of selective sweep windows in purebred broilers. (A-B) Selective sweep results based on ZLSBL and ZHp (for purebred broiler) methods, respectively. The black dash line indicates the top 1% threshold. (C) Mapping of putative sweep windows to known chicken QTLs, the known QTLs were downloaded from chicken QTL database. (D) Putative selected windows and genes in GGA18. (E) LSBL and frequency of missense SNPs in the selected region of GGA18. The upper part is the LSBL result, while the lower part is the frequency distribution of each missense SNP. Only 3 SNPs were detected LSBL significance and frequency difference. (F) The neighbour-joining tree constructed by amino acid sequence of MYH1E in multiple species. The orange background indicated the missense SNP rs740451868. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In broilers, we observed a lead signal on GGA18, which annotated members of the myosin heavy chain (MYH) gene family (Fig. 3A-B, D). MYH1A, MYH1B, MYH1C, etc., are components of myosin and markers of fast-twitch fibers and are involved in the development process of skeletal muscle [54]. Furthermore, we focused on the missense SNPs and calculated the frequency. Three SNPs were obviously different among purebred broilers, local chickens, and GGS (Fig. 3E, supplementary Fig. S11-12). For example, the mutation allele of SNP rs740451868 was fixed in purebred broilers, while the wild allele was fixed in other animals (Fig. 3F). In addition to the MYH gene family, the muscle development-related genes IGF-1, INSR, and SOX6 were detected to harbour the top 1% significant divergent regions (supplementary Fig. S13–15). We also found a list of genes harbouring the top selective sweep windows that were functionally reasonable for domestication and directional breeding in broilers (supplementary table S11). For example, TACR3 and SLC26A8 are involved in spermatogenesis and motility [55], [56]; CPAMD8 and INHBA are associated with eye development; and GLI3 is involved in chicken pigmentation. Additionally, we performed GO enrichment analysis of the gene sets in 391 selective sweep windows in broilers (supplementary Fig. S16, supplementary table S13). Based on the phenotype or physiology process, the GO terms were classified into several clusters, including the development process of the heart (e.g., GO:0008016, GO:0060047, and GO:0060421), muscle functions (e.g., GO:0051155, GO:0014834, and GO:0006941), reproduction (e.g., GO:0022412, GO:0046545, GO:0007292), bone development (GO:0060348), and sensory (GO:1904058, GO:0021618).
Transcription atlas of PSGs in multiple tissues and stages between purebred broilers and local chicken breeds
To further assess the expression abundance of PSGs across tissues in typical broiler Line B and local BJY chickens, we performed RNA sequencing and calculated the count value of each detected gene in six tissues, including breast and thigh muscle, liver, heart, lung, and abdominal fat (Fig. 4A). Based on the whole transcriptome atlas of the two breeds, tissue-specific clusters were presented. Specifically, tissues with similar physiological functions were more likely to cluster together (supplementary Fig. S17-19). For the 163 PSGs, we analysed the expression difference between the two breeds. First, approximately 13.50% to 28.83% of genes were excluded due to low expression levels across tissues (count < 10, supplementary table S14). Differential expression analysis was conducted for those PSGs between Line B and BJY (supplementary table S15). A total of 83 DEGs from 163 PSGs were identified in six tissues (Fig. 4B), while additional genes appeared to function in other tissues or developmental stages. Through enrichment analysis, we found that the tight junction pathway and myosin function (e.g., myosin complex, motor activity) were significantly different between Line B and BJY. These functions and genes were obviously predominant in Line B (supplementary Fig. S20-21). The genes (e.g., MYH1A, MYH1B, and MYH1C) involved in these functions were mainly related to muscle development, especially in breast muscle. Additionally, most putative selected lncRNAs were detected to have zero expression, only 9 of which were expressed in six tissues, but no significant difference was found (supplementary table S16).
Fig. 4.
Transcriptomic atlas of putative selected genes (PSGs) in purebred broiler Line B and local breed BJY chicken. (A) Strategy for DEGs detection from 163 PSGs in six tissues of two breeds. (B) Differential expression profile for 163 PSGs, color indicated the expression fold change in log2 scale, * indicated P < 0.05, ** indicated P < 0.01. The number below the tissues was the DEGs between two chicken breeds. (C) Expression pattern of the tissue-specific genes (TSGs), color indicated the expression fold change in log2 scale, * indicates that the genes were significantly expressed between two breeds in corresponding tissues, including MYH1B, MYH1C, and MYH1G in breast muscle, COL6A2 and BTBD11 in liver. (D) Functional modules detection based on step-by-step method. Each major branch represented a color-coded module that contains a set of highly connected genes. (E) Heatmap between 20 modules and 6 tissues. Pearson correlation coefficients and their associated P values were presented at each box. Red and green indicated that the given tissue had a strong positive and negative correlation relative to all other tissues, respectively. (F) GO enrichment results based on the genes in breast muscle-related modules, the above and below bars represented the GO terms enriched by genes from darkgreen and darkturquoise modules, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Next, we focused on the TSGs identified as presented above from 163 PSGs. A total of 18 and 22 genes were scanned for tissue-specific expression in the two stages, respectively (Fig. 4C, supplementary table S17). For these TSGs, over 50% of genes (9 in D1 and 12 in D42) were highly expressed in muscle tissue (breast and thigh muscle), including TMEM38A and members of the MYH gene family on GGA18. In addition, TPM4, FTCD, DRAM1, and TMEM26 were also specifically expressed in the heart, liver, and lung, respectively. Consistent with the tissue-specific expression profile, some genes (MYH1B, MYH1C, MYH1G) in the MYH gene family were highly expressed in the breast muscle of Line B in D1 and D42 compared to that of BJY (Fig. 4C). In the liver, FTCD and BTBD11 were expressed at lower levels and were specifically expressed in broilers (Fig. 4C). Additionally, we found that COL6A2 was highly expressed in abdominal fat but differentially expressed in the liver. In addition, we also found higher expression of the SOX6 gene in the breast muscle of Line B than in that of BJY (supplementary table S15). In total, each organ of Line B was obviously different from that of BJY, but it can be inferred that the muscle was a major difference between broilers and local chicken breeds at the transcription level. The genes (MYH1A, MYH1B, MYH1C, etc.) in the MYH gene family were identified as functional targets regulating muscle development.
