Abstract
Background:
Head neck squamous cell carcinoma (HNSC) is globally prevalent cancer attributed to tobacco habit. Despite the significant advances in early diagnosis and treatment of HNSC chemo-radio resistance are routinely observed in patients. Aberrant DNA repair mechanisms mainly microhomology mediated DNA end joining (MMEJ) pathway causing deleterious mutations and is implicated in treatment resistance. X-ray cross complimenting group 1 (XRCC1) has recently been shown to play an essential role in MMEJ making XRCC1 a potential therapeutic target to render tumors chemo-radiosensitive. This study analyzes the correlation between the expression level of XRCC1 gene with survival, regulation by miRNA and synthetic lethality partners in HNSCC.
Materials and Methods:
XRCC1 gene expression was evaluated in 520 HNSC patients and 44 of normal tissues using the UALCAN (TCGA) database and its correlation with survival outcome of HNSC patients was analyzed by Kaplan-Meier plot. Infiltration of immune cells in tumors was analyzed by “Tumor-Infiltrating Immune Estimation Resource (TIMER) and promoter methylation status of XRCC1 in samples was analysed by UALCAN. STRING was used to find gene interacting partners of XRCC1.
Results:
XRCC1 was significantly overexpressed in primary tumor of HNSCC and significantly increased with tumor stages and grade and associated with poor survival rate. High XRCC1 expression in HNSC was positively correlated with infiltration level of B cells naïsve, CD4+ and macrophages.
Conclusion:
These results indicate that XRCC1 is a prognostic marker for predicting survival in HNSC patients. Understanding how XRCC1 leads to treatment resistance and modulate immune response can lead to development of targeted therapy.
Key Words: HNSCC, XRCC1, MMEJ, Prognostic biomarker, UALCAN (TCGA) database
Introduction
Head neck squamous cell carcinoma (HNSC) is globally prevalent cancers and common in Indian men and is attributed to tobacco use (Jethwa and Khariwala, 2017). Despite the significant advances in early diagnosis and treatment of HNSC by surgery and concurrent radio-chemotherapy is routinely observed in patients leading to recurrence, morbidity and mortality (Kim, 2017). Mutagenic agents including ionizing radiation can trigger the accumulation of genetic alteration resulting it affect the cell cycle regulation, proliferation, apoptosis and modification in DNA repair. DNA repair mechanism plays an important role in the protection of the genome against carcinogenic agents and preventing the cancer cells altered from continuing the cell cycle and their inappropriate proliferation. DNA repair genes are the key regulators in different metabolic pathways and maintain the structural integrity and functions of DNA. X-ray repair cross-complementing 1 (XRCC1) is the first human gene identified that affects cell sensitivity to ionizing radiation (London, 2015). XRCC1 participates in base excision repair (BER), single-strand break repair (SSBR) and microhomology mediated DNA end joining (MMEJ) to repair the DNA damage induced by ionizing radiation, alkylation and chemical mutagens. Aberrant DNA repair mechanisms causing deleterious mutations are implicated in treatment resistance (Sfeir, 2015; Sharma et al., 2015). Hence biomarkers to assess DNA repair in tumors is warranted for tailoring therapeutic regimen and better prognosis.
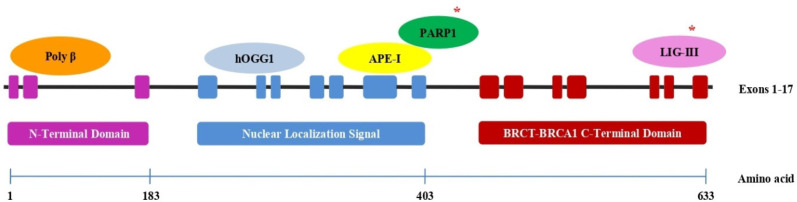
XRCC1 (Figure 1) has recently been shown to play an essential role in DNA repair mechanism mainly MMEJ, making XRCC1 a potential therapeutic target to render tumor chemo-radiosensitive (Sfeir, 2015; Dutta et al., 2017; Ali et al., 2020). The objective of this study was to analyzes the correlation between the expression level of XRCC1 gene with tumor grade, tumor immunity and survival in HNSC using bioinformatics approach.
Figure 1.
XRCC1 Gene Overview and Binding Sites for Other Genes (* indicates proteins of MMEJ repair pathway)
Materials and Methods
Correlation between XRCC1 expression and survival analysis
Gene expression data of 520 HNSC tissues and 44 of normal tissues was downloaded from TGCA RNAseq (UALCAN) publically open database in Jane 2022 (Chandrashekar et al., 2017) and analysed, moreover, methylation of XRCC1 gene was also analysed TCGA tissue samples. We downloaded survival time information from TCGA (including Overall survival and Disease-free survival). These tumor samples were divided into high and low expression group according the median XRCC1 expression value. The survival analysis was done using the Kaplan-Meier plot method (Győrffy et al., 2013) the significant differences determined by a long rank test. P< 0.05 considered as statistically significant for overall survival.
Correlation of the XRCC1 expression with clinical parameters in HNSC
A correlation analysis of the expression levels of XRCC1 with the different clinical stages of cancer (HNSC: stage I, n = 27; stage II, n = 71; stage III, n = 81; stage IV, n = 264) and gender of patients (HNSC: Female, n = 136; Male, n = 383) was performed using the TCGA database. Spearman correlation test was used to analyze the correlation between XRCC1 expression and tumor stages, box plots obtained from TCGA RNAseq database. Then, the correlation between the expression levels of XRCC1 and the gender of patients was analyzed using the Wilcoxon test to identify the significant differences between the two groups, box plots obtained from TCGA RNAseq database (Chandrashekar et al., 2017).
Methylation of XRCC1 with survival of HNSC
Methylation status of XRCC1 gene was analysed using SurvivalMeth publically available database (Zhang et al., 2021). This tool provides comparative analysis of gene methylation level with patient’s survival.
