Abstract
Coronatine (COR) is a plasmid-encoded phytotoxin synthesized by several pathovars of phytopathogenic Pseudomonas syringae. The COR biosynthetic gene cluster in P. syringae pv. glycinea PG4180 is encoded by a 32-kb region which contains both the structural and regulatory genes needed for COR synthesis. The regulatory region contains three genes: corP, corS, and corR. corS is thought to function as a histidine protein kinase, whereas corP and corR show relatedness to response regulators of the two-component regulatory paradigm. In the present study, we investigated whether CorR is a positive activator of COR gene expression. We also studied whether CorR specifically binds the DNA region located upstream of cfl, a gene located at the 5′ end of the gene cluster encoding coronafacic acid, the polyketide portion of COR. Complementation analysis with a corR mutant, PG4180.P2, and transcriptional fusions to a promoterless glucuronidase gene (uidA) indicated that CorR functions as a positive regulator of COR gene expression. Deletion analysis of the 5′ end of the cfl upstream region was used to define the minimal region required for COR gene expression. A 360-bp DNA fragment located over 500 bp upstream from the cfl transcriptional start site was used in DNase I protection assays to define the specific bases bound by CorR. An area extending from −704 to −650 with respect to the cfl transcriptional start site was protected by DNase I footprinting, indicating a rather large area of protection. This area was also conserved in the promoter region for cmaA, which encodes a transcript containing genes for coronamic acid synthesis, another intermediate in the COR biosynthetic pathway. The results obtained in the current study suggest that both the coronafacic acid and the coronamic acid structural genes are controlled by CorR, a positive activator of COR gene expression.
The phytotoxin coronatine (COR) is a plasmid-encoded virulence factor synthesized by Pseudomonas syringae pv. glycinea, a pathogen of soybean (3). The structure of COR can be divided into two distinct parts: (i) coronafacic acid (CFA) is of polyketide origin, and (ii) coronamic acid (CMA) is an ethylcyclopropyl amino acid derived from isoleucine (14, 26, 34). Both CFA and CMA function as distinct intermediates and are secreted by COR-producing strains at low levels (27, 48). The final step in the pathway to COR is presumed to be the ligation of CFA and CMA via amide bond formation.
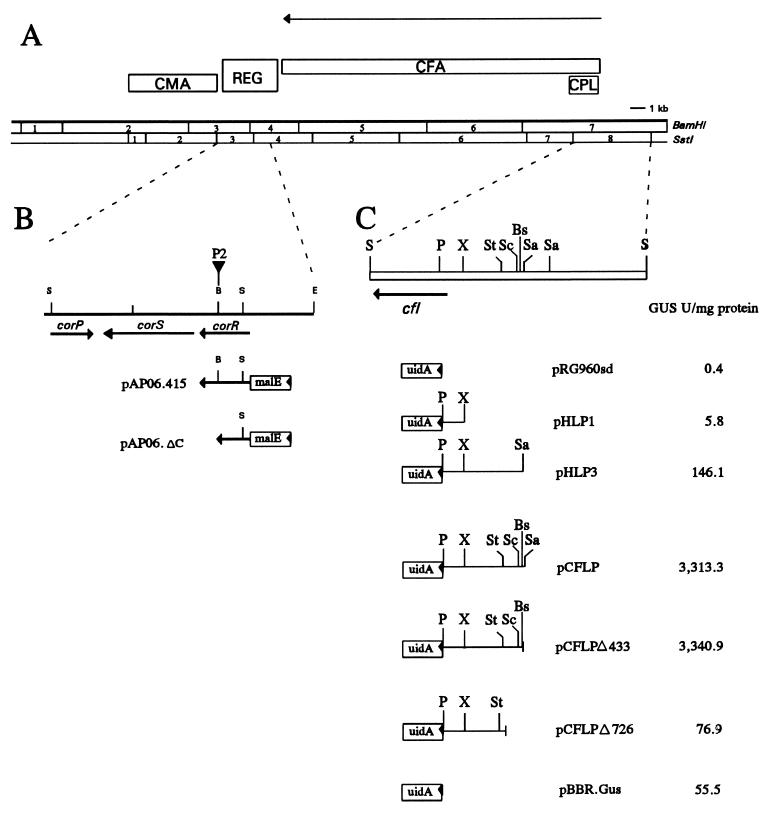
In P. syringae pv. glycinea PG4180, the COR biosynthetic cluster is borne on a 90-kb plasmid designated p4180A (3). Saturation Tn5 mutagenesis, exogenous feeding studies with CFA and CMA, complementation analysis, and nucleotide sequence analysis were used to construct a functional map of the COR biosynthetic region (Fig. 1) (3, 24, 43, 44, 48). Two regions in the COR biosynthetic cluster contain structural genes for CMA and CFA biosynthesis; these are separated by a 3.4-kb regulatory region (Fig. 1) (4). The functional area designated CPL was required for the coupling of CFA and CMA via amide bond formation, a step presumably catalyzed by the cfl gene encoding coronafacate ligase (3, 24).
FIG. 1.
(A) Physical and functional map of the COR biosynthetic gene cluster in P. syringae pv. glycinea PG4180. Functional regions of the COR biosynthetic cluster are shown in the rectangles above the physical map and are abbreviated as follows: CMA, CMA biosynthetic gene cluster; REG, regulatory region; CFA, CFA biosynthetic gene cluster; CPL, coupling region (coupling of CFA and CMA via amide bond formation). The horizontal arrow above the functional map indicates the location of the cfl/CFA operon. (B) Expanded view of the regulatory region that shows the physical location of corP, corR, and corS. The location and orientation of pAP06.415 and pAP06.ΔC, the two malE-corR translational fusions described in the present study, are shown. (C) Physical map of SstI fragment 8 showing the location of the gene encoding coronafacate ligase (cfl) from the translational start site to the translational stop site (arrowhead). The location and orientation of promoter probe constructs used in this study are indicated. Glucuronidase activity for cfl::uidA fusions is shown in the column adjacent to each construct. Restriction enzymes: B, BamHI; Bs, BsrBI; E, EcoRI; P, PstI; S, SstI; Sa, SalI; Sc, ScaI; St, StyI; and X, XbaI.
