Abstract
The staphylococcal accessory regulator (encoded by sarA) is an important global regulator of virulence factor biosynthesis in Staphylococcus aureus. To further characterize its role in virulence determinant production, an sarA knockout mutant was created by insertion of a kanamycin antibiotic resistance cassette into the sarA gene. N-terminal sequencing of exoproteins down-regulated by sarA identified several putative proteases, including a V8 serine protease and a novel metalloprotease, as the major extracellular proteins repressed by sarA. In kinetic studies, the sarA mutation delays the onset of α-hemolysin (encoded by hla) expression and reduces levels of hla to approximately 40% of the parent strain level. Furthermore, SarA plays a role in signal transduction in response to microaerobic growth since levels of hla were much lower in a microaerobic environment than after aerobic growth in the sarA mutant. An exoprotein exhibiting hemolysin activity on sheep blood, and up-regulated by sarA independently of the accessory gene regulator (encoded by agr), was specifically induced microaerobically. Transcriptional gene fusion and Western analysis revealed that sarA up-regulates both toxic shock syndrome toxin 1 gene (tst) expression and staphylococcal enterotoxin B production, respectively. This study demonstrates the role of sarA as a signal transduction regulatory component in response to aeration stimuli and suggests that sarA functions as a major repressor of protease activity. The possible role of proteases as regulators of virulence determinant stability is discussed.
Staphylococcus aureus is a major human pathogen, responsible for a large number of nosocomial infections (66). The pathogenesis of S. aureus has been attributed to its potential to produce a diverse range of extracellular proteins (e.g., hemolysins, toxic shock syndrome toxin 1 [TSST-1], and proteases) and cell wall-associated proteins (e.g., protein A and fibronectin binding protein), many of which are virulence factors (33). The production of secreted exo- and surface proteins is coordinately regulated in a growth phase-dependent manner, occurring preferentially in the post-exponential and log phases of growth, respectively (9, 65). Modulation of virulence determinant biosynthesis also occurs in response to the growth conditions (12, 57, 58), reflecting the ability of S. aureus to adapt and survive in many different environmental niches.
The regulation of virulence determinant production in S. aureus involves several global regulatory loci; of these, sar and agr are the best characterized (15, 45, 54, 56), though other regulators have been described (27, 30). Inactivation of the sar or agr locus results in a pleiotropic decrease in levels of exoproteins and an overproduction of surface proteins, while mutants are less virulent than the parental strain in several animal models (1, 15, 17, 18, 39, 45, 54, 56).
The agr locus consists of two major divergent operons. One operon encodes a unique RNA molecule, RNAIII, responsible for the up-regulation of extracellular protein production and the down-regulation of surface proteins primarily at the transcriptional level (34, 46, 51). This operon has a single promoter, P3 (39). In the opposite direction to RNAIII, the P2 promoter is responsible for expression of RNAII from a four-gene operon, agrBDCA (39). AgrC and AgrA show homology to members of the classical family of two-component sensor and regulator proteins, respectively (39, 50). In addition, agrB and agrD generate a quorum-sensing signalling molecule, a small peptide, which activates expression of RNAIII, and hence agr target genes, in a cell density-dependent manner (7, 35, 36). Mutations in any of the agrBDCA cluster of genes results in lower levels of RNAIII, implying that RNAII products are required for optimal RNAIII expression (50). In the signal transduction pathway, AgrA is believed to bind to environmental concentrations of the autoinducing peptide, transducing this signal via phosphorylation to AgrC, which results in activated AgrC putatively binding to the agr P2 and P3 promoter regions. This leads to increased levels of both RNAII, and hence enhanced levels of the autoinducer molecule itself, and RNAIII, thus ensuring a rapid alteration in virulence gene expression via RNAIII, in response to bacterial population density (35, 50).
The sar operon was first identified by Cheung and coworkers (15) in a Tn917 library screen for fibrinogen binding protein-deficient mutants. The inactivated locus was subsequently found to pleiotropically affect the expression of exoproteins and surface proteins (15, 18). Molecular characterization of the sar operon revealed three overlapping transcripts all encoding SarA, a regulatory DNA binding protein product involved in virulence gene expression (8). Transcriptional and binding studies have shown that the SarA protein binds to the P2 and P3 promoter regions of the agr locus, increasing levels of both RNAII and RNAIII and hence altering the synthesis of virulence factors (13, 31, 47). The mechanism by which S. aureus controls virulence determinant gene expression is therefore complex, involving an interactive, hierarchical regulatory cascade between the products of the sar and agr loci and possibly other components.
To further our understanding of how S. aureus responds to the environment to bring about changes in virulence determinant production, we investigated the role of sarA in the signal transduction pathway in response to environmental stimuli. This effort was facilitated by the creation of a sarA-inactivated mutant. Analysis of the mutant revealed SarA to be a potent repressor of proteases. The role of SarA in the transduction of specific environmental signals was also studied.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani medium and selection with ampicillin (50 μg/ml) where appropriate. S. aureus strains were grown in brain heart infusion (BHI) medium containing erythromycin (5 μg/ml), tetracycline (5 μg/ml), kanamycin (50 μg/ml), neomycin (50 μg/ml), or lincomycin (25 μg/ml) where suitable. All bacteria cultures were grown at 37°C. Phage transduction was performed as described by Novick (49), using φ11 as the transducing phage.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristicsa | Origin (reference) |
|---|---|---|
| S. aureus | ||
| RN4220 | Restriction negative, modification positive | R. Novick 40 |
| 8325-4 | Wild-type strain cured of known prophages | R. Novick 48 |
| PC6911 | agrΔ::tetM Tcr | Laboratory stock 12, 51 |
| PC2429 | sarA+ sarA::km Kmr Eryr | This study |
| PC1839 | sarA::km Kmr | This study |
| PC18391 | sarA::km agrΔ::tetM Kmr Tcr | This study |
| PC322 | hla+ hla::lacZ Eryr | Laboratory stock 12 |
| PC3221 | hla+ hla::lacZ sarA::km Eryr Kmr | This study |
| PC324 | hla+ hla::lacZ agrΔ::tetM Eryr Tcr | Laboratory stock 12 |
| PC3241 | hla+ hla::lacZ sarA::km agrΔ::tetM Eryr Kmr Tcr | This study |
| PC203 | spa+ spa::lacZ Eryr | Laboratory stock 12 |
| PC2031 | spa+ spa::lacZ sarA::km Eryr Kmr | This study |
| PC206 | spa+ spa::lacZ agrΔ::tetM Eryr Tcr | Laboratory stock 12 |
| PC2062 | spa+ spa::lacZ sarA::km agrΔ::tetM Eryr Kmr Tcr | This study |
| PC1072 | tst::lux Tcr | Laboratory stock 12 |
| PC1091 | tst::lux sarA::km Tcr Kmr | This study |
| S6 | Enterotoxin-producing strain | S. Khan 20 |
| PC1700 | agrΔ::tetM in S6 Tcr | This study |
| PC1841 | sarA::km in S6 Kmr | This study |
| PC1845 | sarA::km agrΔ::tetM in S6 Tcr Kmr | This study |
| E. coli DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Promega Corp. |
| Plasmids | ||
| pUBS1 | E. coli cloning vector; Apr | G. Murphy 25 |
| pAZ106 | Promoterless, transcriptional lacZ fusion vector; Apr (E. coli) Eryr (S. aureus) | A. Moir 38 |
| pDG783 | 1.5-kb kanamycin resistance cassette in pSB118; Apr | P. Stragier 29 |
| pPC905 | 1.3-kb SalI-BamHI cut PCR fragment containing the complete sar operon in SalI-BamHI-cut pUBS1; Apr | This study |
| pH4 | 1.5-kb EcoRI fragment from pDG783 containing the kanamycin resistance cassette in EcoRI-cut pPC905; Apr | This study |
| pH4A5 | 3.3-kb PvuII fragment containing sarA::km in SmaI-cut pAZ106; Apr (E. coli) Eryr (S. aureus) | This study |
Abbreviations: Tcr, Kmr, Eryr, and Apr, resistance to tetracycline, kanamycin/neomycin, erythromycin/lincomycin, and ampicillin, respectively.