XRCC1 expression and immune cell infiltration
For the investigation of tumor immune cells in the RNAseq TGCA database, the Tumor Immune Estimate Resource (TIMER) is a freely accessible computational tool (Li et al., 2017). Based on the gene expression profile, a deconvolution method is employed to determine the amount of tumor immune infiltration cells. Eight different types of tumor immune infiltration cells (B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and DCs) were chosen for our investigation, which examined the relationships between the expressions of the XRCC1 gene and TIICs in patients with HNSC. For measuring the Spearman’s correlation, tumor purity was taken into account (Li et al., 2017). Due to the expected negative relationships between highly expressed genes in the invading immune cells and tumor purity. Statistical significance was defined as a p value 0.05.
Gene-function interaction analysis of XRCC1
Virtual screening of XRCC1 associated genes were performed using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) a publically available database (Szklarczyk et al., 2019; Szklarczyk et al., 2021). It is user friendly tool provide hypothetic gene function and functional essays. This tool also predict the function of given query genes, the interactions of genes are in the form of physical, co-expression, prediction, proteins homology, co-localization, genetic and pathway interaction (Szklarczyk et al., 2021).
miRNA regulating XRCC1 expression
The publically available database miRWalk was utilised to find XRCC1 associated miRNAs in humans with predicted and validated miRNA binding sites. The miRNA target site prediction for miRWalk is based on the random-forest approach. The list of all miRNAs associated with XRCC1 displayed on \were arranged as per their expression levels if upregulated or downregulated. The top three upregulated miRNAs were further tested for expression level in HNSC using the publically available mirCancer database (Xie et al., 2013).
Correlation with XRCC1 expression and miR-21
Expression of miR-21 and survival information was obtained from TCGA miRNA database. MiRNAs control the gene expression by suppressing the gene expression. In this study miRNA-21 was over expressed in TCGA samples.
Mutation analysis in XRCC1 and miRNA-21
Somatic mutations in XRCC1 gene were analyzed using the publically COSMIC database (Forbes et al., 2015). The mutations in the miRNA-21 binding site were identified using the database miRNASNP-V3 Liu et al., 2021). Gene and miRNA binding site were predicted by miRWalk database (Stich et al., 2018).
Statistical analysis
Bioinformatics statistical analysis was performed by the online freely available ‘R’ software. Student t-test was used for analysis of expression differences into normal and tumor tissues of TCGA samples. Logrank test was used for comparison of K-M survival curve and genetic alteration prognostic plots. Moreover, P value <0.05 was considered as statistical significant difference.
Results
XRCC1 is significantly overexpressed in HNSC
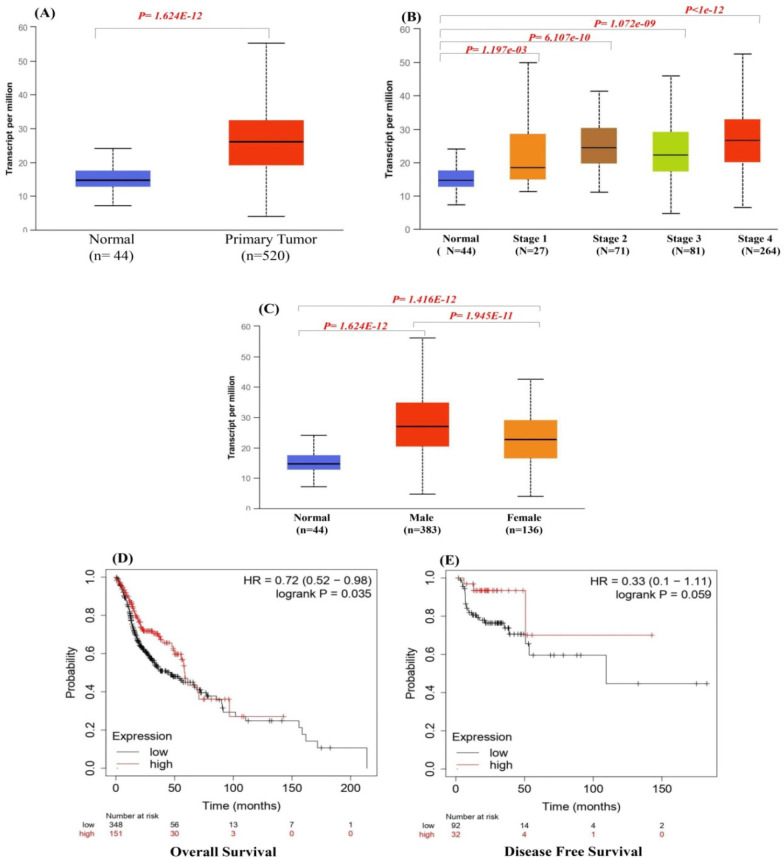
Analysis of the XRCC1 expression level in tumor samples (n = 520) and normal tissue samples (n = 44) indicated that XRCC1 expression level was significantly higher in HNSC tissues than in the normal tissues (P < 0.001) (Figure 2). The higher mRNA expression level of XRCC1 was significantly associated with poor over all patient survival and disease free OS in patients with HNSC (Figure 2D-E).
Figure 2.
(A) Expression of XRCC1 in TCGA samples, Correlation of XRCC1 with clinical factors (B) Correlation between XRCCs expression and tumor stage in HNSC patients (C) Correlation between XRCC1 expression and gender in HNSC patients, (D-E) Overall and Disease Free Survival
Correlation of the expression levels of XRCC1with the clinical parameters of HNSC
Statistically significant differences were observed in the patients with HNSC in the XRCC1 expression (P < 0.05), with a positive correlation between the tumor stage and gene expression (Figure 2B). The highest gene expression was observed in stage IV HNSC (Figure 2C). In addition, mRNA levels of XRCC1in HNSC patients were higher in men than in women (P < 0.05; Figure 2B-C). Overall, these findings imply that the expression levels of XRCC1 are partially correlated with the clinical parameters in HNSC patients.