Sequence analysis of the regulatory region indicated the presence of three genes: corP, corS, and corR (Fig. 1B). The deduced amino acid sequence of corS indicated relatedness to histidine protein kinases which function as environmental sensors, whereas corP and corR showed similarity to response regulators which function as members of two-component regulatory systems (45). Response regulators control the adaptive response in two-component regulatory systems and are characterized by an N-terminal receiver domain which functions as the phosphorylation site and a C-terminal effector domain with a DNA-binding, helix-turn-helix (HTH) motif (32, 33). Both domains are strongly conserved in CorR; CorP, however, contains the highly conserved receiver domain (at least two aspartate residues and a conserved lysine) but lacks the HTH motif. The N-terminal receiver domains of CorR and CorP are almost identical when aligned, suggesting a shared specificity for the same phosphodonor protein(s).
The CFA biosynthetic gene cluster was previously shown to be encoded by a single, 18.8-kb transcript; the cfl gene mapped at the 5′ end of the transcript (23). Previous results indicated that the CFA biosynthetic gene cluster was regulated by corRPS; for example, mutants defective in corR, corP, or corS were defective in CFA biosynthesis, and expression of a cfl::uidA transcriptional fusion required functional copies of each regulatory gene (23).
Both CorR and CorP showed significant sequence relatedness to response regulators in the ROIII group (33), which includes NarL, BvgA, and FixJ. Several of these response regulators function as positive activators of transcription, and some bind to specific target sequences upstream of the promoters they regulate (1, 5, 7). In the present study, we investigated whether CorR is a positive activator of COR gene expression and demonstrated specific binding of this protein to the cfl promoter. We also investigated whether the C-terminal portion of CorR is required for binding.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Pseudomonas strains were routinely cultured on King’s medium B (19) or mannitol-glutamate medium (17) at 24 to 26°C. Escherichia coli DH5α (39) was used as a host in cloning experiments and was cultured in Terrific Broth or Luria-Bertani medium at 37°C (39). Protein contents of cell lysates were determined with the Bio-Rad (Richmond, Calif.) protein assay kit as recommended by the manufacturer. The following antibiotics were added to media in the indicated concentrations (μg/ml): tetracycline, 12.5; kanamycin, 12.5; ampicillin, 40; spectinomycin, 25; streptomycin, 25; chloramphenicol, 12.5; and gentamicin, 2.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli DH5α | 39 | |
| P. syringae pv. glycinea | ||
| PG4180.N9 | COR+; Kmr | 44 |
| PG4180.P2 | COR−; corR::Gmr | 35 |
| Plasmids | ||
| pAP06.415 | Apr Tcr; contains corR; chimeric plasmid constructed from pAP06 and pRK415; 17.7 kb | 35 |
| pAP06.ΔC | Apr Tcr; a 242-bp deletion was introduced into the C terminus of corR; derived from pAP06.415 | This study |
| pMAL-c2 | Apr; ColE1 origin, tac promoter, encodes malE and lacZα; contains factor Xa cleavage site | New England Biolabs |
| pRK415 | Tcr; RK2-derived cloning vector | 18 |
| pBluescript SK(+) | Apr; ColE1 origin, cloning vehicle | Stratagene |
| pRG960sd | Smr Spr; contains promoterless uidA with start codon and Shine-Dalgarno sequence | 46 |
| pHLP1 | Smr Spr; 0.46-kb XbaI-PstI fragment from pHL1 in pRG960sd (XbaI-PstI-uidA) | 24 |
| pHLP3 | Smr Spr; 1.4-kb SalI-PstI fragment from pHL1 in pRG960sd | 23 |
| pBBR1MCS | Cmr; 4.7-kb broad-host-range cloning vector | 20 |
| pCFLP | Cmr; 8-kb promoter probe vector containing the cfl promoter as a PstI-SalI fragment and the uidA gene from pHLP3 in pBBR1MCS | This study |
| pCFLPΔ433 | Cmr; derivative of pCFLP containing a 433-bp deletion from the 5′ end (SalI site) of the cfl promoter | This study |
| pCFLPΔ726 | Cmr; derivative of pCFLP containing a 726-bp deletion from the 5′ end (SalI site) of the cfl promoter | This study |
| pBBR.Gus | Cmr; 6.6-kb promoter probe vector containing the uidA gene from pHLP3 in pBBR1MCS | This study |
| pAPXS9 | Apr; 950-bp XbaI-SalI fragment from pHLP3 in pBluescript SK(+) | This study |
| pAPSS27 | Apr; 278-bp StyI-ScaI fragment from pAPXS9 in pBluescript SK(+) | This study |
| pAPSB36 | Apr; 360-bp StyI-BsrBI fragment from pAPXS9 in pBluescript SK(+) | This study |
DNA procedures.
Electrophoresis, purification of DNA fragments from agarose gels, and small-scale plasmid DNA preparations were performed by standard procedures (39). Selected constructs were transformed into P. syringae by electroporation as described previously (39).
