Abstract
Jingchuvirales is an order of negative-sense RNA viruses with genomes of 9.1–15.3 kb that have been associated with arachnids, barnacles, crustaceans, insects, fish and reptiles in Africa, Asia, Australia, Europe, North America and South America. The jingchuviral genome has two to four open reading frames (ORFs) that encode a glycoprotein (GP), a nucleoprotein (NP), a large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain, and/or proteins of unknown function. Viruses in the order are only known from their genome sequences. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the order Jingchuvirales and on the families Aliusviridae, Chuviridae, Crepuscuviridae, Myriaviridae and Natareviridae, which are available at ictv.global/report/jingchuvirales, ictv.global/report/aliusviridae, ictv.global/report/chuviridae, ictv.global/report/crepuscuviridae, ictv.global/report/myriaviridae and ictv.global/report/natareviridae, respectively.
Keywords: Aliusviridae, aqualaruvirus, charybdivirus, chu-like, Chuviridae, chuvirus, Crepuscuviridae, ICTV Report, Jingchuvirales, Myriaviridae, myriavirus, Natareviridae, obscuruvirus, ollusvirus, taxonomy
Virion
Unknown.
Genome
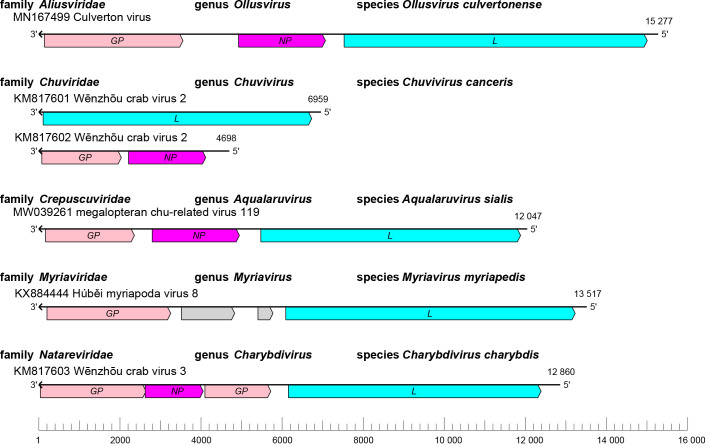
The jingchuviral genome is a nonsegmented linear, nonsegmented circular, bisegmented linear, or bisegmented circular negative-sense RNA of 9.1–15.3 kb containing two to four open reading frames (ORFs) that encode a glycoprotein (GP), a nucleoprotein (NP), a large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain, and/or proteins of unknown function [1–5] (Table 1, Fig. 1).
Table 1.
Characteristics of members of the order Jingchuvirales
|
Example |
Wēnzhōu crab virus 2 (S1: KM817601; S2: KM817602), species Chuvivirus canceris, genus Chuvivirus, family Chuviridae |
|---|---|
|
Virion |
Unknown |
|
Genome |
9.1–15.3 kb of nonsegmented linear, nonsegmented circular, bisegmented linear, or bisegmented circular negative-sense RNA |
|
Replication |
Unknown |
|
Translation |
Unknown |
|
Host range |
Arachnids; barnacles; crustaceans; insects; myriapods; fish; reptiles |
|
Taxonomy |
Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Monjiviricetes; the order includes >4 families, >20 genera and >57 species |
Fig. 1.
Genome organisation of representative jingchuvirals. ORFs are coloured according to the predicted protein function. GP, glycoprotein gene; NP, nucleoprotein gene; L, large protein gene. Grey colour indicates ORFs encoding proteins of unknown function.
Replication
Unknown.
Taxonomy
Current taxonomy: ictv.global/taxonomy. The order Jingchuvirales includes >50 virus species in the families Aliusviridae, Chuviridae, Crepuscuviridae, Myriaviridae and Natareviridae for viruses that infect arachnids, barnacles, crustaceans, insects, fish and reptiles (Fig. 2).
Fig. 2.
Phylogenetic relationships of jingchuvirals. Maximum-likelihood tree (midpoint-rooted) inferred using large protein gene (L) sequences. For details see full ICTV Report. Numbers near nodes on the trees indicate percentage bootstrap values.
Resources
Full ICTV Report on the order Jingchuvirales and the families Aliusviridae, Chuviridae, Crepuscuviridae, Myriaviridae and Natareviridae: ictv.global/report/jingchuvirales, ictv.global/report/aliusviridae, ictv.global/report/chuviridae, ictv.global/report/crepuscuviridae, ictv.global/report/myriaviridae and ictv.global/report/natareviridae, respectively.
Funding information
Production of this Profile, the ICTV Report, and associated resources was supported by the Microbiology Society. This work was supported in part through the Laulima Government Solutions, LLC, prime contract with the US National Institute of Allergy and Infectious Diseases (NIAID) under contract no. HHSN272201800013C. J.H.K. performed this work as an employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC, under contract no. HHSN272201800013C. The content of this publication should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of Health and Human Services or of the institutions and companies affiliated with the authors, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Acknowledgements
We thank Stuart G. Siddell, Elliot J. Lefkowitz, Sead Sabanadzovic, Peter Simmonds, F. Murilo Zerbini, Evelien Adriaenssens, Mart Krupovic, Luisa Rubino, Arvind Varsani (ICTV Report Editors) and Donald B. Smith (Managing Editor, ICTV Report). We thank Anya Crane (Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Fort Detrick, Frederick, MD, USA) for critically editing the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: GP, glycoprotein; L, large protein; NP, nucleoprotein; RdRP, RNA-directed RNA polymerase.
References
- 1.Di Paola N, Dheilly NM, Junglen S, Paraskevopoulou S, Postler TS, et al. Jingchuvirales: a new taxonomical framework for a rapidly expanding order of unusual monjiviricete viruses broadly distributed among arthropod subphyla. Appl Environ Microbiol. 2022;88:e0195421. doi: 10.1128/AEM.01954-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Käfer S, Paraskevopoulou S, Zirkel F, Wieseke N, Donath A, et al. Re-assessing the diversity of negative strand RNA viruses in insects. PLoS Pathog. 2019;15:e1008224. doi: 10.1371/journal.ppat.1008224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife. 2015;4:e05378. doi: 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi M, Lin X-D, Tian J-H, Chen L-J, Chen X, et al. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 5.Wu H, Pang R, Cheng T, Xue L, Zeng H, et al. Abundant and diverse RNA viruses in insects revealed by RNA-Seq analysis: ecological and evolutionary implications. mSystems. 2020;5:e00039-20. doi: 10.1128/mSystems.00039-20. [DOI] [PMC free article] [PubMed] [Google Scholar]