Abstract
Repeat spillover of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) into new hosts has highlighted the critical role of cross-species transmission of coronaviruses and establishment of new reservoirs of virus in pandemic and epizootic spread of coronaviruses. Species particularly susceptible to SARS-CoV-2 spillover include Mustelidae (mink, ferrets and related animals), cricetid rodents (hamsters and related animals), felids (domestic cats and related animals) and white-tailed deer. These predispositions led us to screen British wildlife with sarbecovirus-specific quantitative PCR and pan coronavirus PCR assays for SARS-CoV-2 using samples collected during the human pandemic to establish if widespread spillover was occurring. Fourteen wildlife species (n=402) were tested, including: two red foxes (Vulpes vulpes), 101 badgers (Meles meles), two wild American mink (Neogale vison), 41 pine marten (Martes martes), two weasels (Mustela nivalis), seven stoats (Mustela erminea), 108 water voles (Arvicola amphibius), 39 bank voles (Myodes glareolous), 10 field voles (Microtus agrestis), 15 wood mice (Apodemus sylvaticus), one common shrew (Sorex aranaeus), two pygmy shrews (Sorex minutus), two hedgehogs (Erinaceus europaeus) and 75 Eurasian otters (Lutra lutra). No cases of SARS-CoV-2 were detected in any animals, but a novel minacovirus related to mink and ferret alphacoronaviruses was detected in stoats recently introduced to the Orkney Islands. This group of viruses is of interest due to pathogenicity in ferrets. The impact of this virus on the health of stoat populations remains to be established.
Keywords: Coronavirus, mustelid, cricetid rodent, Minacovirus, stoat, SARS-CoV-2
Data Summary
The Minacovirus sequence assembled from this study has been deposited in the NCBI GenBank database under accession number OP933726. In addition, Illumina read datasets generated have been submitted under Bioproject accession number PRJNA897822, SRA accession numbers SAMN3158039, SAMN31580331 and SAMN31580344, and Biosample accession number SRS1567284850. Supplementary information for this paper is available at https://doi.org/10.6084/m9.figshare.24231709 [1].
Introduction
Coronaviruses are a large and diverse group of enveloped RNA viruses found in diverse vertebrate hosts. Well-studied mammalian hosts, such as humans and domestic dogs, have multiple coronaviruses of several different subfamilies, although most of those of concern in mammals are members of the genera Alphacoronavirus and Betacoronavirus [2]. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic is thought to have arisen via a spillover from horseshoe bats (the natural hosts of the Betacoronavirus subgenus sarbecoviruses), with human infection probably via a ‘liaison host’ such as the raccoon dog (Nyctereutes procyonoides) or Malayan pangolin (Manis javanica) [3–5].
The overwhelming scale of the global human pandemic led to repeated spillovers and onward transmission into other mammalian species such as domestic cats (Felis catus) [6–8], farmed American mink (Neogale vison) [9–13] and Syrian hamsters (Mesocricetus auratus) [14, 15], and the establishment of a new reservoir in North American white-tailed deer (Odocoileus virginianus) [16, 17]. A wide range of other species are either able to be infected experimentally or have been subjects of sporadic case reports of SARS-CoV-2 infection or seroconversion including: cricetid rodents, felids, other small carnivores. mustelids, primates and bats (reviewed in [18, 19]).
SARS-CoV-2 infection in Muridae such as house mice (Mus musculus) and brown rats (Rattus norvegicus) may depend on the virus strain: initial studies with the original (Wuhan) strains of the virus failed to infect them [20, 21] and field studies failed to demonstrate evidence of infection in wild populations (27 Mus musculus and 97 R. norvegicus). Later variants did however cause infection in laboratory studies [21–24] and there have been several subsequent field reports of sporadic infection of rats [25–27].
The present study focused on species of wild animals present in Great Britain that were assessed to be of higher risk for SARS-CoV-2 spillover in 2021, when the study was begun. These included horseshoe bats (the subject of a separate report [28]), mustelids, small carnivores and cricetid rodents.
Thus far, no infection with SARS-CoV-2 has been reported in horseshoe bats (Rhinolophus spp.) in Britain or mainland Europe [28–30] although other related coronaviruses have been detected in these species. Reports in European deer prior to 2022 all failed to detect any exposure [31–33], but 57 % of fallow deer in Dublin, Ireland, seroconverted in early 2022 [34] and sporadic seropositivity in fallow and red deer in Spain in 2021–22 has also been reported [35]. Wild animal surveillance studies in mainland Europe have indicated sporadic detection in wild mustelids, including by quantitative (q)PCR in wild American mink (N. vison), particularly near farmed mink outbreaks, and one otter (L. lutra) [36–38]. Serological evidence of exposure has been described in 3/14 pine martens (Martes martes) and 2/10 badgers (Meles meles) [39], but other studies found no evidence of infection in 48 polecats (Mustela putorius), 163 badgers or cricetid and murid rodents (694 Myodes glareolus, two Microtus arvalis, 27 Mus musculus, 97 R. norvegicus and eight Apodemus species) [40–42].
Methods
Sample collection
A total of 402 samples from 14 wildlife species (Table 1) were collected through a network of wildlife researchers and volunteers engaged in wildlife conservation, monitoring or pest control. The majority of samples were collected during the human coronavirus disease 2019 (Covid-19) pandemic, with the exception of a small number of historical otter samples. The species targeted were primarily mustelids (otters L. lutra, badgers Meles meles, mink, pine martens Martes martes, weasels Mustela nivalis, stoats Mustela erminea) or cricetid rodents (water voles Arvicola amphibius, field voles Microtus agrestis, bank voles Myodes glareolus) with a small number of other species collected opportunistically (hedgehogs Erinaceus europaeus, common shrews Sorex araneus, pygmy shrews Sorex minutus, red foxes Vulpes vulpes, wood mice Apodemus sylvaticus). Ethical approval was granted by the University of Nottingham School of Veterinary Medicine and Science Committee for Animal Research and Ethics (CARE), and the University of Sussex Animal Welfare and Ethical Review Board.
