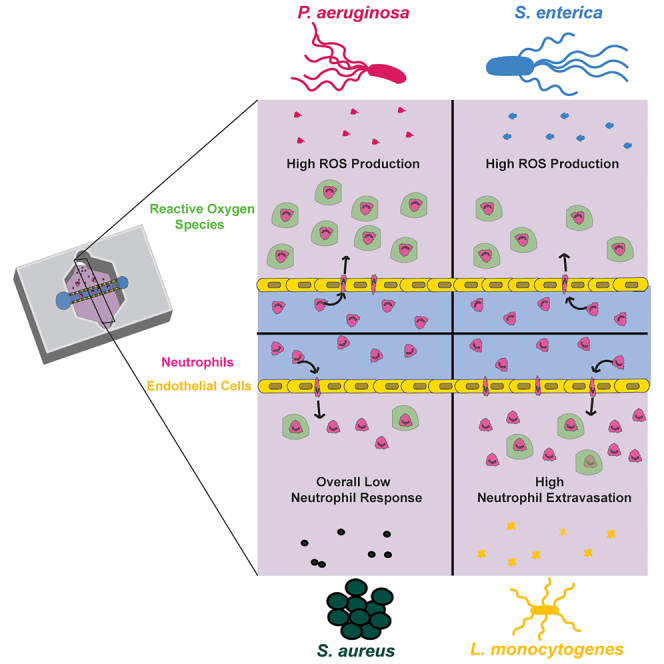
Summary
An efficient neutrophil response is critical for fighting bacterial infections, which remain a significant global health concern; therefore, modulating neutrophil function could be an effective therapeutic approach. While we have a general understanding of how neutrophils respond to bacteria, how neutrophil function differs in response to diverse bacterial infections remains unclear. Here, we use a microfluidic infection-on-a-chip device to investigate the neutrophil response to four bacterial species: Pseudomonas aeruginosa, Salmonella enterica, Listeria monocytogenes, and Staphylococcus aureus. We find enhanced neutrophil extravasation to L. monocytogenes, a limited overall response to S. aureus, and identify IL-6 as universally important for neutrophil extravasation. Furthermore, we demonstrate a higher percentage of neutrophils generate reactive oxygen species (ROS) when combating gram-negative bacteria versus gram-positive bacteria. For all bacterial species, we found the percentage of neutrophils producing ROS increased following extravasation through an endothelium, underscoring the importance of studying neutrophil function in physiologically relevant models.
Subject areas: Immunology, Bacteriology
Graphical abstract
Highlights
-
•
Neutrophils have enhanced extravasation toward L. monocytogenes
-
•
Neutrophils exhibit a unique and limited response to S. aureus
-
•
Gram-negative bacteria elicit more ROS-producing neutrophils than gram-positive bacteria
-
•
Physiologically relevant models can uncover unique immune responses to infection
Immunology; Bacteriology
Introduction
Neutrophils serve an important physiological role as the first line of defense against infection. In a healthy individual, neutrophils are constitutively present in the blood in a quiescent state. Following infection, they become activated by signals from both the pathogen and host cells, including the blood vessel endothelium. Upon activation, neutrophils extravasate through the blood vessel in a process known as transendothelial migration or extravasation. They then migrate through the tissue to reach the site of infection where they perform a variety of antimicrobial functions to clear the infection.1 Not only do bacterial signals activate and direct this neutrophil response, bacteria also release factors that damage neutrophils and interfere with their ability to efficiently clear the infection. This process generally describes the neutrophil response to infection; however, it has also been shown that the extent of the neutrophil recruitment and the specific antimicrobial functions carried out by neutrophils varies in response to distinct pathogen sources.2 While our general understanding of the neutrophil response to infection is well defined, how neutrophils differentially respond to diverse pathogens remains unclear.
Bacterial infections can wreak havoc on the human body, causing tissue damage and, in the most extreme cases, can lead to death due to sepsis or other inflammatory diseases.3,4,5 Bacterial infections remain a major health concern due to their prevalence, particularly in vulnerable, immunosuppressed individuals, and the increase in antibiotic-resistant strains which limits our ability to treat them. Treating bacterial infections by targeting innate immune cells, including neutrophils, could be a more effective and sustainable way to treat infection. However, we first need a more complete understanding of how neutrophils respond to distinct bacterial species and what signals are universally important or unique to individual infections. Bacterial species are typically categorized in two ways, by the presence of a cell wall (gram-negative vs. gram-positive) or by their method of infection (extracellular vs. intracellular).6,7 Different bacterial species have been shown to elicit differing neutrophil responses. An early study showed varying requirements for protein production and CD18 binding for neutrophils transmigrating to sources of Escherichia coli, Staphylococcus aureus, and Streptococcus pneumonia; however, a full understanding of these distinct responses has yet to be achieved.2 Pseudomonas aeruginosa, Salmonella enterica, Listeria monocytogenes, and S. aureus are all bacterial species of special interest to the fields of human health and immunology. Three of these bacterial species (P. aeruginosa, S. enterica, and S. aureus) have been identified as priority pathogens for new antibiotic development by the World Health Organization, as each have multidrug-resistant strains and are common nosocomial infections. As a result, there is a pressing need to develop new therapeutic avenues to treat these infections. Specifically, P. aeruginosa is a common nosocomial, opportunistic pathogen and a leading cause of death in cystic fibrosis patients.8 S. enterica is a leading cause of gastroenteritis with a global estimate of around 94 million cases each year.9 L. monocytogenes is a major foodborne pathogen with particular dangers for pregnant woman via fetal-placental infection.10 S. aureus is frequently implicated worldwide for a role in morbidity and mortality as a cause of sepsis and pneumonia and is the most common multidrug-resistant bacterial species.11,12 A better understanding of the distinct ways these bacterial species lead to neutrophil activation, migration, and effector mechanisms, such as reactive oxygen species (ROS) production, could lead to the development of targeted therapeutics for the treatment of these unique pathogens.
One limitation for studying the neutrophil response has been the development of accurate and relevant models of the human immune system. To overcome this challenge, we recently developed an infection-on-a-chip device to investigate how cell-cell interactions regulate primary human neutrophil extravasation and migration distance. This device is ideal for studying the neutrophil response to infection as it contains important aspects of an in vivo infectious microenvironment including relevant three-dimensional blood vessel architectures, a collagen extracellular matrix, live and intact bacterial species, and primary human cells including neutrophils and endothelial cells. We previously used this device to investigate neutrophil-endothelial cell interactions in the presence of P. aeruginosa and found that endothelial cells are crucial for driving neutrophil extravasation and increasing neutrophil lifetime through production of IL-6 and GM-CSF, respectively.13 In a follow-up study, we found that neutrophil extravasation dynamics in the presence of the opportunistic fungal pathogen Aspergillus fumigatus were different than P. aeruginosa but the role of IL-6 was conserved.14 This indicates the presence of universally required signals for the neutrophil response even when the response varies from pathogen to pathogen. It also demonstrated the ability of our infection-on-a-chip device to elucidate differences in the human neutrophil response to distinct pathogens that mimic in vivo biology. This makes it an ideal platform for interrogating how the human neutrophil response varies under diverse bacterial conditions. While we previously quantified extravasation and distance from the lumen within our device, other important aspects of the neutrophil response could be affected—including interstitial migration or antimicrobial function. It is, therefore, important to consider those as well when developing a holistic understanding of the neutrophil response.
In this study, we used our infection-on-a-chip device to study neutrophil extravasation, migration, and ROS production in response to P. aeruginosa, S. enterica, L. monocytogenes, and S. aureus. We found L. monocytogenes elicited significantly greater neutrophil extravasation compared to the other bacterial species and demonstrated a conserved requirement of IL-6 signaling for neutrophil extravasation across our four bacterial species. Further, we show a significantly higher percentage of neutrophils produce intracellular ROS in response to gram-negative bacteria compared to gram-positive bacteria and the percentage of neutrophils producing intracellular ROS in response to all bacterial species was higher following extravasation through the endothelium. Together, these results show that our infection-on-a-chip device can be used to identify distinct neutrophil responses to diverse pathogens through quantification of extravasation, migration, and ROS production and highlight the importance of using physiologically relevant models for studying human neutrophil function.
Results
Validation of infection-on-a-chip device for diverse bacterial species
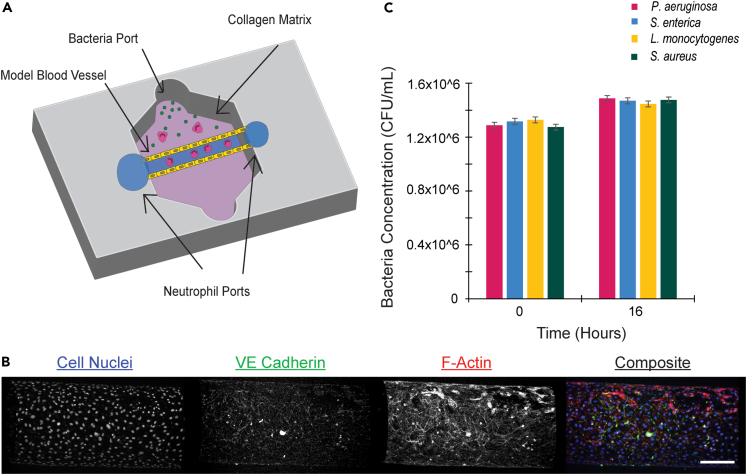
We previously developed an infection-on-a-chip device that incorporates important aspects of the infectious microenvironment including live, intact bacteria, a model blood vessel, and primary human immune cells (Figure 1A).15 This study sought to investigate how neutrophils respond to and interact with diverse bacterial species using this model. Our model, based on the LumeNEXT device, uses endothelial lumens in a collagen matrix as a model blood vessel. To confirm the formation of patent endothelial cell monolayers, we fixed lumens and stained for cell nuclei, actin, and VE-Cadherin (Figure 1B).16 Stained images showed even distribution of cell-nuclei, extensive actin coverage, and tight cell junctions demonstrating a confluent endothelial cell monolayer across the lumen structure within the infection-on-a-chip device.
