Abstract
In recent studies, we have shown that Pseudomonas aeruginosa strains that are acutely cytotoxic in vitro damage the lung epithelium in vivo. Genetic analysis indicated that the factor responsible for acute cytotoxicity was controlled by ExsA and therefore was part of the exoenzyme S regulon. The specific virulence determinant responsible for epithelial damage in vivo and cytotoxicity in vitro was subsequently mapped to the exoU locus. The present studies are focused on a genetic characterization of the exoU locus. Northern blot analyses and complementation experiments indicated that a region downstream of exoU was expressed and that the expression of this region corresponded to increased ExoU secretion. DNA sequence analysis of a region downstream of exoU identified several potential coding regions. One of these open reading frames, SpcU (specific Pseudomonas chaperone for ExoU), encoded a small 15-kDa acidic protein (137 amino acids [pI 4.4]) that possessed a leucine-rich motif associated with the Syc family of cytosolic chaperones for the Yersinia Yops. T7 expression analysis and nickel chromatography of histidine-tagged proteins indicated that ExoU and SpcU associated as a noncovalent complex when coexpressed in Escherichia coli. The association of ExoU and SpcU required amino acids 3 to 123 of ExoU. In P. aeruginosa, ExoU and SpcU are coordinately expressed as an operon that is controlled at the transcriptional level by ExsA.
Pseudomonas aeruginosa is an opportunistic nosocomial pathogen involved in septicemia and/or severe pneumonia in a diverse patient population that includes burn victims, cystic fibrosis patients, and immunocompromised individuals. The numerous virulence factors produced by P. aeruginosa include a group of antihost or effector proteins that are secreted by a type III or contact-dependent secretion pathway (7, 40). The components of the P. aeruginosa pathway include over 20 gene products predicted to make up the type III apparatus and at least four proteins that have been implicated in the translocation of effector proteins (reviewed in reference 7). Translocation is defined as the process by which the bacterium transfers effectors directly into the host cell cytoplasm and is considered to represent a specialized intoxication mechanism (14, 16, 19, 27, 30). Because the P. aeruginosa type III system has been recently discovered, most of the functional characteristics attributed to these proteins are based on the high level of homology with proteins encoded by the Yersinia virulence plasmid pYV. The P. aeruginosa type III secretion apparatus, translocation proteins, and effectors are coordinately controlled by a common transcriptional activator, ExsA, and are collectively referred to as members of the exoenzyme S regulon (7, 8).
Proteins that are unique to the exoenzyme S regulon include a group of secretion substrates that serve as effectors or antihost proteins. These proteins include ExoS, ExoT (7, 39), ExoY (42), and ExoU (5). ExoS and ExoT are ADP-ribosyltransferases that covalently modify several eukaryotic proteins. The primary substrates include members of the small GTP-binding H- and K-ras protein families that serve as regulators of host cell growth and differentiation. The expression of ExoS and ExoT enzymatic activity is absolutely dependent on a eukaryotic cofactor, FAS (factor activating exoenzyme S). FAS is a member of the 14-3-3 family of cofactors that serve as regulators of eukaryotic enzyme activities (11). ExoT is relatively inactive (0.2%) when compared to the ADP-ribosyltransferase activity exhibited by ExoS (39). ExoS has been shown to possess cytotoxic activity, to disrupt Ras-mediated signal transduction pathways in eukaryotic cells, to covalently modify Ras in vivo, and to cause changes in cytoskeletal structure (2, 9, 12, 23, 25). ExoY is a recently described adenylate cyclase secreted by the P. aeruginosa type III system. Translocation of ExoY has been demonstrated, because cells infected with an ExoY-producing P. aeruginosa strain accumulate supraphysiological levels of cyclic AMP (42). Similar to ExoS and ExoT, ExoY requires a eukaryotic cofactor for the stimulation of adenylate cyclase activity. The cofactor stimulating ExoY activity has not been identified, but appears to be distinct from calmodulin (42). The most cytotoxic product of the P. aeruginosa type III pathway appears to be ExoU. ExoU (5) or PepA (15) was discovered because of the acute cytotoxic reaction observed when certain strains of P. aeruginosa were cocultured with MDCK cells. ExoU production is correlated with lung injury in an acute infection model in mice (5). Unlike ExoS, ExoT, or ExoY, an ExoU-associated enzymatic activity has not been identified, and the mechanism of ExoU-mediated acute cytotoxicity has not been defined.
To determine the minimal region of the ExoU locus required for expression and cytotoxicity, we performed a deletion analysis. Preliminary data suggested that a region downstream of the exoU open reading frame (ORF) was required for maximal ExoU secretion from P. aeruginosa. These data were reminiscent of the results observed when mutations are introduced in the Yersinia Syc loci. The Syc proteins are small cytosolic chaperones that bind their cognate Yop effector proteins (4, 32) before or during secretion from the bacterium (6, 35, 36). In this article, we report the observation that SpcU (specific Pseudomonas chaperone for ExoU) affects the extracellular levels and/or secretion of ExoU. SpcU shares several of the common structural properties reported for the cytosolic chaperones of Yersinia (37). SpcU is predicted to encode a small-molecular-mass (14.9-kDa) protein with an acidic pI (4.4) and a conserved C-terminal leucine motif. We show that histidine-tagged recombinant SpcU and ExoU associate in a noncovalent complex that requires an amino-terminal domain within ExoU. exoU and spcU are encoded within the same mRNA and form an operon.