We constructed networks based on weighted gene co-expression network analysis (WGCNA) to detect biological relationships and possible functions of core genes across tissues. Here, a total of 15,620 genes (count > 10) were selected for WGCNA. The soft threshold value β was set to 6 according to the criteria of R2 > 0.85 (supplementary Fig. S20). In total, 20 modules were collected based on the whole transcriptome profile (Fig. 4D-E), each of which comprised 64–5513 genes except for the grey module (supplementary table S18). To obtain tissue-specific modules, the association between 20 modules and 6 tissues was evaluated with the criteria of correlation coefficient r > 0.5 or r < -0.5 and P value < 1.0-10. A total of 10 tissue-specific modules were identified (Fig. 4E). For instance, the darkturquoise and greenyellow modules were highly correlated with breast muscle (r = 0.71, P = 1.0-26; r = 0.71, P = 4.0-27). Based on the GO annotation, we found that the functional enrichments of genes in tissue-specific modules were consistent with tissue function (supplementary table S19). For example, the genes in breast muscle-specific modules (darkturquoise and greenyellow) were enriched in regulating myoblast differentiation and skeletal muscle cell differentiation (Fig. 4F), as well as glycolytic processes and gluconeogenesis. The white and yellow modules correspond to the liver, and the genes within these modules were found to be related to oxidoreductase activity and oxidation–reduction processes, as well as to fatty acid metabolism. Additionally, based on the criteria of GS > 0.2 and MM > 0.8, almost 23% of the genes involved in tissue-specific modules were identified as hub genes related to the six tissues (supplementary table S20). For example, a subset of hub genes, including MYH1A, MYH1C, and MYH1D, were found to have a high correlation and specific expression in breast muscle, which emphasized the role of members of the MYH gene family in breast muscle.
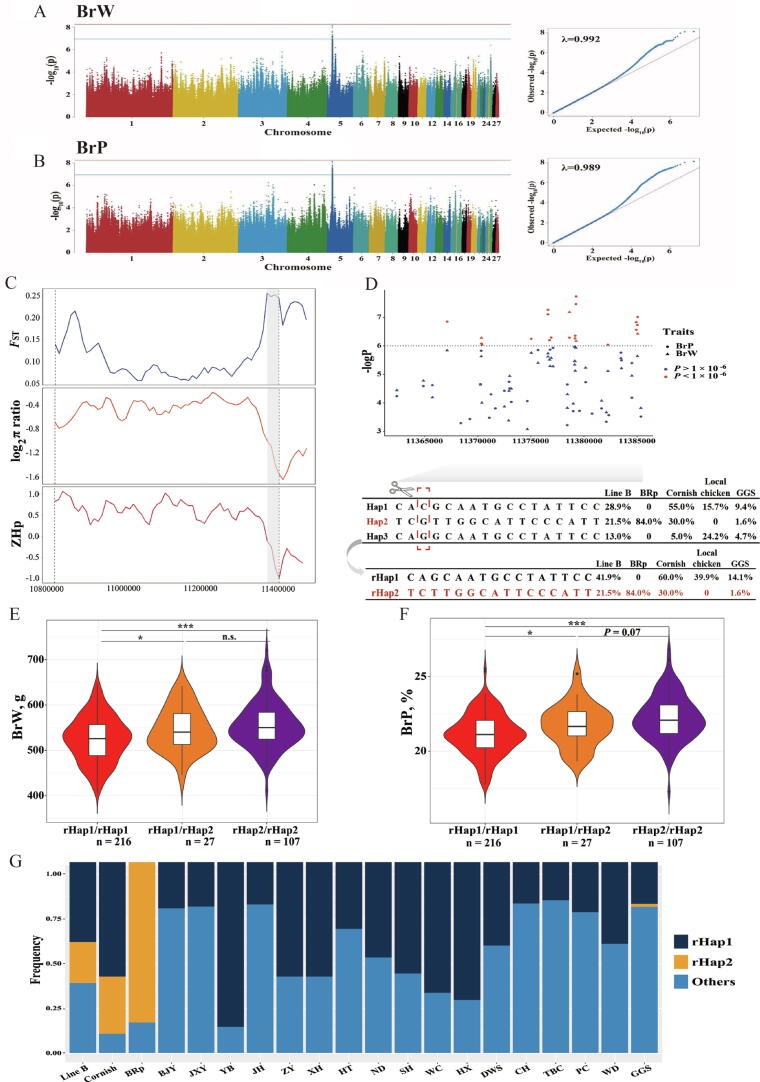
GWAS and fine-mapping for BrW and BrP
Both BrW and BrP are key traits in chicken breeding. The purebred broiler chicken Line B has undergone genomic selection for several generations, and both traits have also changed significantly. Therefore, we aimed to detect the genetic markers associated with them in Line B using the GWAS method. Generation and sex effects were included in the GWAS model due to their significant influence on phenotypes. An ∼ 580 kb region located on GGA5 (10.81–11.39 Mb) showed a significant peak for the two breast muscle traits (Fig. 5A-B). No population stratification was found due to the calculation result of the genomic inflation factor (0.989–0.992) in the Q-Q plot. A total of 18 and 36 SNPs over the suggestive threshold were observed for BrW and BrP, respectively (supplementary table S21). For the lead SNP in the GWAS results of the two traits, the proportions of genetic variance explained by genotype were 13.61% and 9.57%, respectively (supplementary table S21). Within this region, INSC, SOX6 and four noncoding RNAs were identified.
Fig. 5.