Figure 1.
Validation of infection-on-a-chip device
(A) Schematic of infection-on-a-chip device showing primary human neutrophils (pink) inside of an endothelial cell lumen (yellow) migrating to a source of live bacteria (green). Collagen matrix (purple) surrounds the lumen.
(B) Stained images of endothelial cell lumens. Cell nuclei are stained using Hoechst (blue), F-actin is stained with Phalloidin (red) and VE-Cadherin (green) is stained with an anti-CD144 antibody. Scale bar is 100 μm.
(C) Bacterial CFUs at 0 and 16 h for P. aeruginosa, S. enterica, L. monocytogenes, and S. aureus. Data are represented as mean ± SEM.
This study investigated the neutrophil response to four diverse bacterial species: Pseudomonas aeruginosa, Salmonella enterica, Listeria monocytogenes, and Staphylococcus aureus. To study how neutrophils respond to diverse bacterial species without the confounding factor of differential bacterial growth, we designed our devices to include bacteria that were alive but not dividing. We first determined that bacterial growth of each species would remain consistent in EGM-2 media to replicate conditions in the infection-on-a-chip device. We measured the rates at which the four bacterial species grew over the time course of the experiment. Measured colony-forming unit (CFU) values after 16 h showed no significant differences between pathogens (Figure 1C), confirming the bacterial burden remains consistent between species throughout the time course of an experiment. Very little bacterial growth was seen in EGM-2 for any bacterial species. To confirm bacteria were still alive and capable of replicating, bacterial growth in LB broth, for gram-negative bacteria, or BHI broth, for gram-positive bacteria, was compared to bacterial growth in EGM-2. Each bacterial species grew and divided in its respective bacterial broth but not in EGM-2 (Figure S1A). Finally, to determine if bacterial loads remained consistent in the infection-on-a-chip device itself, we determined bacterial CFU values in the collagen after 16 h. We found comparable concentrations of each bacterial species (Figure S1B). Together, these results show our model is well designed for comparing the neutrophil response to distinct bacterial species with the same bacterial burden.
Neutrophils demonstrate increased extravasation toward L. monocytogenes
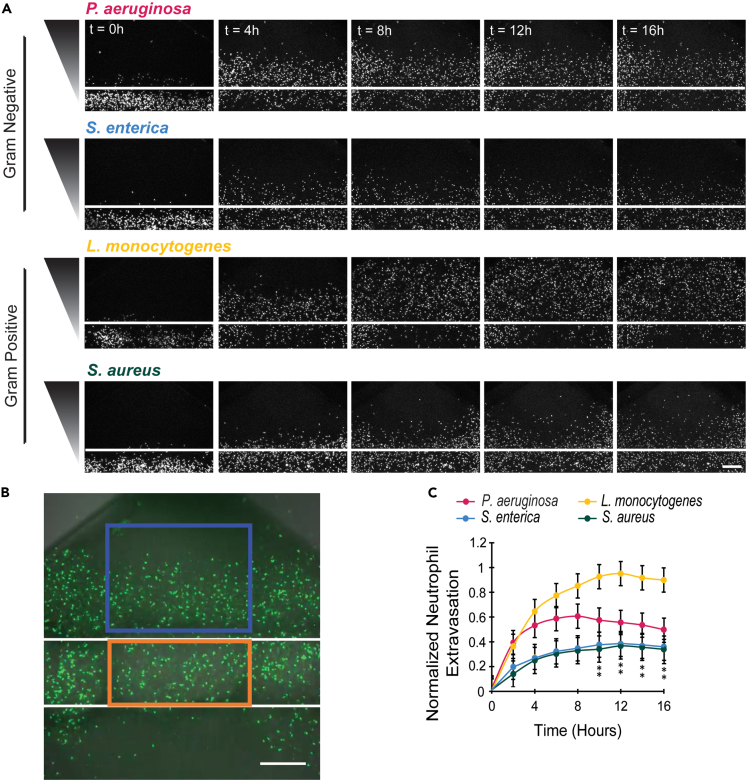
Following activation, neutrophils must cross the blood vessel endothelium via extravasation to reach the tissue and migrate to the site of infection.17 We were first interested in determining how distinct bacterial species influence the extent of neutrophil extravasation. In the infection-on-a-chip device, neutrophils progressively migrated out of the lumen and into the collagen in response to each bacterial species throughout the 16-h experiment (Figure 2A and Video S1); however, more neutrophils migrated out of the lumen in the L. monocytogenes condition. We have previously shown that without a bacterial source, neutrophils do not extravasate out of the lumen.13 To quantify neutrophil extravasation, we counted neutrophils within the region of interest (ROI) outside of the lumen (blue box) at each of the time points and normalized these values to the initial number of neutrophils within the ROI inside of the lumen (orange box) as a loading control (Figures 2B and S2). This quantification confirmed that L. monocytogenes elicits significantly greater neutrophil extravasation compared to the other bacterial species (Figure 2C). Interestingly, the number of extravasated neutrophils plateaus for P. aeruginosa, S. enterica, and S. aureus after 6 to 8 h while continuing until 10 h for L. monocytogenes. These data show a uniquely robust response of neutrophils to L. monocytogenes compared to the other bacterial species. As a control, neutrophil extravasation was also measured in the absence of endothelial cells (Figures S3A–S3C). The results showed similar levels of neutrophil extravasation for each bacterial species, indicating the endothelium plays a significant role in this process.
Figure 2.
Neutrophils have increased extravasation in response to L. monocytogenes
(A) Representative images of neutrophils extravasating out of lumens in response to P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus at 4-h intervals. Neutrophils stained with Calcein AM (white). White line represents the lumen boundary. Bacterial gradient direction shown on left. Scale bar is 100 μm.
(B) Image showing region of interest used for quantification of neutrophil extravasation. Normalized neutrophil extravasation count is determined by dividing the number of neutrophils in the box outside of lumen (blue) by the number of neutrophils in rectangle inside of lumen (orange) at time zero as a loading control. Scale bar is 100 μm.
(C) The number of neutrophils outside the lumen, normalized to the number of neutrophils initially in the lumen was quantified for P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus every 4 h for 16 h. Data quantified from 14 lumens (P. aeruginosa), 13 lumens (S. enterica), 14 lumens (L. monocytogenes), or 12 lumens (S. aureus) across 5 independent experiments. Error bars represent the mean plus SEM. All bacteria were compared to each other at each time point and analyzed with ANOVA. For each condition, estimated marginal means (emmeans) and SEM were calculated and pairwise comparisons were performed with Tukey’s adjustment. Asterisks represent significance of neutrophil extravasation for each bacterial species condition compared to L. monocytogenes condition. P values are labeled as ∗p < 0.05.
Neutrophil migration characteristics in the presence of diverse pathogens
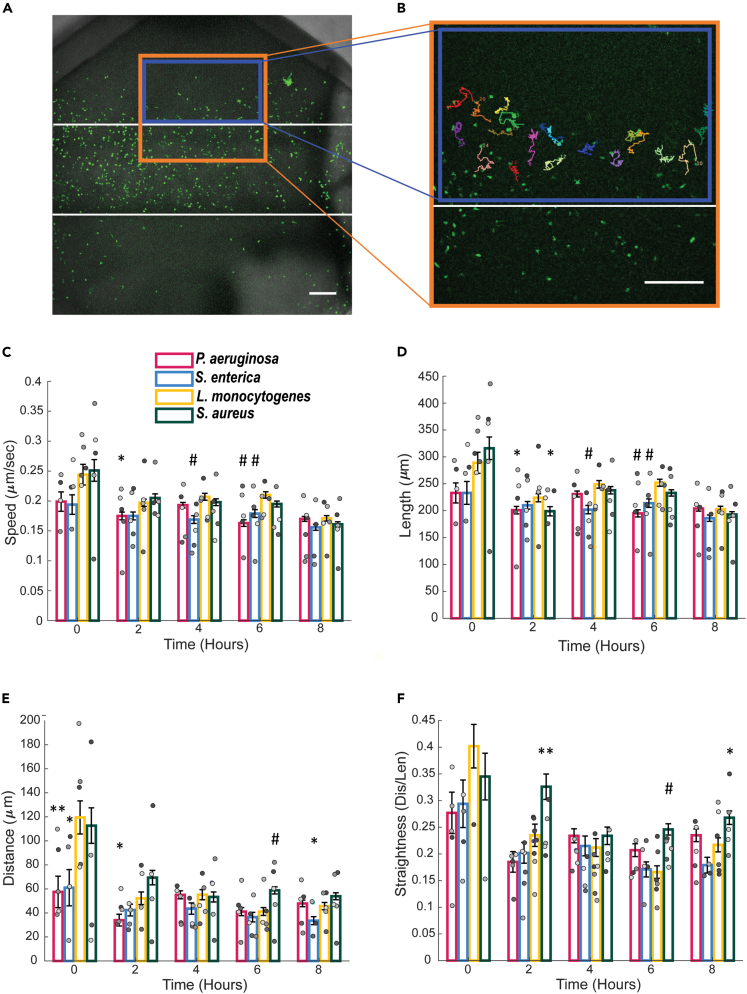
Following extravasation, neutrophils migrate through the tissue to reach sites of infection. We wanted to determine how parameters that describe this migration including speed, directionality, and length differ in response to distinct bacterial species. Only neutrophils in the tissue compartment of the infection-on-a-chip device were tracked, shown as a blue box in Figures 3A and 3B. Overall, no major and persistent differences between bacterial species were found in any of the migration categories quantified. Neutrophils showed increased migratory speed in the presence of L. monocytogenes compared to other bacterial species at the 4- and 6-h time points (Figure 3C). These trends support our extravasation data (Figure 2B) where we found that neutrophils in the presence of L. monocytogenes had greater sustained extravasation compared to the other bacterial species. Additionally, neutrophils in the presence of L. monocytogenes had greater track lengths compared to neutrophils responding to other bacterial species at the 6-h time point (Figure 3D). Neutrophils responding to S. aureus showed increased neutrophil migration distance and straightness (Figures 3E and 3F) between 6 and 8 h, demonstrating a greater ability of neutrophils to directly migrate toward their target. While there were no consistent differences in migration parameters between bacterial species, the results show trends of L. monocytogenes eliciting a faster neutrophil migration phenotype and S. aureus a more directed neutrophil migration phenotype. Interestingly, for each of the bacterial species, there was a significant difference in migration between time points. Specifically, compared to the initial (zero) time point, all subsequent time points have significantly lower values of all migration characteristics for each bacterial species with only a few exceptions (Figure S4). Indicating, there is an initial burst of neutrophil migration in response to each bacterial species as they first extravasate through the endothelium, which qualitatively matches what we see in the migration videos.