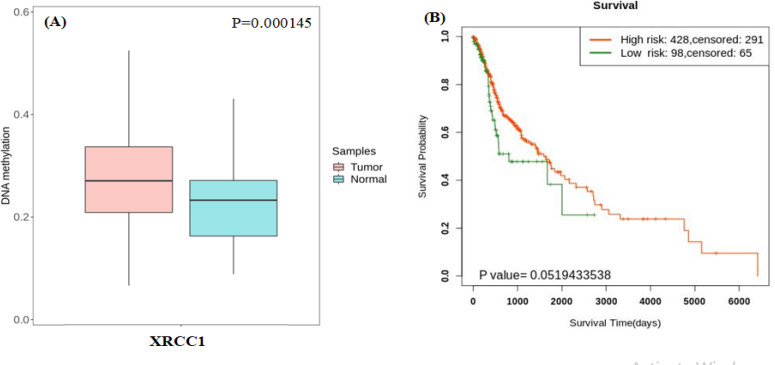
Methylation of XRCC1 with survival of HNSC
Based on our finding suggesting that XRCC1 overepression can be used as potential independent risk factor for patients with HNSC, we further analyzed the methylation sites of XRCC1 using Survival Meth (Li et al., 2019). For Comparison of the methylation levels in HNSC tumor tissues and in normal tissues (Table 1). Then, we divided the samples with differentially methylated sites into high- and low-risk groups and performed survival analysis using the KM method (Győrffy et al., 2013). The results showed that the XRCC1 overexpressing high-risk groups were associated with a poor prognosis (Figure 3), which is consistent with the conclusions of our previous survival analysis.
Table 1.
Methylation Site Differences of XRCCs in HNSC and Normal Tissues
| Gene | Site | Average of tumor | Average of Normal | Delta value | P value |
|---|---|---|---|---|---|
| XRCC1 | cg01880404 | 0.049194 | 0.060121 | -0.01092 | 6.82E-08 |
| cg08112313 | 0.277669 | 0.230684 | 0.046984 | 0.000145 | |
| cg15167433 | 0.042735 | 0.051365 | -0.00862 | 8.24E-05 |
Figure 3.
DNA-Methylation of XRCC1 Gene
Correlation analysis of expression levels of XRCC1and immune cell infiltration
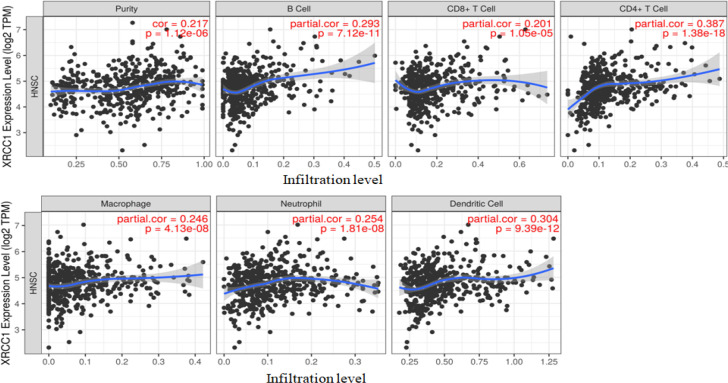
Correlation with expression level of XRCC1 and immune cell infiltration in HNSC was analysed using TIMER (cistrome.shinyapps.io/timer). Expression level and infiltration level were significantly positive correlated in our study (Figure 4). Overexpression of XRCC1 demonstrated that a strong negative correlation with the infiltration level of differentiation CD8+ T cells. Moreover, a CD4+ T cell, B cell, Macrophages, Neutrophils and Dendritic cells shows the significant positive association with the infiltration level (Figure 4).
Figure 4.
Tumor Purity and Immune Cell Infiltration Associated with XRCC1 Expression in Patients with HNSC
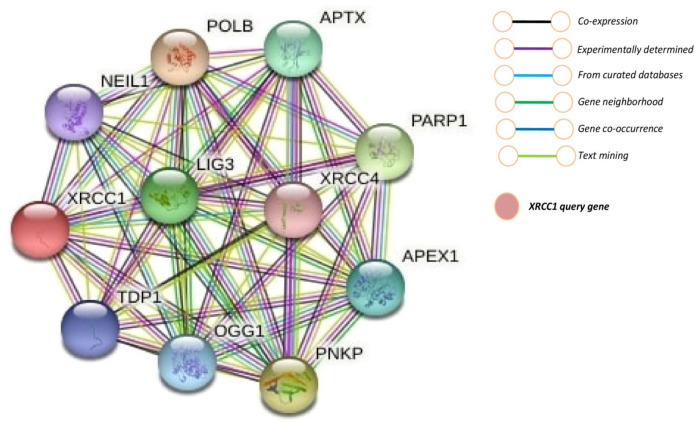
Gene-function interaction analysis of XRCC1
To understand the molecular mechanism of gene expression and their impact on the tumor, we constructed a network of co-expressed genes, including XRCC1 using STRING. The results showed that shows close association with XRCC1 given in Figure 5. LIG3, POLB, NEIL1, TDP1, APTX, PNKP, APEX1, OGG1, and XRCC4 genes were significantly associated with XRCC1. XRCC1shows multiple interactions with LIG3 and POLB, the most significant interactions were physical and pathway interactions.
Figure 5.
Gene Interaction Network Associate with XRCC1 Function
miRNA regulating XRCC1 expression
The miRWalk database search revealed more than twenty five hundreds miRNAs that were associated with XRCC1 which are intact on different position of gene according to the result generated by miRWalk database. We further screen out these miRNAs using mirCancer database, out of 2609 miRNAs only 63 were up/down regulated in HNSC, thus mirCancer database provided the expression profile of cancer related miRNAs which were studied in clinical samples. We were filtered these miRNAs and most over expressed miRNAs i.e. has-mir-21 were select for further analysis.
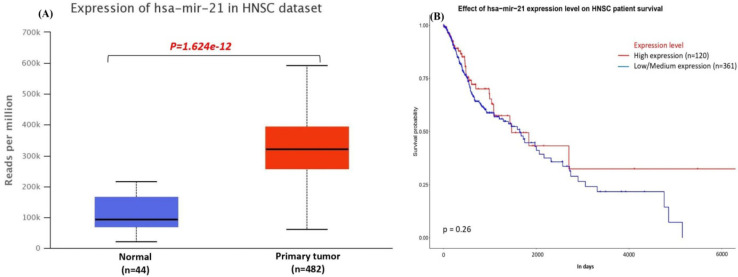
Correlation with XRCC1 expression and miR-21
Expression level of miR-21 was analyzed in TCGA database, as results miR-21 was significantly (P< 0.001) overexpressed in n= 482 tumor tissues of HNSC in TCGA dataset. Moreover, there was no significant (p= 0.26) effect found of miR-21 expression level in HNSC patients survival. That results show the negative correlation because miRNAs regulated the gene expression by suppressing the expression (Figure 6A-B).