GWAS and fine-mapping for BrW and BrP in purebred broiler Line B. (A-B) Manhattan plot and Q-Q plot of association signals for BrW and BrP in Line B, respectively. The red and blue line represented the genomic and suggestive threshold. (C) Results of selective sweep analysis within the GWAS peak. Grey background represented the selected regions. (D) Results for regional GWAS and haplotype inferring. Upper part was regional GWAS results. For the lower part, top haplotypes were firstly inferred with 17 SNPs, bottom refined haplotypes were determined after discarding the SNP within black box, the frequency of each haplotype in various breeds or types of chickens was provided. (E-F) Comparison result for BrW and BrP based on the chickens carrying different refined haplotypes (rHap I and rHap II), respectively. * indicated P < 0.05, *** indicated P < 0.001, n.s. indicated P > 0.05. (G) Refined haplotype frequency distribution among 21 chicken breeds/lines. BRp: broiler paternal line, BJY: Beijing You chicken, CH: Chahua chicken, DWS: Daweishan mini chicken, GGS: Gallus gallus spadiceus, HBM: Huaibei Ma chicken, HT: Hetian chicken, HX: Huxu chicken, JH: Jianghan chicken, JXY: Jingxing Yellow chicken, ND: Ningdu chicken, PC: Piao chicken, SH: Sanhuang chicken, TBC: Tibetan chicken, WC: Wenchang chicken, WD: Wuding chicken, XH: Xianghuang chicken, YB: Yuanbao chicken, ZY: Zhengyang chicken. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Next, we performed selective sweep analysis based on the comparison of Line B with local chicken breeds and GGS to narrow the candidate region (Fig. 5C). Within the GWAS peak, a 30 kb (10.36–10.39 Mb) narrowed region was scanned with an obvious selective signature: FST > 0.25, log2(π broiler/π other) < -1, and ZHp was close to −1. To validate this result, we also performed the same analysis for the three fast-growing purebred broilers, local chicken breeds, and GGS (supplementary Fig. S13). A similar selective signature of reduced pooled heterozygosity (ZHpbroiler < -1.80), decreased nucleotide polymorphism (log2(πbroiler/πother) < -1), and higher population differentiation (FST (broiler vs other) > 0.25) was found in fast-growing purebred broilers, while no evidence was found in local chicken breeds and GGS (ZHplocal > 0, ZHpGGS > 0.5, log2(πlocal/πGGS): 1.02–1.07, FST < 0.05). Therefore, the narrowed 30 kb region was a valid candidate divergent region and affected breast muscle traits.
Within this region, a total of 133 SNPs were detected, and 17 SNPs were found to be related to BrW or BrP (P < 1 × 10-6) (Fig. 5D, supplementary table S22). We inferred haplotypes of these significant SNPs, and three major haplotypes were identified (Fig. 5D). The chickens carrying haplotype II had a more prominent phenotype for breast muscle (supplementary Fig. S23-24). Only one base was different between haplotypes I and III (Fig. 5D), and no phenotypic difference was found between the chickens carrying haplotypes I and III. Therefore, this SNP may not be a causal variant and was not considered further. Therefore, the refined haplotypes were analysed, and two major haplotypes (rHap I and rHap II) were determined (Fig. 5D). Similarly, a significant phenotypic difference was found among the chickens carrying different refined haplotypes (Fig. 5E-F). In addition, the haplotype status among the other 20 breeds was determined (Fig. 5G, supplementary Fig. S25); only fast-growing purebred broilers (Cornish and BRp) were detected as carrying refined haplotype II, and this haplotype was not found in the other 18 breeds (Fig. 5G, supplementary table S23). All the evidence indicated that the refined haplotype II was plausibly the genetic factor in breast muscle traits.
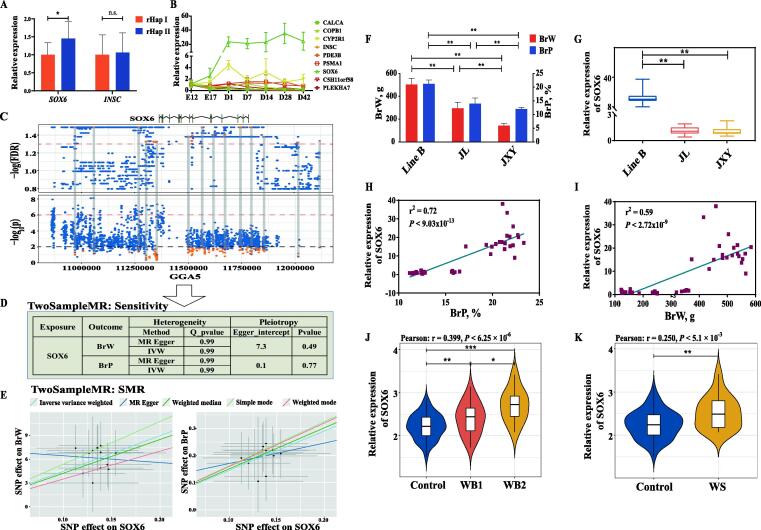
The SOX6 gene is a potential causal gene for BrW and BrP
To identify the causal gene, we first performed RT-PCR for candidate genes in breast muscle. According to the results of the comparison, only SOX6 was differentially expressed at the mRNA and protein levels based on the different refined haplotypes (Fig. 6A, supplementary Fig. S26). While the INSC gene was not differentially expressed, these four noncoding RNAs were not expressed in breast muscle samples. To maximize the possibility of mining the causal genes, we examined the expression of the flanking genes around the candidate region (supplementary Fig. S27). However, no DEGs were found between samples with two refined haplotypes. Additionally, we also determined the expression changes across various developmental stages. SOX6 presented an increasing trend with growth and development processes, while other genes exhibited smaller expression fluctuations (Fig. 6B). Therefore, SOX6 was identified as a potential causal gene affecting BrW and BrP.
Fig. 6.
Expression and correlation analysis for SOX6 and phenotypes in different types of breeds. (A) Effect of refined haplotype on SOX6 and INSC expression. * indicated P < 0.05, n.s. indicated P > 0.05. (B) Expression trend of candidate genes in various developmental stages. E indicated embryo stage; D indicated the days after hatching. (C) Cis-eQTL and GWAS results for BrW in surrounding region of SOX6. The orange dots indicated the SNPs which correlated to SOX6 expression while not to BrW and BrP, the red dot indicated the SNPs were selected after clumping and used in summary-data-based Mendelian randomization (SMR) analysis. (D) Sensitivity analysis in SMR. (E) SMR result for SOX6 on BrW and BrP based on different models. (F) Phenotypic analysis of purebred broiler Line B, local breeds JL (Jinling chicken), and JXY. (G) Relative expression of SOX6 among three breeds. (H-I) Correlation analysis between SOX6 expression and BrP and BrW, respectively. (J-K) The relationship between SOX6 expression level and WB, WS, respectively. WB1 indicated mild WB (score 0), while WB2 indicated moderate WB (score 2), control indicated the normal breast muscle, WS indicated the mild WS (score 1). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
For further verification, we first performed cis-eQTL analysis based on SNPs surrounding SOX6. Significant effects (FDR < 0.05) on SOX6 expression were detected (Fig. 6C), although they did not contribute to narrowing the region or haplotype. Next, we performed SMR to estimate the effect of SOX6 on breast muscle traits. A total of 14 qualified SNPs were included in SMR, and the sensitivity analysis confirmed that no heterogeneity (Q value > 0.05) and pleiotropy (P > 0.05) were present (Fig. 6D, supplementary table S24, Fig. S28-29). The associations between SOX6 and breast muscle traits were not driven by any individual SNP according to the leave-one-out method (supplementary Fig. S30). Then, significant positive effects of SOX6 were detected on BrW (P < 8.20 × 10-13) and BrP (P < 5.34 × 10-14) (Fig. 6E, supplementary table S25), which proved that SOX6 was the causal gene for breast muscle traits.