Figure 3.
Neutrophils migration characteristics in the presence of diverse pathogens
(A and B) The Fiji software plugin MTrackJ was used to track neutrophils. The orange box indicates the area imaged for all migration experiments and the blue box represents the area in which the neutrophils were tracked. Representative images with tracks are shown in B. Scale bar is 100 μm for A and B.
(C–F) Neutrophil migration parameters including speed (C), track length (D), total migration distance (E), and straightness (F) in response to P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus from the cell tracks over 20-min intervals at 2-h time points. Data quantified from 14 lumens (P. aeruginosa), 12 lumens (S. enterica), 13 lumens (L. monocytogenes), or 14 lumens (S. aureus) across 5 independent experiments. Error bars represent the mean plus SEM. All bacteria were compared to each other at each time point and analyzed with ANOVA. For each condition, emmeans and SEM were calculated and pairwise comparisons were performed with Tukey’s adjustment. Asterisks represent significance of neutrophil migration for each bacterial species condition compared to L. monocytogenes condition at that time point. P values are labeled as ∗p < 0.05; ∗∗p <0 .001; #p < 0.0001. Individual data points are displayed with each gray scale color representing a different replicate.
An increased percentage of neutrophils produce ROS in response to gram-negative bacteria, P. aeruginosa, and S. enterica
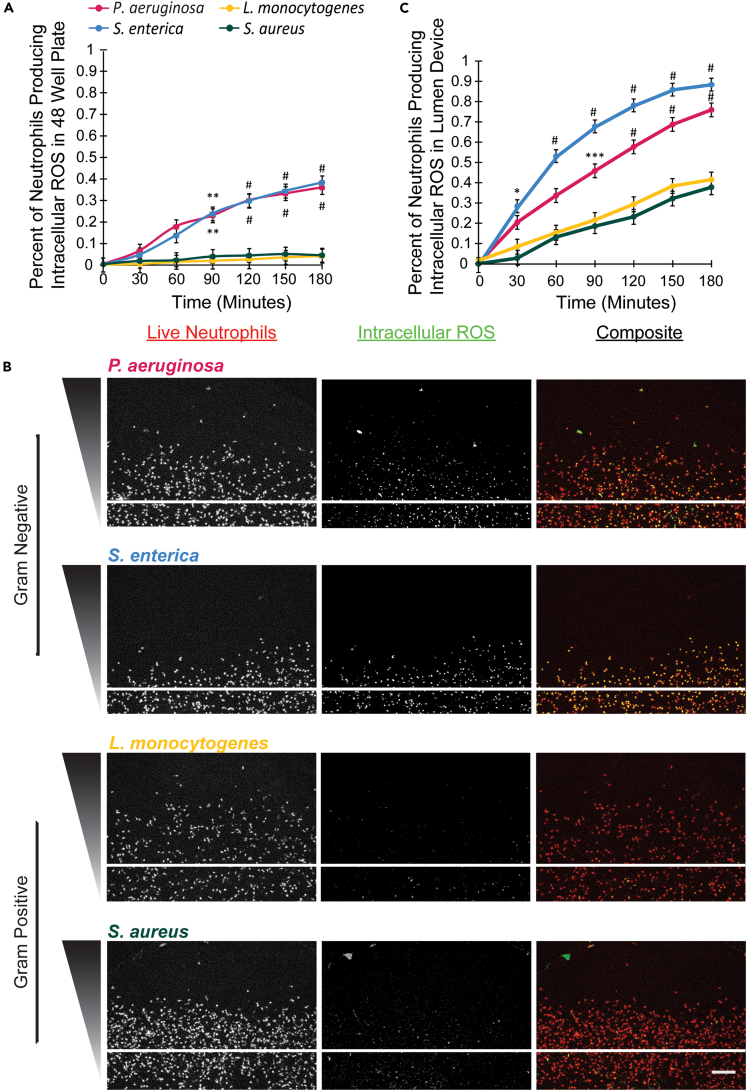
Upon reaching the site of infection, migrating neutrophils encounter the invading pathogen and execute the next step of their function: antimicrobial control. One of the most common antimicrobial effector mechanisms neutrophils use is the production of reactive oxygen species or ROS.18,19,20,21,22,23,24 We, therefore, wanted to determine if the number of neutrophils producing ROS varies in response to distinct bacterial species. We first visualized ROS production by neutrophils seeded in collagen gels in a 48-well plate (Figure S5). We found a significantly higher number of neutrophils producing intracellular ROS in the presence of P. aeruginosa and S. enterica, gram-negative bacteria, compared to neutrophils in the presence of L. monocytogenes and S. aureus, gram-positive bacteria (Figure 4A).
Figure 4.
An increased percentage of neutrophils produce ROS in response to gram-negative bacteria, P. aeruginosa and S. enterica
(A) Neutrophils were seeded in collagen gels in a 48-well plate in the presence of P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus and stained with Calcein AM to visualize all live cells and DHR123 to visualize intracellular ROS production. The percentage of neutrophils producing intracellular ROS in response to P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus was quantified by dividing the number of DHR123-positive neutrophils by the total number of neutrophils (Calcein AM). Data quantified from 3-well plates for each bacterial species across 3 independent experiments. Error bars represent the mean plus SEM. All bacteria were compared to each other at each time point and analyzed with ANOVA. For each condition, emmeans and SEM were calculated and pairwise comparisons were performed with Tukey’s adjustment. Significance is shown with respect to both the L. monocytogenes and S. aureus condition. P values are labeled as ∗∗p <0 .01; #p <0 .0001.
(B) Neutrophils were seeded in the infection-on-a-chip device and stained with DHR123 to visualize intracellular ROS production following extravasation in response to P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus. Representative images showing intracellular ROS production (DHR123) and total neutrophils (Calcein AM) in the infection-on-a-chip device in the presence of P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus. Images were taken every 4 min for 8 h. Images shown are at 3 h after introduction of bacteria. The first column shows all cells stained red with Calcein AM, the second column shows DHR123-positive green, fluorescent ROS producing cells. Scale bar is 100 μm.
(C) The percentage of neutrophil expressing ROS was quantified in response to P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus. Data quantified from 9 lumens for each bacterial species across 3 independent experiments. Error bars represent the mean plus SEM. All bacteria were compared to each other at each time point and analyzed with ANOVA. For each condition, emmeans and SEM were calculated and pairwise comparisons were performed with Tukey’s adjustment. Significance is shown with respect to both the L. monocytogenes and S. aureus condition. P values are labeled as ∗p < 0.05; ∗∗∗p <0 .001; #p <0 .0001.
In vivo, neutrophils typically produce ROS in the tissue following extravasation; therefore, we were interested in determining how neutrophil ROS production would differ between bacterial species following extravasation in the infection-on-a-chip device. We found neutrophils produce intracellular ROS in response to each bacterial species (Figure 4B). In the presence of P. aeruginosa and S. enterica, a significantly greater number of neutrophils produced intracellular ROS as compared to neutrophils in the presence of L. monocytogenes and S. aureus (Figures 4B and 4C). Intriguingly, the results from these experiments also showed a dramatic difference between the number of neutrophils producing ROS following extravasation in the infection-on-a-chip device compared to neutrophils in a well plate. In the infection-on-a-chip device, up to 90% of neutrophils responding to S. enterica and 75% of neutrophils responding to P. aeruginosa were positive for intracellular ROS at 3 h compared to ∼35% in a well plate. Even in the gram-positive bacteria conditions, for which only ∼5% of neutrophils produced intracellular ROS in the well plate, had ∼40% of neutrophils producing intracellular ROS following extravasation through the endothelium. These results demonstrate not only an increase in the number of neutrophils producing intracellular ROS in response to gram-negative bacteria compared to gram-positive bacteria, but also indicate that extravasation through the endothelial lumen enhances the number of neutrophils producing ROS in response to all bacterial species.