Experimental growth conditions.
For transcriptional fusion and protein studies, strains were grown exactly as previously described (12). Microaerobic cultures were grown in a variable-atmosphere (8% O2, 5% CO2, 87% N2) incubator (Don Whitley) with agitation at 37°C. Anaerobic growth at 37°C was conducted in an atmosphere of 10% H2, 10% CO2, and 80% N2.
DNA manipulations.
All molecular biology techniques and recombinant DNA manipulations were carried out as described in reference 61. DNA sequencing was carried out with AmpliTaq DNA polymerase (Applied Biosystems) based on the dye terminator cycle sequencing method and analyzed on an Applied Biosystems automated DNA sequencer.
Hemolysin assay.
α- or β-hemolysin activity was determined on a 10% (vol/vol) rabbit blood (E & O Laboratories, Bonnybridge, United Kingdom) overlay plate or a 5% (vol/vol) sheep blood (TCS Biologicals, Buckingham, United Kingdom) plate. All plates were incubated at 37°C until zones of activity were visible. Assays were performed in triplicate, and the means and standard deviations of activities were calculated. α-Hemolysin levels in bacterial culture supernatants were determined based on the method of Rowe and Welch (60). Bacterial samples (0.5 ml) were centrifuged (5,500 × g, 4°C, 1 min), the supernatant was removed, and phenylmethylsulfonyl fluoride (Sigma) was added to a final concentration of 0.25 mM, and the samples were stored at −20°C; 0.1 ml of supernatant was made up to 1 ml in hemolysin buffer (0.145 M NaCl, 20 mM CaCl2) prior to the addition of 25 μl of defibrinated rabbit blood. After incubation for 15 min at 37°C, we centrifuged the samples (5,500 × g, room temperature, 1 min) and measured the optical density at 543 nm (OD543) of the supernatant. One unit of hemolysin activity is defined as the amount which causes an increase in OD543 of 0.5 per min due to erythrocyte lysis per OD600 unit of original culture.
β-Galactosidase and luciferase assays.
β-Galactosidase (with 4-methylumbelliferyl-β-d-galactopyranoside [MUG] as the substrate) and luciferase activities were measured as previously described (12).
Protein gel analysis.
Samples were prepared, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as previously described (26).
Samples for Western blot analysis were separated by SDS-PAGE on a 10% (wt/vol) gel, then transferred to nitrocellulose, and treated with anti-staphylococcal enterotoxin B (SEB) antibody (Sigma) at a 1:2,000 dilution as described by Burnette (10). Antigen-antibody complexes were detected by reaction with goat anti-rabbit immunoglobulin G (1:30,000) conjugated to alkaline phosphatase (Sigma). The intensity of each band was quantified by densitometry using Bio-Profil electrophoresis gel imagery (Vilber Lourmat).
Zymogram analysis of exoprotein samples for protease activity was carried out as described above except that the sample buffer contained no β-mercaptoethanol and the samples were denatured at room temperature for 15 min. This did not affect the protein profile by comparison to the standard sample preparation protocol. Protein samples were resolved on a 12% (wt/vol) acrylamide Zymogram Ready Gel (Bio-Rad) containing casein, renatured overnight at 37°C, and visualized with Coomassie blue stain according to the manufacturer’s instructions.
N-terminal sequence determination.
After SDS-PAGE, proteins were transferred onto a Protoblot membrane (Applied Biosystems) by electrophoresis and visualized with Coomassie blue according to the manufacturer’s instructions. Protein bands were excised from the membrane and sequenced in an Applied Biosystems 476A protein sequencer.
Construction of an sarA insertionally inactivated mutant.
To study the role of sarA in the regulation of virulence determinant production, an insertionally inactivated sarA mutant was made. Forward and reverse primers corresponding to the 5′ and 3′ ends of the sar operon with added SalI and BamHI restriction sites, respectively (shown in boldface and flanked by GC-rich regions), were designed based on the published sequence (underlined nucleotides) (8). These primers, 5′-ACGCGTCGACGTCGAAAGCGTTGATTTGGGTAGTA-3′ (nucleotides 1 to 21) and 5′-CGCGGATCCGCGAGTGCCATTAGTGCAAAACCT-3′ (complement of nucleotides 1329 to 1349), respectively, were used to PCR amplify the complete sar operon. The 1,349-bp PCR product was cut with SalI-BamHI and cloned into pUBS1 (25) cut with SalI-BamHI to give plasmid pPC905 in E. coli DH5α. A 1,489-bp EcoRI fragment containing a kanamycin resistance cassette of Streptococcus faecalis was excised from pDG783 (29), dephosphorylated, and cloned into the unique EcoRI site internal to the sarA gene in pPC905 cut with EcoRI and dephosphorylated, to give pH4 in E. coli DH5α. Restriction mapping, and DNA sequence analysis using primers internal to sarA and flanking its EcoRI site, was used to confirm insertion of the kanamycin resistance cassette into the sarA gene. A 3,296-bp PvuII fragment containing the inactivated sarA gene and flanking sequences was excised from pH4 and cloned into the vector pAZ106 (38) cut with SmaI and dephosphorylated, giving plasmid pH4A5 in E. coli DH5α. Plasmid DNA of pH4A5 (50 μg) was transformed into S. aureus RN4220 by electroporation (62) and selected on plates containing kanamycin and erythromycin. A kanamycin- and erythromycin-resistant transformant, PC2429, was also checked for neomycin and lincomycin resistance to eliminate transformants which may emerge due to spontaneous chromosomal mutations. The presence of an inactivated and an intact copy of sarA were verified by PCR using sarA internal primers (as described above for pH4 construction) and Southern blot analysis using the pPC905 insert as the probe. A phage lysate of PC2429 was prepared from φ11 stocks, transduced into S. aureus 8325-4, and selected on kanamycin and neomycin plates. Of the 96 transductants recovered, 2 of 65 which were tested were also erythromycin sensitive. Successful integration of the sarA::Km marker into the S. aureus chromosome of one of these transductants, PC1839, was confirmed by PCR using sarA internal primers and Southern blot analysis with the pPC905 insert as the probe.
RESULTS
Construction and phenotypic characterization of an sarA knockout mutant.
To determine the role of sarA in the control of virulence determinant expression, the sarA mutant PC1839 was made by insertion of a kanamycin resistance cassette into the sarA gene in the S. aureus chromosome. A kanamycin resistance cassette was chosen as a selective marker to allow other chromosomal markers carried by reporter gene fusions (12) and regulatory loci mutation to be introduced into PC1839. The plasmid construct pH4A5, containing the inactivated sarA gene, lacks a replicon functional in S. aureus. Hence, when the plasmid was introduced into S. aureus RN4220 by transformation, kanamycin- and erythromycin-resistant colonies which emerge result from a single crossover recombination event between homologous DNA regions of the sar loci on the chromosome and plasmid. It is unlikely that a double crossover will occur due to the poor efficiency of recombination in S. aureus (49). Hence, transformants such as PC2429 contain an intact copy of sarA plus an extra copy of the disrupted sarA gene and are resistant to erythromycin/lincomycin and kanamycin/neomycin. The sarA::Km marker was introduced into the chromosome of S. aureus 8325-4 by transductional outcross (51). A kanamycin-resistant, erythromycin-sensitive colony, PC1839, containing the insertionally inactivated sarA was selected. PCR screening of PC1839 gave a single product corresponding to sarA containing the 1.5-kb cassette (results not shown). Southern blot analysis also confirmed the presence of the cassette when genomic DNA of PC1839 was compared to that of 8325-4, probed with the pPC905 insert. PC1839 (sarA) grew at approximately the same rate and gave the same yield (OD600 of 8 to 10) as the parental strain. As the doubling time during exponential growth was not significantly different from that of the wild type (see Fig. 4 and 5), changes in virulence determinant expression are due to inactivation of sarA rather than changes in growth rate. The mutation is stable and does not require continued antibiotic selection.