Although a firm association has been confirmed between the expression level of SOX6 and BrW and BrP phenotypes in Line B, it is necessary to verify this association in other breeds. Hence, we compared the gene expression and phenotypic changes in the local breeds JXY and JL and purebred broiler Line B (Fig. 6F–G). As expected, lower expression of SOX6 was accompanied by smaller breast size, and SOX6 expression was positively correlated with BrW and BrP among the three breeds (Fig. 6G–I). Furthermore, there was a consistent increase in the protein level of SOX6 in Line B compared to JX and JL (supplementary Fig. S31), which is in line with the mRNA measurement results.
Myopathy is commonly observed in chicken breast muscle. Lake et al. indicated that SOX6 may be a potential functional gene regulating WS and WB based on a preliminary GWAS in chickens [57]. Therefore, we scored the WB and WS and correlated them with SOX6 to investigate the regulatory pattern (supplementary Fig. S32-33). The SOX6 was positively correlated with WB and WS status in Line B (Fig. 6J-K). The relationship between CF and myopathy was proven in a previous report and this study (supplementary Fig. S34-35), and an accelerating effect of SOX6 was found on CF traits (supplementary Fig. S36). Additionally, the drip loss of breast muscle was also associated with SOX6 (supplementary Fig. S37). In summary, we found that SOX6 had a crucial influence on myopathy and meat quality related traits.
Discussion
Domestic chicken is an excellent model for genetic studies of phenotypic evolution [2], [58]. After long-term domestication, an array of distinctive local breeds was developed. These breeds were genetically adapted to new environments and selection, such as the local breeds BJY and JXY, known for their high-quality meat flavour; DWS and CH, characterized by dwarfism; and PC and YB, characterized by the absence of a tail and known for their appearance [8], [24], [59], [60], [61]. However, production efficiency challenges persisted. To address these challenges, the modern poultry breeding system has emerged, and the productivity of chickens has been enhanced significantly by intensive directional selection for production traits [3]. Selection for specific traits in chickens was the crucial factor in the steep increase in productivity, accounting for>90% of the improvement [3]. Although the genetic basis of meat production traits has been studied extensively, the core genes resulting in great phenotypic changes have remained largely unclear. In this study, we conducted a comprehensive investigation of genetic diversity and selective sweeps in fast-growing (modern broiler), slow-growing (local chicken), and wild ancestor (GGS) chickens. A study of selective sweeps revealed a list of 163 protein-coding genes that underwent positive selection and played a pivotal role in domestication and breeding processes in the development of traits related to muscle production, reproduction, digestive ability, disease resistance, and sensory performance. The most significant enrichment terms and mapped known QTLs are related to muscle development and body composition in broilers.
This study provides a large-scale whole-genome sequencing dataset for purebred broilers, especially for Line B, which had been developed over three generations using genomic selection [15]. The phylogenetic analysis showed that Line B shared a similar but not completely identical genetic background with other broilers. Compared to local and wild chickens, a relatively low diversity of 3.8 × 10-3 was observed in Line B, indicating genomic evidence of intensive selection for phenotypic improvement. Although meat production was elevated by intensive selection, it incurred a certain selection “cost” [62], [63], [64]. A higher number of dSNPs and a higher ratio to synonymous SNPs were found in Line B and other broilers compared to GGS and local chicken breeds; this result corroborates the “cost of domestication” hypothesis, similar to the conclusion of Wang et al. [53]. Most harmful alleles were masked in heterozygotes. Here, our results showed that domestication and the current breeding process were accompanied by increasing genetic load, especially in the heterozygous variant load. Remarkably, we found that directional breeding for modern purebred broilers relieved the accumulation of deleterious mutations caused by long-term domestication. These findings revealed the advantages of current breeding systems and strategies in removing latent unfavourable effects from dSNPs while pursuing desired production traits.
In this study, we detected putative sweeps in broilers specifically as described above. Here, we used the yellow skin locus gene (BCO2) [65] and reproduction-related gene (TSHR) [66] as proofs of principle (supplement table S26), demonstrating that our approach can reveal fixed sweeps. The sweep window harbouring the TSHR locus showed a ZLSBL value of 3.15 (top 1.61%) and a ZHp value of −3.88 (top 0.1%). The window covering the BCO2 gene showed a ZLSBL value of 2.48 (top 3%) and a ZHp value of −3.57 (top 0.03%). Similarly, the genes INSR, MYH1s, SOX6, and MYO9B, which are related to skeletal muscle development, were also detected in sweep windows in broilers. In a previous study, Qanbari et al. compared the genomic variants between modern broilers and RJFs and detected a list of putative sweeps with a strict threshold [9]. We found that the most significant overlapping genes included IGF-1, BCO2, and SLC26A8, as well as other genes (NUP37, MAPK13, MAPK14, and PMCH) with ZFST > 6 from a previous report [9]. In the current study, we excluded the putative sweep windows that were also differentiated between local chicken breeds and GGS and reserved the windows harbouring only the top significant signal in broilers, although the significance threshold was relatively relaxed compared to that reported by Qanbari et al. [9]. These results suggested that our findings were plausibly associated with modern intensive selection, especially for meat production traits, which provided new genetic targets in chicken breeding programs, including the SOX6 and some MYH1 genes.