Endothelial cells upregulate expression of IL-6 in response to diverse bacterial species
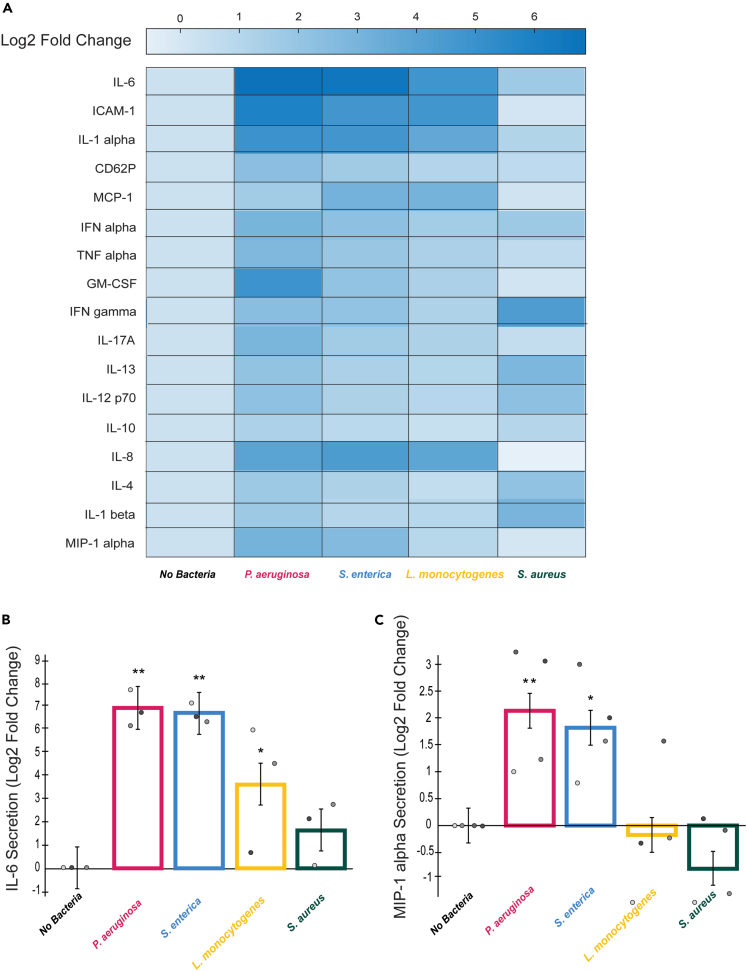
One way in which endothelial cells activate neutrophils and modulate neutrophil function is via the release of pro-inflammatory signals in response to infection.14,13,25,26,27,28,29,30 Given our findings that neutrophil extravasation and intracellular ROS production varies in response to diverse bacterial species, we wanted to determine if this could be due to differential secretion of pro-inflammatory signals by endothelial cells. We used a multiplexed ELISA panel to measure total levels of pro-inflammatory signaling molecules produced by endothelial cells in response to our four bacterial species. Fold changes in endothelial secretion of the different signals in response to each bacterial condition compared to endothelial cells with no bacteria present are shown (Figure 5A). This assay identified two proteins of interest: IL-6 and MIP-1 alpha. IL-6 secretion was increased in response to each bacterial species compared to the control (Figure 5B). The bacteria elicited varying levels of this upregulation, with S. aureus having a more limited increase. Of interest, S. aureus does not elicit significant expression of pro-inflammatory cytokines (e.g., IL-6, IL-8) but does have an increased expression of IL-4 and IL-13, immune regulatory signals. The assay also revealed upregulation of MIP-1 alpha secretion in the presence of P. aeruginosa and S. enterica but not L. monocytogenes and S. aureus compared to the control condition (Figure 5C). This was interesting as neutrophils displayed a greater level of intracellular ROS production in the presence of P. aeruginosa and S. enterica compared to L. monocytogenes and S. aureus.
Figure 5.
Endothelial cells upregulate expression of IL-6 in response to diverse bacterial species
A multiplexed ELISA screen was conducted for endothelial lumen-conditioned media with no bacteria, P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus present.
(A) Log2 fold changes of secreted signals from endothelial cells in the infection-on-a-chip device in the presence of P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus compared to no bacteria condition. Scale ranges from darker blue, higher expression, to lighter blue, lower expression. Factors measured are labeled on the left side of the heatmap.
(B) The levels of IL-6 expressed as a log2 fold change over the no bacteria condition for endothelial cells in the presence of P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus. Data quantified from 12 lumens for each bacterial species across 4 independent experiments. Error bars represent least-squared mean plus SEM. All bacteria were compared to each other at each time point and analyzed with ANOVA. For each condition, emmeans and SEM were calculated and pairwise comparisons were performed with Tukey’s adjustment. Significance is shown with respect to the no bacteria condition. ∗p < 0.05; ∗∗p <0 .01. Individual data points are displayed with each gray scale color representing a different replicate.
(C) The levels of MIP-1 alpha expressed as a log2 fold change over the no bacteria condition for endothelial cells in the presence of P. aeruginosa, S. enterica, L. monocytogenes, or S. aureus. Data quantified from 12 lumens for each bacterial species across 4 independent experiments. Error bars represent least-squared mean plus SEM. All bacteria were compared to each other at each time point and analyzed with ANOVA. For each condition, emmeans and SEM were calculated and pairwise comparisons were performed with Tukey’s adjustment. Significance is shown with respect to the no bacteria condition. ∗p < 0.05; ∗∗p <0 .01. Individual data points are displayed with each gray scale color representing a different replicate.
To determine if MIP-1 alpha plays a role in neutrophil ROS production, we blocked MIP-1 alpha using a blocking antibody and measured neutrophil intracellular ROS expression but saw no difference between the blocking condition and the control condition (data not shown).
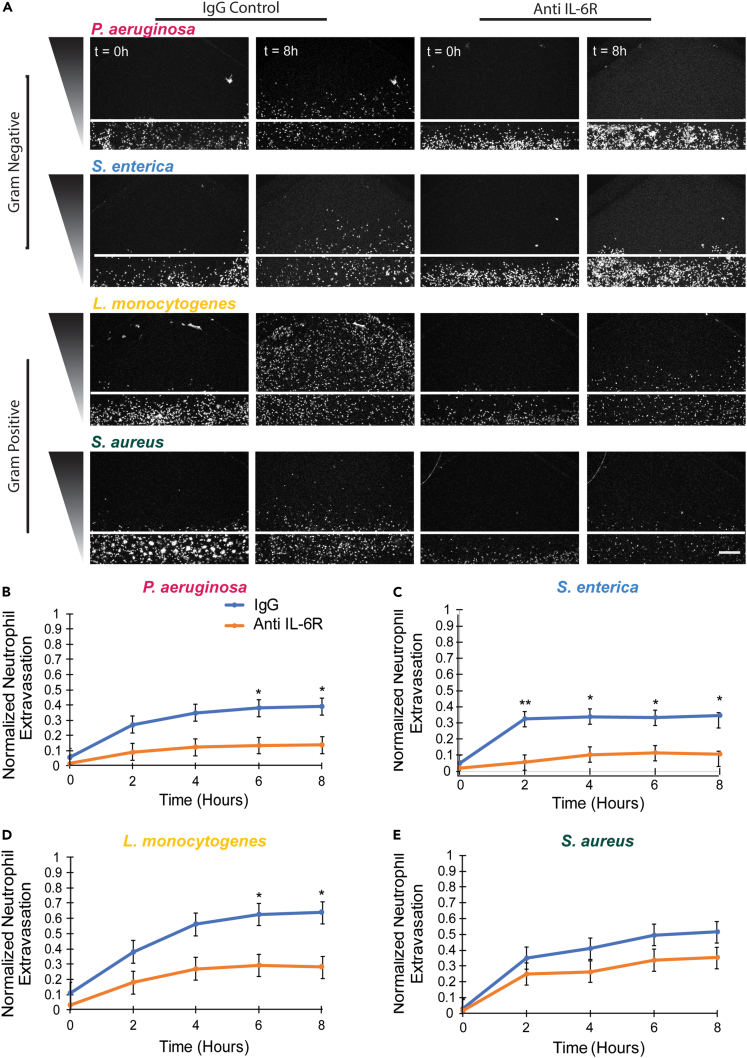
IL-6 is necessary for neutrophil extravasation in response to diverse bacterial species
We have previously shown that IL-6 secretion by the endothelium enhances neutrophil extravasation in response to P. aeruginosa and Aspergillus fumigatus.13,14 Here, we found increased secretion of IL-6 in response to all four bacterial species. Therefore, we hypothesized that IL-6 signaling was critical for neutrophil extravasation in response to each bacterial species. To test this hypothesis, we used an IL-6 receptor blocking antibody to inhibit IL-6 signaling. We observed a reduced number of extravasating neutrophils compared to a control IgG antibody for each bacterial species (Figure 6A). Quantification of neutrophil extravasation showed consistently lower levels of neutrophil extravasation in the absence of IL-6 signaling compared to the control condition in response to each of the bacteria (Figure 6B–6E, Videos S2, S3, S4, S5, and Figures S6A–S6D). The decrease in neutrophil extravasation did not reach significance in response to S. aureus but there was a trend toward lower extravasation. Of note, S. aureus elicited the lowest level of IL-6 secretion from endothelial cells (Figure 5). Together, these results indicate a conserved requirement for IL-6 signaling for neutrophil extravasation in response to diverse bacterial species.
Figure 6.
IL-6 is required for neutrophil extravasation in response to diverse bacterial species
(A) Representative images of neutrophils migrating out of endothelial lumens at 0 and 8 h in the presence of a control IgG antibody (two left columns) or an IL-6 receptor blocking antibody (two right columns). White line represents the lumen boundary. Scale bar is 100 μm.
(B–E) The number of neutrophils outside the lumen, normalized to the number of neutrophils initially in the lumen was quantified for (B) P. aeruginosa, (C) S. enterica, (D) L. monocytogenes, or (E) S. aureus every 2 h for 8 h in the presence of either an IgG control antibody or IL-6 receptor blocking antibody. Data quantified from 9 lumens for each bacterial species and each antibody condition across 3 independent experiments. Error bars represent the mean plus SEM. All bacteria were compared to each other at each time point and analyzed with ANOVA. For each condition, emmeans and SEM were calculated and pairwise comparisons were performed with Tukey’s adjustment. Asterisks represent significance between IL-6 receptor blocking antibody condition and IgG control condition. P values are labeled as ∗p < 0.05; ∗∗p <0 .01.