FIG. 4.
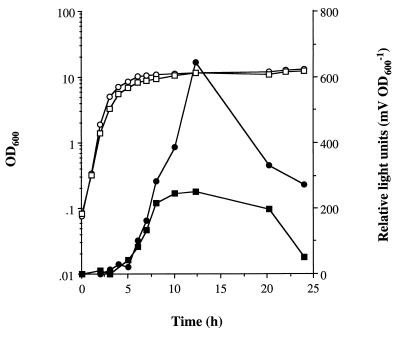
Role of sarA in the regulation of tst::lux expression during aerobic growth. S. aureus PC1072 (tst::lux) (○, •) and PC1091 (sarA tst::lux) (□, ■) were grown in BHI at 37°C with shaking at 250 rpm as described in Materials and Methods. Bacterial growth was measured by OD600 (○, □), and fusion expression was determined by luciferase activity (•, ■).
FIG. 5.
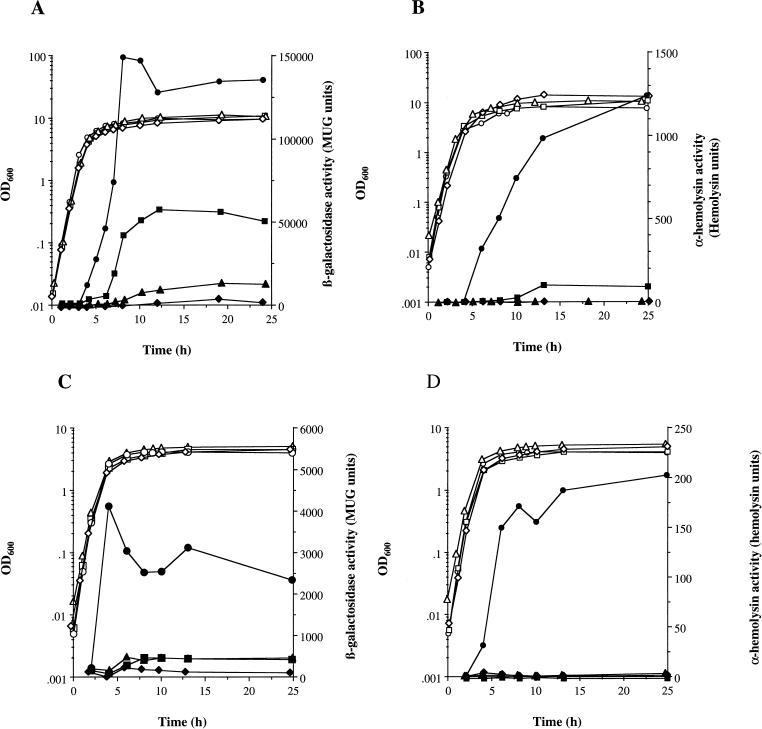
Role of sarA and agr in the regulation of hla::lacZ expression and Hla activity in response to environmental conditions. S. aureus PC322 (hla::lacZ) (○, •), PC3221 (sarA hla::lacZ) (□, ■), PC324 (agr hla::lacZ) (▵, ▴), and PC3241 (sarA agr hla::lacZ) (◊, ⧫) were grown in BHI at 37°C as described in Materials and Methods. (A and B) Aerobic growth in a shaking water bath at 250 rpm; (C and D) microaerobic growth on a shaking platform in a cabinet containing 8% O2, 5% CO2, and 87% N2. Bacterial growth was measured by OD600 (○, □, ▵, ◊). (A) hla::lacZ expression was determined by measuring β-galactosidase activity (•, ■, ▴, ⧫); (B) Hla activity in cell-free supernatants (•, ■, ▴, ⧫) was determined as described in Materials and Methods.
Identification of sarA-regulated exoproteins.
The effect of the sarA mutation on total exoprotein production in an 8325-4 background was examined during the growth cycle by SDS-PAGE analysis (Fig. 1A). The mutation resulted in a pleiotropic alteration in the extracellular protein profile compared to the wild-type strain as has been previously noted (18). It has been previously shown that most exoproteins are preferentially expressed in a growth phase-dependent manner during the transition between late post-exponential (T = 6 h [Fig. 1A]) and stationary phases (T = 8 h [Fig. 1A]) of growth (18). Several exoproteins present in the parental strain are missing or reduced in PC1839 (sarA) and are hence up-regulated by SarA. Figure 1A also shows many exoproteins which overaccumulate in PC1839 (sarA) compared to the parent and therefore are negatively regulated by SarA. To further identify the SarA-regulated proteins, several of these exoproteins were N-terminally sequenced (Table 2).
FIG. 1.
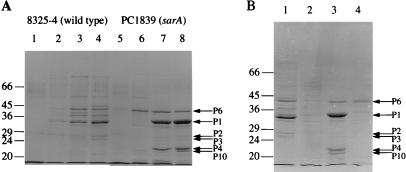
Effects of regulatory mutations on exoprotein production in S. aureus SDS–10% (wt/vol) polyacrylamide gels show total extracellular protein production during growth as described in Materials and Methods. (A) S. aureus 8325-4 lanes 1 to 4 and PC1839 (sarA) (lanes 5 to 8) at T = 2, 4, 6, and 8 h, corresponding to log, early and late post-exponential, and stationary phases of growth, respectively. (B) S. aureus 8325-4 (lane 1), PC6911 (agr) (lane 2), PC1839 (sarA) (lane 3), and PC18391 (sarA agr) (lane 4), all in stationary phase (T = 8 h). All lanes contain exoproteins from the equivalent of 0.05 OD600 units of original culture. Representative sarA-regulated proteins are indicated by arrows. The molecular masses of protein standards (in kilodaltons) are shown.
TABLE 2.
N-terminal sequences of PC1839 (sarA) exoproteins
| Designation | Approximate size (kDa) | Sequence | Homologya | Accession no. (reference) |
|---|---|---|---|---|
| P6 | 40 | AAETGKGKGVLGDTKDININ | 85% identity to metalloprotease (SepPI) of S. epidermidis | P4314863 |
| P1 | 35 | VILPNNDRHQI | 100% identity to V8 serine protease of S. aureus | P0418811, S2175869 |
| P2 | 28 | ENNVTKIKDTNIFPYTGVVAFFSASGF | 88 and 67% identities to novel antigens ORF-1 and ORF-2 of S. aureus, respectively | U60589 and U6352959, respectively |
| P3 | 26 | ENNVKQITNTNVAPYNGVTV | 55% identity to both novel antigens ORF-1 and ORF-2 of S. aureus | U60589 and U6352959, respectively |
| P4 | 22 | DQVQYENTLKNFKIREQQFD | 100% identity to ORFX 3′ of V8 serine protease of S. aureus | W. Hufnagle 32 |
Homology at the amino acid level to proteins in the SWISSPROT and EMBL databases.
In PC1839 (sarA), a 35-kDa protein (designated P1) is the major product in the extracellular fluid and appears preferentially during the stationary phase of growth (T = 6 and 8 h) (Fig. 1A). In 8325-4, it is likely to correspond to the minor band directly above the most prominent protein in the 8325-4 profile (Fig. 1A, lane 4), which is α-hemolysin (Hla; 33 kDa) (28). The identity of α-hemolysin was verified by N-terminal sequencing (results not shown). The intensity of the 35-kDa exoprotein in PC1839 (sarA) is at least 10-fold higher than in 8325-4, and hence the protein is strongly negatively regulated by SarA. This regulation may occur at the transcriptional and/or posttranscriptional levels. In Fig. 1B, the same protein (P1) is missing or greatly reduced in both PC6911 (agr) and PC18391 (sarA agr) compared to 8325-4, implying that its production is positively controlled by agr independently of sarA. N-terminal sequence analysis of the 35-kDa protein (P1) revealed 100% identity in an 11-amino-acid overlap to both a 29.0-kDa staphylococcal serine protease (also called V8 protease) (11) and a 31.3-kDa glutamic acid-specific endopeptidase (69) from two different S. aureus strains (Table 2). The P1 sequence aligns exactly with residue 69 of each protease, the first amino acid of the mature polypeptide.