Based on the QTL annotation (meat production, fatness, CO2 partial pressure) and GO enrichment (heart development) results of selected regions (Fig. 3C, supplementary Fig. S16, table S13), muscle development, cardiopulmonary function and health condition, and fatness related metabolism were significantly changed in purebred broilers. Correspondingly, the breast and thigh muscle, heart and lung tissues, and liver and abdominal fat tissues were applied to transcriptomic study. According to transcriptomic evidence of six tissues from Line B and JXY, we identified a list of hub genes for the development of different tissues. The genes within the corresponding modules were annotated to related biological functions of various organs, which is consistent with the results in cattle [67], sheep [68], and local chicken breeds [69]. Based on the empirical threshold of hub genes and TSGs, we found that the expression of a subset of hub genes (MYH1A, MYH1C, MYH1D, etc.) was muscle specific. The MYH1s are the marker genes of fast-type muscle fibers, which is especially consistent with the function of breast muscle. For the 163 PSGs, we found 18 and 22 TSGs, some of which were differentially expressed between Line B and BJY. For instance, FTCD is formiminotransferase cyclodeaminase and involved in autoimmune hepatitis [70]. This gene was expressed at lower levels in the liver of Line B, which may be correlated with the low incidence of hepatic diseases, such as fatty liver syndrome, in fast-growing chickens [71]. In addition, consistent with previous results, members of the MYH gene family were highly expressed in the breast muscle of Line B. Hence, it is reasonable to conclude that breast muscle is the major distinction between modern broilers and local chicken breeds or GGS based on the genomic and transcriptomic evidence presented here.
Breast muscle weight and size are crucial in modern chicken breeding programs. Chickens living in wild environments have smaller breast muscles, which could help them adapt to the stressful environment and natural enemies [72]. For example, GGSs have small body sizes and weights and weak flying abilities, which are beneficial for migrating and avoiding natural disasters. However, these characteristics are a severe disadvantage in modern aviculture. After decades of directional breeding, the BrP of modern broilers can reach 25% (breast weight to dressed weight) [73], [74]. This produces a plump carcass, the appearance of which is preferred over that of local breeds in the chicken market. For Line B, the breeding period is relatively short compared to breeding periods of other commercial breeds (Cobb, Cornish, etc.); in this line, the BrP reached 21.5% (breast weight to body weight) in the most recent generation [15]. Combining the results of GWAS and selective sweep analysis, we found that the genomic region GGA5: 11.36–11.39 Mb appeared to be a plausible candidate region and was significantly associated with BrW and BrP and genetically differentiated between purebred broilers and local chicken breeds or GGS. Within this region, we detected two refined haplotypes affecting SOX6 expression. The frequency of the dominant haplotype (corresponding to higher BrW and BrP) was higher in Line B, Cornish, and BRp, while it was extremely low in local chicken breeds and GGS. It is practical to perform BLUP-given genetic architecture (BLUP|GA) by constructing a GA matrix based on the haplotype information and incorporating it into the G matrix. This method has been proven to have a higher prediction accuracy [75], [76]. These results support available genetic markers for improving breast muscle traits in modern chicken breeding programs.
SOX6 is a well-known factor determining the conversion of muscle fibre types in animals [77]. In a report by Quiat et al., SOX6 was enriched in fast myofibers and directly repressed the transcription of slow myofiber-enriched genes [78]. However, Zhang et al. reported that overexpression of SOX6 in leg muscle unexpectedly increased the transcription of the slow myofiber marker gene MYH7B, as well as the fast myofiber marker genes (MYH1A, Tnnc2, Tnni2, etc.) [78]. For breast muscle, the myogenesis and development of satellite cells could be promoted by overexpression of SOX6, which also led to the increased expression of MYH1A and decreased expression of MYH7B [78]. In total, these findings suggest a stimulating function for breast muscle weight due to the higher fibre diameter in the fast-twitch group compared to the slow-twitch group. For broiler Line B, fast-twitch myofibers are the major component of breast muscle, as well as for other commercial broilers. Remarkably, this gene spanned a significant selective signature (π ratio < 0.3) in purebred broilers, indicating that SOX6 was selected for over the breeding history, which is consistent with previous reports on chickens [79], [80]. The higher expression in Line B compared to the slow-growing line supported this conjecture. In addition, Mendelian randomization is a solid approach that is applied to the use of genetic variants to address causality [81]. To prove the causal effect of SOX6 on BrW and BrP, we performed the SMR method integrating cis-eQTL and GWAS results and proved the causal relationship.
In addition, myopathy (WS and WB) is caused by the rapid growth of breast muscle. It is contradictory to modern human demand, and the ‘antagonism’ relationship could be a more anticipated outcome for breast muscle development and myopathy. It has been predicted that the SOX6 locus might be correlated with muscle myopathy in the Cobb breed [57], as well as MYH1s [82]. Here, an increasing effect of SOX6 on WB and WS was discovered, which was consistent with a previous assumption [57]. Notably, both SOX6 and MYH1s harboured top selection signatures in purebred broilers, and MYH1s were directly regulated by the transcription factor SOX6 in muscle tissue. Hence, it is logical to infer that the SOX6-MYH1 axis is the regulatory core of breast muscle development, and this axis could act as a genetic target to regulate the balance between higher breast yield and improved meat quality in modern chicken breeding programs.
The productivity of modern broilers far exceeds that of their ancestor GGS and that of local chicken breeds; however, the increase in meat production efficiency is gradually slowing, although the production performance is still improving. Therefore, there is still great potential for development before reaching their growth limit. Here, the SOX6 gene was reported to harbour some copy number variants and binding sites for microRNAs (miRNAs) [83], [84], [85], as well as some motifs regulating muscle-related genes as a transcription factor [86], which provide plentiful genetic materials for use in genetic breeding aimed at enhancing breast yield and production efficiency in high-performance broilers or local chicken breeds.
Conclusions
In conclusion, we generated a large-scale atlas comprising the genomic variants and transcriptional changes in multiple tissues and stages related to muscle development in fast-growing purebred broilers and slow-growing local chicken breeds, providing sufficient and valuable information for exploring the genetic basis of meat production traits. Breast muscle was the major difference between purebred broilers and local chicken breeds or GGS based on genomic and transcriptomic evidence. PSGs, TSGs, and DEGs contributed to a better understanding of the evolutionary differences in different types of chickens. The SOX6–MYH1s axis and causal gene SOX6 for BrW, BrP, and myopathy will facilitate selection aimed at high meat yield in chickens.