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli TG1 was used as a host strain for M13 bacteriophage propagation (22). For genetic manipulations, Pseudomonas strains were cultivated in Luria-Bertani (LB) broth and plated on Vogel-Bonner medium (34). E. coli DH5α and BL21(DE3) were cultured in LB broth or agar. For ExoU production, P. aeruginosa strains were cultivated in a deferrated dialysate of Trypticase soy broth containing 10 mM nitrilotriacetic acid as previously described (17). Bacteria containing plasmids were grown in the presence of the appropriate antibiotic(s) at a final concentration of 100 μg of ampicillin, 34 μg of chloramphenicol (E. coli), 400 μg of carbenicillin, and 100 μg of tetracycline (P. aeruginosa) per ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant properties or genotypea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| BL21(DE3) | hsd-5 gal (λ cIts857 indl Sam7 nin5 lacUV5-T7 gene1) | 33 |
| BL21(DE3) pLysS | pLysS (Cmr) | 33 |
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 | 22 |
| thi-1 relA1 | ||
| SM10 | thi thr leu tonA lacY supE recA::Rp4-2Tc::Mu Km | 39 |
| P. aeruginosa | ||
| PA103 | Wild type, exoU+exoT+ exoS | 5, 8 |
| PAK | Wild type, exoU exoT+ exoS+ | 5, 8 |
| PA103ΔexoU | exoU nonpolar mutant | This study |
| PA103exoU::Tn5Tc | exoU polar mutant, mini-Tn5Tc insertion | 5 |
| Plasmids | ||
| pUC18, pUC19 | Wild type, Apr | 22 |
| pUCP19 | Wild type, E. coli-P. aeruginosa shuttle vector, Apr | 28 |
| pUC18exoU656 | Contains the exoU promoter and 656 aa of ExoU | This study |
| pET16b | N-terminal His tag fusion expression vector, Apr | Novagen |
| pET23b | C-terminal His tag fusion expression vector, Apr | Novagen |
| pET16bΔEcoRI | Deletion of EcoRI site of pET16b | J. T. Barbieri |
| pETexoS | pET16bΔEcoRI encoding His-ExoS | 20 |
| pETexoU656 | pET16bΔEcoRI encoding 656 aa of His-ExoU | This study |
| pETexoU | pET16bΔEcoRI encoding 687 aa of His-ExoU, | This study |
| pETexoUspcU | pET16bΔEcoRI encoding His-ExoU and SpcU | This study |
| pET23exoUspcU | pET23b encoding ExoU and SpcU-His | This study |
| pET23Δ3–123exoUspcU | pET23b encoding an in-frame deletion (Δ3–123) of ExoU and SpcU-His | This study |
| pUCPNexoU | pUCP19 encoding only ExoU with the native promoter and stop sequences | This study |
| pUCPexoUEcoRV | pUCP19 containing exoUspcU, as a 2.8-kb EcoRV fragment | This study |
| pUCPexoU(Bam6.5) | pUCP19 containing a 6.5-kb BamHI fragment with exoU and flanking downstream sequences | 5 |
| pNOT19 | Allelic replacement vector | 29 |
| pNOTexoU | pNOT19 containing a 6.5-kb fragment with exoU and additional downstream sequences | This study |
| pNOTΔexoU | pNOT19 containing an in-frame deletion of aa 3–674 of ExoU | This study |
| pMOB3 | pHSS21; Kmr CmrsacBR oriT as a NotI cartridge | 29 |
| pMOBΔexoU | pNOTΔexoU with the NotI cartridge from pMOB3 | This study |
aa, amino acids.
DNA methods.
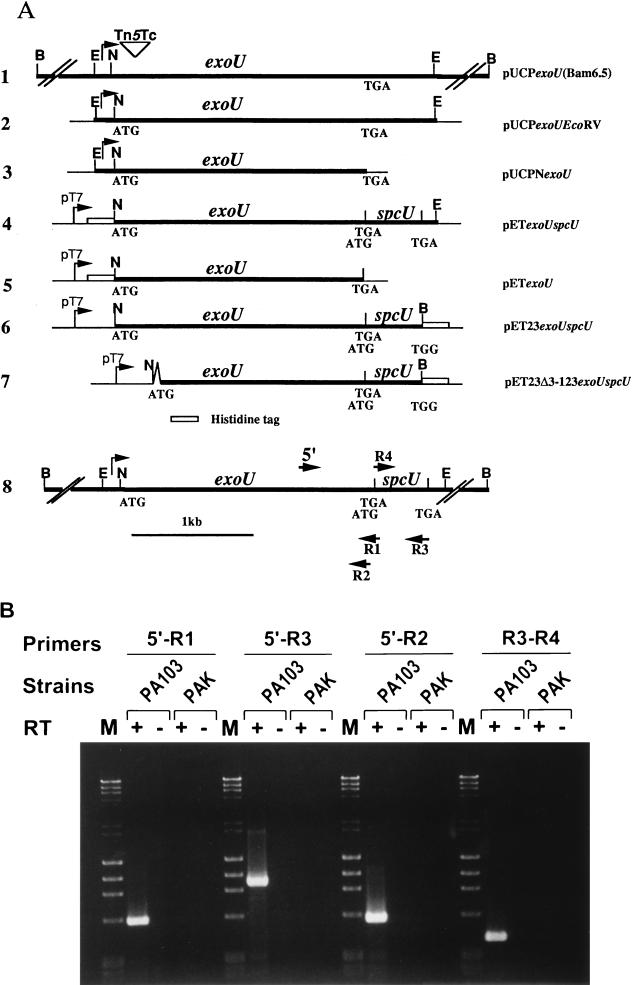
Plasmid DNA was isolated by standard alkaline lysis procedures (22) or by the use of Qiagen spin or Midi-Prep columns (Qiagen, Inc., Santa Clarita, Calif.). DNA fragments were purified from agarose gels by using a Qiaquick gel extraction kit (Qiagen, Inc.). Transformation of E. coli and P. aeruginosa was performed as previously described (22, 24). PCRs were performed with Deep Vent polymerase (New England Biolabs, Beverly, Mass.) in an Amplitron II thermocycler (Thermolyne, Barnstead, Dubuque, Iowa). A 2-min preincubation at 98°C was followed by 30 cycles consisting of a 1-min denaturation step; annealing for 1 min at variable temperature, depending on the primer; and extension at 72°C for 1 min. A 5-min extension step at 72°C at the end of the cycling period was performed. All primers were obtained from Operon Technologies, Inc. (Alameda, Calif.) and the location of relevant primers for reverse transcriptase-PCR (RT-PCR) is shown in Fig. 4A.
FIG. 4.
ExoU locus constructs and amplification of the exoU-spcU transcript by RT-PCR. (A) Schematic representation of the location of the exoU and spcU ORFs within different plasmid constructs used in complementation analyses in P. aeruginosa (constructs, lines 1 to 3), protein expression and purification in E. coli (constructs, lines 4 to 7), and mapping of the mRNA 3′ end (line 8). Double slash marks indicate regions that are not drawn to scale. The chromosomal insertion of the polar Tn5 insertion within the 5′ coding region of exoU is illustrated by an inverted triangle (line 1). Corresponding constructs include pUCPexoU(Bam6.5) (line 1), pUCPexoUEcoRV (line 2), pUCPNexoU (line 3), pETexoUspcU (line 4), pETexoU (line 5), pET23exoUspcU (line 6), and pET23Δ3–123exoUspcU (line 7). N, NsiI; E, EcoRV; B, BamHI. An inverted V in the construct on line 7 represents the deletion of amino acids 3 to 123 of ExoU. Line 8, map of the ExoU locus with the approximate locations of primers used for PCR amplification. Total RNAs from P. aeruginosa strains PA103 (exoU+) and PAK (exoU) were used for the first-strand cDNA synthesis in the presence (+) or absence (−) of Superscript II RT. RT-PCR was performed as described in Materials and Methods. (B) Products of the RT-PCRs were analyzed on a 1.2% agarose gel. PA103 (exoU+) and PAK (exoU) strains are indicated over the brackets, and the primer pairs used for each reaction are underlined. M, molecular weight markers. + and −, reaction mixtures containing (+) or not containing (−) RT.