Table 1.
Species and sample type screened for SARS-CoV-2
|
Species |
Oral swab |
Rectal swab |
Lung sample |
Faecal sample |
Total no. of animals |
Sample collection dates |
|---|---|---|---|---|---|---|
|
Mustelids |
||||||
|
Otter |
– |
– |
75 |
– |
75 |
Jan 2020–Apr 2022 (51 samples) Apr 2017–Dec 2019 (25 samples) |
|
Badger |
– |
– |
101 |
– |
101 |
Apr 2021–Apr 2022 |
|
Mink |
1 |
1 |
– |
– |
2 |
Aug 2021–Jun 2022 |
|
Pine marten |
– |
– |
– |
41 |
41 |
Mar 2020–Oct 2021 |
|
Weasel |
– |
– |
2 |
– |
2 |
Sep 2021 |
|
Stoat |
7 |
7 |
– |
– |
7 |
Dec 2021 |
|
Cricetid rodents |
||||||
|
Water vole |
– |
– |
– |
108 |
108 |
Aug–Nov 2021 |
|
Bank vole |
– |
– |
7 |
32 |
39 |
Jan–Feb 2022 |
|
Field vole |
– |
– |
5 |
5 |
10 |
Nov 2021–Feb 2022 |
|
Other species |
||||||
|
Wood mouse |
– |
– |
– |
10 |
10 |
Jul 2021–Feb 2022 |
|
Common shrew |
– |
– |
– |
1 |
1 |
Jan 2022 |
|
Pygmy shrew |
– |
– |
– |
2 |
2 |
Jan 2022 |
|
Hedgehog |
– |
– |
– |
2 |
2 |
Nov 2022 |
|
Red fox |
2 |
2 |
– |
– |
2 |
Apr 2022 |
|
Total |
402 |
Lung samples were taken from postmortem cadavers submitted to the Cardiff Otter Project (otters) or badgers found dead and submitted for tuberculosis monitoring to the University of Nottingham (badgers tested for covid were all confirmed culture-negative for Mycobacterium tuberculosis complex infections). Mink and stoat samples (oronasal and rectal swabs from cadavers) were provided from programmes for invasive species control and were from animals that were live trapped and euthanized (mink) or lethally trapped (stoats). Rodent samples were faecal samples from animals live trapped in Longworth or Elliot traps for population monitoring (faecal samples taken from traps or latrine/burrow sites), and from a small number of captive water voles from a licensed breeding and release programme. A small number of lung samples were taken from animals found dead. Weasel, hedgehog and shrew samples were from a small number of animals caught as bycatch in Longworth or Elliot traps. Pine marten samples were all faecal samples (scat) from environmental monitoring. Samples were collected from a variety of British locations (Supplementary Information, available in the online version of this article). Collectors were solicited by social media, notices in game and hunting organization newsletters, letters in the Veterinary Record and contact networks of wildlife organizations working with study participants and were provided with sampling packs including gloves, collection and shipping material and instructions. Samples were collected into RNAlater before either storage at −20 °C or direct shipping to The University of Nottingham, depending on the capacity of the collectors.
RNA extraction, reverse transcriptase (RT) and RNA-dependent RNA polymerase (RDRP) gene coronaviruses generic conventional PCR and envelope gene sarbecovirus-specific real-time PCR
RNA extraction from lung tissue, faecal samples, rectal and oronasal swabs, and cell culture supernatant as a positive control was carried out using the Macherey-Nagel RNA tissue extraction kit as per the manufacturer’s instructions. The Wuhan SARS-CoV-2 strain positive control sample used throughout this study was kindly donated by Dr Christopher Coleman (Division of Infection, Immunity and Microbes, School of Life Sciences, University of Nottingham, UK). Reverse transcription was performed in two steps, using M-MLV-RT and random hexamer primers (Promega) as per the manufacturer’s instructions. All cDNA products were stored at −20 °C for conventional PCR.
A generic pan-coronavirus PCR assay [43] was used to amplify a 440 bp fragment of the coronavirus RDRP gene using Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs cat. no.: M0493S). Primers were: F: GGTTGGGACTATCCTAAGTGTGA and R: CCATCATCAGATAGAATCATCATA. PCR products were purified using the Nucleospin extract II kit (Macherey-Nagel) according to the manufacturer’s instructions and were Sanger sequenced (Eurofins UK).
Real-time PCR was carried out using the Promega GoTaq Probe 1-Step RT-qPCR System (Promega) with Sarbecovirus-specific envelope gene primers as previously described [44]. Primers and probes were: F: ACAGGTACGTTAATAGTTAATAGCGT, Probe: FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ and R: ATATTGCAGCAGTACGCACACA.
RNA and cDNA quality control was assessed via partial amplification of 108 bp of the beta actin gene using a published conventional PCR protocol [45]. Primers were F: CAGCACAATGAAGATCAAGATCATC and R: CGGACTCATCGTACTCCTGCTT
High-throughput sequencing and genome analyses
RNA sequencing was performed on positive samples by Novogene, using the Illumina NovaSeq 6000 platform. Quality filtering and trimming to remove adapters, duplicates and low-quality reads was achieved using fastp v0.23.1 [46]. Kraken2 v2.1.2 was used for taxonomic classification of paired-end reads against the Kraken2 viral Refseq database [47] (retrieved 9 June 2022). Reads were assembled using the coronaSPAdes option in SPAdes genome assembler v3.15.4 [48] using default parameters. While CheckV v1.0.1, a fully automated command-line pipeline, was used for identification and quality assessment of contigs, contigs were also queried against the NCBI custom blastn (v2.12.0) viral database [49] (retrieved 3 July 2022).
Assembled contigs classified and assessed as complete alphacoronavirus genomes were indexed and extracted for downstream analysis using the SAMtools v1.16.1 faidx option [50]. Assembled genomes were annotated in Geneious Prime (v.2022.2.2) using NCBI coronavirus reference sequences for minacoviruses and tegacoviruses.