A 40-kDa exoprotein repressed by sarA is preferentially produced during the post-exponential phase of growth in PC1839 (sarA) (Fig. 1A, P6). Interestingly, in the sarA mutant, production of this exoprotein increases 2 h earlier during the post-exponential phase of growth (T = 4 h) (Fig. 1A, lane 6) compared to the appearance of most of the other exoproteins (T = 6 and 8 h). P6 is not apparently present in 8325-4, although a slightly larger protein is suggesting that the presence of P6 is repressed by SarA (Fig. 1B, lanes 1 and 3). P6 is not present in PC6911 (agr) but is present in PC1839 (sarA agr) (Fig. 1B, lanes 2 and 4), which shows that the presence of P6 is repressed by SarA but independently of agr. The P6 N-terminal sequence revealed 85% identity over 20 amino acids to an extracellular elastase precursor from Staphylococcus epidermidis (63), also called metalloprotease or SepPI. SepPI, which requires zinc for its proteolytic activity, and P6 are more distantly related to various Bacillus proteases (reference 63 and results not shown). P6 shows homology starting at the first residue of the mature form of the extracellular elastase of S. epidermidis.
Two exoproteins with estimated sizes of 28 and 26 kDa (P2 and P3) are produced at low levels in cultures of PC1839 (sarA) (T = 6 and 8 h) (Fig. 1A, lanes 7 and 8). P2 and P3 are present in the parental strain (8325-4) but are absent from PC6911 (agr) and PC18391 (sarA agr) (Fig. 1B). These proteins are thus apparently not strongly regulated by SarA but up-regulated by agr. N-terminal sequencing of P2 and P3 revealed 88 and 55% best identity over 27 and 20 amino acids, respectively, to a 26-kDa novel antigen called ORF-1, and also 67% and 55% identity, respectively, to a second 26-kDa novel antigen, called ORF-2 (59), both from an enterotoxin-producing S. aureus strain (Table 2). ORF-1 and ORF-2 show 61.5% identity to each other over their entire lengths. The functions of these novel antigens are unknown. However, the antigens show highest homology (approximately 30% over the entire lengths of both proteins) to V8 serine protease of S. aureus and thus can be added to the multitude of proteases so far identified in the sarA mutant of 8325-4. ORF-1 and ORF-2, like V8 serine protease, also showed approximately 25 to 30% identity over their entire lengths to both the 26.9-kDa exfoliative toxin A (ETA) of S. aureus (accession no. M17347) (41, 53) and the 27.3-kDa exfoliative toxin B (ETB) (accession no. M17348) (41).
Two proteins of approximately 22 and 21 kDa are present in PC1839 (sarA) but absent in both wild-type 8325-4 and agr mutant PC6911 (Fig. 1B). Hence, these exoproteins are strongly repressed by sarA apparently independently of agr. N-terminal and sequence analysis of the 22-kDa exoprotein (P4) did not show homology to any proteins in established databases (Table 2). However, P4 showed 100% identity over 20 amino acids to a recently identified S. aureus protein, ORFX, located immediately 3′ of V8 protease (32). The N terminus of the 21-kDa protein (P10) did not have any significant homologues (results not shown).
Protease activity analysis.
To examine the proteolytic activity of exoproteins regulated by sarA, total extracellular proteins were analyzed on a renaturing gel with casein as the substrate. PC1839 (sarA) show an intense zone of clearing indicative of protease activity at 35 kDa which corresponds to V8 serine protease activity (Fig. 2, lane 3). It is absent in 8325-4 (even though the sample represents 10-fold more OD600 units), which confirms its down-regulation by sarA (Fig. 1). A second band of activity in PC1839 (sarA) runs at approximately 23 kDa (Fig. 2, lane 3). Comparison of the relative intensities of likely V8 serine protease activity in 8325-4 and PC1839 (sarA) (Fig. 2 and results not shown) suggests there is approximately 1,000-fold more V8 protease activity in PC1839 (sarA) than in 8325-4. Apparent protease activity is missing in PC6911 (agr) and is lower in PC18391 (sarA agr) than in PC1839 (sarA), consistent with SarA negatively regulating V8 serine protease production and agr positively regulating V8 serine protease, as previously observed on the Coomassie blue-stained gel after SDS-PAGE (Fig. 1B). Even in PC18391 (sarA agr), there is still approximately 100-fold more activity than in 8325-4 (Fig. 2 and results not shown).
FIG. 2.
Zymogram analysis of protease activity in exoproteins of S. aureus. Lane 1, S. aureus 8325-4; lane 2, PC6911 (agr); lane 3, PC1839 (sarA); lane 4, PC18391 (sarA agr). Exoproteins are from the equivalents of 0.05 (lanes 1 and 2) and 0.005 (lanes 3 and 4) OD600 units of original culture. Stationary-phase samples (T = 18 h) were prepared and separated on a 12% (wt/vol) polyacrylamide gel with casein as the substrate, and the gel was renatured as described in Materials and Methods. Proteolytic activity can be seen as a zone of clearing on the gel, indicated by arrows. The molecular masses of protein standards (in kilodaltons) are shown.
Several additional protease bands were visible at approximate sizes of 28, 20, 19, 70, and 38 kDa, in order of decreasing activity, when the zymogram gel was overloaded with supernatant samples from the sarA mutant (results not shown). Some of these may correspond to the putative proteases identified by N-terminal sequencing of exoproteins in PC1839 (sarA) (Table 2).
Role of SarA in regulation of SEB and TSST-1.
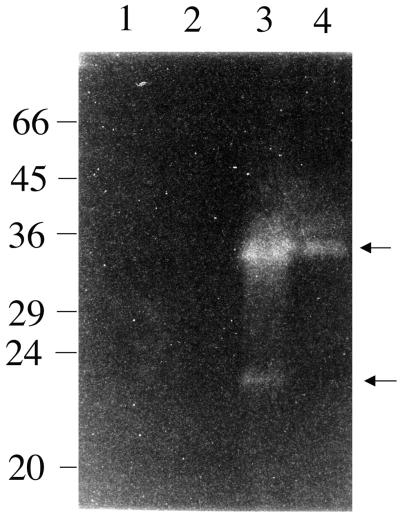
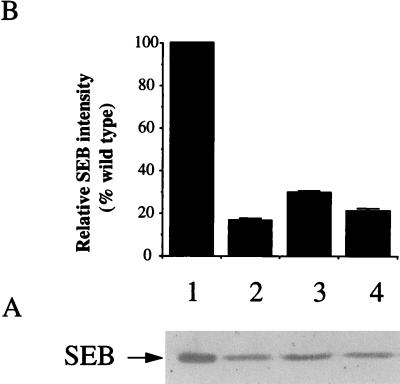
The set of regulatory mutations was introduced into S. aureus S6, an enterotoxin-producing strain. SEB is the major exoprotein produced by S6 and is known to be up-regulated by agr (20). The role of sarA in the regulation of SEB was examined by Western blot analysis (Fig. 3). In stationary-phase cultures (T = 18 h), the 28-kDa SEB band was reduced in PC1841 (sarA) to approximately 30% of the S6 level. PC1700 (agr) and PC1845 (sarA agr) had 16 and 20%, respectively, of the wild-type level of SEB.
FIG. 3.
Western blot analysis of SEB production. (A) Western blot of total extracellular proteins of S. aureus S6 (lane 1), PC1700 (agr) (lane 2) PC1841 (sarA) (lane 3), and PC1845 (sarA agr) (lane 4). All lanes contain exoproteins from the equivalent of 0.05 OD600 units of original culture (T = 18 h). Proteins were separated by SDS-PAGE on a 10% (wt/vol) gel, transferred onto a nitrocellulose membrane (BDH), and probed with a 1:2,000 dilution of antibodies to SEB (Sigma) prior to colorimetric detection as described in Materials and Methods. (B) The relative intensity of each protein band quantified by densitometry (mean of three experiments plus standard deviation).