Construction of transcriptional fusions.
pCFLP, which contains a cfl::uidA transcriptional fusion in pBBR1MCS, was constructed using pHLP3 as a source of the cfl promoter and the uidA gene. pHLP3 was digested with SalI and SstI to release the 3.27-kb fragment containing the cfl promoter and the uidA gene. This fragment was then ligated into pBBR1MCS digested with SalI and SstI, and the ligation mixture was transformed into E. coli DH5α. Transformants were selected on LB agar containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and chloramphenicol; pCFLP contained the cfl::uidA fusion in the transcriptionally active orientation. In another experiment, the uidA gene was excised from pHLP3 with PstI and SstI and directionally cloned into the polylinker site of pBBR1MCS to create pBBR.Gus.
Deletion analysis.
Exonuclease III (ExoIII) was used to determine the minimal size of the cfl promoter. pCFLP was digested with SalI and SphI, which generate ExoIII-sensitive and ExoIII-resistant sites, respectively. Staggered deletions in the cfl promoter region were generated using the protocols supplied with the Erase-a-Base kit (Promega, Madison, Wis.). Transcriptional fusions were then transformed into PG4180.N9 and assayed for glucuronidase activity as described below.
Glucuronidase assays.
Transcriptional activity was initially assayed by spotting bacterial suspensions (A600 of 0.1) onto mannitol-glutamate plates containing 20 μg of X-Gluc (5-bromo-4-chloro-indolyl glucuronide) per ml, followed by incubation at 18°C for 2 to 3 days. Glucuronidase activity was quantified by fluorometric analysis of cells grown for 48 h in 10 ml of Hoitink-Sinden medium optimized for COR production (HSC) at 18°C (30, 31). Fluorescence was monitored with a Fluoroscan II version 4.0 microplate reader (ICN Biomedicals, Inc., Costa Mesa, Calif.) in 96-well microtiter plates. Glucuronidase activity was expressed in units per milligram of protein, with 1 U being equivalent to 1 nmol of methylumbelliferone formed per min. The values presented for glucuronidase activity represent the average of two experiments with three replicates per experiment.
Production and purification of fusion proteins.
Overproduction of fusion proteins was first evaluated in E. coli DH5α containing selected constructs. Cells were grown at 37°C in Terrific Broth to an optical density at 600 nm (OD600) of 0.4 to 0.5, induced with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and incubated an additional 3 h. Aliquots of cells (1 ml) were removed before and after induction, pelleted by centrifugation, resuspended in lysis buffer (39), and incubated on ice for 30 min. The cell suspension was then sonicated as described previously (38) and centrifuged at 14,000 × g for 20 min at 4°C. The pellet was discarded, and the supernatant (which contained the soluble fraction of the crude extract) was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% polyacrylamide gel (39).
For routine analysis, fusion proteins were overproduced in P. syringae pv. glycinea PG4180.N9 by growing the cells at 18°C in TB to an OD600 of 0.4 to 0.5, inducing them with 5.0 mM IPTG, and incubating them an additional 6 h. Aliquots of cells (1 ml) were removed before and after the induction period, and total cellular proteins were analyzed as described above. Fusion proteins were purified from P. syringae by pelleting 100 ml of IPTG-induced cells, suspending them in 8 ml of lysis buffer, and then incubating them on ice for 30 min. The soluble fraction of the crude cellular extract was recovered as described above and applied to the amylose resin as described previously (38).
Maltose binding protein (MBP)-CorR fusion proteins used in gel retardation and DNase I footprinting analyses were isolated from PG4180.N9 cells (30 ml) grown as described above but for 15 h after induction with 5.0 mM IPTG. Subsequent steps were performed at 0 to 4°C in TEDG buffer (50 mM Tris [pH 7.5], 0.5 mM EDTA, 2 mM dithiothreitol, 10% [wt/vol] glycerol). Cells were harvested by centrifugation (5,000 × g, 1 min), supernatants were discarded, and cells were washed in 20 ml of TEDG buffer and collected by centrifugation. Cells were then resuspended in 10 ml of TEDG and lysed by sonication. Lysates were centrifuged at 23,000 × g for 10 min at 4°C; supernatants were then collected and transferred into a clean centrifuge tube to which amylose resin (50 μl) was added. The mixture was then centrifuged for 10 s, and the supernatant was discarded. The resin was washed with 1 ml of column buffer (20 mM Tris-HCl, 0.2 M NaCl, 1 mM EDTA, 10 mM mercaptoethanol, and 1 mM sodium azide), and centrifuged for 10 s. Then, 100 μl of 10 mM maltose was added to elute the fusion protein, and the reaction mixture was centrifuged for 10 s. The supernatant was used in all subsequent assays.
The concentration of MBP-CorR was evaluated by loading different volumes of the fusion protein to a polyacrylamide gel containing known amounts of bovine serum albumin. The gel was analyzed, and MBP-CorR concentrations were determined by using the Bio-Rad PhosphorImager system, the GS-700 densitometer, and the Molecular Analyst software (version 2.1).
Gel shift assays.
To facilitate end labeling with [α-32P]dATP, DNA fragments used for gel retardation were subcloned into pBluescript SK(+) and excised with enzymes which generate 5′ overhanging ends. DNA fragments were then separated from vector DNA on 5% polyacrylamide gels and end labeled with [α-32P]dATP (39).
Gel retardation assays were performed by incubating 20 nM purified fusion protein with 2,000 cpm of end-labeled probe in binding buffer containing 10 mM Tris-HCl (pH 7.5), 10 mM KCl, 1 mM EDTA (pH 8.0), 1 mM dithiothreitol, 10% glycerol, and 1 μg of poly(dI-dC). After 20 min on ice, 2 μl of loading buffer (binding buffer supplemented with 0.4% bromophenol blue and 1% glycerol) was added, and the samples were loaded onto a 5% polyacrylamide gel. After electrophoresis, the gels were either dried and autoradiographed or scanned with a Bio-Rad GS-525 Molecular Imager.