Plasmid construction.
For expression and purification of recombinant ExoU and/or SpcU in E. coli, pET16b and pET23b vectors were used (Novagen). pET16b and pET23b each encode histidine tags (10 and 6 amino acids, respectively) which were cloned in frame with either ExoU (N-terminal tag) or SpcU (C-terminal tag). DNA fragments containing the coding regions for exoU and spcU were either excised as restriction fragments from subclones of pUCPexoU(Bam6.5), pUCPexoUEcoRV, and pUC18exoU656 or as fragments isolated from PCRs. After being subcloned in M13 vectors, all amplified fragments were subjected to nucleotide sequence analysis with an ALF automated sequencer and reagents (Pharmacia, Piscataway, N.J.) to confirm the fidelity of the Deep Vent polymerase.
For expression and complementation studies in P. aeruginosa, plasmids containing exoU on a 6.5-kb BamHI fragment [pUCPexoU(Bam6.5)], exoU and spcU on a 2.8-kb EcoRV fragment (pUCPexoUEcoRV), or the exoU ORF alone (pUCPNexoU) were constructed in the E. coli-P. aeruginosa shuttle vector pUCP19 (28). Fragments were obtained by standard techniques through partial restriction endonuclease digestion or PCR amplification. All constructs were transcribed by the native exoU promoter region, which is controlled by the transcriptional activator ExsA.
To construct a nonpolar mutation in exoU without disrupting spcU, a flanking region upstream and including the start codon of ExoU was cloned in the gene replacement vector, pNOT19, as a HindIII-NsiI fragment. A tetracycline cartridge was inserted into the HindIII site to serve as a selectable marker for transconjugants. This marker was located outside the region needed for homologous recombination into the chromosome. To provide downstream flanking sequences, a region was amplified that included the last 14 codons of ExoU and 738 bp of downstream DNA. Primers used for amplification contained 5′ NsiI and 3′ EcoRI sites to ligate into the 5′NsiI site of exoU and a 3′ EcoRI site of the vector. This strategy deleted 672 codons of ExoU and provided an in-frame stop codon for the predicted 13-amino-acid peptide. The deletion was confirmed by nucleotide sequence analysis. The NotI mobilization cartridge (29) was added as a NotI fragment to complete the construct. Conjugation, selection of transconjugants, and resolution of plasmid sequences from the chromosome were performed as previously described (29, 39). The chromosomal exoU deletion in strain PA103 (PA103ΔexoU) was confirmed by Southern blot analysis with both internal and downstream probes (data not shown).
Complementation of PA103exoU::Tn5Tc or PA103ΔexoU.
Complementation plasmids were transformed into PA103exoU::Tn5Tc or PA103ΔexoU by the MgCl2 method (24). Transformants were grown under inducing conditions for maximal expression of ExoU and SpcU (5, 8). Cultures were harvested at the stationary phase and normalized to identical optical density readings at 540 nm. Cellular lysates and concentrated supernatant fractions were prepared as previously described (8).
Nucleotide sequence analysis.
DNA was subcloned in M13mp18 and -19 according to standard protocols (22), and DNA sequencing was performed with an ALF automated sequencer (Pharmacia) and fluoresceinated primers (Operon Technologies). Analysis of sequences was performed with Genetics Computer Group software (Madison, Wis.).
RNA isolation, RT-PCR, and Northern blot analysis.
Total RNA was isolated from PA103, PA103exoU::Tn5Tc, and PA103exoU::Tn5Tc/pUCPexoU(Bam6.5) from cultures grown under inducing conditions (13). Northern blot analysis was performed as previously described (41) with a 2-kb EcoRV restriction fragment and a 466-bp XhoI restriction fragment to detect messages specific for exoU and downstream loci, respectively.
To map the 3′ end of the exoU locus mRNA, a combination of primers were used with RT-PCR amplification of cDNA. P. aeruginosa PA103 (exoU+) and PAK (exoU) were grown under inducing conditions, and total RNA was isolated from mid-log-phase bacterial cultures (13). Random primers (50 ng) and Superscript II RT (Life Technologies) were used for first-strand cDNA synthesis according to the manufacturer’s recommendations. Ten percent of the first-strand reaction mixture was used as a template for specific amplification with primers R1-4 and 5′ (Fig. 4A).
Expression, purification, and quantitation of recombinant proteins in E. coli.
Plasmids containing histidine-tagged versions of ExoU and/or SpcU were transformed into BL21(DE3) pLysS. After overnight growth, strains were diluted 50-fold into 800 ml of LB broth with ampicillin and chloramphenicol. After 2 h of growth at 30°C, expression of recombinant proteins was induced with 0.75 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and incubation was continued for an additional 2 h. Cellular extracts were prepared as described previously (39), filtered, and loaded onto a 3-ml nickel-nitrilotriacetic acid-agarose column (Qiagen). For the pET16b series (10-histidine amino-terminal leader), the column was washed with 30 ml of binding buffer (wash 1) followed by binding buffer containing 60 mM imidazole (wash 2). Bound proteins were eluted with binding buffer containing 0.5 M imidazole and collected as 2-ml fractions. For the pET23b series (a six-histidine carboxyl-terminal tag), wash 2 contained 25 mM imidazole, and bound proteins were eluted with 250 mM imidazole.