Phylogenetic analysis
Complete coronavirus genomes, and extracted RDRP, spike and nucleocapsid nucleotide sequences from alphacoronavirus genomes assembled in this study, and a total of 22 reference alphacoronavirus genomes (all Minacovirus full genomes available and a selection of Refseq or well-characterized full-length isolates of tegacoviruses with the reference sequence of porcine epidemic diarrhoea virus, PEDV, as an outgroup, Supplementary Information) were downloaded from NCBI, and aligned using Mafft v7.490 [51]. Maximum likelihood phylogenetic trees were reconstructed based on complete coronavirus genomes, and four different genes using IQ-TREE v2.0.7 [52], with 1000 ultrafast bootstrap approximations using UFBoot2 within IQ-TREE v2.0.7 to evaluate branch support [53]. The ModelFinder function within IQ-TREE was used to select the best-fitting nucleotide substitution model for phylogenetic reconstruction [54]. Phylogenetic trees were visualized and annotated in FigTree v1.4.4 (https://github.com/rambaut/figtree/). Using the same approach trees were also similarly reconstructed for individual genes (25 spike and nucleocapsid genes as the S genes of this group of viruses are known to recombine) and for all available Minacovirus partial gene fragments of RDRP and spike (where there are a lot more sequences available than for other parts of the genome).
Results
No animal sample tested positive on the Sarbecovirus-specific E gene qPCR.
Four (out of seven) stoat rectal swab samples (57 %) tested positive on the pancoronavirus PCR. Sanger sequencing of PCR products indicated that these were alphacoronaviruses of the Minacovirus group. No oral swab samples tested positive from the same animals. All stoat samples in this study were from the same population, and were sourced from the Orkney Islands stoat eradication programme (https://www.nature.scot/professional-advice/land-and-sea-management/managing-wildlife/orkney-native-wildlife-project) and are from the same recently introduced and heavily bottlenecked population. All except one sample in this study were from adult animals with a mix of males and females. Positive samples were from one adult male, one juvenile female and two adult females.
Illumina sequencing taxonomic classification, and genome assembly
Taxonomic classification using Kraken2 identified reads assigned to other viral operational taxonomic units, but only reads classified to the Coronaviridae viral family are reported in this study. De novo assembly of datasets from the stoats yielded one full-length coronavirus contig of 28.1 kb (100 % quality, 99.9 % completeness) and two partial contigs (7.5 kb, 27 % quality, 26.6 % completeness; and 27.9 kb, 99 % quality and 99.1 % completeness) from a further two samples. The remaining sample did not yield any coronavirus contigs. The sequences had closest homology to alphacoronaviruses of the Minacovirus group. The complete full genome sequence of the most complete contig has been deposited in GenBank (accession number OP933726).
Genome annotation and organization
Genome annotation demonstrated a typical Alphacoronavirus genome organization consisting of 5′ and 3′ UTRs, a large ORF1ab, encoding 16 non-structural peptides (nsp1-16) making up about two-thirds of the viral genome, and genes encoding four structural proteins: the spike (S), membrane (M), envelope (E) and nucleocapsid (N) (Fig. 1). Accessory proteins included ORFs with homology to ORF3c and 7b of feline coronavirus isolates. A number of smaller potential ORFs within these regions were also present, possibly corresponding to other accessory proteins with homology to the ORF3 or 7 of other Minacovirus and Tegacovirus isolates.
Fig. 1.
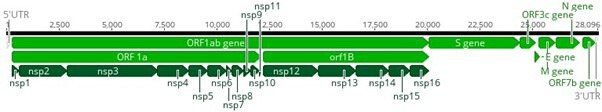
Genome organization of the stoat sequences derived from this study. ORFs are shown in light green, UTRs are shown in grey and regions with homology to non-structural proteins derived from feline coronavirus ORF1ab are marked in dark green.
Phylogenetic analysis
Results from the maximum likelihood phylogenetic trees drawn using all available complete Minacovirus genomes and selected reference sequences for tegacoviruses demonstrated that the stoat sequence is most closely clustered with ferret isolates (Fig. 2). This relationship is consistent across analysis of individual genes (spike, nucleocapsid full genes) and in larger phylogenetic trees of all partial fragments of Minacovirus RDRP and spike available in the NCBI database (Supplementary Information).
Fig. 2.
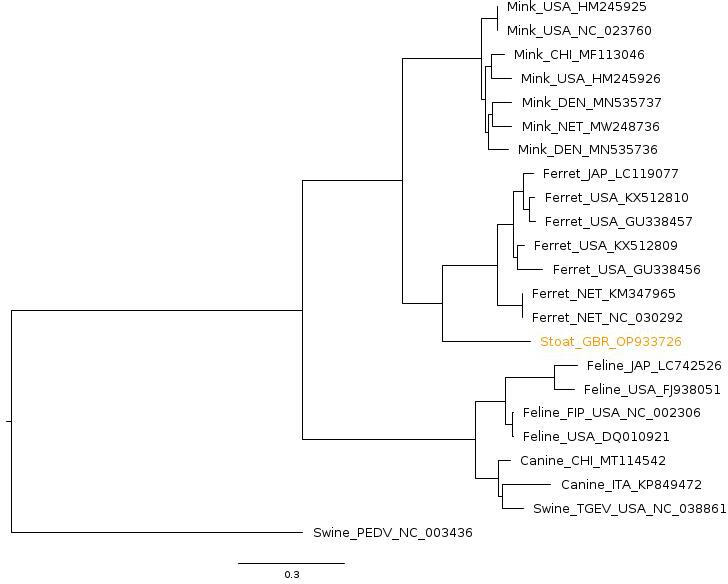
Maximum likelihood phylogenetic tree of full genomes of minacoviruses and tegacoviruses constructed with 1000 bootstrap approximation, rooted on the PEDV coronavirus reference sequence. Twenty-three full genomes were included. The sequence from this study is marked in orange. Sequences are named with species of origin, name of virus (if applicable) and a three-letter code for geographical origin (GBR=Great Britain, USA=United States of American, CHI=China, JAP=Japan, NET=Netherlands, DEN=Denmark) and GenBank ID. Bootstrap values are shown on nodes, the scale bar represents the number of substitutions per site.