DNase I footprinting.
Footprinting was carried out on both strands of a DNA fragment containing a 360-bp fragment isolated from the cfl upstream region. pHLP3 was digested with XbaI and SalI, and the 950-bp fragment was subcloned in pBluescript SK(+) to generate pAPXS9. Plasmid pAPXS9 was then digested with StyI and SalI, and the 778-bp fragment containing the coding strand was isolated by electrophoresis (5% polyacrylamide) and electroelution. This fragment was then digested with BsrBI, and the 360-bp StyI/BsrBI fragment was cloned into the SmaI site of pBluescript SK(+), resulting in pAPSB36. Plasmid pAPSB36 was then digested with BamHI and EcoRI (sites flanking SmaI in pBluescript), and the 381-bp BamHI/EcoRI fragment was isolated as described above. Both the coding and noncoding fragments (1 μg each) were end labeled with [α-32P]dCTP and [α-32P]dGTP (for coding strand) and [α-32P]dATP (for noncoding strand) with Klenow polymerase. Reactions were incubated at 37°C for 30 min, and unincorporated deoxynucleoside triphosphates were removed by chromatography through Sephadex G-50 columns (39).
For DNase I footprinting experiments, the procedure of Leblanc and Moss (21) was used. Labeled DNA fragments (ca. 300,000 cpm) were incubated for 30 to 40 min on ice with various amounts of MBP-CorR, binding buffer (21), and 1 μg of poly(dI-dC) in a final volume of 10 μl. After the binding reaction, 75 U of RNase-free DNase I (Gibco BRL) was added to the mixture, and the reaction proceeded for 1 min at room temperature (25°C). The reaction was then terminated by the addition of 5 μl of loading buffer (7 M urea, 0.05% xylene cyanol, and 0.05% bromophenol blue). Samples were heated to 100°C briefly and then loaded (15,000 cpm/lane) onto 8% polyacrylamide gels.
A G+A sequencing ladder was prepared by cleaving end-labeled DNA (300,000 cpm) with 1 M pyridine formate (pH 2.0). The ladder was prepared as described previously (21), and the cleaved DNA (15,000 cpm/lane) was loaded onto the gel used for DNase I footprinting.
Analysis of the C-terminal region of CorR.
The corR mutant PG4180.P2 contains a gentamicin resistance (Gmr) cassette cloned into the BamHI site of corR (35). Insertion of the Gmr cassette at this position disrupts the HTH motif at the C terminus of CorR. Our previous analysis indicated that PG4180.P2 was totally defective in the biosynthesis of COR, indicating that the carboxyl terminus of CorR is required for functional activity. The importance of the C-terminal region of CorR was investigated in the present study by constructing a mutant fusion protein designated MBP::CorRΔC. The truncated version of CorR was constructed by digesting pAP06.415 with BamHI, excising the fragment which contains the C-terminal region, and religating this construct to produce pAP06.ΔC (Fig. 1B). This construct was used to investigate the role of the C-terminal portion of CorR in transcriptional activation and DNA binding.
Detection of COR.
P. syringae strains were grown at 18°C in HSC medium, and supernatants were analyzed for COR production by high-pressure liquid chromatography 7 days after inoculation (30). When the objective was to determine the effect of a selected fusion protein on COR production, cells were induced with 5 mM IPTG 24 h after inoculation into HSC medium. Each strain was inoculated to three replicate aliquots (10 ml) of HSC medium for evaluation of COR production, and each experiment was repeated.
RESULTS
CorR is a positive regulator of COR gene expression.
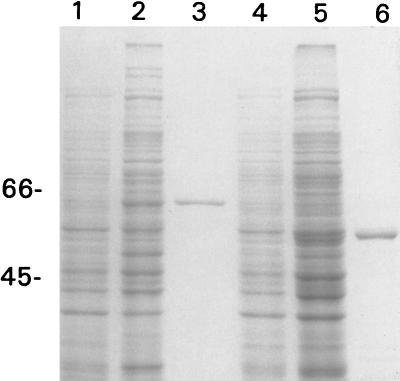
We previously showed that the tac promoter and the lac repressor (encoded by lacIq) could be used in P. syringae for the controllable production of translational fusions to MBP (35). The construction of pAP06.415, a chimeric plasmid consisting of pRK415 and a malE-corR fusion in pMal-c2, was described previously (35). When PG4180.N9(pAP06.415) cells were induced with IPTG, a 64-kDa protein was observed which corresponds to the predicted size of a fusion protein consisting of MBP (42.7 kDa) and CorR (21.5 kDa) (Fig. 2, lane 2). Furthermore, this fusion protein could be partially purified from PG4180.N9 by affinity chromatography on an amylose column (Fig. 2, lane 3).
FIG. 2.
SDS-PAGE analysis of P. syringae pv. glycinea PG4180.N9 containing pAP06.415 and pAP06.ΔC. Lanes 1, 2, 4, and 5 show total cellular proteins from the following strains: lane 1, PG4180.N9(pAP06.415), uninduced; lane 2, PG4180.N9(pAP06.415), induced with IPTG; lane 4, PG4180.N9(pAP06.ΔC), uninduced; and lane 5, PG4180.N9(pAP06.ΔC), induced with IPTG. Lanes 3 and 6 show the MBP-CorR and MBP-CorRΔC fusions which were isolated from induced cells of PG4180.N9(pAP06.415) and PG4180.N9(pAP06.ΔC), respectively, and were purified by affinity chromatography on amylose resin. The migration of the molecular weight markers is shown on the left; numbers indicate the molecular mass in kilodaltons.