Figure 6.
(A) expression of miR-21 in HNSC (B) effect of miR-21 expression on HNSC patients survival
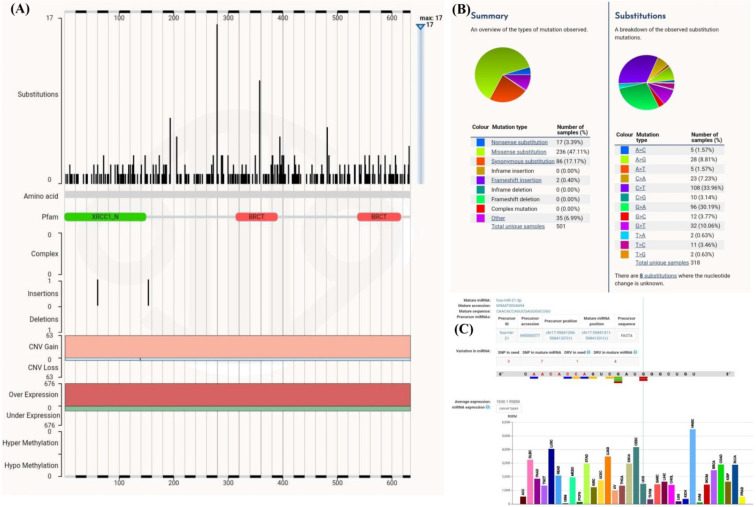
Mutation analysis in XRCC1 and miRNA-21
Using COSMIC database, we found that there are more than 500 mutations in XRCC1 coding region that are reported (Figure 7A-B). Amongst these mutations there are three SNPs in mature miR 21 seed region that is responsible for binding the miR21. These mutations in the miR21 binding site are found in HNSC (Figure 7C). Moreover, predictive binding of site of miRNA-21 in human XRCC1 is on CDC region, and there are huge numbers of mutation present that affect the XRCC1 regulation by miRNA-21.
Figure 7.
(A-B) mutations in XRCC1 coding region, (C) mutations in mature miRNA-21
Discussion
Ionizing radiation and alkalytic agents play crucial role in tumor recurrence by affecting the genetic stability. The DNA repair genes are closely associated with tumorigenesis (Borrego-Soto and Ortiz-Lopez, 2015). XRCC1 is an important member of XRCC family which plays an essential role in DNA repair mechanism (Brem, 2005; London, 2020). XRCC1 is a key protein of single strand break repair, but recent studies shows it is also an important member of double strand break repair (DSBR) mechanism (Charbonnel et al., 2011; Sharma et al., 2015; Dutta et al., 2017; Eckelmann et al., 2020). XRCC1 is a scaffold protein assembled the group of protein which will repair the gap, and bind with PARP-1 and 2, LIG1 and 3, Polyβ etc. XRCC1-LIG3 complex complete the ligation process (London, 2015; 2020; Tang and Caglayan, 2021). Most of the clinical work related to XRCC1 gene in cancer has focused on gene polymorphisms in XRCC1 i.e. Arg399Gln is a genetic susceptibility factor for the HNSC and other cancer (Dutta et al., 2020; Sobiahe et al., 2020; Kabzinski et al., 2021).
In this study using bioinformatics approach we report that XRCC1 is over expressed in HNSC and this result in poor overall survival. These results are similar with previous studies (Ang et al., 2011; Moreira et al., 2020; Wang et al., 2021) though different from (Yadav et al., 2011). High rate of recurrence and metastases in patients is a major concern affecting morbidity and mortality in HNSC. The reason for the metastasis and recurrence of the disease may be due to the interaction of the surrounding tissues and immune cells that make up the tumor microenvironment (Perri et al., 2020; Wang et al., 2021). We reported that high level of XRCC1 expression showed a significant negative correlation with the infiltration of the cluster of differentiation CD8+ T cells and positive correlation with the infiltration of the cluster of differentiation CD4+ T cells, B cell, Macrophages, Neutrophils and Dendritic cells (Fan et al., 2021).
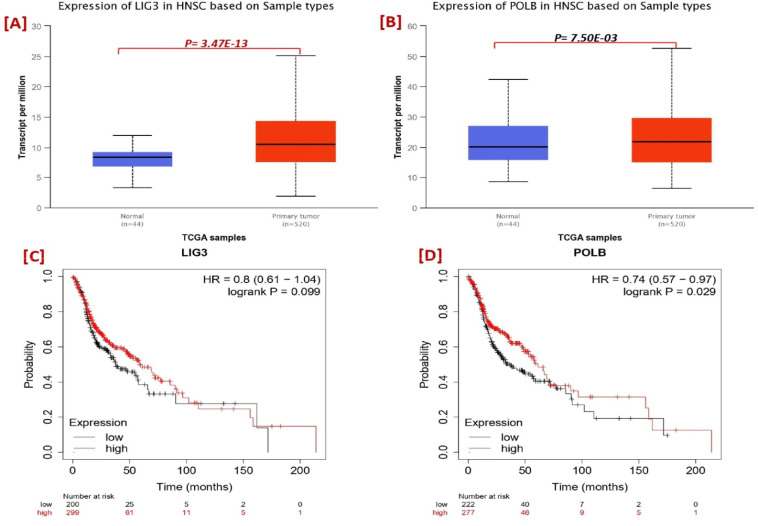
It is very useful to understanding the molecular mechanisms of their intrinsic associations with other protein and their collective impact on the tumor. We constructed a network of co-expressed genes and proteins, including XRCC1 using the STRING publically available database (Szklarczyk et al., 2019; 2021). On visual analysis of protein and gene and 10 genes identified that closely interacted with XRCC1 (Figure 5). Protein- protein network shows the physical, co-expression, prediction, co-localization, genetic and pathway interaction. XRCC1 had multiple interactions with LIG3 and POLB; the most significant interactions were physical interactions and pathway. LIG3 and POLB genes are the DNA repair proteins that plays distinct role in DNA repair pathway. For understanding the association of LIG3 and POLB with XRCC1 we further investigate the expression and survival of these genes, we found that LIG3 was significantly overexpress in HNSC patient (Figure 8A) and may affect the overall survival (Figure 8C). Although, there was no significant association were found between POLB and XRCC1 expression and survival (Figure 8B, D).