To investigate the role of sarA in the control of TSST-1 gene (tst) expression, the sarA mutation was introduced into a previously constructed tst::lux fusion (12) to create strain PC1091 (sarA tst::lux). The effect of sarA on tst levels as measured by luciferase activity was determined (Fig. 4). In PC1072 (tst::lux), tst expression rapidly increases postexponentially at 5 h and reaches a maximum of approximately 600 relative light units at T = 12 h. In PC1091 (sarA tst::lux), there was an approximate twofold reduction in tst levels during the stationary phase (T = 10 to 20 h) compared to PC1072 (tst::lux). There was no apparent difference in the lag period prior to tst expression between the wild-type and sarA backgrounds.
Kinetics of virulence determinant expression in an sarA mutant.
SarA has been shown to regulate both spa and hla expression (14, 18). In epistasis experiments, the effect of sarA on the kinetics of expression of hla and spa was examined to determine the role of sarA in the hierarchical regulation of these virulence determinants during aerobic growth. The previously constructed (12) lacZ transcriptional reporter gene fusions hla::lacZ and spa::lacZ were introduced into the sarA and sarA agr backgrounds of 8325-4, and the effects of these mutations on virulence determinant gene expression was measured by changes in β-galactosidase levels. In PC322 (hla::lacZ), hla expression rapidly increases at 3 h of growth, reaching a maximum of 146,000 MUG units in the post-exponential phase (T = 10 h) and remains high at T = 12 to 24 h (130,000 MUG units) (Fig. 5A). In PC1839 (sarA hla::lacZ), there is an additional 3-h lag prior to a sudden increase in hla transcription at 6 h compared to PC322 (hla::lacZ). Thereafter, aerobic levels rose only to about 40% of the parental level during the stationary phase of growth (T = 12 to 24 h) (Fig. 5A). Expression of hla in PC324 (agr hla::lacZ) was reduced to approximately 8% of the level in PC322 (hla::lacZ) and was further diminished in PC3241 (sarA agr hla::lacZ), to approximately 3% of the parental level after 18 h of growth.
To determine any posttranscriptional regulation of α-hemolysin production, hla::lacZ expression and α-hemolysin protein (Hla) activity were compared. Specific Hla activity in PC322 (hla::lacZ) began to increase as the cells entered post-exponential phase (T = 4 h) and continued to rise into the stationary phase of growth (T = 13 to 24 h) (Fig. 5B). In PC3221 (sarA hla::lacZ), Hla levels were not measurable until 6 to 8 h into growth, implying an additional 2- to 4-h lag period prior to the onset of Hla activity compared to the parent (Fig. 5B). Thereafter, Hla activity rose to only 10% of the parental PC322 (hla::lacZ) level at T = 13 h (Fig. 5B), compared to 40% of the parental level when hla::lacZ expression was determined (Fig. 5A). Hla activity in PC324 (agr hla::lacZ) and PC3241 (agr sarA hla::lacZ) was <1% of the activity in PC322 (hla::lacZ) (Fig. 5B).
Expression of spa::lacZ occurs optimally during late exponential phase in a wild-type background (PC203; T = 3 h), (2). spa levels are dramatically (23- 24-, and 20-fold) increased compared to PC203 (spa::lacZ) in PC2031 (sarA spa::lacZ), PC206 (agr spa::lacZ), and PC2062 (sarA agr spa::lacZ), respectively, at T = 3 h (results not shown) which is consistent with previous studies (14).
Role of SarA in the response to environmental signals.
To determine the role of sarA in the transduction of specific environmental signals which affect virulence determinant expression, we studied the effects of the sarA mutation on spa and hla expression under different growth conditions. Initial studies using our sarA::lacZ fusion suggested that sarA levels may be responsive to aeration levels (12a). To examine the role of sarA in the transduction of this environmental signal, the levels of expression of both hla and spa were compared in wild-type and sarA backgrounds under aerobic and microaerobic growth conditions as described in Materials and Methods. During microaerobic growth, hla expression in PC322 (hla::lacZ) increased between T = 2 h and 4 h, rapidly reaching a maximum of 4,200 MUG units at late log phase (T = 4 h) (Fig. 5C). Microaerobic expression of hla in PC3221 (sarA hla::lacZ) resulted in a dramatic reduction in hla::lacZ expression to only 17 and 14% of PC322 (hla::lacZ) levels in stationary phase (T = 10 and 13 h, respectively) (Fig. 5C). Expression of hla was reduced to similar levels: 19 and 14% of PC322 (hla::lacZ) levels in PC324 (agr hla::lacZ) at T = 10 and 13 h, respectively (Fig. 5C). In PC3241 (sarA agr hla::lacZ), hla expression was further diminished to 8 and 5% of the level in parent PC322 strain at the same time points. When α-hemolysin protein activity was concurrently measured from the same culture samples, the specific Hla activity was only <1% of the parental level in sarA and agr mutant backgrounds during growth (Fig. 5D).
Comparison of microaerobic and aerobic hla::lacZ expression and Hla activity (Fig. 5) showed that both microaerobic expression and activity were significantly reduced in the wild-type background (PC322), to <15% of aerobic levels (T = 24 h). Most interestingly, under aerobic conditions hla expression was approximately 40% in the sarA mutant (PC3221) compared to the wild type (PC322) (Fig. 5A), while under microaerobic conditions the sarA mutant (PC3221) showed only 14% of parental expression during late stationary phase (T = 13 to 24 h), which was equivalent to expression in the agr mutant (PC324) (Fig. 5C). A similar pattern was observed for Hla activity (Fig. 5B and D). Aerobically, Hla levels in the SarA mutant were reduced to 10% of the parental level during stationary phase (Fig. 5B, T = 13 to 24 h), while microaerobically, Hla activity was only <1% of the parental level at the same stage of growth (Fig. 5D). This finding suggests that sarA is important in the regulation of hla expression and stability under different aeration conditions.
The role of sarA in the regulation of spa expression during microaerobic growth was determined. Microaerobic spa expression at T = 4 h was 80-fold higher in PC2031 (sarA spa::lacZ) than in PC203 (spa::lacZ), while in PC206 (agr spa::lacZ) and PC2062 (sarA agr spa::lacZ), spa levels were 40- to 60-fold higher than in PC203 (results not shown).
The effect of sarA on virulence determinant gene expression in response to other environmental signals was also studied. The presence of 1 M NaCl was previously shown to dramatically repress levels of hla and spa (12, 52). To determine whether the effect of NaCl was mediated via sarA, the fusion strains in wild-type and sarA backgrounds were grown in BHI supplemented with 1 M NaCl. Fusion expression of hla and spa was similarly repressed in both parental and sarA mutant strains in the presence of 1 M NaCl (results not shown). The effect of divalent cation availability on spa expression was also examined. The addition of a sub-growth-inhibitory concentration of the metal ion chelator EDTA (0.5 mM), EGTA (0.5 mM), or 2,2′-dipyrydyl (0.4 mM) to BHI repressed spa levels to various extents (12), but this occurred independently of SarA (results not shown).
Induction of a hemolysin positively regulated by sarA in microaerobic and anaerobic conditions.
To further identify components which may be regulated by sarA in an aeration-dependent manner, hemolysin production was measured under aerobic, microaerobic, and anaerobic conditions. On solid media incubated aerobically (18 h), microaerobically (18 h), and anaerobically (2 days), zones of hemolytic activity were observed in wild-type 8325-4 and sarA mutant (PC1839) strains when assayed for α- and β-hemolysin (results not shown). No activity was observed in the agr (PC6911) or sarA agr (PC18391) mutant strain aerobically. Surprisingly, a zone of hemolysis (approximately 10 to 20% of that in 8325-4) was noted in the agr mutant (PC6911), but only on sheep blood plates and specifically in a microaerobic or anaerobic environment. The agr-independent, aeration-regulated activity requires SarA, as it is absent in PC18391 (sarA agr).