Discussion
In this study, we used our recently developed infection-on-a-chip device to investigate the neutrophil response to diverse pathogens by quantifying neutrophil extravasation, interstitial migration, and ROS production. We found neutrophils exhibited enhanced extravasation in response to L. monocytogenes compared to the other bacterial species. Additionally, we demonstrated a universal role for IL-6 in mediating neutrophil extravasation to diverse bacterial species. Overall, our interstitial migration data showed similar results across bacteria but higher migration parameters early in infection for each bacterial species. Finally, we saw an increased number of neutrophils producing ROS when responding to gram-negative bacteria compared to neutrophils responding to gram-positive bacteria and, for all bacterial species, more neutrophils produced ROS following extravasation through an endothelium than when studied in a well plate. We also saw an overall lack of neutrophil response to S. aureus, consistent with its ability to inhibit neutrophil function. Together, these results demonstrate that our infection-on-a-chip device can be used to identify distinct neutrophil responses to diverse pathogens through quantification of extravasation, migration, and ROS production and highlight the importance of using physiologically relevant models for studying human neutrophil function.
It is known that neutrophils play a critical role in the early immune response to L. monocytogenes. Neutrophils in mice rapidly accumulate at the site of L. monocytogenes infection and mice depleted of neutrophils are shown to have severe listeriosis in the liver.31,32 Additionally, the neutrophil response to L. monocytogenes has been shown to be regulated at least in part by soluble signals released from the endothelium.33 Our investigation of neutrophil extravasation using our infection-on-a-chip device demonstrated neutrophils responding to L. monocytogenes had significantly greater extravasation than neutrophils responding to the other bacterial species. The particularly important role of neutrophils in the immune response to L. monocytogenes could explain the more robust neutrophil response we found in the presence of this bacterial species. Both Salmonella enterica and S. aureus elicited significantly lower neutrophil extravasation in comparison to L. monocytogenes. Both bacteria have previously described mechanisms in which they can suppress neutrophil function and evade neutrophil clearance. S. enterica has been shown to alter regulation of neutrophil ribosomal genes associated with cell-cycle arrest, apoptosis, and innate immunity in a way that promotes its survival.34 S. aureus has been previously shown to inhibit the neutrophil response in multiple ways including decreasing overall neutrophil extravasation. S. aureus releases toxins and membrane-associated proteins that block chemokine and cytokine signaling and decrease neutrophil extravasation.35,36 Our infection-on-a-chip device was able to replicate this known ability of S. enterica and S. aureus to decrease neutrophil extravasation while highlighting the strong neutrophil response to L. monocytogenes.
We found the percentage of neutrophils producing intracellular ROS was dependent on the infecting bacterial species. Specifically, a greater percentage of neutrophils produce intracellular ROS in the presence of P. aeruginosa and S. enterica, gram-negative bacteria, compared to in the presence of L. monocytogenes and S. aureus, gram-positive bacteria. S. aureus has previously been shown to decrease neutrophil ROS production via the staphylococcal superantigen-like 7 protein.37 Additionally, S. aureus release of SaeR/S-regulated factors reduces neutrophil ROS production.38 While the differences in ROS production we observed were correlated to the gram-negative or gram-positive categorization, it is unclear if this distinction is directly connected to the presence of a cell wall. In general, though, distinctions in the neutrophil response to infection between gram-negative and gram-positive bacteria have been previously shown, including in the context of ROS production where CD137 has been shown to increase ROS production in neutrophils responding to gram-positive bacteria but decreases ROS production in neutrophils responding to gram-negative bacteria.39 One possible explanation could be the activation of neutrophil ROS production by lipopolysaccharide (LPS), a gram-negative-specific cell wall component. Previous work has shown a role of LPS in inducing neutrophil ROS production via NADPH oxidation pathways.40 Another possibility could be gram-negative bacteria producing more ROS due to activation by IL-6 as gram-negative bacteria elicited greater levels of IL-6 secretion from endothelial cells in our devices. IL-6 signaling and ROS production have been linked in other cell types, including nerve cells and islet beta cells.41,42 Furthermore, evidence exists that neutrophil ROS and IL-6 participate in a positive feedback loop that potentiates ROS production.43 In general, the connection between IL-6 and ROS production in neutrophils remains unclear and further work is needed to clarify their relationship in bacterial infections.
Not only did we find distinct intracellular ROS production between bacteria, but we also found a universal increase in intracellular ROS following extravasation through an endothelium compared to neutrophils seeded in a dish. This indicates a role of the endothelium in activating neutrophil ROS production in the presence of diverse bacterial species and highlights the importance of using physiologically relevant models of the infectious microenvironment for studying neutrophil function. Neutrophil and endothelial cell interactions play an important role in neutrophil recruitment upon infection.44 Specifically, we previously demonstrated endothelial cells regulate neutrophil extravasation and lifetime via secretion of IL-6 and GM-CSF, respectively.13 In addition to our ROS data, results from our present study show a consistent role of the endothelium in activating neutrophil function in response to diverse bacterial species with a conserved requirement of IL-6 for the neutrophil response to bacteria.
The role of IL-6 in the neutrophil response to infection has been previously investigated. IL-6 is known to modulate neutrophil function by binding to an IL-6 receptor on neutrophils and activating downstream signaling effects through the STAT3 signaling pathway.45 IL-6 activation of neutrophils has been shown to increase their migration toward IL-8.46 There are several IL-6 receptor blocking drugs, such as tocilizumab and sarilumab, on the market that have been widely studied and effectively used to reduce inflammation and improve patient outcomes for diseases such as rheumatoid arthritis and recently COVID-19.47,48 Clinical studies have shown these drugs do not increase infection rates in patients indicating the presence of other immune pathways to make up for reduced levels of neutrophil extravasation.
A consistent trend we observed throughout our study was an overall more limited neutrophil response to S. aureus in comparison to the other bacterial species. Neutrophil extravasation, migration, ROS production, and endothelium signal secretion of known pro-inflammatory signals, including IL-6 and IL-8, are consistently lower for neutrophils responding to S. aureus. Additionally, endothelial secretion of signals associated with a dampened neutrophil response, including IL-4 and IL-13, were upregulated in the presence of S. aureus compared to the other bacterial species.49 As previously discussed in the case of neutrophil ROS production, S. aureus is capable of hampering the neutrophil response in the body. S. aureus inhibits neutrophil rolling, an important step in the extravasation process, through release of SSL5.50 Additionally, S. aureus targets another step of neutrophil extravasation, neutrophil adherence to endothelial cells, through release of extracellular adherence protein.51 S. aureus also inhibits neutrophil migration toward some chemokines including fMLP and C5a.52 The capability of our infection-on-a-chip device to accurately model how S. aureus slows the neutrophil response during infection provides the intriguing possibility of using our model to better study mechanisms of S. aureus inhibition of neutrophils and to eventually use it for screening potential drug targets against this harmful pathogen.
Our findings reveal diverse neutrophil responses to different bacterial species, raising intriguing questions about the underlying mechanisms. Specifically, we observed a robust neutrophil response to L. monocytogenes compared to the other bacterial species studied; however, different causes could underlie this differential response. For example, given the reduced number of ROS-producing cells in response to L. monocytogenes, it is possible that the neutrophils are not as readily able to clear L. monocytogenes compared to the other bacterial species, leading to a more sustained response. This could also account for the increased neutrophil migration in response to L. monocytogenes. Alternatively, the endothelial response to L. monocytogenes could differ compared to the other bacterial species, especially if we consider the possibility for temporal differences. The multiplexed ELISA provided us a snapshot of endothelial protein secretion, but these levels could vary over time, leading to differential neutrophil activation. One potential factor this study does not investigate is neutrophil LTB4 production. LTB4 is a secondary chemoattractant released by neutrophils that plays an important role in initiating the inflammatory response.53 Future studies should investigate whether LTB4 production varies in the presence of diverse bacteria and the role this has on the variability of the neutrophil response. We also observed distinct levels of ROS production in response to gram-positive versus gram-negative bacteria, both with and without extravasation through an endothelium. This indicates that differences in bacteria-neutrophil interactions contribute to the prevalence of ROS producing neutrophils, but we do not have a clear understanding of what is driving this distinction. Interestingly, following extravasation through the endothelium, ROS production increases for all bacterial species, indicating the endothelial cells provide an important activation cue but we have yet to identify this signal. There are many factors, including phagocytosis and binding of pathogen-associated molecular patterns (PAMPs), such as bacterial peptides, to neutrophil receptors, that influence neutrophil ROS production. The levels of phagocytosis or PAMPs may vary between bacteria conditions leading to differences in neutrophil ROS production. The fact that neutrophils produce more ROS following extravasation suggests that the differences shown are a result of a neutrophil factor rather than a bacterial factor, pointing to phagocytosis. Future studies will investigate neutrophil phagocytosis of these bacteria in our system to better answer this question. This study emphasizes the ongoing importance of investigating neutrophil function in physiologically relevant human systems, which holds great potential for future advancements in human health.