Compliance with Ethics requirements
All Institutional and National Guidelines for the care and use of chickens were exactly followed.
CRediT authorship contribution statement
Xiaodong Tan: Conceptualization, Methodology, Software, Data curation, Visualization, Writing – review & editing. Ranran Liu: Conceptualization, Methodology, Data curation, Writing – review & editing. Di Zhao: Investigation, Software. Zhengxiao He: Investigation, Software. Wei Li: Investigation, Software. Maiqing Zheng: Investigation, Data curation. Qinghe Li: Investigation, Software. Qiao Wang: Investigation, Software. Dawei Liu: Investigation. Furong Feng: Investigation. Dan Zhu: Investigation. Guiping Zhao: Conceptualization, Methodology, Writing – review & editing. Jie Wen: Conceptualization, Methodology, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was funded by grants from the National Key Research and Development Program of China (2022YFD1301600), the Agricultural Science and Technology Innovation Program (CAAS-ZDRW202005), and the China Agriculture Research System (CARS-41).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.02.016.
Contributor Information
Guiping Zhao, Email: zhaoguiping@caas.cn.
Jie Wen, Email: wenjie@caas.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.FAOSTAT. [cited 2022 Oct 10] Available from: https://www.fao.org/faostat/en/#data/QCL.
- 2.Darwin C. The variation of animals and plants under domestication. D. Appleton; 1894. [Google Scholar]
- 3.Muir W.M., Wong G.K., Zhang Y., Wang J., Groenen M.A., Crooijmans R.P., et al. Genome-wide assessment of worldwide chicken SNP genetic diversity indicates significant absence of rare alleles in commercial breeds. Proc Natl Acad Sci U S A. 2008;105(45):17312–17317. doi: 10.1073/pnas.0806569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubin C.J., Zody M.C., Eriksson J., Meadows J.R., Sherwood E., Webster M.T., et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464(7288):587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- 5.Zuidhof M.J., Schneider B.L., Carney V.L., Korver D.R., Robinson F.E. Growth, efficiency, and yield of commercial broilers from 1957, 1978, and 2005. Poult Sci. 2014;93(12):2970–2982. doi: 10.3382/ps.2014-04291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooke M., Birkhead T.R. Cambridge University Press; 1991. Cambridge encyclopedia of ornithology. [Google Scholar]
- 7.IPCC, 2021: Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kingdom and New York, NY, USA, In press, doi:10.1017/9781009157896.
- 8.Wang M.S., Otecko N.O., Wang S., Wu D.D., Yang M.M., Xu Y.L., et al. An Evolutionary Genomic Perspective on the Breeding of Dwarf Chickens. Mol Biol Evol. 2017;34(12):3081–3088. doi: 10.1093/molbev/msx227. [DOI] [PubMed] [Google Scholar]
- 9.Qanbari S., Rubin C.J., Maqbool K., Weigend S., Weigend A., Geibel J., et al. Genetics of adaptation in modern chicken. PLoS Genet. 2019;15(4):e1007989. doi: 10.1371/journal.pgen.1007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X., Sun J., Zhao G., Li W., Tan X., Zheng M., et al. Identification of Major Loci and Candidate Genes for Meat Production-Related Traits in Broilers. Front Genet. 2021;12 doi: 10.3389/fgene.2021.645107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K., Hu H., Tian Y., Li J., Scheben A., Zhang C., et al. The Chicken Pan-Genome Reveals Gene Content Variation and a Promoter Region Deletion in IGF2BP1 Affecting Body Size. Mol Biol Evol. 2021;38(11):5066–5081. doi: 10.1093/molbev/msab231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Z., Li M., Cheng H., Fan W., Yuan Z., Gao Q., et al. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat Commun. 2018;9(1):2648. doi: 10.1038/s41467-018-04868-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutter N.B., Bustamante C.D., Chase K., Gray M.M., Zhao K., Zhu L., et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316(5821):112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan X., Liu L., Liu X., Cui H., Liu R., Zhao G., et al. Large-Scale Whole Genome Sequencing Study Reveals Genetic Architecture and Key Variants for Breast Muscle Weight in Native Chickens. Genes (Basel) 2021;13(1):3. doi: 10.3390/genes13010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan X., Liu R., Li W., Zheng M., Zhu D., Liu D., et al. Assessment the effect of genomic selection and detection of selective signature in broilers. Poult Sci. 2022;101(6) doi: 10.1016/j.psj.2022.101856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong F., Zhao G., He Z., Sun J., Wang X., Liu D., et al. Serum Creatine Kinase as a Biomarker to Predict Wooden Breast in vivo for Chicken Breeding. Front Physiol. 2021;12 doi: 10.3389/fphys.2021.711711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong F., Bai L., He Z., Sun J., Tan X., Zhao D., et al. Integrated metabolomics and lipidomics evaluate the alterations of flavor precursors in chicken breast muscle with white striping symptom. Front Physiol. 2022;13:1079667. doi: 10.3389/fphys.2022.1079667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu R., Kong F., Xing S., He Z., Bai L., Sun J., et al. Dominant changes in the breast muscle lipid profiles of broiler chickens with wooden breast syndrome revealed by lipidomics analyses. J Anim Sci Biotechnol. 2022;13(1):93. doi: 10.1186/s40104-022-00743-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riggs M.R., Hauck R., Baker-Cook B.I., Osborne R.C., Pal A., Terra M.T.B., et al. Meat quality of broiler chickens processed using electrical and controlled atmosphere stunning systems. Poult Sci. 2022;102(3) doi: 10.1016/j.psj.2022.102422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W., Liu R., Zheng M., Feng F., Liu D., Guo Y., et al. New insights into the associations among feed efficiency, metabolizable efficiency traits and related QTL regions in broiler chickens. J Anim Sci Biotechnol. 2020;11:65. doi: 10.1186/s40104-020-00469-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulfah M., Kawahara-Miki R., Farajalllah A., Muladno M., Dorshorst B., Martin A., et al. Genetic features of red and green junglefowls and relationship with Indonesian native chickens Sumatera and Kedu Hitam. BMC Genomics. 2016;17(1):1–9. doi: 10.1186/s12864-016-2652-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q., Jia Y., Wang Y., Jiang Z., Zhou X., Zhang Z., et al. Evolution of cis-and trans-regulatory divergence in the chicken genome between two contrasting breeds analyzed using three tissue types at one-day-old. BMC Genomics. 2019;20(1):1–10. doi: 10.1186/s12864-019-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X., Otecko N.O., Peng M., Weng Z., Li W., Chen J., et al. Genome-wide genetic structure and selection signatures for color in 10 traditional Chinese yellow-feathered chicken breeds. BMC Genomics. 2020;21(1):1–12. doi: 10.1186/s12864-020-6736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M.S., Huo Y.X., Li Y., Otecko N.O., Su L.Y., Xu H.B., et al. Comparative population genomics reveals genetic basis underlying body size of domestic chickens. J Mol Cell Biol. 2016;8(6):542–552. doi: 10.1093/jmcb/mjw044. [DOI] [PubMed] [Google Scholar]
- 25.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997 2013. doi: doi.org/10.48550/arXiv.1303.3997.