Denaturing chromatography was performed to determine the binding characteristics of histidine-tagged ExoU or SpcU. Histidine-tagged proteins were first isolated under nondenaturing conditions by nickel chromatography as described above. The peak fraction containing the eluted tagged protein and other associated peptide(s) was diluted in a solution of urea to obtain a final concentration of 6 M. The fraction was further dialyzed in either binding buffer (His-ExoU) or phosphate-buffered saline (SpcU-His) containing 6 M urea. Samples were subsequently reapplied to the nickel column in the presence of urea. The column was washed and eluted with buffers containing 6 M urea. Fractions were collected, and samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (13.5% polyacrylamide) and Western blot analyses (8). Blots were probed with rabbit anti-ExoU or monoclonal anti-histidine tag antibodies (Qiagen) with a secondary rabbit anti immunoglobulin G (IgG) (Sigma Chemical Co.). Bound IgG was detected with 125I-protein A and autoradiography. Proteins were quantitated with the bicinchonic acid protein kit (Pierce, Rockford, Ill.) or by densitometry of Coomassie-stained polyacrylamide gels with bovine serum albumin as a standard.
Nucleotide sequence accession number.
The accession number for the nucleotide sequence reported for spcU is U97065.
RESULTS
Complementation of P. aeruginosa PA103exoU::Tn5Tc.
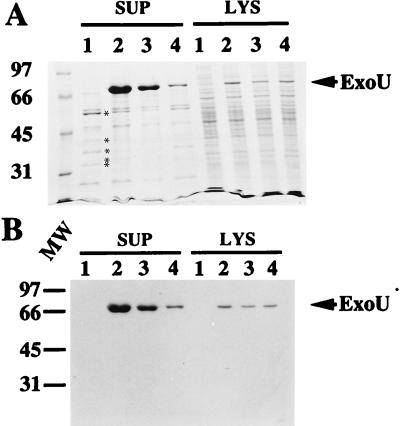
Previous studies had shown that the PA103exoU::Tn5Tc mutant strain, when complemented in trans with a plasmid containing exoU on a 6.5-kb BamHI fragment, produced extracellular ExoU, was cytotoxic in vitro, and resulted in lung injury in vivo (5). To map the minimal region required for ExoU expression and secretion, we constructed 3′ deletions of pUCPexoU(Bam6.5) (Table 1 and Fig. 4A) and performed complementation analyses with PA103exoU::Tn5Tc. Comparisons of equivalent amounts of total protein from the supernatants and lysates of complemented strains indicated that the amount of extracellular ExoU varied with the length of flanking downstream sequence (Fig. 1A and B, lanes 2 to 4, SUP) in a specific manner. The levels of ExoU produced in the lysates appeared similar in all complemented strains (Fig. 1A and B, lanes 2 to 4, LYS). These data suggested that sequences downstream of exoU were not required for synthesis but appeared essential for maximal expression of ExoU in P. aeruginosa supernatants.
FIG. 1.
Complementation of PA103exoU::Tn5Tc. (A) SDS-polyacrylamide gel (11% acrylamide), stained with Coomassie blue R250, of protein profiles of P. aeruginosa PA103exoU::Tn5Tc (a polar insertion within exoU, host strain) containing different plasmid constructs of the ExoU locus. All strains were grown under inducing conditions for expression of the exoenzyme S regulon. Equivalent amounts (normalized to the culture optical density at 540 nm) of either extracellular fractions (supernatant [SUP]) or lysate (LYS) preparations were loaded in each lane. Lanes: 1, P. aeruginosa PA103exoU::Tn5Tc; 2, PA103exoU::Tn5Tc complemented in trans with pUCPexoU(Bam 6.5); 3, the host strain with pUCPexoUEcoRV; and 4, the host strain with pUCPNexoU. The positions of other type III-secreted products of the exoenzyme S regulon are shown by asterisks in panel A (in descending order, ExoT, PopB, PopD, PopN, and PcrV). (B) Western Blot analysis of a duplicate gel probed with polyclonal anti-ExoU antibodies. The positions of ExoU (arrow) and molecular weight (MW) standards (left [thousands]) are indicated.
One hypothesis to account for these observations is that an ORF(s) located downstream of exoU was involved in ExoU secretion. To test this hypothesis, total RNA was isolated from strains cultured under inducing conditions for ExoU expression and subjected to Northern blot analysis. Two probes were used to detect mRNA. One probe corresponded to the exoU promoter region and a majority of the ORF, and the second probe was located immediately downstream (103 bp) of the exoU stop codon. Both probes hybridized to a 2.4-kb message (data not shown). These data indicated that the exoU mRNA was approximately 400 bp larger than expected and may contain another ORF.
Nucleotide sequence analysis of the exoU downstream region.
Downstream sequences flanking exoU were subjected to restriction mapping analysis and subcloned into M13 vectors for sequencing. The G+C content of the region downstream of exoU was significantly lower (56%) than that of the P. aeruginosa genome sequences (67%), which is consistent with the lower observed G+C content of exoU (5). Translation of the nucleotide sequence identified three overlapping ORFs encoding predicted proteins with molecular masses of 14.9, 8.0, and 4.6 kDa. The first ORF possessed a start codon that overlapped the stop codon of exoU and a putative ribosome binding site (located 5 bp upstream of the start codon). This ORF, now termed spcU (specific Pseudomonas chaperone for ExoU), encoded a protein of 137 amino acids, with a predicted molecular mass of 14.9 kDa and an acidic pI of 4.4. The physical properties of SpcU are consistent with the characteristics of type III cytosolic chaperones (35, 37). We performed BESTFIT and Pile-Up alignments (Fig. 2) with Orf1 of P. aeruginosa (postulated chaperone for ExoS and ExoT), SycE (chaperone for YopE), SycH (chaperone for YopH), and SycT (chaperone for YopT). Pile-up analysis (Fig. 2) indicated that the C-terminal region of SpcU aligned with a common leucine-rich motif found within the chaperone family, as first defined by Cornelis and coworkers (18, 35, 37).
FIG. 2.
Pileup alignment of the leucine repeat motif from Syc and Syc-like chaperones. Amino acids 84 to 115 of SpcU were aligned to amino acids 85 to 116 of Orf1 (putative chaperone for ExoS or ExoT) (41), amino acids 87 to 118 of SycE (37), amino acids 89 to 120 of SycH (35), and amino acids 81 to 112 of SycT (18). Common amino acids that conform to the consensus sequence are presented in shaded boxes. A consensus sequence, which includes SpcU as part of the alignment, is shown. A consensus sequence, as determined by Iriarte and Cornelis (18), which includes a leucine-rich motif, is indicated for comparison.
Complementation of a nonpolar exoU mutation in strain PA103.