Discussion
This study found no evidence of widespread circulation of SARS-CoV-2 in our opportunistically gathered British wild carnivores and cricetid rodents in 2021–22. The study was designed to detect an incidence rate of 5 % based on our previous studies of alphacoronaviruses in British rodents [55] with a target of 73 samples per species calculated with the online Epitools sample size calculator [56]. For some species including otters and badgers, for which there were ongoing post-mortem studies of found dead (largely road kill) animals [57–60], this sample size target was readily achieved. This target was also achieved for water voles facilitated by a network of wildlife monitoring and captive breeding for re-introduction [61]. However, we were reliant on volunteer submitters for other species and did not achieve the target levels of fresh samples for hedgehogs, foxes, martens and other species. We were also restricted in the type of sample collected for most species as it was primarily non-invasive (faeces or post-mortem rectal and oronasal swabs) that could be collected by network members. There remains a possibility that our study did not detect low-level circulation of SARS-CoV-2 in the species tested if infection is spatially restricted, that we targeted the wrong tissue sample type or that our samples were too degraded to detect virus with RNA-based methods. This is inherent to the design of a study such as this one seeking to provide the basis for a preliminary assessment with scope to implement a spatially stratified study.
The detection of another coronavirus in this study, however, indicates that the methods used and sample preservation were adequate for viral detection, at least at high prevalence, even in small populations of samples. Our study is also in line with other European wildlife studies indicating absence of widespread SARS-CoV-2 circulation in wild small carnivores and rodents, including wild American mink [38–42, 62, 63]. Detection of SARS-CoV-2 in one Eurasian river otter in lung tissue and nasal swabs and detection of a Eurasian badger-specific coronavirus in lung tissue from similar post-mortem monitoring programmes indicates that lungs are an appropriate target tissue for coronavirus monitoring in those species [37, 42]. SARS-CoV-2 is also readily detected in faecal samples in laboratory studies of rodents and carnivore species as well as field studies of farmed mink [64–67], indicating faecal sample screening is also an appropriate sample for coronavirus monitoring. Faecal samples or rectal swabs are also among the easiest and most acceptable samples for volunteer submitters to collect safely, promoting their use in disease surveillance.
Serological testing for SARS-CoV-2 seroconversion would have been a useful addition to this study as PCR-based testing used in our study can only detect current viral nucleic acid shedding whereas serology can detect prior exposure to the virus and give a longer term picture of virus exposure. The limitations of volunteer sample submission meant that for many species blood, tissue or body fluids (such as pleural effusion) from which reasonable antibody recovery could be expected were unavailable. Commercial pan-species serology tests for SARS-CoV-2 are available and have been used in other studies of wildlife exposure to SARS-CoV-2 [31, 33, 34, 68] . However, these kits are not validated for all species and we do not know what the cross-reactivity is with some of the more divergent coronaviruses detected recently in European wildlife [42]. The results of these tests are usually confirmed with virus neutralization assays with false positives a common finding [39, 69]. These caveats make serology testing for an expected low case rate hard to interpret and this work remains for follow-up studies.
The stoat alphacoronavirus reported in this study is a novel finding. The stoats all came from an eradication programme on the Orkney Islands, an archipelago off the northeastern tip of mainland Great Britain. They are not native to the islands having been first found in 2010, and cause considerable negative impact on breeding bird populations [70]. Despite the small number of samples collected (seven) more than half of them (four) were PCR positive on rectal swabs, suggesting that the prevalence of this virus in this population is high. Wider studies of this population and mainland populations would be warranted to gauge the prevalence of the virus in the larger (and source) populations.
The novel virus we identified in Orkney stoats is a member of the Minacovirus subgroup of the genus Alphacoronavirus, and clusters closely to mink and ferret coronaviruses. The ferret and mink viruses have a well-described pathogenicity, causing diarrhoea and sometimes a systemic disease syndrome, similar to feline infectious peritonitis in cats [71–79]. Wider studies of the pathogenesis of this virus in stoats and the impact of this on the species would be warranted. There are multiple reports of the ferret viruses in this group recombining with each other [73, 75, 80] and multiple reports of farmed mink infected with SARS-CoV-2 also being co-infected with minacoviruses at a high prevalence rate [13, 81, 82]. While recombination between SARS-CoV-2 and minacoviruses has not been observed it is a possibility that warrants monitoring, particularly in farmed mink outbreaks.
Of interest, the tegacoviruses (the most closely related alphacoronavirus clade to the minacoviruses) are known for readily recombining and jumping host species, with canine, feline and porcine recombinants in this group reported in multiple separate events [83–88]. Canine coronaviruses have also been reported multiple times in multiple locations in people with respiratory disease [89–91], making this group of viruses of concern for recombination and cross-species transmission potential and warranting monitoring.
Sequences in the Tegacovirus group have also been reported from raccoon dogs [88, 92], suggesting that there could be cross-species transmission between raccoon dogs, domestic cats, dogs, pigs and people. This is of particular concern as raccoon dogs are known to be able to be infected with and transmit SARS-CoV-2 and are one of the main suspects for the origin of the SARS-CoV-2 outbreak in humans [3, 93, 94].
The Minacovirus sequences isolated to date are all associated with mustelids of the genus Mustela, subfamily Mustelinae (ferrets, mink, stoats). However, there are relatively few studies of coronaviruses in mustelids. That is beginning to be rectified with the publication of SARS-CoV-2 monitoring studies, with a possible Gammacoronavirus identified in Chinese ferret badgers (Melogale moschata) and, most recently, isolates of a possibly new genus, Epsiloncoronavirus, in Italian badgers [42, 95]. The potential host and geographical ranges of these viruses remain unknown.
Overall this study adds to a growing picture of a lack of widespread SARS-CoV-2 circulation in wild European mammals, other than fallow deer. However, a novel alpha coronavirus of the Minacovirus sub-genus has been reported in an island population of stoats, adding much needed information on alphacoronavirus diversity in mustelids. The wider disease impacts and epidemiology of this virus in this species is, however, unknown and requires further study.