- 27.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10(2) doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falush D., Wirth T., Linz B., Pritchard J.K., Stephens M., Kidd M., et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S., Nei M., Dudley J., Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9(4):299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C., Dong S.S., Xu J.Y., He W.M., Yang T.L. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35(10):1786–1788. doi: 10.1093/bioinformatics/bty875. [DOI] [PubMed] [Google Scholar]
- 36.Terhorst J., Kamm J.A., Song Y.S. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet. 2017;49(2):303–309. doi: 10.1038/ng.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam K., Mugal C., Nabholz B., Schielzeth H., Wolf J.B., Backström N., et al. Molecular evolution of genes in avian genomes. Genome Biol. 2010;11(6):R68. doi: 10.1186/gb-2010-11-6-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H., Patterson N., Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20(3):393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovaka S., Zimin A.V., Pertea G.M., Razaghi R., Salzberg S.L. and Pertea M Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019;20(1):278. doi: 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.She X., Rohl C.A., Castle J.C., Kulkarni A.V., Johnson J.M., Chen R. Definition, conservation and epigenetics of housekeeping and tissue-enriched genes. BMC Genomics. 2009;10:269. doi: 10.1186/1471-2164-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nature Genet. 2012;44(7):821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aslam M.L., Carraro R., Bestin A., Cariou S., Sonesson A.K., Bruant J.S., et al. Genetics of resistance to photobacteriosis in gilthead sea bream (Sparus aurata) using 2b-RAD sequencing. BMC Genet. 2018;19(1):43. doi: 10.1186/s12863-018-0631-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilmour A., Gogel B., Cullis B., Welham S., Thompson R. VSN international ltd; Hemel hempstead: 2015. ASReml user guide release 4.1 structural specification. [Google Scholar]
- 47.Turner S.D. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv. 2014 doi: 10.21105/joss.00731. [DOI] [Google Scholar]
- 48.Zheng D., Li N., Hou R., Zhang X., Wu L., Sundquist J., et al. Glucagon-like peptide-1 receptor agonists and diabetic retinopathy: nationwide cohort and Mendelian randomization studies. BMC Med. 2023;21(1):40. doi: 10.1186/s12916-023-02753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shabalin A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28(10):1353–2138. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Xie X., Yang Y., Ren Q., Ding X., Bao P., Yan B., et al. Accumulation of deleterious mutations in the domestic yak genome. Anim Genet. 2018;49(5):384–392. doi: 10.1111/age.12703. [DOI] [PubMed] [Google Scholar]
- 53.Wang M.S., Zhang J.J., Guo X., Li M., Meyer R., Ashari H., et al. Large-scale genomic analysis reveals the genetic cost of chicken domestication. BMC Biol. 2021;19(1):1–16. doi: 10.1186/s12915-021-01052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z., Han S., Shen X., Wang Y., Cui C., He H., et al. The landscape of DNA methylation associated with the transcriptomic network in layers and broilers generates insight into embryonic muscle development in chicken. Int J Biol Sci. 2019;15(7):1404–1418. doi: 10.7150/ijbs.35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geng D, Yang X, Wang R, Deng S, Li L, Hu X, et al. A novel stopgain mutation c.G992A (p.W331X) in TACR3 gene was identified in nonobstructive azoospermia by targeted next-generation sequencing. J Clin Lab Anal 2019; 33(3): e22700. doi: 10.1002/jcla.22700. [DOI] [PMC free article] [PubMed]
- 56.Dirami T., Rode B., Jollivet M., Da Silva N., Escalier D., Gaitch N., et al. Missense mutations in SLC26A8, encoding a sperm-specific activator of CFTR, are associated with human asthenozoospermia. Am J Hum Genet. 2013;92(5):760–766. doi: 10.1016/j.ajhg.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lake J.A., Dekkers J.C.M., Abasht B. Genetic basis and identification of candidate genes for wooden breast and white striping in commercial broiler chickens. Sci Rep. 2021;11(1):6785. doi: 10.1038/s41598-021-86176-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang M.S., Thakur M., Peng M.S., Jiang Y., Frantz L.A.F., Li M., et al. 863 genomes reveal the origin and domestication of chicken. Cell Res. 2020;30(8):693–701. doi: 10.1038/s41422-020-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao G.P., Chen J.L., Zheng M.Q., Wen J., Zhang Y. Correlated responses to selection for increased intramuscular fat in a Chinese quality chicken line. Poult Sci. 2007;86(11):2309–2314. doi: 10.1093/ps/86.11.2309. [DOI] [PubMed] [Google Scholar]
- 60.Liu R., Wang H., Liu J., Wang J., Zheng M., Tan X., et al. Uncovering the embryonic development-related proteome and metabolome signatures in breast muscle and intramuscular fat of fast-and slow-growing chickens. BMC Genomics. 2017;18(1):816. doi: 10.1186/s12864-017-4150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y.M., Khederzadeh S., Li S.R., Otieno Otecko N., Irwin D.M., Thakur M., et al. Integrating Genomic and Transcriptomic Data to Reveal Genetic Mechanisms Underlying Piao Chicken Rumpless Trait. Genom Proteom Bioinf. 2021;9(5):787–799. doi: 10.1016/j.gpb.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Larson G., Fuller D.Q. The evolution of animal domestication. Annu Rev Ecol Evol Syst. 2014;45:115–136. doi: 10.1146/annurev-ecolsys-110512-135813. [DOI] [Google Scholar]
- 63.Frantz L.A., Larson G.A. Hybrid Communities. Routledge; 2018. genetic perspective on the domestication continuum; pp. 23–37. [Google Scholar]