Figure 8.
Expression and Survival Analysis of LIG3 and POLB
Scaffold protein XRCC1 play diverse role in base excision repair (BER), SSBR and MMEJ also know as alt-NHEJ. XRCC1 is consists of three terminal domains including N- terminal domain which is binds to DNA strands break. The C-terminal domain is the central domainconsisting of BRCT-I binds with poly (ADPribose) polymerase 1 (PARP1) and BRCT-II binds with to Ligase III (LIG3) (London, 2015; 2020). Moreover, XRCC1 also interact with other proteins like, OGG1, ATM, APE1, POLB, LIG1, MRE11 and BRCA1 etc., these proteins can be the synthetic lethality partner of XRCC1 (Table 2). Synthetic lethality therapeutically exploits the inter-gene relationship where the loss of function of either of two related genes is non-lethal, but loses of both causes cell death (Nijman et al., 2011). PARP1 overexpression in different cancer associated with poor clinical outcome (Rojo et al., 2012; Liu et al., 2016; Li et al., 2018). XRCC1-PARP1 interaction was well defined in BER and SSBR, PARP1 has bind on C-terminal domain of XRCC1 gene. It is noted that high XRCC1 or high PARP1 protein level was associated with aggressive phenotype and significantly linked with poor overall survival (Ali et al., 2020). Moreover, pre-clinical gene therapy was selectively toxic in XRCC1 deficient or platinum sensitive ovarian cancer cells; Ali et al., (2020) concluded that XRCC1 deficient ovarian cancer cells suitable for synthetic lethality targeting using PARP1 inhibition. In addition, XRCC1 deficiency/mutation can also hyper-activate PARP1 (Hoch et al., 2017).
Table 2.
Synthetic Lethality Partner of XRCC1
| S N | Synthetic Lethality Partner | Expression | Type of Cancer | Reference |
|---|---|---|---|---|
| 1 | PARP1 | Up regulate | Ovarian Cancer | [7] |
| 2 | LIG1 | Up regulate | Ovarian Cancer | [40] |
| 3 | BRCA2 | Down regulated | In vitro | [22] |
| 4 | ATM | Up regulate | Breast Cancer | [40] |
| 5 | ATR | Up regulate | Breast Cancer | [41] |
| 6 | Wee1 | Up regulate | Breast Cancer | [40] |
| 7 | MRE11 | Up regulate | Oral Cancer | [42] |
Double strand breaks (DSBs) repair mechanism is still not fully understood but it can be repaired by different pathways including homologous recombination (HR) and non-homologous end joining (NHEJ) (Scrully et al., 2019). Homologous recombination (HR) is an error-free repair mechanism uses a homologous template for DSBs repair (Sfeir, 2015; Wang et al., 2017) and it is only activated when cells enter S/G2 cell cycle phase because this pathway required cyclin-dependent kinases (CDKs) for promoting end resection for its activation. Although, classical nonhomologous end joining (C-NHEJ) pathway relies on Ku70/Ku80 and ligates DSB ends without a template and it is activated throughout the cell cycle. Some time DNA double strand breaks repair by another alternative- NHEJ which is called alt-NHEJ or Microhomology Mediated End Joining (MMEJ), this is highly error prone pathway and may associated with aggressive tumor phenotype, recurrence and resistant to therapy. The mechanism of MMEJ activation itself a big research question for scientific community and still under investigation. MMEJ required the proteins which are used in single strand repair mechanism, common are LIG3, POLB, PARP1 and XRCC1.
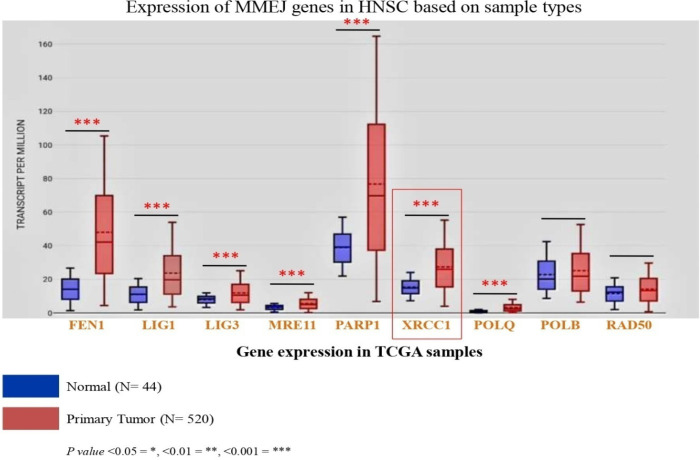
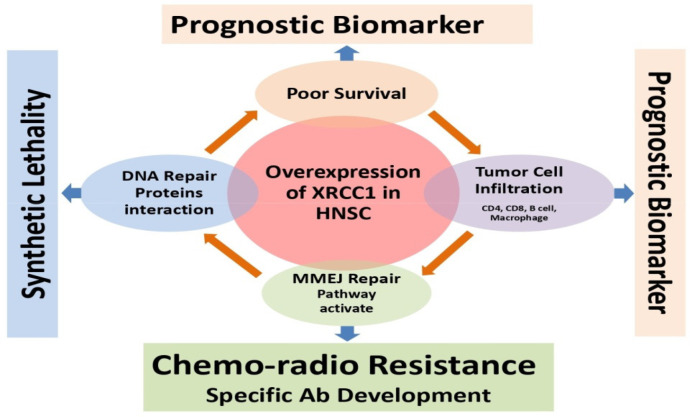
Along with XRCC1, FEN1, LIG1 and 3, PARP1, MRE11, NBS1 POLQ, POLB, RAD50 and Polθ plays a distinct role in MMEJ repair pathway. Expression of these genes in HNSC cancer patients decides the choice of repair pathways selection. We have analyzed the expression of above mention genes in TCGA RNAseq database; we found that most of them are overexpressed in TCGA HNSC tumor tissues (Figure 9). Furthermore, we analyze the survival of TCGA HNSC patients in reference to MMEJ pathway proteins, we found that XRCC1, POLQ and LIG1 are significantly affected the overall survival of HNSC patients (Table 3). In addition, XRCC1could be a biomarker for prognosis and prediction of chemo-radio resistance in head neck cancer patient (Figure 10).