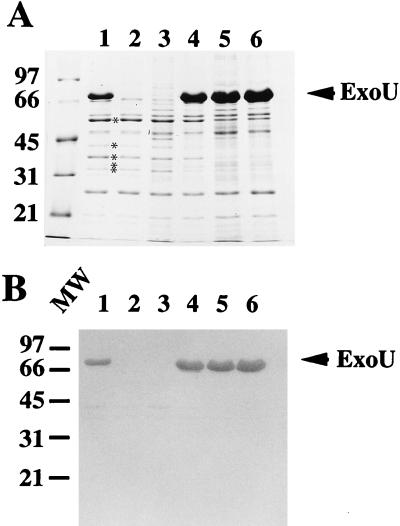
To determine if SpcU was required for ExoU secretion, a nonpolar deletion in exoU was introduced into the chromosome of strain PA103 by standard allelic replacement procedures (29, 39). This strain, PA103ΔexoU, no longer produced ExoU (Fig. 3, lanes 2 and 3) but was able to synthesize and secrete other proteins of the exoenzyme S regulon (PopB, PopD, PopN, PcrV, and ExoT). Southern blot analysis using internal and downstream probes for the exoU locus confirmed the deletion of exoU and demonstrated that the region downstream of exoU was intact (data not shown). The complementation analysis was repeated with the series of deletion plasmids used in the initial complementation experiments of the polar mutant PA103exoU::Tn5Tc. No differences were detected in the amount of extracellular ExoU when PA103ΔexoU was complemented with constructs that differed in terms of the length of included downstream sequences (Fig. 3). These data are consistent with the hypothesis that an ORF located downstream of exoU affects the extracellular levels and/or secretion of ExoU. The ability of ExoU to mediate cytotoxicity was also assessed in the nonpolar mutant. In tissue culture assays using trypan blue staining to detect dead cells, PA103ΔexoU was noncytotoxic (data not shown). The cytotoxic phenotype was restored with a plasmid containing only the exoU ORF, pUCPNexoU (data not shown). This analysis indicates that ExoU expression alone is responsible for the observed acute cytotoxic phenotype of strain PA103.
FIG. 3.
Complementation of PA103ΔexoU. (A) SDS-PAGE analysis (11% acrylamide; Coomassie-stained gel) of equivalent amounts of extracellular proteins from P. aeruginosa strains grown under inducing conditions for the exoenzyme S regulon. Lanes: 1, strain PA103; 2, PA103ΔexoU (a nonpolar deletion in exoU); 3, PA103ΔexoU containing the vector control plasmid pUCP19; 4, PA103ΔexoUpUCPNexoU; 5, PA103ΔexoUpUCPexoUEcoRV; 6, PA103ΔexoUpUCPexoU(Bam 6.5). The positions of other type III-secreted products of the exoenzyme S regulon are shown by asterisks in panel A (in descending order, ExoT, PopB, PopD, PopN, and PcrV). (B) Western blot of a duplicate gel probed with polyclonal anti-ExoU antibodies. The positions of ExoU (arrow) and molecular weight (MW) standards (left [thousands]) are indicated.
The exoU locus is organized as an operon.
Our initial results with Northern blot analysis suggested that the exoU locus encoded an mRNA containing both exoU and spcU coding regions. Since Northern blot analysis is relatively insensitive and it is difficult to accurately interpret the size of the message, we used RT-PCR to confirm the operon organization of the exoU locus. PCR primers were designed to differentiate between one unique or two separate transcripts (Fig. 4A). Total RNA was isolated from a strain containing (PA103) and a strain not containing (PAK) the exoU locus (5). Amplified products were detected only from RNA isolated from PA103 and only in reaction mixtures containing RT. These controls show that the cDNA products detected were specific to exoU mRNA. The size of the detected cDNA products was consistent with the calculated fragments based on a single mRNA encoding both exoU and spcU (Fig. 4B). We conclude that the exoU locus is organized as an operon encoding exoU and spcU.
Association of recombinant ExoU and SpcU.
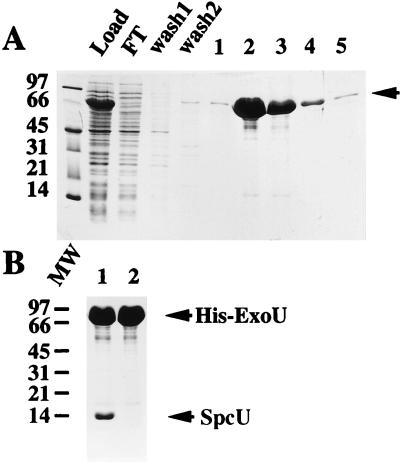
To investigate whether ExoU would bind to the postulated SpcU chaperone, exoU and/or spcU was expressed from pET16b in E. coli (Fig. 4A). When the exoU ORF was expressed (pETexoU) and subjected to nickel chromatography, His-ExoU was specifically eluted by imidazole (Fig. 5A). An identical experiment was repeated with pETexoUspcU, and the peak fractions between the two experiments were compared by SDS-PAGE (Fig. 5B). When spcU was also present in the construct, a protein with the predicted molecular mass of SpcU coeluted with His-ExoU (Fig. 5B, lane 1). To test for nonspecific binding to the nickel columns, pT7-7exoUspcU was constructed. In this construct, ExoU and SpcU were coexpressed as native proteins without histidine tags. After chromatography, neither protein bound to the nickel columns, eliminating possible nonspecific associations of ExoU and SpcU with the matrix (data not shown). These preliminary results suggested that SpcU bound noncovalently to ExoU. To confirm the identity of the SpcU ORF, the 14-kDa band, isolated from peak fractions containing His-ExoU (Fig. 5B, lane 1), was transferred to Immobilon polyvinylidene difluoride and subjected to N-terminal amino acid sequence analysis. The amino terminus (MIDTXLAQXGLR) of the 14-kDa protein was identical (12 amino acids) to the predicted amino terminus of SpcU.
FIG. 5.
Nickel chromatography, under native conditions, of recombinant amino-terminally-tagged ExoU. (A) SDS-PAGE (13.5% polyacrylamide stained with Coomassie blue R250) analysis of the cell lysate from host strain E. coli BL21(DE3) pLysS with pETexoU (N-terminal 10-histidine tag). Protein profiles of the cell lysate are shown as the material loaded onto a nickel column (Load), the flowthrough from the column (FT), wash fractions 1 and 2, and the elution fractions 1 to 5 in the presence of high concentrations of imidazole. The arrow denotes the position of His-ExoU. (B) SDS-PAGE analysis of peak eluate fractions from nickel chromatography of two expression constructs. Lane 1 contains the peak eluate fraction from an expression experiment using the pETexoUspcU construct to produce recombinant protein (His-ExoU and SpcU). Lane 2 contains a similar fraction from the expression of pETexoU (His-ExoU). Molecular weight (MW) standards (left [thousands]) and recombinant proteins (arrows) are indicated.