DISCUSSION
SarA has been previously shown to be a pleiotropic regulator of virulence determinant production in S. aureus (14, 15, 18). To further characterize the SarA regulon and to determine its role in the transduction of specific environmental signals, the sarA gene was disrupted by insertional inactivation using a kanamycin antibiotic resistance cassette. α-Hemolysin expression was diminished in the sarA mutant of S. aureus 8325-4, but interestingly, under our specific growth conditions, apparent transcription as measured by the use of an hla::lacZ fusion was reduced only to approximately 40% of the parental level. Our results are significantly different from those of Cheung and Ying (18), who found that α-hemolysin expression was almost totally abolished in a SarA mutant in an 8325-4-related strain background. Environmental conditions have huge effects on virulence determinant production (12). The discrepancy in results may therefore be due to different culture conditions, as these have not been fully described previously (15, 18).
SarA is involved in the temporal regulation of hla expression, since in the sarA mutant expression occurred 3 h later than in the parent (Fig. 5A). This occurred at the transcriptional level, as in both wild-type and sarA backgrounds, the times of onset of hla expression and activity were identical, implying that there is no additional translational control of hla timing by SarA. Previously, Vandenesch et al. (65) demonstrated that hla transcription could not be induced early in the growth cycle by RNAIII expressed on a plasmid, suggesting that additional, unknown signals independent of RNAIII were involved in its control. Our results indicate sarA is also needed to stimulate hla expression during the post-exponential phase, possibly by the accumulation of an activator up-regulated by sarA. The temporal delay in hla expression in the SarA mutant may explain why previous work detected no hla expression in an sarA mutant, as samples for Northern blot analysis were apparently taken only into early stationary phase (18).
The role of sarA in the regulation of SEB was also investigated. SEB, like Hla, is positively regulated by agr at the mRNA level, though host factors other than RNAIII have been implicated in its control (20). Western blot analysis showed that SEB expression is subject to positive control by sarA (Fig. 3). Transcriptional analysis using tst::lux reporter gene fusions demonstrated that sarA is also involved in the up-regulation of tst during the post-exponential phase, possibly via agr. Unlike the case for agr (39, 56), however, inactivating SarA only partially attenuates the expression of TSST-1 and Hla, suggesting that SarA may have a secondary role in the up-regulation of the production of these secreted proteins compared to agr.
Comparison of the exoprotein profiles of the various mutants in an isogenic background allowed the identification of a number of SarA-regulated proteins. Several proteins which are repressed by SarA were visualized. Sequence analysis of the exoproteins present in the supernatant of the sarA mutant surprisingly revealed a range of extracellular proteins exhibiting homology to known proteases, suggesting that sarA is a pleiotropic repressor of proteases in S. aureus. Enhanced proteolytic activity alone does not explain generally reduced levels of toxins in the sarA mutant, since hla is also controlled at the level of expression. The overproduction of protease in the sarA mutant may, however, have an important role in the posttranslational regulation of exoprotein and surface protein stability. The discrepancy between hla expression and Hla activity (Fig. 5A and B) could be due to digestion of the mature protein by SarA-regulated protease activity. Proteases may be involved in the modification of cell surface proteins, determining the relative amounts of cell-bound or extracellular surface proteins during infection. Recent evidence has suggested that the role of extracellular proteases may be to alter bacterial surface protein properties and assist the release the bacteria from attachment sites and, hence, dissemination during infection (44). Furthermore, the lack of secreted protease activity in a clinical isolate of S. aureus may explain why it showed high Hla production despite exhibiting normal levels of RNAIII and reduced levels of hla transcript compared to 8325-4 (42).
Staphylococcal proteases have been well characterized (3), though their possible roles in bacterial pathogenesis are poorly understood. In S. aureus 8325-4, we have identified V8 serine protease and a likely metalloprotease as the major exoproteins repressed by sarA. The production of V8 serine protease is growth phase dependent and subject to positive regulation by agr and SarA at the transcriptional level (34, 39, 42a). Gross protease activity on fibrin plates has also been shown to be enhanced in a sarA mutant (18).
Our studies have shown that the synthesis of V8 serine protease is complex and involves positive regulation by agr and negative regulation by sarA. Hence, two opposing but interactive regulatory pathways are implicated in the biosynthesis of this protease. In addition, there is an interdependence between the regulatory elements since in a sarA mutant V8 protease production reaches its highest level only when an intact agr is present (Fig. 1B). Most interestingly, located immediately 3′ and transcribed in the same operon as V8 serine protease in S. aureus 8325-4 is ORFX, encoding a protein of unknown function (32). P4 is an SarA-repressed protein which requires agr for its expression. The N-terminal sequence of P4 show 100% identity with ORFX 3′ of V8 protease, consistent with it being in an operon with V8 protease whose expression is repressed by SarA. The role of ORFX 3′ of V8 protease is unknown. A putative 40-kDa metalloprotease, P6, was also identified as being repressed by SarA in strain 8325-4. The putative metalloprotease of 8325-4 has previously been shown to be up-regulated by agr (34). Two other putative serine proteases were identified during this study as exoproteins P2 and P3, both of which require agr for production but are only slightly up-regulated by SarA. Interestingly, P2 and P3 also show relatedness to ETA and ETB, recognized virulence determinants controlled by agr (53). Both ETA and ETB are serine proteases (6, 21).
The staphylococcal metalloproteases are produced throughout the growth cycle but are mainly present after the exponential phase (2–5). In contrast, expression of serine protease of S. aureus V8 has been reported to occur exclusively in the post-exponential phase, consistent with the exoprotein being up-regulated by agr (9). Our data suggest that in the 8325-4 background, most of the proteases are mainly produced during the post-exponential phase of growth (Fig. 1A). Interestingly, in the sarA mutant, and in contrast to V8 serine protease synthesis, the putative 40-kDa metalloprotease is induced earlier during growth, suggesting that sarA is involved in suppressing the log-phase production of the metalloprotease. We have shown that SarA also affects the timing and level of expression of hla during the growth cycle, but unlike the case for the metalloprotease, expression of hla, which is up-regulated by sarA, is delayed when sarA is absent. In S. aureus V8, a role for the staphylococcal metalloprotease as an activator of a precursor of an inactive serine protease produced by the same bacterium has been suggested (24). Furthermore, a metalloprotease from Staphylococcus hyicus was recently shown to be involved in lipase processing in a growth phase-dependent manner (5). Hence, the presence of the putative metalloprotease during exponential growth in S. aureus 8325-4 culture supernatant prior to the appearance of the V8 serine protease is consistent with a possible role for metalloprotease in the processing of V8 serine protease when sarA expression or activity is suppressed. This finding suggests that there is an additional signal, possibly environmental, involved in regulating sarA in the hierarchical production of proteases.
This study has demonstrated that SarA is involved in the signal transduction pathway in response to the availability of O2 and/or CO2. Microaerobically, levels of Hla and toxins are generally diminished, possibly due to a decrease in growth yield, and thus reduced quorum-sensing potential compared to aerobic growth, although a specific regulatory mechanism may also account for this (52). The involvement of SarA in the up-regulation of both α- and β-hemolysin production in response to low O2 or high CO2 levels was shown by the greater attenuation of hla expression in an sarA mutant than in the parent after microaerobic compared to aerobic growth. This occurs at the level of hla expression. Furthermore, an unknown sarA-regulated hemolysin specifically induced under microaerobic conditions independently of agr was also identified in 8325-4. We are currently investigating the identity of this hemolysin in a β-hemolysin mutant background. Microaerobic conditions have previously been reported to increase expression of Hla in different S. aureus strains (52, 67). Our studies showed no significant increase in hla transcription or activity under microaerobic compared to aerobic conditions (12). However, our results do indicate that hla expression may be induced early in growth in a lower-oxygen environment since hla transcription occurs during the late log phase under microaerobic conditions but during post-exponential phase during aerobic growth.