Figure 9.
Expression of MMEJ Pathway Genes in TCGA Samples
Table 3.
Overall Survivals of MMEJ Genes of TCGA RNAseq HNC Samples
| S N | MMEJ Genes | Expression | Hazard ratio (HR) | Logrank P value |
|---|---|---|---|---|
| 1 | XRCC1 | Overexpress | 0.72 (0.52- 0.98) | 0.035* |
| 2 | LIG1 | Overexpress | 0.67 (0.48-0.92) | 0.013* |
| 3 | POLQ | Overexpress | 0.71 (0.52-0.95) | 0.022* |
| 4 | FEN1 | Overexpress | 1.29 (0.98-1.69) | 0.065 |
| 5 | LIG3 | Overexpress | 0.80 (0.61-1.04) | 0.099 |
| 6 | PARP1 | Overexpress | 0.78 (0.57-1.05) | 0.100 |
| 7 | MRE11 | Overexpress | 0.75 (0.56-1.00) | 0.052 |
| 8 | POLB | No Significant expression | 0.74 (0.57-0.97) | 0.029* |
| 9 | RAD50 | No Significant expression | 1.32 (0.99-1.75) | 0.060 |
Figure 10.
Translational Potential of XRCC1 as a Prognostic Biomarker
In conclusion, these results provide evidence for using XRCC1 expression in tumor as prognostic marker for predicting survival in HNSC patients. Understanding how XRCC1 leads to treatment resistance and modulate immune response can lead to development of targeted therapy.
Author Contribution Statement
SSA: Drafting the manuscript, data search and analysis, and conceptualizing the MS; SL: Data search and analysis on miR-21; MG: Providing critical inputs for clinical utility of the findings, drafting the manuscript; RC: Providing critical inputs on the search strategy and bioinformatics analysis, drafting the manuscript; AK: Conceptualizing the MS, training SSA and SL for data search and analysis validation of results. Drafting and reviewing the manuscript.
Acknowledgements
We would like to thank the Department of Biochemistry, All India Institute of Medical Sciences (AIIMS) Bhopal (M.P.) India for infrastructure and computational support.
Funding statement
This study is approved and financially supported by the Indian Council of Medical Research New Delhi, India for ICMR- Research Associateship (45/01/2020-HUM /BMS) to SSA under the mentorship of AK.
Ethical issue
This study is a part of ICMR-RA ship, and approved by the Institutional Ethical Committee (IHEC) AIIMS Bhopal, India (Ref. No. IHEC-LOP/2021/EF0224).
Data availability
Appendix
The data used in this study is drawn from the publically available domain and is thus permitted for this study. ULCAN-TCGA (The University of ALabama at Birmingham CANcer data analysis Portal- The Cancer Genome Atlas): This user friendly tool provides access to TCGA, MET500, CPTAC and CBTTC cancer OMICS database and is a good platform for identifying for novel biomarkers, to perform in silico validation of potential genes and provides information on miRNAs expression and patients survival.
Date of use: 30 June 2022
URL: https://ualcan.path.uab.edu/analysis.html
K-M plotter (Kaplan-Meier Plotter): The Kaplan Meier plotter can investigate the relationship between all gene expression (mRNA, miRNA, protein) and survival in over thirty thousand samples from 21 different tumour types, including HNSC, lung, breast, stomach, and colon cancer, and myeloma. The major goal of the tool is to generate and assess survival biomarkers.
Date of use: 2 July 2022
URL: https://kmplot.com/analysis/
SurvivalMeth: A comprehensive web-based automated service for investigating the impact of DNA methylation-related functional elements (DMFEs) on prognosis in various cancer types. It incorporates a variety of combinations, such as single DMFEs, multiple DMFEs, and clinical data, to perform detailed survival analysis and visualization on preupload data, and customised DNA methylation profiles of DMFEs from various diseases to be analysed.
Date of use: 2 July 2022
URL: http://bio-bigdata.hrbmu.edu.cn/survivalmeth/
TIMER (Tumor Immune Estimate Resource): It is a novel statistical web based resource designed for the systematic evaluation of the clinical impact of t immune cells namely B cell, CD4+ T cell, CD8+ T cell, neutrophil, macrophage and dendritic cell that are abundant in tumor microenvironment in different cancers. TIMER provides survival analysis of given immune cells in desired cancer type and correlation analysis of immune cells with the expression of selected gene.
Date of use: 5 July 2022
URL: http://timer.cistrome.org/
STRING (Search Tool for the Retrieval of Interacting Genes/Proteins): This is an online utility tool analyses interaction between proteins and their functional associations along with visualization of partial protein interaction networks and executes gene set enrichment analysis for the entire input.
Date of use: 5 July 2022
URL: https://string-db.org/
miRWalk: This resource offers validated and anticipatory details regarding miRNA-binding locations spanning the 3’UTR, 5’UTR, and CDC region a specified gene of interest across various species, including humans. The method employed for forecasting miRNA target sites relies on a random forest-based technique, specifically implemented through the TarPmiR software. This tool thoroughly scans the entire gene sequence.
Date of use: 30 June 2022
URL: http://mirwalk.umm.uni-heidelberg.de/
COSMIC (Catalogue of Somatic Mutation in Cancer): This is a comprehensive manually curated and descriptive database that provides detailed information of effects of somatic mutations including non-coding mutations, gene fusions, alterations in gene copy numbers, and mutations responsible for drug resistance in human cancer.
Date of use: 10 Oct 2022
URL: https://cancer.sanger.ac.uk/cosmic
miRNASNP-V3: The miRNASNP-v3 investigates relationships between miRNA-associated SNPs and diseases; analyses impact of SNPs on miRNA-target interactions, expression and their structure; in different cancers, performs functional enrichment analysis to discern miRNA target gain/loss due to SNPs; scrutinizes the links between drug sensitivity and miRNA expression.
