Abstract
Discoviridae is a family of negative-sense RNA viruses with genomes of 6.2–9.7 kb that have been associated with fungi and stramenopiles. The discovirid genome consists of three monocistronic RNA segments with open reading frames (ORFs) that encode a nucleoprotein (NP), a nonstructural protein (Ns), and a large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Discoviridae, which is available at ictv.global/report/discoviridae.
Keywords: Discoviridae, ICTV Report, orthodiscovirus, taxonomy
Virion
Unknown.
Genome
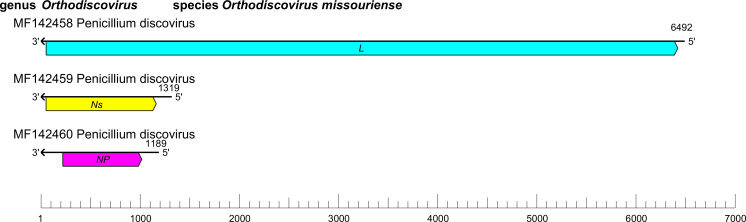
The genome of discovirids comprises three RNA segments (small [S], medium [M], and large [L]) of linear negative-sense RNA with a total length of 6.2–9.7 kb (S segment: about 1.1–1.2 kb; M segment: about 1.7–1.9 kb; and L segment: about 3.4–6.5 kb) (Table 1). Each segment contains at least one ORF that encodes either an NP, an Ns or an L protein containing an RNA-directed RNA polymerase (RdRP) domain [1–3] (Fig. 1).
Table 1.
Characteristics of members of the family Discoviridae
|
Example |
Penicillium discovirus (S: MF142460; M: MF142459; L: MF142458), species Orthodiscovirus missouriense, genus Orthodiscovirus |
|---|---|
|
Virion |
Unknown |
|
Genome |
6.2–9.7 kb of trisegmented negative-sense RNA |
|
Replication |
Unknown |
|
Translation |
Unknown |
|
Host range |
Peronosporaceaen stramenopiles, eurotiomycete and sordariomycete fungi |
|
Taxonomy |
Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Ellioviricetes, order Bunyavirales; the family includes the genus Orthodiscovirus and several species |
Fig. 1.
Genome organisation of Penicillium discovirus. ORFs are indicated as boxes, coloured according to the predicted protein function (L, large protein gene; NP, nucleoprotein gene; Ns, nonstructural protein gene).
Replication
Unknown.
Taxonomy
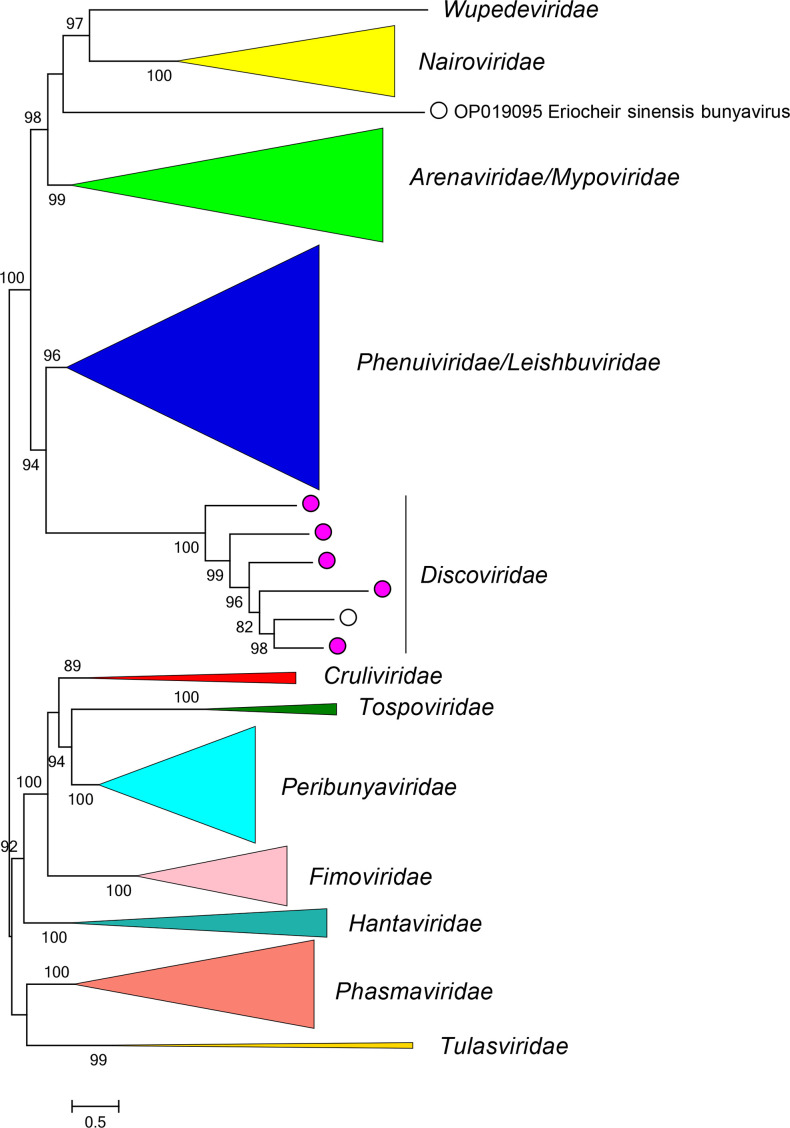
Current taxonomy: ictv.global/taxonomy. Discovirids are most closely related to arenavirids, leishbuvirids, mypovirids, nairovirids, phenuivirids, and wupedevirids [4] (Fig. 2). The family includes the genus Orthodiscovirus and >4 species; numerous discovirus-like sequences have also been found in sequence-read archives [5]. Like most other bunyavirals, Discovirids (i) have multisegmented, negative-sense single-stranded RNA genomes; (ii) encode proteins with high sequence identity to proteins of other bunyavirals, (iii) and have five conserved motifs (A–E) in their RdRP domain.
Fig. 2.
Phylogenetic relationships of viruses in the family Discoviridae. Branches for other families are collapsed. Numbers at nodes indicate bootstrap support >70 %. Full details of the virus sequences and methods used are available in the full ICTV Report on the family Discoviridae.
Resources