- 64.Larson G., Burger J. A population genetics view of animal domestication. Trends Genet. 2013;29(4):197–205. doi: 10.1016/j.tig.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 65.Eriksson J., Larson G., Gunnarsson U., Bed'Hom B., Tixier-Boichard M., Strömstedt L., et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008;4(2):e1000010. doi: 10.1371/journal.pgen.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rubin CJ, Zody MC, Eriksson J, Meadows JR, Sherwood E, Webster MT, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 2010; 464(7288): 587-591. doi: 10.1371/journal.pgen.1000010. [DOI] [PubMed]
- 67.Zhang T., Wang T., Niu Q., Xu L., Chen Y., Gao X., et al. Transcriptional atlas analysis from multiple tissues reveals the expression specificity patterns in beef cattle. BMC Biol. 2022;20(1):79. doi: 10.1186/s12915-022-01269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sabino M., Carmelo V.A.O., Mazzoni G., Cappelli K., Capomaccio S., Ajmone-Marsan P., et al. Gene co-expression networks in liver and muscle transcriptome reveal sex-specific gene expression in lambs fed with a mix of essential oils. BMC Genomics. 2018;19(1):236. doi: 10.1186/s12864-018-4632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xing S., Liu R., Zhao G., Groenen M.A.M., Madsen O., Liu L., et al. Time Course Transcriptomic Study Reveals the Gene Regulation During Liver Development and the Correlation With Abdominal Fat Weight in Chicken. Front Genet. 2021;12 doi: 10.3389/fgene.2021.723519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Terziroli Beretta-Piccoli B., Mieli-Vergani G., Vergani D. Autoimmmune hepatitis. Cell Mol Immunol. 2022;19(2):158–176. doi: 10.1038/s41423-021-00768-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Z., Li Q., Liu R., Zhao G., Zhang Y., Zheng M., et al. Expression and methylation of microsomal triglyceride transfer protein and acetyl-CoA carboxylase are associated with fatty liver syndrome in chicken. Poult Sci. 2016;95(6):1387–1395. doi: 10.3382/ps/pew040. [DOI] [PubMed] [Google Scholar]
- 72.Desta T. Phenotypic characteristic of junglefowl and chicken. Worlds Poult Sci J. 2019;75(1):69–82. doi: 10.1017/S0043933918000752. [DOI] [Google Scholar]
- 73.Agostini P., Santos R., Khan D., Siebert D., van der Aar P. The optimum valine: lysine ratios on performance and carcass traits of male broilers based on different regression approaches. Poult Sci. 2019;98(3):1310–1320. doi: 10.3382/ps/pey454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walk C.L., Rao S.V.R. High doses of phytase on growth performance and apparent ileal amino acid digestibility of broilers fed diets with graded concentrations of digestible lysine. J Anim Sci. 2019;97(2):698–713. doi: 10.1093/jas/sky441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morgante F., Huang W., Maltecca C., Mackay T.F.C. Effect of genetic architecture on the prediction accuracy of quantitative traits in samples of unrelated individuals. Heredity (Edinb) 2018;120(6):500–514. doi: 10.1038/s41437-017-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Z., Erbe M., He J., Ober U., Gao N., Zhang H., et al. Accuracy of whole-genome prediction using a genetic architecture-enhanced variance-covariance matrix. 2015;5(4):615–627. doi: 10.1534/g3.114.016261. G3 (Bethesda. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quiat D., Voelker K.A., Pei J., Grishin N.V., Grange R.W., Bassel-Duby R., et al. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proc Natl Acad Sci U S A. 2011;108(25):10196–10201. doi: 10.1073/pnas.1107413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Z., Lin S., Luo W., Ren T., Huang X., Li W., et al. Sox6 Differentially Regulates Inherited Myogenic Abilities and Muscle Fiber Types of Satellite Cells Derived from Fast- and Slow-Type Muscles. Int J Mol Sci. 2022;23(19) doi: 10.3390/ijms231911327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu Z., Sun C., Qu L., Wang K. and Yang N Genome-Wide Detection of Selective Signatures in Chicken through High Density SNPs. PLoS One. 2016;11(11):e0166146. doi: 10.1371/journal.pone.0166146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu W., Lee W.R. and Abasht B Detection of genomic signatures of recent selection in commercial broiler chickens. BMC Genet. 2016;17(1):122. doi: 10.1186/s12863-016-0430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanderson E., Glymour M.M., Holmes M.V., Kang H., Morrison J., Munafò M.R., et al. Mendelian randomization Nat Rev Methods Primers. 2022;2(1):6. doi: 10.1038/s43586-021-00092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pampouille E., Berri C., Boitard S., Hennequet-Antier C., Beauclercq S.A., Godet E., et al. Mapping QTL for white striping in relation to breast muscle yield and meat quality traits in broiler chickens. BMC Genomics. 2018;19(1):202. doi: 10.1186/s12864-018-4598-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin S., Lin X., Zhang Z., Jiang M., Rao Y., Nie Q., et al. Copy Number Variation in SOX6 Contributes 3to Chicken Muscle Development. Genes. 2018;9(1) doi: 10.3390/genes9010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X.Y., Chen X.L., Huang Z.Q., Chen D.W., Yu B., He J., et al. MicroRNA-499-5p regulates porcine myofiber specification by controlling Sox6 expression. Animal. 2017;11(12):2268–2274. doi: 10.1017/s1751731117001008. [DOI] [PubMed] [Google Scholar]
- 85.Liu Y., Zhang M., Shan Y., Ji G., Ju X., Tu Y., et al. miRNA-mRNA network regulation in the skeletal muscle fiber phenotype of chickens revealed by integrated analysis of miRNAome and transcriptome. Sci Rep. 2020;10(1):10619. doi: 10.1038/s41598-020-67482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.An C.I., Dong Y., Hagiwara N. Genome-wide mapping of Sox6 binding sites in skeletal muscle reveals both direct and indirect regulation of muscle terminal differentiation by Sox6. BMC Dev Biol. 2011;11:59. doi: 10.1186/1471-213x-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.