Limitations of the study
While the infection-on-a-chip device used for this work contains many of the important physiological components of the human vasculature and infectious microenvironment, it certainly does not completely encapsulate every element of an in vivo system. This device does not contain any other immune cell types beyond neutrophils, such as tissue macrophages and tissue dendritic cells. Furthermore, it uses endothelial cells as the vasculature model but no other components of the vasculature are included. As a result, there is additional immune cell and vasculature signaling not represented in this system. Additionally, collagen is the only protein used as the extracellular matrix (ECM) source where other proteins in addition to collagen make up the ECM in the body. The study spans the range of gram-negative, gram-positive, intracellular, and extracellular bacteria but excludes some common bacterial pathogens (Escherichia coli, for example) and does not test multiple strains of each bacterial species. Variations may occur in the neutrophil response to differing bacterial species and clinical strains versus lab strains. Finally, the study uses bacteria that are alive but not dividing to eliminate confounding factors due to differences in bacterial growth dynamics. This is not completely representative of an in vivo infection but still provides a significant advantage over studies that use uniform formylated peptides or lipopolysaccharides to mimic bacteria instead of including live pathogens. Future experiments will address these limitations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial strains | ||
| Pseudomonas aeruginosa | Takeya K, Amako K. 1966 | K |
| Salmonella enterica | Hoiseth and Stocker, 1981 | SL1344 |
| Listeria monocytogenes | Edman et al. 1968 | 10403 |
| Staphylococcus aureus | Los Angeles County isolate, LAC | USA300 |
| Chemicals, peptides, and recombinant proteins | ||
| Endothelial Growth Media 2 | Lonza | NC9525043 |
| Trypsin EDTA | ATCC | 50189662FP |
| Trypsin Neutralizing Solution | ATCC | 50189663FP |
| Lennox L Broth | Gibco | 10855021 |
| Brain Heart Infusion Broth | Sigma Aldrich | 53286 |
| Lennox L Agar | Invitrogen | 22700–025 |
| Brain Heart Infusion Agar | Research Products International | B11500 |
| Streptomycin | Sigma Aldrich | S1277 |
| Two polydimethylsiloxane | Electron Microscopy Sciences | 24236–10 |
| Polyethyleneimine | Sigma Aldrich | 03880 |
| GA | Sigma Aldrich | G6257 |
| Type 1 Rat Tail Collagen | Corning | 354249 |
| Paraformaldehyde | Thermo Scientific | AAJ19943K2 |
| Phosphate Buffered Saline | Thermo Scientific | BP2944-100 |
| Hoechst | Sigma Aldrich | 23491-45-4 |
| Phalloidin | abcam | ab176757 |
| VE-Cadherin | Miltenyi Biotech | 130-123-688 |
| Dihydrorhodamine-123 | Thermo Scientific | D23806 |
| IL-6 receptor blocking antibody | R&D Systems | MAB227; RRID: AB_2127908 |
| IgG antibody | R&D Systems | HAF007; RRID: AB_357234 |
| MIP-1 alpha receptor blocking antibody | R&D Systems | AF-270-SP; RRID: AB_354436 |
| IgG antibody | R&D Systems | AB-108-C; RRID: AB_354267 |
| Critical commercial assays | ||
| Neutrophil Isolation Kit | Miltenyi Biotec | 130-104-434 |
| Erythrocyte Depletion Kit | Miltenyi Biotec | 130-098-196 |
| ProcartaPlex Human Inflammation Panel | Invitrogen | EPX200-12185-901 |
| Experimental models: Cell lines | ||
| Human Umbilical Vein Endothelial Cells | Promocell | 50-305-964 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Laurel Hind (laurel.hind@colorado.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Human participants
Primary human neutrophils were isolated from whole blood samples obtained from healthy donors. Informed consent was obtained from all donors at the time of the blood draw in accordance with our Institutional Review Board approved protocol. All donors were between the ages of 18–65. Information on sex/gender, ancestry, race, ethnicity, and socioeconomic status was not collected in accordance with our approved protocol.
Cell lines
Pooled Human Umbilical Vein Endothelial Cells (HUVEC, 50-305-964, Promocell GmbH C12203, Heidelerg, Germany) were maintained in Endothelial Growth Media (EGM-2, NC9525043, Lonza Walkersville CC3162, Basel, Switzerland) until they reached 80% confluence. These cells were used from passages 2 to 7. Media was changed every two days. The cells were detached using Trypsin EDTA (0.05% Trypsin and 0.02% EDTA in phosphate buffered saline without calcium or magnesium)(50189662FP, ATCC, Mannassas, Virginia) and Trypsin Neutralizing Solution (5% FBS in phosphate buffered saline without calcium and magnesium)(50189663FP, ATCC, Mannassas, Virginia). They were than split and reseeded at a concentration of 375,000 cells/15 mL.
Microbe strains
Four bacterial species were used in this study: Pseudomonas aeruginosa (strain K), Salmonella enterica (serovar Typhimurium strain SL1344), Listeria monocytogenes (serotype 1/2a strain 10403s) and Staphylococcus aureus (strain USA300 LAC). Agar plates were made for growing each of these bacterial species. Lennox L (LB) agar (22700-025, Invitrogen, Waltham, Massachusetts) plates and LB broth (10855021, Gibco, Waltham, Massachusetts) were used with P. aeruginosa. LB agar plates with 40 μg/mL streptomycin (S1277, Sigma Aldrich, St. Louis, Missouri) were used with S. enterica. Brain Heart Infusion (BHI, B11500, Research Products International, Mount Prospect, Illinois) agar plates and BHI broth (53286, Sigma Aldrich, St. Louis, Missouri) were used with both L. monocytogenes and S. aureus. Plates were steaked using the quadrant streaking method.
Method details
Microfluidic device fabrication
Microfluidic devices were fabricated as previously described.16 The devices were formed by patterning two polydimethylsiloxane (PDMS, 24236-10, Electron Microscopy Sciences, Hatfield, Pennsylvania) layers from two different SU-8 silicon masters (MicroChem). The two distinct layers were then stacked and aligned and a PDMS rod was placed inside the center chamber. The aligned devices were bonded to a glass-bottom 50 mm dish (P50G-1.5-30-F, Mattek, Ashland, Massachusetts) using oxygen plasma from a PE25-JW Plasma Etcher (Plasma Etch, Carson City, Nevada) to create the final devices.
Device and collagen preparation
Infection-on-a-Chip devices were prepared following the same protocol outlined in previous publications.54 First, a 2% polyethyleneimine (PEI) solution in deionized water (03880, Sigma Aldrich, St. Louis, Missouri) was pipetted into the device and incubated for 10 min. Then a 0.4% glutaraldehyde (GA) solution in deionized water (G6257, Sigma Aldrich, St. Louis, Missouri) was pipetted into the device and incubated for 30 min. The devices were then washed three times with water. Type I Rat Tail Collagen (354249, Corning, Corning, New York) was neutralized to a desired pH of 7.2 at a concentration of 4 mg/mL. The collagen solution was added into the devices where it polymerized around the PDMS rod in the central chamber. After polymerization, the PDMS rod was removed leaving behind the hollow lumen structure. The lumens were then seeded with HUVECs at a concentration of 20,000 cells/μL and incubated overnight on a rotator. HUVECs were allowed to grow and form a monolayer with twice daily media changes for two days.
Lumen staining protocol
Fully formed HUVEC lumens were incubated with 4% paraformaldehyde (PFA) (AAJ19943K2, Thermo Scientific, Waltham, Massachusetts) in phosphate buffered saline (PBS, BP2944-100, Thermo Scientific, Waltham, Massachusetts) for 10 min at room temperature while on a rocker. Next, the lumens were incubated with 0.005 mg/mL Hoescht (23491-45-4, Sigma Aldrich, St. Louis, Missouri), Phalloidin (1:200, ab176757, abcam, Cambridge, UK), and VE-Cadherin (1:120, 130-123-688, Miltenyi Biotec, Bergisch Gladbach, Germany) overnight at 4°C. Single time point z-stacks were taken to image the entire lumen.
Neutrophil isolation
Primary human neutrophils were isolated from whole blood samples using the MACSxpress Whole Blood Neutrophil Isolation Kit (130-104-434, Miltenyi Biotec, Bergisch Gladbach, Germany) and the erythrocyte depletion kit (130-098-196, Miltenyi Biotec, Bergisch Gladbach, Germany), according to the manufacturer’s instructions. Informed consent was obtained from all donors at the time of the blood draw in accordance with our Institutional Review Board approved protocol. Neutrophils were stained with Calcein AM at a concentration of 10 nM (C3100MP, Thermo Scientific, Waltham, Massachusetts).
Bacteria preparation
Sixteen hours before an experiment, a single colony from a streaked plate was obtained using an inoculation loop and then added into a 5 mL broth solution: LB broth for P. aeruginosa and S. enterica and BHI broth for L. monocytogenes and S. aureus. At the time of the experiment, 1 mL of this solution was diluted with 4 mL of additional broth and incubated for 75 min. Then, 1 mL of the bacterial solution was spun down at 16,000xg for 1 min and resuspended in 100 μL of EGM-2. A 1:100 solution was created by diluting in EGM-2 and the OD was measured. The volume of the solution is adjusted to a final concentration of 1.25 × 106 CFU/mL.
Colony forming unit (CFU) to optical density (OD) relationships for each bacterial species were determined by creating serial dilutions of the bacterial cultures, measuring the OD of each dilution, and counting CFUs for each dilution. The linear relationship between the CFUs and OD was then determined by plotting these points and forming a line of best fit.
Bacterial species growth analysis
Bacterial species were grown as described in the bacteria preparation section. To obtain initial CFUs, bacteria solutions were plated onto agar and incubated for 16 h at 37°C. CFUs were then counted. To determine bacterial growth over the time course of the experiment, bacteria solutions were grown in EGM-2 for 16 h at 37°C, plated onto agar, and grown for 16 h at 37°C. CFUs were then counted.
To compare bacterial growth in EGM-2 to bacterial growth in LB/BHI broth, a single bacterial colony was added into 5 mL of either EGM-2 or LB/BHI broth and then incubated for 16 h. The number of CFUs was then measured in each condition.
Bacterial viability analysis
Bacterial species were added into the Infection-on-a-Chip device as previously described with neutrophils present and incubated at 37°C for 16 h. The top layer of the device was then peeled off and 5 μL of 8 mg/mL collagenase into each device and the devices were incubated for 30 min at 37°C. Collagenase solutions were then removed and added to bacterial agar plates. Plates were incubated at 37°C for 16 h and then individual colonies were counted.
Neutrophil extravasation
Approximately 4 μL of neutrophils at a concentration of 7 × 106 cells/mL were added into each lumen. Three microliters of bacterial species at a concentration of 1.25 × 106 CFU/mL were added to the top port of the device.
Neutrophil extravasation analysis
Experiments were analyzed by counting the number of neutrophils outside of the lumen in a region of interest (600 pixels by 900 pixels) directly above the lumen edge and dividing by the initial number of cells inside a region of interest (300 pixels by 900 pixels) in the center of the lumen to get normalized values (Figure 2B). Images were taken every 10 min for 16 h. Multiple 10 μm z-stacks were imaged to capture all cells present in the device.