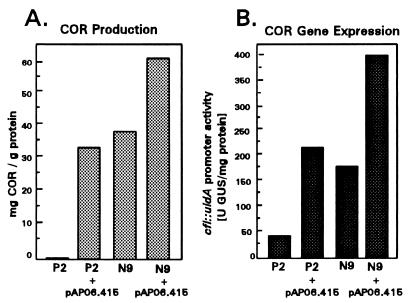
To assess the function of the MBP-CorR fusion in vivo, we investigated whether pAP06.415 could restore COR production to PG4180.P2, a corR mutant completely defective in COR production (35). PG4180.P2(pAP06.415) cells produced 31 mg of COR/g of protein, a level comparable to the COR-producing PG4180.N9, indicating that the MBP-CorR fusion was fully functional in vivo (Fig. 3A). We also investigated whether the overproduction of MBP-CorR in PG4180.N9 could increase COR production; PG4180.N9(pAP06.415) cells induced with IPTG produced 58.9 mg of COR/g of protein, a quantity significantly higher (P = 0.05) than the amount produced by the wild-type PG4180.N9 (Fig. 3A).
FIG. 3.
Effect of genetic background on COR production (A) and the transcriptional activity of the cfl::uidA transcriptional fusion contained in pHLP3 (B). Abbreviations: P2, PG4180.P2, a corR mutant of P. syringae pv. glycinea; N9, PG4180.N9, a COR-producing strain of P. syringae pv. glycinea. MBP-CorR was overproduced by introducing pAP06.415 and adding 5 mM IPTG as described in the text.
These results suggested that CorR might function as a positive regulator of COR gene expression. This hypothesis was investigated by overproducing MBP-CorR in PG4180.P2 and PG4180.N9 containing pHLP3; the construct pHLP3 contains the cfl promoter fused to a promoterless glucuronidase gene (uidA) (23). When induced with IPTG, PG4180.P2(pAP06.415, pHLP3) produced 213 U of glucuronidase/mg of protein, a level approximately fivefold higher than for PG4180.P2(pHLP3) (Fig. 3B). Furthermore, overproduction of MBP-CorR in PG4180.N9(pHLP3, pAP06.415) resulted in 395 U of glucuronidase/mg of protein, a level significantly higher than glucuronidase activity in PG4180.N9(pHLP3) (Fig. 3B). These results confirmed the role of CorR as a positive activator of the cfl transcript.
Analysis of the cfl promoter region.
Previous work indicated that a 311-bp region upstream of the cfl transcriptional start site contained promoter activity (24). This was shown with pHLP1 (Fig. 1C), a construct containing the 311-bp region upstream of the promoterless uidA gene in pRG960sd. In a subsequent study, Liyanage et al. (23) showed that PG4180(pHLP3) produced approximately 30 times more glucuronidase than PG4180(pHLP1) (Fig. 1C). Since pHLP3 contained an additional 950 bp that are not present in pHLP1 (Fig. 1C), this result indicated that additional DNA upstream of the XbaI site was needed for a full level of transcriptional activation.
One explanation for the differential level of transcriptional activation between pHLP1 and pHLP3 was the possible occurrence of a CorR-binding site in pHLP3 which was absent in pHLP1. Therefore, we initially examined two fragments for their ability to bind purified MBP-CorR. These were the 460-bp PstI-XbaI fragment contained in pHLP1 and the 950-bp XbaI-SalI fragment contained in pHLP3. When the 460-bp PstI-XbaI fragment was used as target DNA, no gel retardation was observed regardless of the amount of MBP-CorR utilized (data not shown). In contrast, migration of the 950-bp XbaI-SalI fragment was retarded when incubated with 150 and 200 ng of MBP-CorR (data not shown). The specificity of complex formation between MBP-CorR and the 950-bp XbaI-SalI fragment was investigated by adding increasing amounts of the unlabeled XbaI-SalI fragment to the reaction mixture. When cold fragment was added as a competitor in amounts of 125 ng or higher, binding was either significantly reduced or completely abolished. In comparison, when poly(dI-dC) was added to the reaction mixture, binding was either not affected or was reduced only slightly (data not shown). These results indicate that MBP-CorR specifically binds the 950-bp XbaI-SalI fragment.
To determine the minimum sequence necessary for cfl expression and MBP-CorR binding, we constructed a series of deletions from the 5′ (SalI) end of the cfl upstream DNA. A new construct, pCFLP (Fig. 1C), was designed for this purpose since the pBBR1MCS polylinker in pCFLP was more amenable to deletion analysis than was the multicloning site in pRG960sd, the vector used for construction of pHLP3. Although pCFLP and pHLP3 contain the same cfl::uidA fusion, pCFLP was 21 times more active when assayed for glucuronidase activity (Fig. 1C); this discrepancy is most likely due to the higher copy number of pBBR1MCS. Two deletion derivatives of pCFLP, pCFLPΔ433 and pCFLPΔ726, proved useful for delineating the cfl promoter region; sequence analysis indicated that these two constructs lacked 433 and 726 bp of DNA downstream of the SalI site, respectively. PG4180.N9(pCFLPΔ433) (Fig. 1C) retained the full level of glucuronidase activity exhibited by PG4180.N9(pCFLP), suggesting that a 433-bp region downstream of the SalI site was dispensable for promoter activity. However, glucuronidase activity in PG4180.N9(pCFLPΔ726) was 43-fold lower than in PG4180.N9(pCFLPΔ433), demonstrating that deletion of an additional 293 bp from the 5′ end of pCFLPΔ433 virtually eliminated cfl promoter activity (Fig. 1C).