Funding information
This work was funded by the Biotechnology and Biosciences Research Council (BBSRC) grant number BB/W009501/1. During this period, the Otter Project was supported by funding from the Environment Agency, and by the Waterloo Foundation. The badger post-mortem study was funded by DEFRA as part of their ongoing tuberculosis monitoring.
Acknowledgements
We are very grateful to the many volunteers who collected samples for this study including Holly Broadhurst, Emma Bryce, Rachel Ewans, Martina Sekirnik, Ellen Bielinksi, Cristobal Castillo, Max Kampmann, John Bruce, Andrea Sartorius, the RSPB Orkney, RSPCA Oak and Furrows, the Vincent Wildlife Trust and Northumberland Wildlife Trust. Thanks also go to the many individuals and organizations who contribute to the otter collection network, including the Environment Agency, Natural Resources Wales, Wildlife Trusts, Local County Ecologists, Trunk Road Agencies, and local mammal groups.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Ethical approval was granted by the University of Nottingham School of Veterinary Medicine and Science Committee for Animal Research and Ethics (CARE), and the University of Sussex Animal Welfare and Ethical Review Board.
Footnotes
Abbreviations: RDRP, RNA-dependent RNA polymerase; RT-qPCR, reverse transcriptase quantitative PCR; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
References
- 1.Apaa T, Withers AJ, MacKenzie L, Staley C, Dessi N, et al. Lack of detection of SARS-Cov-2 in British wildlife in 2020-21 and first description of a stoat (Mustela erminea) Minacovirus . Microbiology Society. Dataset. 2023 doi: 10.6084/m9.figshare.24231709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo PCY, Lau SKP, Lam CSF, Lau CCY, Tsang AKL, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crits-Christoph A, Gangavarapu K, Pekar JE, Moshiri N, Singh R, et al. Genetic Evidence of Susceptible Wildlife in SARS-CoV-2 Positive Samples at the Huanan Wholesale Seafood Market Wuhan: Analysis and Interpretation of Data Released by the Chinese Center for Disease Control. Zenodo; 2023. [Google Scholar]
- 4.Holmes EC, Goldstein SA, Rasmussen AL, Robertson DL, Crits-Christoph A, et al. The origins of SARS-CoV-2: a critical review. Cell. 2021;184:4848–4856. doi: 10.1016/j.cell.2021.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu MQ, Lin HF, Li J, Chen Y, Luo Y, et al. A SARS-CoV-2-related virus from malayan pangolin causes lung infection without severe disease in human ACE2-transgenic mice. J Virol. 2023;97:e0171922. doi: 10.1128/jvi.01719-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goletic S, Goletic T, Softic A, Zahirovic A, Rukavina D, et al. The evidence of SARS-CoV-2 human-to-pets transmission in household settings in Bosnia and Herzegovina. Front Genet. 2022;13:839205. doi: 10.3389/fgene.2022.839205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosie MJ, Hofmann-Lehmann R, Hartmann K, Egberink H, Truyen U, et al. Anthropogenic infection of cats during the 2020 COVID-19 pandemic. Viruses. 2021;13:185. doi: 10.3390/v13020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jairak W, Chamsai E, Udom K, Charoenkul K, Chaiyawong S, et al. SARS-CoV-2 delta variant infection in domestic dogs and cats, Thailand. Sci Rep. 2022;12:8403. doi: 10.1038/s41598-022-12468-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domańska-Blicharz K, Orłowska A, Smreczak M, Niemczuk K, Iwan E, et al. Mink SARS-CoV-2 infection in Poland - short communication. J Vet Res. 2021;65:1–5. doi: 10.2478/jvetres-2021-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckstrand CD, Baldwin TJ, Rood KA, Clayton MJ, Lott JK, et al. An outbreak of SARS-CoV-2 with high mortality in mink (Neovison vison) on multiple Utah farms. PLoS Pathog. 2021;17:e1009952. doi: 10.1371/journal.ppat.1009952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammer AS, Quaade ML, Rasmussen TB, Fonager J, Rasmussen M, et al. SARS-CoV-2 transmission between mink (neovison vison) and humans, Denmark. Emerg Infect Dis. 2021;27:547–551. doi: 10.3201/eid2702.203794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Putintseva YA, Bondar EI, Simonov EP, Sharov VV, Oreshkova NV, et al. Siberian larch (Larix sibirica Ledeb.) mitochondrial genome assembled using both short and long nucleotide sequence reads is currently the largest known mitogenome. BMC Genomics. 2020;21:654. doi: 10.1186/s12864-020-07061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wasniewski M, Boué F, Richomme C, Simon-Lorière E, Van der Werf S, et al. Investigations on SARS-CoV-2 and other coronaviruses in mink farms in France at the end of the first year of COVID-19 pandemic. bioRxiv. 2023:2023.02.02.526749. doi: 10.1101/2023.02.02.526749. [DOI] [PMC free article] [PubMed]
- 14.Kok K-H, Wong S-C, Chan W-M, Wen L, Chu AW-H, et al. Co-circulation of two SARS-CoV-2 variant strains within imported pet hamsters in Hong Kong. Emerg Microbes Infect. 2022;11:689–698. doi: 10.1080/22221751.2022.2040922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yen H-L, Sit THC, Brackman CJ, Chuk SSY, Gu H, et al. Transmission of SARS-CoV-2 delta variant (AY.127) from pet hamsters to humans, leading to onward human-to-human transmission: a case study. Lancet. 2022;399:1070–1078. doi: 10.1016/S0140-6736(22)00326-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hale VL, Dennis PM, McBride DS, Nolting JM, Madden C, et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature. 2022;602:481–486. doi: 10.1038/s41586-021-04353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuchipudi SV, Surendran-Nair M, Ruden RM, Yon M, Nissly RH, et al. Multiple spillovers from humans and onward transmission of SARS-CoV-2 in white-tailed deer. Proc Natl Acad Sci U S A. 2022;119:e2121644119. doi: 10.1073/pnas.2121644119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuchipudi SV, Tan C, van Dorp L, Lichtveld M, Pickering B, et al. Coordinated surveillance is essential to monitor and mitigate the evolutionary impacts of SARS-CoV-2 spillover and circulation in animal hosts. Nat Ecol Evol. 2023;7:956–959. doi: 10.1038/s41559-023-02082-0. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen SS, Alvarez J, Bicout DJ, Calistri P, Canali E, et al. SARS-CoV-2 in animals: susceptibility of animal species, risk for animal and public health, monitoring, prevention and control. EFSA J. 2023;21:e07822. doi: 10.2903/j.efsa.2023.7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinnon KH, Leist SR, Schäfer A, Edwards CE, Martinez DR, et al. A mouse-adapted SARS-CoV-2 model for the evaluation of COVID-19 medical countermeasures. Microbiology. 2020 doi: 10.1101/2020.05.06.081497. [DOI]