Neutrophil migration analysis
Lumens for neutrophil migration experiments were set up in the same manner as extravasation experiments. Only neutrophils initially outside of the lumen were tracked. Speed, distance migrated (the length of a line drawn from the start point to the endpoint of the cell track), length of tracks, and straightness of migration (the distance migrated divided by the length of the track) were quantified. Cells were tracked using MTrackJ software in FIJI and then quantification was done using a custom MATLAB script. Images were taken every 30 s for 20 min at 2 h intervals for a total of 8 h of elapsed time. Multiple 10 μm z-stacks were imaged to capture all cells present in the device.
ROS production
Intracellular ROS production by neutrophils was measured both in collagen in a 48-well plate and in the Infection-on-a-Chip device. Dihydrorhodamine-123 (DHR-123, D23806, Thermo Fisher Scientific, Waltham, Massachusetts) was used to label intracellular ROS. Neutrophils were seeded in 4 mg/mL collagen in a 48-well plate. 10 μM DHR-123 was pipetted into the well and incubated for 30 min at 37°C. Bacterial species were then introduced into the system. Images were taken every 4 min for 3 h. Multiple 10 μm z-stacks were imaged to capture all cells present throughout the collagen. In the Infection-on-a-Chip device, the lumen was preincubated with 10 μM DHR-123 for 30 min at 37°C. Neutrophils were then added to the system in the continued presence of 10 μM DHR-123. Images were taken every 4 min for 8 h. Multiple 10 μm z-stacks were imaged to capture all cells present throughout the device.
Image acquisition
All imaging was conducted using a Nikon A1R HD25 Laser Scanning Confocal Microscope built on the Nikon Ti2-E Inverted Microscope System, a Nikon 10x/0.45 (NA) objective, a fully automated stage and Nikon elements acquisition software.
Multiplex ELISA assay
A Luminex Multiplex assay, ProcartaPlex Human Inflammation Panel (EPX200-12185-901, Invitrogen, Waltham, Massachusetts), was used to measure protein secretion from endothelial cells in the Infection-on-a-Chip device. The Infection-on-a-Chip devices were set up as described above with endothelial lumens and bacteria but without neutrophils. The devices were incubated for 16 h at 37°C. Media from 6 lumens was collected for each condition. Collected media was pooled and centrifuged to remove any cellular debris. Data shown is from a total of 4 replicates. Collected samples were frozen and stored at −20°C until the assay was conducted. Protein levels in the supernatants were then measured to determine protein secretion from endothelial cells under the different infection conditions compared to endothelial expression in the absence of any pathogen. Heat maps were created using MATLAB and are displayed as a Log2 fold change over the condition with no pathogen present.
Antibody blocking
Infection-on-a-Chip devices were preincubated with either 10 μg/mL IL-6 receptor blocking antibody (MAB227, R&D Systems, Minneapolis, Minnesota) or 10 μg/mL IgG antibody (HAF007, R&D Systems, Minneapolis, Minnesota) as a control. Antibodies were also added to the neutrophil solution and to the devices at the start of the experiment. Extravasation of neutrophils was quantified and analyzed in the same manner as previously described.
Infection-on-a-Chip devices were preincubated with either 10 μg/mL MIP-1 alpha receptor blocking antibody (AF-270-SP, R&D Systems, Minneapolis, Minnesota) or 10 μg/mL IgG antibody (AB-108-C, R&D Systems, Minneapolis, Minnesota) as a control. Antibodies were also added to the neutrophil solution and the devices at the start of the experiment. Intracellular ROS production of neutrophils was quantified and analyzed in the same manner as previously described.
Quantification and statistical analysis
For all experiments, data was pooled from three or more independent replicates. Statistical analysis was conducted using R. While the data were pooled, the statistical analysis accounts for variability within and between replicates and considers independent replicates as individual entities. All bacteria were compared to each other at each time point and analyzed with analysis of variance (ANOVA). For each condition, estimated marginal means (emmeans) and standard error (SEM) were calculated and pairwise comparisons were performed with Tukey’s adjustment. p values are labeled as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Acknowledgments
This work was supported by the National Institutes of Health through R35 GM1 46737A.
Author contributions
I.M.R., C.J.C., E.L.G., and L.E.H. designed the research and wrote the manuscript; I.M.R. and E.L.G. performed the experiments; I.M.R., C.J.C., E.L.G., E.N., K.A.P., and L.E.H. analyzed and interpreted the data.
Declaration of interests
None to declare.
Published: December 2, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108627.
Supplemental information
References
- 1.Richardson I.M., Calo C.J., Hind L.E. Microphysiological Systems for Studying Cellular Crosstalk During the Neutrophil Response to Infection. Front. Immunol. 2021;12:661537. doi: 10.3389/fimmu.2021.661537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moreland J.G., Bailey G., Nauseef W.M., Weiss J.P. Organism-Specific Neutrophil-Endothelial Cell Interactions in Response to Escherichia coli , Streptococcus pneumoniae , and Staphylococcus aureus. J. Immunol. 2004;172:426–432. doi: 10.4049/jimmunol.172.1.426. [DOI] [PubMed] [Google Scholar]
- 3.Angus D.C., van der Poll T. Severe Sepsis and Septic Shock. N. Engl. J. Med. 2013;369:840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 4.Xie J., Van Hoecke L., Vandenbroucke R.E. The Impact of Systemic Inflammation on Alzheimer’s Disease Pathology. Front. Immunol. 2021;12:796867. doi: 10.3389/fimmu.2021.796867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez-García M., Hernández-Lemus E. Periodontal Inflammation and Systemic Diseases: An Overview. Front. Physiol. 2021;12:709438. doi: 10.3389/fphys.2021.709438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silhavy T.J., Kahne D., Walker S. The Bacterial Cell Envelope. Cold Spring Harbor Perspect. Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pizarro-Cerdá, J., and Cossart, P. Listeria Monocytogenes: Cell Biology of Invasion and Intracellular Growth [DOI] [PMC free article] [PubMed]
- 8.Pang Z., Raudonis R., Glick B.R., Lin T.-J., Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019;37:177–192. doi: 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 9.Balasubramanian R., Im J., Lee J.-S., Jeon H.J., Mogeni O.D., Kim J.H., Rakotozandrindrainy R., Baker S., Marks F. The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum. Vaccines Immunother. 2019;15:1421–1426. doi: 10.1080/21645515.2018.1504717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lecuit M. Listeria monocytogenes , a model in infection biology. Cell Microbiol. 2020;22:e13186. doi: 10.1111/cmi.13186. [DOI] [PubMed] [Google Scholar]
- 11.Cheung G.Y.C., Bae J.S., Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12:547–569. doi: 10.1080/21505594.2021.1878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Assis L.M., Nedeljković M., Dessen A. New strategies for targeting and treatment of multi-drug resistant Staphylococcus aureus. Drug Resist. Updates. 2017;31:1–14. doi: 10.1016/j.drup.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Hind L.E., Ingram P.N., Beebe D.J., Huttenlocher A. Interaction with an endothelial lumen increases neutrophil lifetime and motility in response to P aeruginosa. Blood. 2018;132:1818–1828. doi: 10.1182/blood-2018-05-848465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hind L.E., Giese M.A., Schoen T.J., Beebe D.J., Keller N., Huttenlocher A. Immune Cell Paracrine Signaling Drives the Neutrophil Response to A. fumigatus in an Infection-on-a-Chip Model. Cell. Mol. Bioeng. 2021;14:133–145. doi: 10.1007/s12195-020-00655-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bischel L.L., Sung K.E., Jiménez-Torres J.A., Mader B., Keely P.J., Beebe D.J. The importance of being a lumen. FASEB J. 2014;28:4583–4590. doi: 10.1096/fj.13-243733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiménez-Torres J.A., Peery S.L., Sung K.E., Beebe D.J. LumeNEXT: A Practical Method to Pattern Luminal Structures in ECM Gels. Adv. Healthcare Mater. 2016;5:198–204. doi: 10.1002/adhm.201500608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filippi M.-D. Neutrophil transendothelial migration: updates and new perspectives. Blood. 2019;133:2149–2158. doi: 10.1182/blood-2018-12-844605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen G.T., Green E.R., Mecsas J. Neutrophils to the ROScue: Mechanisms of NADPH Oxidase Activation and Bacterial Resistance. Front. Cell. Infect. Microbiol. 2017;7:373. doi: 10.3389/fcimb.2017.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veenith T., Martin H., Le Breuilly M., Whitehouse T., Gao-Smith F., Duggal N., Lord J.M., Mian R., Sarphie D., Moss P. High generation of reactive oxygen species from neutrophils in patients with severe COVID-19. Sci. Rep. 2022;12:10484. doi: 10.1038/s41598-022-13825-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlgren C., Karlsson A., Bylund J. Intracellular Neutrophil Oxidants: From Laboratory Curiosity to Clinical Reality. J. Immunol. 2019;202:3127–3134. doi: 10.4049/jimmunol.1900235. [DOI] [PubMed] [Google Scholar]