Hence, sarA is a key component in the signal transduction pathway of hla expression in response to a microaerobic signal. However, the exact function of SarA and how it regulates virulence determinant expression in S. aureus are unknown. Environmental stimuli such as anaerobiosis and osmolytes are known to alter DNA supercoiling properties of target promoters (23). However, our studies indicate that DNA supercoiling does not have a major role in sarA expression or the environmental regulation of virulence determinant genes (12). Morfeldt et al. (47) speculated that SarA may function homologously to the integration host factor of E. coli by affecting the DNA bending of the target promoters. Binding studies by the same group demonstrated that a cytoplasmic protein purified, and later identified as SarA, binds to two sites within the agr promoter region affecting both RNAII and RNAIII transcription (47). Our study has shown SarA mediates the up-regulation of hla expression in response to microaerobic growth. As this effect probably occurs independently of RNAIII, this observation suggests that SarA or a putative complex containing SarA binds directly at the hla promoter or via an additional regulatory protein. Regions upstream of known agr-regulated genes such as seb, eta, and etb have been predicted to serve as binding sites for regulatory proteins (43, 53). More recently, Chien and Cheung (19) have mapped the SarA binding site to a 29-bp sequence within the agr P2-P3 regulatory region by footprinting experiments. This region is unusual in that it is very AT rich (89.6% over 29 bp) (19). Analysis of the control regions of SarA-regulated genes reveals similar AT-abundant sequences (82.8 to 96.6% over 29 bases) (Fig. 6). The significance of these regions in the SarA-mediated regulation of virulence gene expression remains to be established experimentally.
FIG. 6.
Nucleotide alignment of upstream regions of SarA-regulated genes of S. aureus. Alignments are based on homology of the noncoding regions 5′ of SarA-controlled genes to the proposed 29-bp SarA binding site (19) of the agr P2-P3 regulatory region (accession no. X52543 [50]). Best identity (58.6 to 72.4% identity over 29 bp to the agr sequence) was observed in AT-rich regions of tst (accession no. U93688), spa (accession no. J01786 [64]), hlb (accession no. X13404 [55]), seb (accession no. M11118 [37]), V8 protease (accession no. P04188 [11]), and hla (accession no. X01645 [28]). The distances of the hypothetical SarA binding site from transcriptional start (TS) sites are indicated where known. The 3′ ends of the tst, hlb, V8 protease, and hla sequences are located 145, 61, 9, and 270 bp upstream of their translational start sites. Nucleotides showing >50% identity over seven sequences are boxed in black. A predicted consensus sequence is indicated by upper- and lowercase letters representing 100 and >50% nucleotide identity, respectively, over the seven sequences.
Recently, a modulating effect on SarA expression by putative ORF-3 and ORF-4 located immediately upstream of sarA has been proposed (8). Studies suggest that all three sar transcripts are required for restoring RNAII and RNAIII to wild-type levels in an sarA mutant, possibly by posttranslational interaction (13). In addition, ORF-3 was shown to be necessary for maximal, RNAIII-independent repression of spa expression mediated by SarA (14). Interestingly, an alternate sigma factor, ςB, was identified in S. aureus (68) and shown to interact with the sar operon in vitro (22). Therefore, this positive transcriptional regulation may also have a function in the hierarchical regulation of virulence determinant gene expression.
This study on the role of SarA in virulence determinant production in S. aureus has revealed SarA to be a major repressor of protease activity and a signal transducer which responds to O2 or CO2 levels. Regulation of protease activity in response to external stimuli may allow the organism to react quickly to alterations in its environment. Thus, SarA may not only regulate virulence determinant gene expression directly but also control surface and exoprotein stability by way of protease activity. This would allow the organism to rapidly change its surface array of virulence determinants, which is part of the innate ability of S. aureus to be a such a successful pathogen and inhabit a wide range of niches.
ACKNOWLEDGMENTS
We thank Saleem Khan and Richard Novick for providing strains used in this study, Wendy Hufnagle for sharing results prior to publication, and Jodi Lindsay for helpful discussions.
This research program was supported with funds from MAFF (P.F.C.) and the Royal Society (S.J.F.).
ADDENDUM IN PROOF
The sequence of staphopain (a thiol protease) has recently appeared in the databases (Swiss-Prot accession no. P81297). It shows 47% identity to ORFX over 172 amino acids. Thus, ORFX itself may be another SarA-regulated protease.
REFERENCES
- 1.Abdelnour A, Arvidson S, Bremell T, Rydén C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61:3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvidson S. Studies on extracellular proteolytic enzymes from Staphylococcus aureus. II. Isolation and characterisation of an EDTA-sensitive protease. Biochim Biophys Acta. 1973;302:149–157. doi: 10.1016/0005-2744(73)90017-x. [DOI] [PubMed] [Google Scholar]
- 3.Arvidson S O. Extracellular enzymes from Staphylococcus aureus. In: Easmon C S F, Adlam C, editors. Staphylococci and staphylococcal infections. Vol. 2. London, England: Academic Press Inc.; 1983. pp. 745–808. [Google Scholar]
- 4.Ayora S, Götz F. Genetic and biochemical properties of an extracellular neutral metalloprotease from Staphylococcus hyicus subsp. hyicus. Mol Gen Genet. 1994;242:421–430. doi: 10.1007/BF00281792. [DOI] [PubMed] [Google Scholar]
- 5.Ayora S, Lindgren P-E, Götz F. Biochemical properties of a novel metalloprotease from Staphylococcus hyicus subsp. hyicus involved in extracellular lipase processing. J Bacteriol. 1994;176:3218–3223. doi: 10.1128/jb.176.11.3218-3223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey C J, Smith T P. The reactive serine residue of epidermolytic toxin A. Biochem J. 1990;269:535–537. doi: 10.1042/bj2690535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balaban N, Novick R P. Autocrine regulation of toxin synthesis by Staphylococcus aureus. Proc Natl Acad Sci USA. 1995;92:1619–1623. doi: 10.1073/pnas.92.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bayer M G, Heinrichs J H, Cheung A L. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Björklind A, Arvidson A. Mutants of Staphylococcus aureus affected in the regulation of exoprotein synthesis. FEMS Microbiol Lett. 1980;7:203–206. [Google Scholar]
- 10.Burnette W M. Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 11.Carmona C, Gray G L. Nucleotide sequence of the serine protease gene of Staphylococcus aureus, strain V8. Nucleic Acids Res. 1987;15:6757. doi: 10.1093/nar/15.16.6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan P F, Foster S J. The role of environmental factors in the regulation of virulence determinant expression in Staphylococcus aureus 8325-4. Microbiology. 1998;144:2469–2479. doi: 10.1099/00221287-144-9-2469. [DOI] [PubMed] [Google Scholar]
- 12a.Chan, P. F., and S. J. Foster. Unpublished data.