- 21.Shuai H, Chan JF-W, Yuen TT-T, Yoon C, Hu J-C, et al. Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine. 2021;73:103643. doi: 10.1016/j.ebiom.2021.103643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu H, Chen Q, Yang G, He L, Fan H, et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halfmann PJ, Iida S, Iwatsuki-Horimoto K, Maemura T, Kiso M, et al. SARS-CoV-2 omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C, Cui H, Li E, Guo Z, Wang T, et al. The SARS-CoV-2 B.1.351 variant can transmit in rats but not in mice. Front Immunol. 2022;13:869809. doi: 10.3389/fimmu.2022.869809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher AM, Airey G, Liu Y, Gemmell M, Thomas J, et al. The ecology of viruses in urban rodents with a focus on SARS-CoV-2. Emerg Microbes Infect. 2023;12:2217940. doi: 10.1080/22221751.2023.2217940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson SJ, Kotwa JD, Jeeves S, Himsworth C, Pearl D, et al. Surveillance for SARS-CoV-2 in Norway rats (Rattus norvegicus) from Southern Ontario. Transbound Emerg Dis. 2023 doi: 10.22541/au.166214344.47276029/v1. [DOI] [Google Scholar]
- 27.Wang Y, Lenoch J, Kohler D, DeLiberto TJ, Tang CY, et al. SARS-CoV-2 exposure in Norway rats (Rattus norvegicus) from New York city. mBio. 2023;14:e0362122. doi: 10.1128/mbio.03621-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apaa T, Withers AJ, Staley C, Blanchard A, Bennett M, et al. Sarbecoviruses of British horseshoe bats; sequence variation and epidemiology. J Gen Virol. 2023;104 doi: 10.1099/jgv.0.001859. [DOI] [PubMed] [Google Scholar]
- 29.Orłowska A, Smreczak M, Thor K, Niedbalska M, Pawelec D, et al. The genetic characterization of the first detected bat coronaviruses in Poland revealed SARS-related types and alphacoronaviruses. Viruses. 2022;14:1914. doi: 10.3390/v14091914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sander AL, Moreira-Soto A, Yordanov S, Toplak I, Balboni A, et al. Genomic determinants of Furin cleavage in diverse European SARS-related bat coronaviruses. Commun Biol. 2022;5:491. doi: 10.1038/s42003-022-03421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holding M, Otter AD, Dowall S, Takumi K, Hicks B, et al. Screening of wild deer populations for exposure to SARS-CoV-2 in the United Kingdom, 2020-2021. Transbound Emerg Dis. 2022;69:e3244–e3249. doi: 10.1111/tbed.14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreira-Soto A, Walzer C, Czirják GÁ, Richter MH, Marino SF, et al. Serological evidence that SARS-CoV-2 has not emerged in deer in Germany or Austria during the COVID-19 pandemic. Microorganisms. 2022;10:748. doi: 10.3390/microorganisms10040748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wernike K, Fischer L, Holsteg M, Aebischer A, Petrov A, et al. Serological screening in wild ruminants in Germany, 2021/2022: No evidence of SARS‐CoV‐2, bluetongue virus or pestivirus spread but high seroprevalences against Schmallenberg virus. Transbounding Emerging Dis. 2022;69:e3289–e3296. doi: 10.1111/tbed.14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purves K, Brown H, Haverty R, Ryan A, Griffin LL, et al. First Eurasian cases of SARS-CoV-2 seropositivity in a free-ranging urban population of wild fallow deer. bioRxiv. 2023 doi: 10.1101/2023.07.07.547941. [DOI] [Google Scholar]
- 35.Encinas P, Escalera A, Aydillo T, Iglesias I, Nelson MI, et al. SARS-CoV-2 neutralizing antibodies in free-ranging fallow deer (Dama dama) and red deer (Cervus elaphus) in Suburban and rural areas in Spain. Transbound Emerg Dis. 2023;2023:1–11. doi: 10.1155/2023/3324790. [DOI] [Google Scholar]
- 36.Aguiló-Gisbert J, Padilla-Blanco M, Lizana V, Maiques E, Muñoz-Baquero M, et al. First description of SARS-Cov-2 infection in two feral. Animals. 2021;11 doi: 10.3390/ani11051422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padilla-Blanco M, Aguiló-Gisbert J, Rubio V, Lizana V, Chillida-Martínez E, et al. The finding of the severe acute respiratory syndrome coronavirus (SARS-CoV-2) in a wild Eurasian river otter (Lutra lutra) highlights the need for viral surveillance in wild mustelids. Front Vet Sci. 2022;9:826991. doi: 10.3389/fvets.2022.826991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sikkema RS, Begeman L, Janssen R, Wolters WJ, Geurtsvankessel C, et al. Risks of SARS-CoV-2 transmission between free-ranging animals and captive mink in the Netherlands. Transbound Emerg Dis. 2022;69:3339–3349. doi: 10.1111/tbed.14686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davoust B, Guérin P, Orain N, Fligny C, Flirden F, et al. Evidence of antibodies against SARS‐CoV‐2 in wild mustelids from Brittany (France) Transbounding Emerging Dis. 2022;69:e3400–e3407. doi: 10.1111/tbed.14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carmona G, Burgos T, Barrientos R, Martin-Garcia S, Muñoz C, et al. Lack of SARS-CoV-2 RNA evidence in the lungs from wild European polecats (Mustela putorius) from Spain. Eur J Wildl Res. 2023;69:33. doi: 10.1007/s10344-023-01662-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wernike K, Drewes S, Mehl C, Hesse C, Imholt C, et al. No Evidence for the presence of SARS-CoV-2 in bank voles and other rodents in Germany, 2020-2022. Pathogens. 2022;11:1112. doi: 10.3390/pathogens11101112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zamperin G, Festa F, Palumbo E, Quaranta E, Monne I, et al. Discovery of a coronavirus in the Eurasian badger (Meles meles) belonging to a putative new genus. Infect Genet Evol. 2023;109:105406. doi: 10.1016/j.meegid.2023.105406. [DOI] [PubMed] [Google Scholar]