Full ICTV Report on the family Discoviridae: ictv.global/report/discoviridae.
Funding information
Production of this Profile, the ICTV Report, and associated resources was supported by the Microbiology Society. This work was supported in part through the Laulima Government Solutions, LLC, prime contract with the U.S. National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272201800013C. J.H.K. performed this work as an employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC, under Contract No. HHSN272201800013C. The content of this publication should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Health and Human Services, including the Centres for Disease Control and Prevention, or of the institutions and companies affiliated with the authors, nor does mention of trade names, commercial products, or organisations imply endorsement by the U.S. Government. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centres for Disease Control and Prevention.
Acknowledgements
We thank Stuart G. Siddell, Elliot J. Lefkowitz, Sead Sabanadzovic, Peter Simmonds, F. Murilo Zerbini, Evelien Adriaenssens, Mart Krupovic, Luisa Rubino, Arvind Varsani (ICTV Report Editors), and Donald B. Smith (Managing Editor, ICTV Report). We thank Anya Crane (Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Fort Detrick, Frederick, MD, USA) for critically editing the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: L, large; M, medium; NP, nucleoprotein; Ns, nonstructural protein; RdRP, RNA-directed RNA polymerase; S, small.
References
- 1.Chiapello M, Rodríguez-Romero J, Ayllón MA, Turina M. Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol. 2020;6:veaa058. doi: 10.1093/ve/veaa058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nerva L, Turina M, Zanzotto A, Gardiman M, Gaiotti F, et al. Isolation, molecular characterization and virome analysis of culturable wood fungal endophytes in esca symptomatic and asymptomatic grapevine plants. Environ Microbiol. 2019;21:2886–2904. doi: 10.1111/1462-2920.14651. [DOI] [PubMed] [Google Scholar]
- 3.Nerva L, Forgia M, Ciuffo M, Chitarra W, Chiapello M, et al. The mycovirome of a fungal collection from the sea cucumber Holothuria polii . Virus Res. 2019;273:197737. doi: 10.1016/j.virusres.2019.197737. [DOI] [PubMed] [Google Scholar]
- 4.Huang P, Zhang X, Ame KH, Shui Y, Xu Z, et al. Genomic and phylogenetic characterization of a bunya-like virus from the freshwater Chinese mitten crab Eriocheir sinensis . Acta Virol. 2019;63:433–438. doi: 10.4149/av_2019_410. [DOI] [PubMed] [Google Scholar]
- 5.Neri U, Wolf YI, Roux S, Camargo AP, Lee B, et al. Expansion of the global RNA virome reveals diverse clades of bacteriophages. Cell. 2022;185:4023–4037. doi: 10.1016/j.cell.2022.08.023. [DOI] [PubMed] [Google Scholar]