Date of use: 12 Oct 2022
URL: http://bioinfo.life.hust.edu.cn/miRNASNP/#!/
Declaration of competing interest
The authors declare that they have no competing interests.
References
- Ali R, Alabdullah M, Alblihy A, et al. PARP1 blockade is synthetically lethal in XRCC1 deficient sporadic epithelial ovarian cancers. Cancer Lett. 2020;469:124–33. doi: 10.1016/j.canlet.2019.10.035. [DOI] [PubMed] [Google Scholar]
- Ali R, Alblihy A, Toss MS. XRCC1 deficient triple negative breast cancers are sensitive to ATR, ATM and Wee1 inhibitor either alone or in combination with olaparib. Ther Adv Med Oncol. 2021;21:1758835920974201. doi: 10.1177/1758835920974201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Subhi N, Ali R, Abdel-Fatah T, et al. Targeting ataxia telangiectasia-mutated- and Rad3-related kinase (ATR) in PTEN-deficient breast cancers for personalized therapy. Breast Cancer Res Treat. 2018;169:277–86. doi: 10.1007/s10549-018-4683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang MK, Patel MR, Yin XY, et al. High XRCC1 protein expression is associated with poorer survival in patients with head and neck squamous cell carcinoma. Clin Cancer Res. 2011;17:6542–52. doi: 10.1158/1078-0432.CCR-10-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrego-Soto G, Ortiz-López R, Rojas-Martínez A. Ionizing radiation-induced DNA injury and damage detection in patients with breast cancer. Genet Mol Biol. 2015;38:420–32. doi: 10.1590/S1415-475738420150019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R, Hall J. XRCC1 is required for DNA single-strand break repair in human cells. Nucleic Acids Res. 2005;33:2512–20. doi: 10.1093/nar/gki543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnel C, Allain E, Gallego ME, et al. Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair (Amst) 2011;10:611–9. doi: 10.1016/j.dnarep.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Dutta A, Eckelmann B, Adhikari S, et al. Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Res. 2017;45:2585–99. doi: 10.1093/nar/gkw1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Abarna R, Shubham M, et al. Anbalagan, Effect of Arg399Gln single-nucleotide polymorphism in XRCC1 gene on survival rate of Indian squamous cell head-and-neck cancer patients. J Cancer Res Therapy. 2020;16:551–8. doi: 10.4103/jcrt.JCRT_476_18. [DOI] [PubMed] [Google Scholar]
- Eckelmann BJ, Bacolla A, Wang H, et al. XRCC1 promotes replication restart, nascent fork degradation and mutagenic DNA repair in BRCA2-deficient cells. NAR Cancer. 2020;2:zcaa013. doi: 10.1093/narcan/zcaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Gao Z, Li X, et al. Gene expression and prognosis of x-ray repair cross-complementing family members in non-small cell lung cancer. Bioengineered. 2021;12:6210–228. doi: 10.1080/21655979.2021.1964193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;18:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch NC, Hanzlikova H, Rulten SL, et al. XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature. 2017;541:87–91. doi: 10.1038/nature20790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jethwa AR, Khariwala SS. Tobacco-related carcinogenesis in head and neck cancer. Cancer Metastasis Rev. 2017;35:411–23. doi: 10.1007/s10555-017-9689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabzinski J, Maczynska M, Kaczmarczyk D, et al. Influence of Arg399Gln, Arg280His and Arg194Trp XRCC1 gene polymorphisms of Base Excision Repair pathway on the level of 8-oxo-guanine and risk of head and neck cancer in the Polish population. Cancer Biomark. 2021;32:317–26. doi: 10.3233/CBM-203163. [DOI] [PubMed] [Google Scholar]
- Kim YS. Reirradiation of head and neck cancer in the era of intensity-modulated radiotherapy: patient selection, practical aspects, and current evidence. Radiat Oncol J. 2017;35:1–15. doi: 10.3857/roj.2017.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:108–10. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li C, Jin J, et al. High PARP-1 expression predicts poor survival in acute myeloid leukemia and PARP-1 inhibitor and SAHA-bendamustine hybrid inhibitor combination treatment synergistically enhances anti-tumor effects. EBio Med. 2018;38:47–56. doi: 10.1016/j.ebiom.2018.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CJ, Fu X, Xia M, et al. miRNASNP-v3: a comprehensive database for SNPs and disease-related variations in miRNAs and miRNA targets. Nucleic Acids Res. 2021;49:1276–81. doi: 10.1093/nar/gkaa783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang Y, Zhao Y, et al. High PARP-1 expression is associated with tumor invasion and poor prognosis in gastric cancer. Oncol Lett. 2016;12:3825–35. doi: 10.3892/ol.2016.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London RE. The structural basis of XRCC1-mediated DNA repair. DNA Repair (Amst) 2015;30:90–103. doi: 10.1016/j.dnarep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London RE. XRCC1 - Strategies for coordinating and assembling a versatile DNA damage response. DNA Repair (Amst) 2020;93:102917. doi: 10.1016/j.dnarep.2020.102917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira DG, Morais EF, Santos HB, et al. Freitas, Immunohistochemical expression of DNA repair proteins in oral tongue and lower lip squamous cell carcinoma. Braz Oral Res. 2020;34:e101. doi: 10.1590/1807-3107bor-2020.vol34.0101. [DOI] [PubMed] [Google Scholar]
- Nijman SM. Synthetic lethality: general principles, utility and detection using genetic screens in human cells. FEBS Lett. 2011;585:1–6. doi: 10.1016/j.febslet.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri F, Ionna F, Longo F, et al. Immune response against Head and Neck Cancer: Biological mechanisms and implication on Therapy. Translational Oncol. 2020;13:262–74. doi: 10.1016/j.tranon.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo F, García-Parra J, Zazo S, et al. Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer. Ann Oncol. 2012;23:1156–64. doi: 10.1093/annonc/mdr361. [DOI] [PubMed] [Google Scholar]