- 43.Woo PCY, Lau SKP, Chu C, Chan K, Tsoi H, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer M, Freuling CM, Müller T, Wegelt A, Kooi EA, et al. Molecular double-check strategy for the identification and characterization of European Lyssaviruses. J Virol Methods. 2014;203:23–32. doi: 10.1016/j.jviromet.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 46.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meleshko D, Hajirasouliha I, Korobeynikov A. coronaSPAdes: from biosynthetic gene clusters to RNA viral assemblies. Bioinformatics. 2021;38:1–8. doi: 10.1093/bioinformatics/btab597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 50.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsoleridis T, Onianwa O, Horncastle E, Dayman E, Zhu M, et al. Discovery of novel alphacoronaviruses in European rodents and shrews. Viruses. 2016;8:84. doi: 10.3390/v8030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Epitools Epitools sample size calculation. 2020. https://epitools.ausvet.com.au/samplesize?page=SampleSize
- 57.O’Rourke E, Hynes J, Losada S, Barber JL, Pereira MG, et al. Anthropogenic drivers of variation in concentrations of perfluoroalkyl substances in otters (Lutra lutra) from England and Wales. Environ Sci Technol. 2022;56:1675–1687. doi: 10.1021/acs.est.1c05410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandoval Barron E, Swift B, Chantrey J, Christley R, Gardner R, et al. A study of tuberculosis in road traffic-killed badgers on the edge of the British bovine TB epidemic area. Sci Rep. 2018;8:17206. doi: 10.1038/s41598-018-35652-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swift BMC, Barron ES, Christley R, Corbetta D, Grau-Roma L, et al. Tuberculosis in badgers where the bovine tuberculosis epidemic is expanding in cattle in England. Sci Rep. 2021;11:20995. doi: 10.1038/s41598-021-00473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas NE, Hailer F, Bruford MW, Chadwick EA. Country-wide genetic monitoring over 21 years reveals lag in genetic recovery despite spatial connectivity in an expanding carnivore (Eurasian otter, Lutra lutra) population. Evol Appl. 2022;15:2125–2141. doi: 10.1111/eva.13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirkland C, Farré M. Mitochondrial genome evolution, genetic diversity, and population structure in British water voles (Arvicola amphibius) Genes. 2021;12:138. doi: 10.3390/genes12020138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keller M, Peter N, Holicki CM, Schantz AV, Ziegler U, et al. SARS-CoV-2 and West Nile virus prevalence studies in raccoons and raccoon dogs from Germany. Viruses. 2022;14:2559. doi: 10.3390/v14112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Villanueva-Saz S, Giner J, Palomar AM, Gómez MA, Põdra M, et al. No evidence of SARS-CoV-2 infection in wild mink (Mustela lutreola and neogale vison) from Northern Spain during the first two years of pandemic. Animals. 2022;12:1971. doi: 10.3390/ani12151971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adney DR, Lovaglio J, Schulz JE, Yinda CK, Avanzato VA, et al. Severe acute respiratory disease in American mink experimentally infected with SARS-CoV-2. JCI Insight. 2022;7:e159573. doi: 10.1172/jci.insight.159573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Griffin BD, Chan M, Tailor N, Mendoza EJ, Leung A, et al. SARS-CoV-2 infection and transmission in the North American deer mouse. Nat Commun. 2021;12:3612. doi: 10.1038/s41467-021-23848-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolters WJ, de Rooij MMT, Molenaar RJ, de Rond J, Vernooij JCM, et al. Manifestation of SARS-CoV-2 infections in mink related to host-, virus- and farm-associated factors, The Netherlands 2020. Viruses. 2022;14:1754. doi: 10.3390/v14081754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wurtzer S, Lacote S, Murri S, Marianneau P, Monchatre-Leroy E, et al. Reduction in SARS-CoV-2 virus infectivity in human and hamster feces. Viruses. 2022;14:1777. doi: 10.3390/v14081777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vandegrift KJ, Yon M, Surendran Nair M, Gontu A, Ramasamy S, et al. SARS-CoV-2 omicron (B.1.1.529) infection of wild white-tailed deer in New York city. Viruses. 2022;14:2770. doi: 10.3390/v14122770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fusco G, Cardillo L, Levante M, Brandi S, Picazio G, et al. First serological evidence of SARS-CoV-2 natural infection in small ruminants : brief report. Vet Res Commun. 2023;47:1741–1748. doi: 10.1007/s11259-022-10044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Project ONW. Orkney Native Wilife Project. 2023. https://www.orkneynativewildlife.org.uk/
- 71.Autieri CR, Miller CL, Scott KE, Kilgore A, Papscoe VA, et al. Systemic coronaviral disease in 5 ferrets. Comp Med. 2015;65:508–516. [PMC free article] [PubMed] [Google Scholar]
- 72.Doria-Torra G, Vidaña B, Ramis A, Amarilla SP, Martínez J. Coronavirus infection in ferrets: antigen distribution and inflammatory response. Vet Pathol. 2016;53:1180–1186. doi: 10.1177/0300985816634809. [DOI] [PubMed] [Google Scholar]