Gel retardation assays.
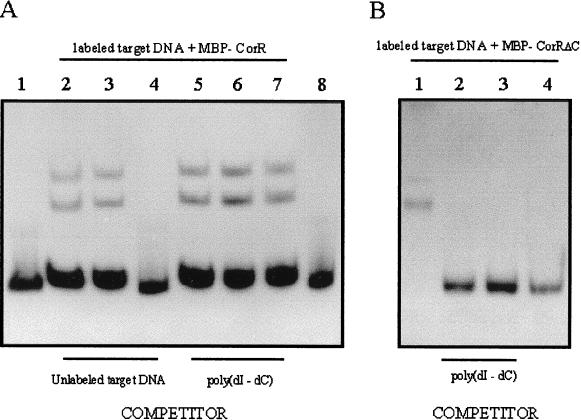
The results described above suggested that the 278-bp region located between the StyI and ScaI sites in the cfl promoter might bind MBP-CorR. This fragment was isolated from pAPSS27, and competition assays were conducted with MBP-CorR, the unlabeled StyI-ScaI fragment, and poly(dI-dC). When MBP-CorR was omitted from the reaction, no gel retardation was observed (Fig. 4A, lanes 1 and 8). Gel retardation was observed when MBP-CorR was incubated with the labeled StyI-ScaI fragment (Fig. 4A, lane 2). Although migration was not inhibited when 1 ng of cold, unlabeled fragment was added to the reaction mixture (Fig. 4A, lane 3), complete inhibition was observed with 20 ng of the unlabeled StyI-ScaI fragment (Fig. 4A, lane 4). The addition of 0, 50, and 100 ng of poly(dI-dC), which was used as a nonspecific competitor, did not inhibit gel retardation (Fig. 4A, lanes 5 to 7). These results indicated that the 278-bp StyI-ScaI fragment specifically binds MBP-CorR.
FIG. 4.
(A) Competition assays with the MBP-CorR fusion and the 278-bp StyI-ScaI fragment. Lanes 1 and 8 show approximately 20 ng of end-labeled target DNA (278-bp StyI-ScaI fragment) and 0 ng of MBP-CorR. Lanes 2 to 7 contain the target DNA fragment and 40 ng of purified MBP-CorR. Specific inhibition of binding was investigated by adding the following amounts of unlabeled target fragment: lane 2, 0 ng; lane 3, 1 ng; and lane 4, 20 ng. The addition of the nonspecific competitor poly(dI-dC) is shown in lanes 5 to 7, which contain 0, 50, and 100 ng of poly(dI-dC), respectively. (B) Competition assays with MBP-CorRΔC fusions and the 278-bp StyI-SacI fragment. Lanes 1 to 3 contain 20 ng of target DNA and 40 ng of MBP-CorRΔC, as well as 0, 100, and 250 ng of poly(dI-dC), respectively. Lane 4 contains 20 ng of end-labeled target DNA (278-bp StyI-ScaI fragment) and 0 ng of MBP-CorRΔC.
DNase I protection assays.
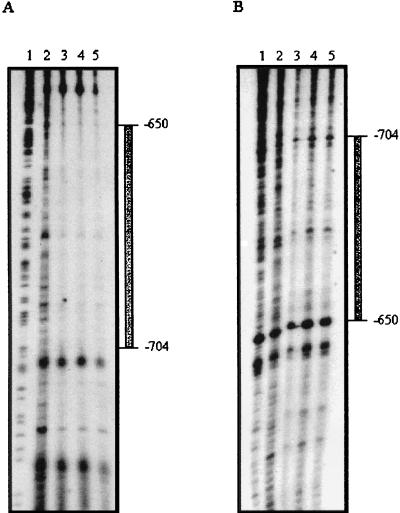
DNase I footprint experiments were performed to establish the precise location of the CorR-binding site within the cfl promoter. The StyI-BsrBI fragment contained in plasmid pAPSB36 was used to examine the upper and lower strands of the cfl promoter, respectively. Comparison of the sequence patterns produced in the absence or presence of MBP-CorR demonstrated protected regions of 54 bp on both the top and bottom strands (Fig. 5). The binding sites extended from position −704 to −650 on the top strand and from position −650 to −704 on the bottom strand relative to the cfl transcriptional start site (Fig. 5). These results were reproduced in several different gels, indicating a fairly large region of protection on both strands.
FIG. 5.
DNase I footprinting of the cfl promoter region contained in the 360-bp StyI-BsrBI fragment with the MBP-CorR fusion protein. DNase I protection assays of the upper strand (A) and lower strand (B) were carried out as described in Materials and Methods. Lanes: 1, G+A sequencing ladder; 2, DNase I only; 3, DNase I plus 20 nM MBP-CorR; 4, DNase I plus 40 nM MBP-CorR; and 5, DNase I plus 80 nM MBP-CorR.
Role of the C-terminal region of CorR in DNA binding.
DH5α(pAP06.ΔC) cells induced with IPTG produced a 55-kDa fusion protein, which is consistent with the deletion of approximately 8.8 kDa from the C terminus of CorR (data not shown). Introduction of this construct into PG4180.N9 and induction with IPTG produced similar results; a 55-kDa fusion protein was produced in the induced cells (Fig. 2, lane 5) and could be purified on amylose resin (Fig. 2, lane 6).