- 13.Cheung A L, Bayer M G, Heinrichs J H. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J Bacteriol. 1997;179:3963–3971. doi: 10.1128/jb.179.12.3963-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung A L, Eberhardt K, Heinrichs J H. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect Immun. 1997;65:2243–2249. doi: 10.1128/iai.65.6.2243-2249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung A L, Projan S J. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J Bacteriol. 1994;176:4168–4172. doi: 10.1128/jb.176.13.4168-4172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung A L, Yeaman M R, Sullam P M, Witt M D, Bayer A S. Role of the sar locus of Staphylococcus aureus in induction of endocarditis in rabbits. Infect Immun. 1994;62:1710–1725. doi: 10.1128/iai.62.5.1719-1725.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung A L, Ying P. Regulation of α- and β-hemolysins by the sar locus of Staphylococcus aureus. J Bacteriol. 1994;176:580–585. doi: 10.1128/jb.176.3.580-585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chien Y-T, Cheung A L. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J Biol Chem. 1998;273:2645–2652. doi: 10.1074/jbc.273.5.2645. [DOI] [PubMed] [Google Scholar]
- 20.Compagnone-Post P, Malyankar U, Khan S A. Role of host factors in the regulation of the enterotoxin B gene. J Bacteriol. 1991;173:1827–1830. doi: 10.1128/jb.173.5.1827-1830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dancer S J, Garratt R, Saldanha J, Jhoti H, Evans R. The epidermolytic toxins are serine proteases. FEBS Lett. 1990;268:129–132. doi: 10.1016/0014-5793(90)80990-z. [DOI] [PubMed] [Google Scholar]
- 22.Deora R, Tseng T, Misra T K. Alternative transcription factor ςSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J Bacteriol. 1997;179:6355–6359. doi: 10.1128/jb.179.20.6355-6359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorman C J. DNA supercoiling and environmental regulation of gene expression in pathogenic bacteria. Infect Immun. 1991;59:745–749. doi: 10.1128/iai.59.3.745-749.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drapeau G R. Role of a metalloprotease in activation of the precursor of staphylococcal protease. J Bacteriol. 1978;136:607–613. doi: 10.1128/jb.136.2.607-613.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foster S J. Cloning, expression, sequence analysis and biochemical characterisation of an autolytic amidase of Bacillus subtilis 168 trpC2. J Gen Microbiol. 1991;137:1987–1998. doi: 10.1099/00221287-137-8-1987. [DOI] [PubMed] [Google Scholar]
- 26.Foster S J. Analysis of the autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1992;174:464–470. doi: 10.1128/jb.174.2.464-470.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giraudo A T, Raspanti C G, Calzolari A, Nagel R. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can J Microbiol. 1994;40:677–681. doi: 10.1139/m94-107. [DOI] [PubMed] [Google Scholar]
- 28.Gray G S, Kehoe M. Primary sequence of the α-toxin gene from Staphylococcus aureus Wood 46. Infect Immun. 1984;46:615–618. doi: 10.1128/iai.46.2.615-618.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guérout-Fleury A-M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 30.Hart M E, Smeltzer M S, Iandolo J J. The extracellular protein regulator (xpr) affects exoprotein and agr mRNA levels in Staphylococcus aureus. J Bacteriol. 1993;175:7875–7879. doi: 10.1128/jb.175.24.7875-7879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinrichs J H, Bayer M G, Cheung A L. Characterization of the sar locus and its interactions with agr in Staphylococcus aureus. J Bacteriol. 1996;178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hufnagle, W. Personal communication.
- 33.Iandolo J J. The genetics of staphylococcal toxins and virulence factors. In: Iglewski B H, Clark V L, editors. Molecular basis of bacterial pathogenesis. New York, N.Y: Academic Press; 1990. pp. 399–426. [Google Scholar]
- 34.Janzon L, Arvidson S. The role of the δ-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji G, Beavis R, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 37.Jones C L, Khan S A. Nucleotide sequence of the enterotoxin B gene from Staphylococcus aureus. J Bacteriol. 1986;166:29–33. doi: 10.1128/jb.166.1.29-33.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kemp E H, Sammons R L, Moir A, Sun D, Setlow P. Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis 168. J Bacteriol. 1991;173:4646–4652. doi: 10.1128/jb.173.15.4646-4652.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kornblum J, Kreiswirth B N, Projan S J, Ross H, Novick R P. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 373–402. [Google Scholar]
- 40.Kreiswirth B, Löfdahl S, Betley M J, O’Reilly M, Schleivert M P, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 41.Lee C Y, Schmidt J J, Johnson-Winegar A D, Spero L, Iandolo J J. Sequence determination and comparison of the exfoliative toxin A and toxin B genes from Staphylococcus aureus. J Bacteriol. 1987;169:3904–3909. doi: 10.1128/jb.169.9.3904-3909.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, Arvidson S, Mölby R. Variation in the agr-dependent expression of α-toxin and protein A among clinical isolates of Staphylococcus aureus from patients with septicaemia. FEMS Microbiol Lett. 1997;152:155–161. doi: 10.1111/j.1574-6968.1997.tb10422.x. [DOI] [PubMed] [Google Scholar]
- 42a.Lindsay, J. A., and S. J. Foster. Unpublished data.
- 43.Mahmood R, Khan S A. Role of upstream sequences in the expression of the staphylococcal enterotoxin B gene. J Biol Chem. 1990;265:4652–4656. [PubMed] [Google Scholar]
- 44.McGavin M J, Zahradka C, Rice K, Scott J E. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect Immun. 1997;65:2621–2628. doi: 10.1128/iai.65.7.2621-2628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morfeldt E, Janzon L, Arvidson S, Löfdahl S. Cloning of a chromosomal locus (exp) which regulates the expression of several exoprotein genes in Staphylococcus aureus. Mol Gen Genet. 1988;211:435–440. doi: 10.1007/BF00425697. [DOI] [PubMed] [Google Scholar]
- 46.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 48.Novick R P. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 49.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 50.Novick R P, Projan S J, Kornblum J, Ross H F, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 51.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohlsen K, Koller K-P, Hacker J. Analysis of expression of the alpha-toxin gene (hla) of Staphylococcus aureus by using a chromosomally encoded hla::lacZ gene fusion. Infect Immun. 1997;65:3606–3614. doi: 10.1128/iai.65.9.3606-3614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Toole P W, Foster T J. Nucleotide sequence of the epidermolytic toxin A gene of Staphylococcus aureus. J Bacteriol. 1987;169:3910–3915. doi: 10.1128/jb.169.9.3910-3915.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng H-L, Novick R P, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Projan S J, Kornblum J, Kreiswirth B, Moghazeh S L, Eisner W, Novick R P. Nucleotide sequence: the β-hemolysin gene of Staphylococcus aureus. Nucleic Acids Res. 1989;17:3305. doi: 10.1093/nar/17.8.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Recsei P, Kreiswirth B, O’Reilly M, Schlievert P, Gruss A, Novick R P. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 57.Regassa L B, Betley M J. Alkaline pH decreases expression of the accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1992;174:5095–5100. doi: 10.1128/jb.174.15.5095-5100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Regassa L B, Novick R P, Betley M J. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect Immun. 1992;60:3381–3388. doi: 10.1128/iai.60.8.3381-3388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reineck K, Renneberg J, Diamant M, Gutschik E, Bendtzen K. Molecular cloning and expression of a novel Staphylococcus aureus antigen. Biochim Biophys Acta. 1997;1350:128–132. doi: 10.1016/s0167-4781(96)00216-3. [DOI] [PubMed] [Google Scholar]
- 60.Rowe G E, Welch R A. Assays of hemolytic toxins. Methods Enzymol. 1994;235:657–667. doi: 10.1016/0076-6879(94)35179-1. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 62.Schenk S, Laddaga R A. Improved methods for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;94:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 63.Teufel P, Götz F. Characterization of an extracellular metalloprotease with elastase activity from Staphylococcus epidermidis. J Bacteriol. 1993;175:4218–4224. doi: 10.1128/jb.175.13.4218-4224.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uhlén M, Guss B, Nilsson B, Gatenbeck S, Philipson L, Lindeberg M. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J Biol Chem. 1984;259:1695–1702. [PubMed] [Google Scholar]
- 65.Vandenesch F, Kornblum J, Novick R P. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J Bacteriol. 1991;173:6313–6320. doi: 10.1128/jb.173.20.6313-6320.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waldvogel F A. Staphylococcus aureus (including toxic shock syndrome) In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 4th ed. New York, N.Y: Churchill Livingstone; 1995. pp. 1754–1777. [Google Scholar]
- 67.Wiseman G M. The hemolysins of Staphylococcus aureus. Bacteriol Rev. 1975;39:317–344. doi: 10.1128/br.39.4.317-344.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshikawa K, Tsuzuki H, Fujiwara T, Nakamura E, Iwamoto H, Matsumoto K, Shin M, Yoshida N, Teraoka H. Purification, characterization and gene cloning of a novel glutamic acid-specific endopeptidase from Staphylococcus aureus ATCC 12600. Biochim Biophys Acta. 1992;1121:221–228. doi: 10.1016/0167-4838(92)90358-k. [DOI] [PubMed] [Google Scholar]