Association of recombinant ExoU and SpcU under denaturing conditions.
If the association between ExoU and SpcU was noncovalent, then chromatography under denaturing conditions should dissociate this interaction. The peak elution fraction from an affinity purification of pETexoUspcU was collected in a solution of urea (6 M final concentration). After dialysis to remove imidazole, this fraction was subjected to a second nickel chromatography in the presence of 6 M urea. Under these conditions, SpcU was predominantly detected in the flowthrough and wash fractions (Fig. 6, lanes FT, wash 1). Some His-ExoU (approximately 12% of the total His-ExoU load) was detected in these fractions, but the majority (88%) of the protein was eluted with imidazole (Fig. 6A, lanes 1 to 3). Western blot analysis with an antiserum specific for ExoU confirmed the approximate values derived from densitometry of Coomassie-stained gels (Fig. 6B). These data indicate that the association between His-ExoU and SpcU is noncovalent and can be disrupted by exposure to the chaotropic reagent, urea.
FIG. 6.
Denaturing nickel chromatography of His-ExoU. An N-terminal histidine-tagged ExoU construct also encoding an untagged version of SpcU (pETexoUspcU) was expressed in E. coli BL21(DE3) pLysS. The peak eluate fraction from a chromatography performed under nondenaturing conditions (Fig. 5B, lane 1) was collected in a urea solution (final concentration of 6 M). After dialysis to eliminate imidazole, the denatured sample was loaded onto a nickel column equilibrated in urea. (A) SDS-PAGE analysis (13.5% polyacrylamide) of the column fractions which include the load, flowthrough (FT), wash fractions 1 and 2, and elution fractions (in the presence of a high concentration of imidazole) 1 to 5. (B) Western blot analysis of a duplicate gel probed with polyclonal antiserum specific for ExoU. Bound IgG was detected by 125I-protein A. His-ExoU and SpcU (arrows) and molecular weight (MW) standards (left [thousands]) are indicated.
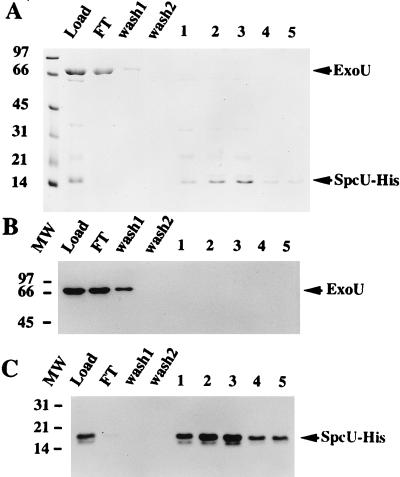
The converse experiment was performed to determine if a C-terminal, polyhistidine-tagged SpcU (SpcU-His) could associate with ExoU under native and denaturing nickel chromatography conditions. The exoUspcU operon was constructed such that the stop codon of spcU was eliminated to fit in frame with the C-terminal polyhistidine tag of pET23b. Under native chromatography conditions, SpcU-His and ExoU coeluted (Fig. 7A, B, and C, load lanes). The elution fractions from native chromatography were dialyzed to remove imidazole, brought to 6 M urea, and loaded onto a second nickel column equilibrated with 6 M urea. Under these binding conditions, ExoU was detected in the flowthrough and wash fractions (Fig. 7A and B, lanes FT and wash 1), while SpcU was detected in the fractions eluted with imidazole (Fig. 7A, lanes 1 to 5). To provide a more sensitive assay for ExoU and SpcU-His, the same fractions were subjected to Western blot analysis with either a rabbit antiserum to ExoU or a monoclonal antibody specific for the histidine-tagged portion of the fusion protein (Fig. 7B and C). These data clearly show that under denaturing conditions, ExoU is present in the load, flowthrough, and wash 1 fractions, while SpcU-His is present in the load and the eluted fractions. Taken together, the results from native and denaturing nickel chromatography, with both His-ExoU and SpcU-His, indicate that ExoU and SpcU specifically associate in a noncovalent complex.
FIG. 7.
Denaturing nickel chromatography of SpcU-His. A C-terminal six-histidine-tagged SpcU-His fusion construct also encoding an untagged version of ExoU was expressed from E. coli BL21(DE3) pET23exoUspcU. The eluate fractions from a native chromatography of the cell lysate were pooled, dialyzed to eliminate imidazole, brought to 6 M urea, and loaded onto a nickel column equilibrated in urea. (A) SDS-PAGE (13.5% polyacrylamide) analysis of the protein profiles of the different fractions: load, flowthrough (FT), wash fractions (wash 1 and 2), and elution fractions 1 to 5. (B) Western blot analysis of a duplicate gel (as in panel A) probed with polyclonal antiserum specific for ExoU. (C) Western blot analysis of a duplicate gel (as in panel A) probed with a primary monoclonal antibody specific for the histidine tag and a secondary rabbit anti-mouse IgG. Bound IgG was detected in blots B and C by using 125I-protein A. Arrows on the right indicate the migration of ExoU and SpcU-His. The molecular weight (MW) markers are indicated on the left (thousands).
Chaperone binding domain of ExoU.
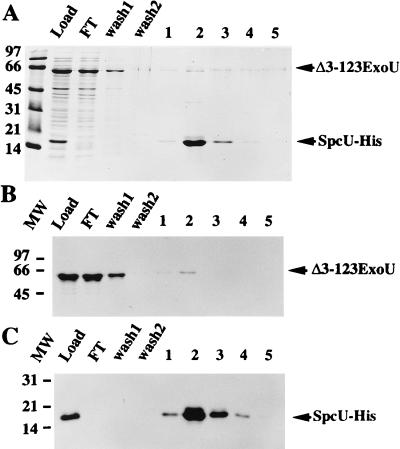
Since many of the type III chaperones bind to the amino-terminal domain of their cognate protein (21, 30, 31), we constructed an in-frame amino-terminal deletion (residues 3 to 123) within ExoU and investigated whether this recombinant derivative interacted with SpcU. Expression of both SpcU-His and Δ3–123ExoU was detected in the load fraction from an extract prepared from pET23Δ3–123exoUspcU (Fig. 8A, load lane). Based on densitometry of a Coomassie-stained gel, approximately 90% of Δ3–123ExoU was detected in the flowthrough and wash 1 fractions. SpcU-His was eluted from this column by using imidazole (Fig. 8A, lanes 2 and 3). Western blot analysis with an antiserum to ExoU (Fig. 8B) or a monoclonal antibody to the histidine tag (Fig. 8C) confirmed the results obtained with the protein profile shown in Fig. 8A. We concluded that amino acids 3 to 123 of ExoU are involved in the interaction between ExoU and SpcU.