- Scully R, Panday A, Elango R, et al. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol. 2019;20:698–714. doi: 10.1038/s41580-019-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Symington LS. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci. 2015;40:701–14. doi: 10.1016/j.tibs.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Javadekar S, Pandey M, et al. Homology and enzymatic requirements of microhomology-dependent alternative end joining. Cell Death Dis. 2015;6:e1697. doi: 10.1038/cddis.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobiahe A, Hijazi E, Al-Ameer HJ, et al. Arg399Gln XRCC1 Polymorphism and Risk of Squamous Cell Carcinoma of the Head and Neck in Jordanian Patients. Asian Pac J Cancer Prev. 2020;21:663–5. doi: 10.31557/APJCP.2020.21.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sticht C, Torre DL, Parveen A, et al. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One. 2018;18:e0206239. doi: 10.1371/journal.pone.0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:10800. doi: 10.1093/nar/gkab835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Çağlayan M. The scaffold protein XRCC1 stabilizes the formation of polβ/gap DNA and ligase IIIα/nick DNA complexes in base excision repair. J Biol Chem. 2021;297:101025. doi: 10.1016/j.jbc.2021.101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhang M, Cheng M, et al. Tumor microenvironment in head and neck squamous cell carcinoma: Functions and regulatory mechanisms. Cancer Lett. 2021;507:55–69. doi: 10.1016/j.canlet.2021.03.009. [DOI] [PubMed] [Google Scholar]
- Wang H, Xu X. Microhomology-mediated end joining: new players join the team. Cell Biosci. 2017;7:6. doi: 10.1186/s13578-017-0136-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Chen YK, Lo S, et al. MRE11 promotes oral cancer progression through RUNX2/CXCR4/AKT/FOXA2 signaling in a nuclease-independent manner. Oncogene. 2021;40:3510–32. doi: 10.1038/s41388-021-01698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Fang PT, Su CW, et al. Excision repair cross-complementing group 2 upregulation is a potential predictive biomarker for oral squamous cell carcinoma recurrence. Oncol Lett. 2021;21:450. doi: 10.3892/ol.2021.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B, Ding Q, Han H, et al. miRCancer: a microRNA-cancer association database constructed by text mining on literature. Bioinformatics. 2013;29:638–44. doi: 10.1093/bioinformatics/btt014. [DOI] [PubMed] [Google Scholar]
- Yadav A, Kumar B, Teknos TN, et al. Sorafenib enhances the antitumor effects of chemoradiation treatment by downregulating ERCC-1 and XRCC-1 DNA repair proteins. Mol Cancer Ther. 2011;10:1241–51. doi: 10.1158/1535-7163.MCT-11-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhao N, Zhang X, et al. SurvivalMeth: a web server to investigate the effect of DNA methylation-related functional elements on prognosis. Brief Bioinform. 2021;20:bbaa162. doi: 10.1093/bib/bbaa162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Appendix
The data used in this study is drawn from the publically available domain and is thus permitted for this study. ULCAN-TCGA (The University of ALabama at Birmingham CANcer data analysis Portal- The Cancer Genome Atlas): This user friendly tool provides access to TCGA, MET500, CPTAC and CBTTC cancer OMICS database and is a good platform for identifying for novel biomarkers, to perform in silico validation of potential genes and provides information on miRNAs expression and patients survival.
Date of use: 30 June 2022
URL: https://ualcan.path.uab.edu/analysis.html
K-M plotter (Kaplan-Meier Plotter): The Kaplan Meier plotter can investigate the relationship between all gene expression (mRNA, miRNA, protein) and survival in over thirty thousand samples from 21 different tumour types, including HNSC, lung, breast, stomach, and colon cancer, and myeloma. The major goal of the tool is to generate and assess survival biomarkers.
Date of use: 2 July 2022
URL: https://kmplot.com/analysis/
SurvivalMeth: A comprehensive web-based automated service for investigating the impact of DNA methylation-related functional elements (DMFEs) on prognosis in various cancer types. It incorporates a variety of combinations, such as single DMFEs, multiple DMFEs, and clinical data, to perform detailed survival analysis and visualization on preupload data, and customised DNA methylation profiles of DMFEs from various diseases to be analysed.
Date of use: 2 July 2022
URL: http://bio-bigdata.hrbmu.edu.cn/survivalmeth/
TIMER (Tumor Immune Estimate Resource): It is a novel statistical web based resource designed for the systematic evaluation of the clinical impact of t immune cells namely B cell, CD4+ T cell, CD8+ T cell, neutrophil, macrophage and dendritic cell that are abundant in tumor microenvironment in different cancers. TIMER provides survival analysis of given immune cells in desired cancer type and correlation analysis of immune cells with the expression of selected gene.
Date of use: 5 July 2022
URL: http://timer.cistrome.org/
STRING (Search Tool for the Retrieval of Interacting Genes/Proteins): This is an online utility tool analyses interaction between proteins and their functional associations along with visualization of partial protein interaction networks and executes gene set enrichment analysis for the entire input.
Date of use: 5 July 2022
URL: https://string-db.org/
miRWalk: This resource offers validated and anticipatory details regarding miRNA-binding locations spanning the 3’UTR, 5’UTR, and CDC region a specified gene of interest across various species, including humans. The method employed for forecasting miRNA target sites relies on a random forest-based technique, specifically implemented through the TarPmiR software. This tool thoroughly scans the entire gene sequence.
Date of use: 30 June 2022
URL: http://mirwalk.umm.uni-heidelberg.de/
COSMIC (Catalogue of Somatic Mutation in Cancer): This is a comprehensive manually curated and descriptive database that provides detailed information of effects of somatic mutations including non-coding mutations, gene fusions, alterations in gene copy numbers, and mutations responsible for drug resistance in human cancer.
Date of use: 10 Oct 2022
URL: https://cancer.sanger.ac.uk/cosmic
miRNASNP-V3: The miRNASNP-v3 investigates relationships between miRNA-associated SNPs and diseases; analyses impact of SNPs on miRNA-target interactions, expression and their structure; in different cancers, performs functional enrichment analysis to discern miRNA target gain/loss due to SNPs; scrutinizes the links between drug sensitivity and miRNA expression.
Date of use: 12 Oct 2022
URL: http://bioinfo.life.hust.edu.cn/miRNASNP/#!/