- 21.Zhang T., Jiang J., Liu J., Xu L., Duan S., Sun L., Zhao W., Qian F. MK2 Is Required for Neutrophil-Derived ROS Production and Inflammatory Bowel Disease. Front. Med. 2020;7:207. doi: 10.3389/fmed.2020.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J., Liu J., Li A. Roles of neutrophil reactive oxygen species (ROS) generation in organ function impairment in sepsis. J. Zhejiang Univ. Sci. B. 2022;23:437–450. doi: 10.1631/jzus.B2101075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Z., Yang W., Parkitny L., Gibson S.A., Lee K.S., Collins F., Deshane J.S., Cheng W., Weinmann A.S., Wei H., et al. Deficiency of Socs3 leads to brain-targeted experimental autoimmune encephalomyelitis via enhanced neutrophil activation and ROS production. JCI Insight. 2019;4:e126520. doi: 10.1172/jci.insight.126520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun L., Wu Q., Nie Y., Cheng N., Wang R., Wang G., Zhang D., He H., Ye R.D., Qian F. A Role for MK2 in Enhancing Neutrophil-Derived ROS Production and Aggravating Liver Ischemia/Reperfusion Injury. Front. Immunol. 2018;9:2610. doi: 10.3389/fimmu.2018.02610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Liu Y. Neutrophil-Induced Liver Injury and Interactions Between Neutrophils and Liver Sinusoidal Endothelial Cells. Inflammation. 2021;44:1246–1262. doi: 10.1007/s10753-021-01442-x. [DOI] [PubMed] [Google Scholar]
- 26.Alves-Filho J.C., Marcel Silva Melo B., Ryffel B. MMP-9 Mediates Cross-Talk between Neutrophils and Endothelial Cells in Psoriasis. J. Invest. Dermatol. 2021;141:716–718. doi: 10.1016/j.jid.2020.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Wu X., Newbold M.A., Gao Z., Haynes C.L. A versatile microfluidic platform for the study of cellular interactions between endothelial cells and neutrophils. Biochim. Biophys. Acta Gen. Subj. 2017;1861:1122–1130. doi: 10.1016/j.bbagen.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt E.P., Lee W.L., Zemans R.L., Yamashita C., Downey G.P. On, Around, and Through: Neutrophil-Endothelial Interactions in Innate Immunity. Physiology. 2011;26:334–347. doi: 10.1152/physiol.00011.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X., Dong H., Wang M., Gao Y., Zhang T., Hu G., Duan H., Mu X. IL-1α-induced microvascular endothelial cells promote neutrophil killing by increasing MMP-9 concentration and lysozyme activity. Immunol. Res. 2016;64:133–142. doi: 10.1007/s12026-015-8731-4. [DOI] [PubMed] [Google Scholar]
- 30.Marki A., Ley K. Leaking chemokines confuse neutrophils. J. Clin. Invest. 2020;130:2177–2179. doi: 10.1172/JCI136259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G., Lin A., Han Q., Zhao H., Tian Z., Zhang J. IFN-γ protects from apoptotic neutrophil-mediated tissue injury during acute Listeria monocytogenes infection. Eur. J. Immunol. 2018;48:1470–1480. doi: 10.1002/eji.201847491. [DOI] [PubMed] [Google Scholar]
- 32.Conlan J.W., North R.J. Neutrophils are essential for early anti-Listeria defense in the liver, but not in the spleen or peritoneal cavity, as revealed by a granulocyte-depleting monoclonal antibody. J. Exp. Med. 1994;179:259–268. doi: 10.1084/jem.179.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drevets D.A. Listeria monocytogenes infection of cultured endothelial cells stimulates neutrophil adhesion and adhesion molecule expression. J. Immunol. 1997;158:5305–5313. [PubMed] [Google Scholar]
- 34.Huang T., Jiang C., Yang M., Xiao H., Huang X., Wu L., Yao M. Salmonella enterica serovar Typhimurium inhibits the innate immune response and promotes apoptosis in a ribosomal/TRP53-dependent manner in swine neutrophils. Vet. Res. 2020;51:105. doi: 10.1186/s13567-020-00828-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenlee-Wacker M., DeLeo F.R., Nauseef W.M. How methicillin-resistant Staphylococcus aureus evade neutrophil killing. Curr. Opin. Hematol. 2015;22:30–35. doi: 10.1097/MOH.0000000000000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zwack E.E., Chen Z., Devlin J.C., Li Z., Zheng X., Weinstock A., Lacey K.A., Fisher E.A., Fenyö D., Ruggles K.V., et al. Staphylococcus aureus induces a muted host response in human blood that blunts the recruitment of neutrophils. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2123017119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bestebroer J., Aerts P.C., Rooijakkers S.H.M., Pandey M.K., Köhl J., Van Strijp J.A.G., De Haas C.J.C. Functional basis for complement evasion by staphylococcal superantigen-like 7: SSL7 inhibits C5a generation. Cell Microbiol. 2010;12:1506–1516. doi: 10.1111/j.1462-5822.2010.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guerra F.E., Addison C.B., De Jong N.W.M., Azzolino J., Pallister K.B., van Strijp J.A.G., Voyich J.M., Voyich J.M. Staphylococcus aureus SaeR/S-regulated factors reduce human neutrophil reactive oxygen species production. J. Leukoc. Biol. 2016;100:1005–1010. doi: 10.1189/jlb.4VMAB0316-100RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen Q.-T., Nguyen T.-H.T., Ju S.-A., Lee Y.-S., Han S.H., Lee S.-C., Kwon B.S., Yu R., Kim G.Y., Lee B.J., Kim B.S. CD137 Expressed on Neutrophils Plays Dual Roles in Antibacterial Responses against Gram-Positive and Gram-Negative Bacterial Infections. Infect. Immun. 2013;81:2168–2177. doi: 10.1128/IAI.00115-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu M., Bedouhene S., Hurtado-Nedelec M., Pintard C., Dang P.M.C., Yu S., El-Benna J. The Prolyl Isomerase Pin1 Controls Lipopolysaccharide-Induced Priming of NADPH Oxidase in Human Neutrophils. Front. Immunol. 2019;10:2567. doi: 10.3389/fimmu.2019.02567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J., Liu Y., Chen J., Hu C., Teng M., Jiao K., Shen Z., Zhu D., Yue J., Li Z., Li Y. The ROS-mediated activation of IL-6/STAT3 signaling pathway is involved in the 27-hydroxycholesterol-induced cellular senescence in nerve cells. Toxicol. Vitro. 2017;45:10–18. doi: 10.1016/j.tiv.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 42.Marasco M.R., Conteh A.M., Reissaus C.A., Mirmira R.G., Linnemann A.K. Interleukin-6 Reduces B-Cell Oxidative Stress by Linking Autophagy with the Antioxidant Response. Diabetes. 2018;67 doi: 10.2337/db17-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., Luo F., Zhao H. Paraquat-Induced Reactive Oxygen Species Inhibit Neutrophil Apoptosis via a p38 MAPK/NF-κB–IL-6/TNF-α Positive-Feedback Circuit. PLoS One. 2014;9:e93837. doi: 10.1371/journal.pone.0093837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillipson M., Kubes P. The neutrophil in vascular inflammation. Nat. Med. 2011;17:1381–1390. doi: 10.1038/nm.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fielding C.A., McLoughlin R.M., McLeod L., Colmont C.S., Najdovska M., Grail D., Ernst M., Jones S.A., Topley N., Jenkins B.J. IL-6 Regulates Neutrophil Trafficking during Acute Inflammation via STAT3. J. Immunol. 2008;181:2189–2195. doi: 10.4049/jimmunol.181.3.2189. [DOI] [PubMed] [Google Scholar]
- 46.Wright H.L., Cross A.L., Edwards S.W., Moots R.J. Effects of IL-6 and IL-6 blockade on neutrophil function in vitro and in vivo. Rheumatology. 2014;53:1321–1331. doi: 10.1093/rheumatology/keu035. [DOI] [PubMed] [Google Scholar]
- 47.Angriman F., Ferreyro B.L., Burry L., Fan E., Ferguson N.D., Husain S., Keshavjee S.H., Lupia E., Munshi L., Renzi S., et al. Interleukin-6 receptor blockade in patients with COVID-19: placing clinical trials into context. Lancet Respir. Med. 2021;9:655–664. doi: 10.1016/S2213-2600(21)00139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maini R.N., Taylor P.C., Szechinski J., Pavelka K., Bröll J., Balint G., Emery P., Raemen F., Petersen J., Smolen J., et al. Double-blind randomized controlled clinical trial of the interleukin-6 receptor antagonist, tocilizumab, in European patients with rheumatoid arthritis who had an incomplete response to methotrexate. Arthritis Rheum. 2006;54:2817–2829. doi: 10.1002/art.22033. [DOI] [PubMed] [Google Scholar]
- 49.Egholm C., Heeb L.E.M., Impellizzieri D., Boyman O. The Regulatory Effects of Interleukin-4 Receptor Signaling on Neutrophils in Type 2 Immune Responses. Front. Immunol. 2019;10:2507. doi: 10.3389/fimmu.2019.02507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bestebroer J., Poppelier M.J.J.G., Ulfman L.H., Lenting P.J., Denis C.V., Van Kessel K.P.M., Van Strijp J.A.G., De Haas C.J.C. Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectin–mediated neutrophil rolling. Blood. 2007;109:2936–2943. doi: 10.1182/blood-2006-06-015461. [DOI] [PubMed] [Google Scholar]
- 51.Chavakis T., Hussain M., Kanse S.M., Peters G., Bretzel R.G., Flock J.-I., Herrmann M., Preissner K.T. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nat. Med. 2002;8:687–693. doi: 10.1038/nm728. [DOI] [PubMed] [Google Scholar]
- 52.Veldkamp K.E., Heezius H.C., Verhoef J., van Strijp J.A., van Kessel K.P. Modulation of Neutrophil Chemokine Receptors by Staphylococcus aureus Supernate. Infect. Immun. 2000;68:5908–5913. doi: 10.1128/iai.68.10.5908-5913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afonso P.V., Janka-Junttila M., Lee Y.J., McCann C.P., Oliver C.M., Aamer K.A., Losert W., Cicerone M.T., Parent C.A. LTB4 Is a Signal-Relay Molecule during Neutrophil Chemotaxis. Dev. Cell. 2012;22:1079–1091. doi: 10.1016/j.devcel.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ingram P.N., Hind L.E., Jiminez-Torres J.A., Huttenlocher A., Beebe D.J. An Accessible Organotypic Microvessel Model Using iPSC-Derived Endothelium. Adv. Healthcare Mater. 2018;7:1700497. doi: 10.1002/adhm.201700497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.