FIG. 8.
Amino acids 3 to 123 of ExoU are required to bind to SpcU-His. The cell lysate from E. coli BL21(DE3)pLysS pET23Δ3–123exoUspcU was subjected to nickel affinity chromatography under native conditions. This construct encodes an amino-terminal deletion derivative of ExoU and SpcU-His. (A) SDS-PAGE analysis (13.5% polyacrylamide) of the protein profiles of the different fractions: load, flowthrough (FT), wash fractions 1 and 2, and elution fractions 1 to 5. (B) Western blot analysis of a duplicate gel (as shown in panel A) probed with polyclonal antiserum specific for ExoU. (C) Western blot analysis of a duplicate gel (as shown in panel A) probed with a primary monoclonal antibody specific for the histidine tag and a secondary rabbit anti-mouse IgG. Bound IgG was detected by 125I-protein A. Arrows on the right indicate the migration of Δ3–123ExoU and SpcU-His. The molecular weight (MW) markers are indicated on the left (thousands).
DISCUSSION
Our initial studies of exoU focused on the expression of the protein relative to the cytotoxicity and lung injury caused by P. aeruginosa (5). Previous analyses indicated that the exoU locus was part of a larger DNA element that may encode other virulence determinants. We noted that the G+C content of the region was significantly lower than that of the P. aeruginosa genome and mapped 5′ insertion-like sequences (5). With these data, we initiated a complementation analysis to further characterize the locus. The complementation profile of a nonpolar chromosomal exoU deletion mutant confirmed the presence of a coding region, located downstream of exoU, which appeared to influence the amount of extracellular ExoU detected. Nucleotide sequence analysis of the region immediately downstream of exoU identified three ORFs, only one of which, SpcU, possessed the common physical characteristics and leucine motif of type III chaperones (35, 37). The identity of the SpcU ORF was confirmed by amino-terminal sequence analysis of a 14-kDa protein that coeluted with His-ExoU. Binding studies indicated that SpcU associated specifically with full-length ExoU but did not associate with an amino-terminal deletion (Δ3–123) derivative of ExoU. The binding of SpcU and ExoU was noncovalent and could be disrupted by using 6 M urea. These experiments, however, do not exclude the possibility that an E. coli-derived protein may mediate the interaction between ExoU and SpcU. Taken together, our results support the hypothesis that SpcU functions as a chaperone for ExoU.
Although the chaperone predicted to bind ExoS and possibly ExoT (Orf1) was reported previously, this is the first report of the binding of a P. aeruginosa type III-encoded chaperone to a secretion substrate. The postulated functions of the type III chaperones are based on the results of studies investigating the mechanism of Yop secretion in Yersinia. Functions that have been attributed to type III chaperones include increasing the cytosolic stability of the Yops (3, 10), preventing the premature association of the Yops with the translocator proteins YopB and YopD (38), targeting of the Yop effectors to the secretion and/or translocation apparatus (3, 19, 21), and preventing cytosolic Yop aggregation (38). Recombinant ExoU, when produced as an amino-terminal histidine-tagged protein, appears to form fibular structures in the absence of SpcU (data not shown). These data suggest that SpcU may function in part by keeping ExoU from aggregating in the P. aeruginosa cytoplasm before the type III apparatus secretes it. ExoU appears to be synthesized as a relatively stable intracellular product, because complementation of the polar insertion in PA103exoU::Tn5Tc with plasmids containing or not containing spcU resulted in similar amounts of protein in bacterial lysates. Interestingly, in the absence of SpcU, small amounts of ExoU were detected in the supernatant fraction. These results suggest that a small amount of protein may be released upon lysis, that SpcU is not absolutely required for secretion, or that another chaperone may be able to partially substitute for the SpcU function. The former hypothesis is consistent with the observation that YopE and YopH appear to have two distinct secretion domains, only one of which involves the chaperone binding domain (3, 21, 30, 38). The most likely candidate for a functional SpcU substitute may be the putative chaperone for ExoS or ExoT, Orf1 (41). Although strain PA103 does not possess exoS, hybridization analysis indicates that this strain possesses a chromosomal copy of orfl (data not shown). We observed in earlier work that ExoS, ExoT, and ExoU all possessed an identical amino-terminal sequence (six amino acids) and suggested that this motif may be related to secretion (5). Domain mapping experiments also determined that the first 99 amino acids of ExoS were required for secretion and that this domain contained a region that encoded the aggregation properties of the molecule (20, 41). When ExoS and ExoU are aligned, a region encompassing 37 amino-terminal amino acids (18 to 53, ExoS; and 4 to 40, ExoU) demonstrates 31% identity with two areas of conserved residues. These data indicate that there may be some cross talk between the Orf1 and SpcU chaperones, perhaps as a protective mechanism to prevent toxic buildup of intracellular type III secretion aggregates.
The exoU locus appears to consist of exoU and spcU and probably does not include further sequences. The sizes of the message on Northern blots obtained with both exoU and spcU probes were identical and appear to correspond to the additive length of both coding regions. RT-PCR analysis demonstrated that the two coding regions were physically associated on the same message. It is unclear at this time why the addition of further downstream sequences appears to enhance ExoU production. Other factors could be encoded downstream of exoU or spcU that either may stabilize ExoU or the operon transcript or may enhance exoU translation. There is some precedence for the hypothesis that a separately encoded product may enhance exoU translation. Genetic studies of exoS expression indicate that the regulatory protein ExsC is involved in either ExoS stability or translation (13). As in Yersinia, these observations suggest that together transcription, translation, and secretion may be important parameters in expression of the P. aeruginosa type III effector proteins (1, 26).
ACKNOWLEDGMENTS
We thank Hai Yuan Sun for assistance with the nucleotide sequence analysis. Amino acid sequence analysis was carried out at the Protein and Nucleic Acid Shared Facility, Medical College of Wisconsin.