The StyI-ScaI fragment obtained from pAPSS27 was used to investigate the DNA binding ability of an MBP fusion lacking the carboxyl terminus of CorR (MBP-CorRΔC). In this experiment, some retardation was observed when MBP-CorRΔC was incubated with the labeled DNA fragment (Fig. 4B, lane 1). However, this interaction was shown to be weak since the addition of poly(dI-dC) completely inhibited gel retardation (Fig. 4B, lanes 3 and 4). These results suggest that the C-terminal portion of CorR is required for specific binding to the cfl upstream region.
DISCUSSION
CorR was previously shown to be required for transcription of the cfl/CFA operon (23). Furthermore, the relatedness of CorR to response regulators in the ROIII group suggested that CorR might function as a positive activator of COR gene expression and bind to the cfl promoter region. The experiments described here confirmed these hypotheses and demonstrated the DNA-binding ability of CorR and the specific region bound by CorR in the cfl upstream region.
Numerous reports exist where MBP translational fusions have been used to investigate the DNA binding function of regulatory proteins (5, 10, 22). Translational fusions to MBP often increase the solubility of regulatory proteins and have little or no effect on protein function in vivo (6, 10, 11, 22). In the present study, the production of COR and the transcriptional activation of cfl were restored to the corR mutant PG4180.P2 by pAP06.415, a construct containing a malE-corR translational fusion. Since the fusion protein was active in vivo, MBP-CorR was used to facilitate purification of CorR for subsequent DNA binding studies.
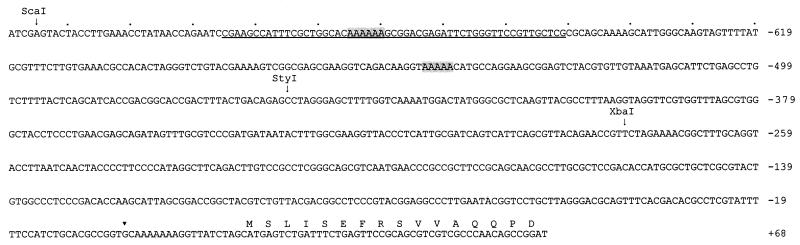
The area protected by MBP-CorR was located at position −704 to −650 with respect to the cfl transcriptional start site. Other regulatory proteins have been shown to bind regions far upstream relative to the start point for transcription (13, 16, 28, 37). In several of these interactions, DNA bending is thought to occur, and this may facilitate interaction between the regulatory protein and RNA polymerase at the transcriptional start site (9, 13). Several accessory elements are known to induce DNA bending, including the cyclic AMP receptor protein and histone-like proteins, such as integration host factor (36). The nucleotide sequence of the region bound by MBP-CorR showed no evidence that specific DNA bending proteins were involved. However, this region did contain two poly(A) tracts at positions −684 to −679 and positions −552 to −548 (Fig. 6). Deoxyadenylate tracts are known to introduce curvature into DNA sequences and may facilitate the interaction between regulatory proteins and the RNA polymerase complex (12, 41). It is also important to note that the A+T content in the region protected by MBP-CorR was 50%, a figure substantially higher than the 37% A+T in the cfl coding region. The high percentage of A+T in the protected region may facilitate transcriptional activation and DNA bending, a hypothesis suggested for A+T-rich regions bound by other transcriptional activators (8, 25).
FIG. 6.
Sequence of the cfl upstream region (GenBank accession number U09027). Underscored bases represent the nucleotides protected by DNase I. Shaded regions indicate poly(A) tracts; the inverted triangle shows the transcriptional start site for cfl. Numbers on the right represent the nucleotide position relative to the cfl transcriptional start site.
Many response regulators in the two-component paradigm contain two distinct regions, a receiver domain containing conserved aspartyl and lysine residues and an output domain containing an HTH motif (33). PG4180.P2 contains a Gmr cassette within corR in a location which disrupts the output domain of the translational product. Although PG4180.P2 was completely defective in COR production, transcriptional activity in PG4180.P2(pHLP3) was not completely abolished (Fig. 3B). These results indicate that the receiver domain is still functional and that some transcriptional activation can occur in the absence of the output domain. A deletion derivative of CorR, MBP-CorRΔC, was overproduced and was used in gel retardation studies; these experiments showed that removal of the predicted output domain of CorR reduced specific binding to the cfl promoter. The C-terminal region of CorR contains a well-defined HTH domain that may be responsible for specific binding to thermosensitive promoters in the COR gene cluster. The present study did not address whether the HTH motif in CorR functions as part of a larger DNA-binding domain as it does in some response regulators (5, 29).
The cmaA promoter, which transcribes the structural genes for CMA biosynthesis (43), also requires corR for functional activity (45). Progressive subcloning in pRG960sd indicated that a 265-bp region located at positions −721 to −456 with respect to the cmaA transcriptional start site was required for cmaA transcriptional activity (43). When the cfl upstream region protected by DNase I was aligned with the 5′ end of the cmaA promoter (43), a 40% identity was observed over a 60-bp region (Fig. 7). The high percentage similarity between the two promoter regions and the conserved location with respect to the transcriptional start sites suggest that CorR may bind to this region in the cmaA promoter. This hypothesis is currently being investigated and will provide further insight into the regulation of thermosensitive transcripts in the COR gene cluster.
FIG. 7.
Alignment of the cfl upstream region protected by DNase I with the cmaA promoter-regulatory region. Shaded bases indicate regions of nucleotide identity; dashes were inserted to maximize the alignment. Numbers on the right indicate the nucleotide position relative to the transcriptional start sites for cmaA (upper sequence) and cfl (lower sequence).