- 73.Lamers MM, Smits SL, Hundie GB, Provacia LB, Koopmans M, et al. Naturally occurring recombination in ferret coronaviruses revealed by complete genome characterization. J Gen Virol. 2016;97:2180–2186. doi: 10.1099/jgv.0.000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lescano J, Quevedo M, Gonzales-Viera O, Luna L, Keel MK, et al. First case of systemic coronavirus infection in a domestic ferret (Mustela putorius furo) in Peru. Transbound Emerg Dis. 2015;62:581–585. doi: 10.1111/tbed.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Minami S, Kuroda Y, Terada Y, Yonemitsu K, Van Nguyen D, et al. Detection of novel ferret coronaviruses and evidence of recombination among ferret coronaviruses. Virus Genes. 2016;52:858–862. doi: 10.1007/s11262-016-1365-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Terada Y, Minami S, Noguchi K, Mahmoud HYAH, Shimoda H, et al. Genetic characterization of coronaviruses from domestic ferrets, Japan. Emerg Infect Dis. 2014;20:284–287. doi: 10.3201/eid2002.130543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vlasova AN, Halpin R, Wang S, Ghedin E, Spiro DJ, et al. Molecular characterization of a new species in the genus Alphacoronavirus associated with mink epizootic catarrhal gastroenteritis. J Gen Virol. 2011;92:1369–1379. doi: 10.1099/vir.0.025353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wills SE, Beaufrère HH, Brisson BA, Fraser RS, Smith DA. Pancreatitis and systemic coronavirus infection in a ferret (Mustela putorius furo) Comp Med. 2018;68:208–211. doi: 10.30802/AALAS-CM-17-000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wise AG, Kiupel M, Garner MM, Clark AK, Maes RK. Comparative sequence analysis of the distal one-third of the genomes of a systemic and an enteric ferret coronavirus. Virus Res. 2010;149:42–50. doi: 10.1016/j.virusres.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Y. Genetic diversity and potential recombination between ferret coronaviruses from European and American lineages. J Infect. 2020;80:350–371. doi: 10.1016/j.jinf.2020.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ip HS, Griffin KM, Messer JD, Winzeler ME, Shriner SA, et al. An opportunistic survey reveals an unexpected coronavirus diversity hotspot in North America. Viruses. 2021;13:2016. doi: 10.3390/v13102016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kwok KTT, de Rooij MMT, Sinartio FF, Smit LAM, Koopmans MPG, et al. Genome sequence of a Minacovirus strain from a farmed mink in The Netherlands. Microbiol Resour Announc. 2021;10:e01451-20. doi: 10.1128/MRA.01451-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen S, Liu D, Tian J, Kang H, Guo D, et al. Molecular characterization of HLJ-073, a recombinant canine coronavirus strain from China with an ORF3abc deletion. Arch Virol. 2019;164:2159–2164. doi: 10.1007/s00705-019-04296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Decaro N, Mari V, Elia G, Addie DD, Camero M, et al. Recombinant canine coronaviruses in dogs, Europe. Emerg Infect Dis. 2010;16:41–47. doi: 10.3201/eid1601.090726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Licitra BN, Duhamel GE, Whittaker GR. Canine enteric coronaviruses: emerging viral pathogens with distinct recombinant spike proteins. Viruses. 2014;6:3363–3376. doi: 10.3390/v6083363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ntafis V, Mari V, Decaro N, Papanastassopoulou M, Papaioannou N, et al. Isolation, tissue distribution and molecular characterization of two recombinant canine coronavirus strains. Vet Microbiol. 2011;151:238–244. doi: 10.1016/j.vetmic.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pratelli A, Tempesta M, Elia G, Martella V, Decaro N, et al. The knotty biology of canine coronavirus: a worrying model of coronaviruses’ danger. Res Vet Sci. 2022;144:190–195. doi: 10.1016/j.rvsc.2021.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y, Ma G, Lu C, Wen H. Detection of canine coronaviruses genotype I and II in raised Canidae animals in China. Berl Munch Tierarztl Wochenschr. 2006;119:35–39. [PubMed] [Google Scholar]
- 89.Lednicky JA, Tagliamonte MS, White SK, Blohm GM, Alam MM, et al. Isolation of a novel recombinant canine coronavirus from a visitor to Haiti: further evidence of transmission of coronaviruses of zoonotic origin to humans. Clin Infect Dis. 2022;75:e1184–e1187. doi: 10.1093/cid/ciab924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vlasova AN, Diaz A, Damtie D, Xiu L, Toh T-H, et al. Novel canine coronavirus Isolated from a hospitalized patient with Pneumonia in East Malaysia. Clin Infect Dis. 2022;74:446–454. doi: 10.1093/cid/ciab456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vlasova AN, Toh T-H, Lee JS-Y, Poovorawan Y, Davis P, et al. Animal alphacoronaviruses found in human patients with acute respiratory illness in different countries. Emerg Microbes Infect. 2022;11:699–702. doi: 10.1080/22221751.2022.2040341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang W, Tian JH, Chen X, Hu RX, Lin XD, et al. Coronaviruses in wild animals sampled in and around Wuhan at the beginning of COVID-19 emergence. Virus Evol. 2022;8:veac046. doi: 10.1093/ve/veac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Freuling CM, Breithaupt A, Müller T, Sehl J, Balkema-Buschmann A, et al. Susceptibility of raccoon dogs for experimental SARS-CoV-2 infection. Emerg Infect Dis. 2020;26:2982–2985. doi: 10.3201/eid2612.203733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rao SS, Parthasarathy K, Sounderrajan V, Neelagandan K, Anbazhagan P, et al. Susceptibility of SARS coronavirus-2 infection in domestic and wild animals: a systematic review. 3 Biotech. 2023;13:5. doi: 10.1007/s13205-022-03416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dong BQ, Liu W, Fan XH, Vijaykrishna D, Tang XC, et al. Detection of a novel and highly divergent coronavirus from asian leopard cats and Chinese ferret badgers in Southern China. J Virol. 2007;81:6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]