This work was supported by grants AI31665 and AI01289 (to D.W.F.) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
REFERENCES
- 1.Anderson D, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 2.Bette-Bobillo P, Giro P, Sainte-Marie J, Vidal M. Exoenzyme S from P. aeruginosa ADP ribosylates rab4 and inhibits transferrin recycling in SLO-permeabilized reticulocytes. Biochem Biophys Res Commun. 1998;244:336–341. doi: 10.1006/bbrc.1998.8263. [DOI] [PubMed] [Google Scholar]
- 3.Cheng L W, Anderson D M, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol Microbiol. 1997;24:757–765. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 5.Finck-Barbançon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish J P, Fleiszig S M J, Wu C, Mende-Mueller L, Frank D W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 6.Forsberg Å, Wolf-Watz H. Genetic analysis of the yopE region of Yersinia spp.: identification of a novel conserved locus, yerA, regulating YopE expression. J Bacteriol. 1990;172:1547–1555. doi: 10.1128/jb.172.3.1547-1555.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank D W. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 8.Frank D W, Nair G, Schweizer H P. Construction and characterization of chromosomal insertional mutations of the Pseudomonas aeruginosa exoenzyme S trans-regulatory locus. Infect Immun. 1994;62:554–563. doi: 10.1128/iai.62.2.554-563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frithz-Lindsten E, Du Y, Rosqvist R, Forsberg Å. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol. 1997;25:1125–1139. doi: 10.1046/j.1365-2958.1997.5411905.x. [DOI] [PubMed] [Google Scholar]
- 10.Frithz-Lindsten E, Rosqvist R, Johansson L, Forsberg Å. The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensible for targeting to the secretion loci. Mol Microbiol. 1995;16:635–647. doi: 10.1111/j.1365-2958.1995.tb02426.x. [DOI] [PubMed] [Google Scholar]
- 11.Fu H, Coburn J, Collier R J. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc Natl Acad Sci USA. 1993;90:2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganesan A K, Frank D W, Misra R P, Schmidt G, Barbieri J T. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J Biol Chem. 1998;273:7332–7337. doi: 10.1074/jbc.273.13.7332. [DOI] [PubMed] [Google Scholar]
- 13.Goranson J, Hovey A K, Frank D W. Functional analysis of exsC and exsB in regulation of exoenzyme S production by Pseudomonas aeruginosa. J Bacteriol. 1997;179:1646–1654. doi: 10.1128/jb.179.5.1646-1654.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakansson S, Schesser K, Persson C, Galyov E E, Rosqvist R, Homble F, Wolf-Watz H. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 15.Hauser A R, Kang P J, Engel J N. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol. 1998;27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 16.Holmstrom A, Pettersson J, Rosqvist R, Hakansson S, Tafazoli F, Fallman M, Magnusson K E, Wolf-Watz H, Forsberg Å. YopK of Yersinia pseudotuberculosis controls translocation of Yop effectors across the eukaryotic cell membrane. Mol Microbiol. 1997;24:73–91. doi: 10.1046/j.1365-2958.1997.3211681.x. [DOI] [PubMed] [Google Scholar]
- 17.Iglewski B H, Sadoff J, Bjorn M J, Maxwell E S. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyl transferase distinct from toxin A. Proc Natl Acad Sci USA. 1978;75:3211–3215. doi: 10.1073/pnas.75.7.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iriarte M, Cornelis G R. YopT, a new Yersinia Yop effector protein, affects the cytoskeleton of host cells. Mol Microbiol. 1998;29:915–929. doi: 10.1046/j.1365-2958.1998.00992.x. [DOI] [PubMed] [Google Scholar]
- 19.Iriarte M, Sory M P, Boland A, Boyd A P, Mills S D, Lambermont I, Cornelis G R. TyeA, a protein involved in control of Yop release and in translocation of Yersinia Yop effectors. EMBO J. 1998;17:1907–1918. doi: 10.1093/emboj/17.7.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knight D A, Finck-Barbançon V, Kulich S M, Barbieri J T. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1995;63:3182–3186. doi: 10.1128/iai.63.8.3182-3186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee V T, Anderson D M, Schneewind O. Targeting of Yersinia Yop proteins into the cytosol of Hela cells: one step translocation of YopE across bacterial and eukaryotic membranes is dependent on SycE chaperone. Mol Microbiol. 1998;28:593–601. doi: 10.1046/j.1365-2958.1998.00822.x. [DOI] [PubMed] [Google Scholar]
- 22.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 23.McGuffie E M, Frank D W, Vincent T S, Olson J C. Modification of Ras in eukaryotic cells by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1998;66:2607–2613. doi: 10.1128/iai.66.6.2607-2613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen R H, Debusscher G, McCombie W R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson J C, McGuffie E M, Frank D W. Effects of differential expression of the 49-kilodalton exoenzyme S by Pseudomonas aeruginosa on cultured eukaryotic cells. Infect Immun. 1997;65:248–256. doi: 10.1128/iai.65.1.248-256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettersson J, Nordfelth R, Dubinia E, Bergman T, Gustafsson M, Magnusson K E, Wolf-Watz H. Modulation of virulence factor expression by pathogen target cell contact. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 27.Rosqvist R, Magnusson K E, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 29.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-based vectors and a family of cassettes containing portable mob sites and the counterselectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 30.Sory M P, Boland A, Lambermont I, Cornelis G R. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sory M P, Cornelis G R. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 32.Straley S C, Skryzpek E, Plano G V, Bliska J B. Yops of Yersinia spp. pathogenic for humans. Infect Immun. 1993;61:3105–3110. doi: 10.1128/iai.61.8.3105-3110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 34.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 35.Wattiau P, Bernier B, Deslee P, Michiels T, Cornelis G R. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sci USA. 1994;91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wattiau P, Cornelis G R. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol Microbiol. 1993;8:123–131. doi: 10.1111/j.1365-2958.1993.tb01209.x. [DOI] [PubMed] [Google Scholar]
- 37.Wattiau P, Woestyn S, Cornelis G R. Customized secretion chaperones in pathogenic bacteria. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 38.Woestyn S, Sory M P, Boland A, Lequenne O, Cornelis G R. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol Microbiol. 1996;20:1261–1271. doi: 10.1111/j.1365-2958.1996.tb02645.x. [DOI] [PubMed] [Google Scholar]
- 39.Yahr T L, Barbieri J T, Frank D W. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J Bacteriol. 1996;178:1412–1419. doi: 10.1128/jb.178.5.1412-1419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 41.Yahr T L, Hovey A K, Kulich S M, Frank D W. Transcriptional analysis of the Pseudomonas aeruginosa exoenzyme S structural gene. J Bacteriol. 1995;177:1169–1178. doi: 10.1128/jb.177.5.1169-1178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]