In many instances, phosphorylated response regulators bind more efficiently to their target sequences than does the nonphosphorylated form (2, 40, 47). However, nonphosphorylated regulators often retain some level of DNA binding ability (5, 15, 42). The phosphorylation state of MBP-CorR was not investigated in the present study; however, the fusion was purified from PG4180.N9, which contains a functional copy of the cognate histidine protein kinase, CorS. We are currently investigating whether autophosphorylation of CorS and subsequent phosphotransfer to CorP and CorR is correlated with COR gene induction.
ACKNOWLEDGMENTS
We thank F. Alarcón-Chaidez for help with the graphics; M. Ullrich for stimulating discussions; and V. Rangaswamy, L. Keith, and F. Alarcón-Chaidez for reviewing the manuscript.
This work was supported by the Oklahoma Agricultural Experiment Station and by NSF grant MCB-9603618.
REFERENCES
- 1.Agron P G, Monson E K, Ditta G S, Helinski D R. Oxygen regulation of expression of nitrogen fixation genes in Rhizobium meliloti. Res Microbiol. 1994;145:454–459. doi: 10.1016/0923-2508(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 2.Arthur M, Depardieu F, Gerbaud G, Galmand M, Leclercq R, Courvalin P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J Bacteriol. 1997;179:97–106. doi: 10.1128/jb.179.1.97-106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender C L, Liyanage H, Palmer D, Ullrich M, Young S, Mitchell R. Characterization of the genes controlling biosynthesis of the polyketide phytotoxin coronatine including conjugation between coronafacic and coronamic acid. Gene. 1993;133:31–38. doi: 10.1016/0378-1119(93)90221-n. [DOI] [PubMed] [Google Scholar]
- 4.Bender C, Palmer D, Peñaloza-Vázquez A, Rangaswamy V, Ullrich M. Biosynthesis of coronatine, a thermoregulated phytotoxin produced by the phytopathogen Pseudomonas syringae. Arch Microbiol. 1996;166:71–75. [Google Scholar]
- 5.Boucher P E, Menozzi F D, Locht C. The modular architecture of bacterial response regulators. Insights into the activation mechanism of the BvgA transactivator of Bordetella pertussis. J Mol Biol. 1994;241:363–377. doi: 10.1006/jmbi.1994.1513. [DOI] [PubMed] [Google Scholar]
- 6.Darwin A J, Stewart V. Nitrate and nitrite regulation of the Fnr-dependent aeg-46.5 promoter of Escherichia coli K-12 is mediated by competition between homologous response regulators (NarL and NarP) for a common DNA-binding site. J Mol Biol. 1995;251:15–29. doi: 10.1006/jmbi.1995.0412. [DOI] [PubMed] [Google Scholar]
- 7.Dong X-R, Li S-F, DeMoss J A. Upstream sequence elements required for NarL-mediated activation of transcription from the narGHJI promoter of Escherichia coli. J Biol Chem. 1992;267:14122–14128. [PubMed] [Google Scholar]
- 8.Foster-Hartnett D, Cullen P J, Monika E M, Kranz R G. A new type of NtrC transcriptional activator. J Bacteriol. 1994;176:6175–6187. doi: 10.1128/jb.176.20.6175-6187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiwara S, Zielinski N A, Chakrabarty A M. Enhancer-like activity of AlgR1-binding site in alginate gene activation: position, orientational, and sequence specificity. J Bacteriol. 1993;175:5452–5459. doi: 10.1128/jb.175.17.5452-5459.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grob P, Guiney D G. In vitro binding of the Salmonella dublin virulence plasmid regulatory protein SpvR to the promoter regions of spvA and spvR. J Bacteriol. 1996;178:1813–1820. doi: 10.1128/jb.178.7.1813-1820.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han D C, Winans S C. A mutation in the receiver domain of the Agrobacterium tumefaciens transcriptional regulator VirG increases its affinity for operator DNA. Mol Microbiol. 1994;12:23–30. doi: 10.1111/j.1365-2958.1994.tb00991.x. [DOI] [PubMed] [Google Scholar]
- 12.Harrington R E. DNA curving and bending in protein-DNA recognition. Mol Microbiol. 1992;6:2549–2555. doi: 10.1111/j.1365-2958.1992.tb01431.x. [DOI] [PubMed] [Google Scholar]
- 13.Huang K-J, Schieberl J L, Igo M M. A distant upstream site involved in the negative regulation of the Escherichia coli ompF gene. J Bacteriol. 1994;176:1309–1315. doi: 10.1128/jb.176.5.1309-1315.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ichihara A, Shiraishi K, Sato H, Sakamura S, Nishiyama K, Sakai R, Furusaki A, Matsumoto T. The structure of coronatine. J Am Chem Soc. 1977;99:636–637. [Google Scholar]
- 15.Jin S, Roitsch T, Christie P J, Nester E W. The regulatory VirG protein specifically binds to a cis-acting regulatory sequence involved in transcriptional activation of Agrobacterium tumefaciens virulence genes. J Bacteriol. 1990;172:531–537. doi: 10.1128/jb.172.2.531-537.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato J, Chakrabarty A M. Purification of the regulatory protein AlgR1 and its binding in the far upstream region of the algD promoter in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1991;88:1760–1764. doi: 10.1073/pnas.88.5.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keane P J, Kerr A, New P B. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust J Biol Sci. 1970;23:585–595. [Google Scholar]
- 18.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 19.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 20.Kovach M E, Phillips R W, Elzer P H, Roop III R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 21.Leblanc B, Moss T. DNase I footprinting. Methods Mol Biol. 1994;30:1–10. doi: 10.1385/0-89603-256-6:1. [DOI] [PubMed] [Google Scholar]
- 22.Lee H-S, Berger D K, Kustu S. Activity of purified NIFA, a transcriptional activator of nitrogen fixation genes. Proc Natl Acad Sci USA. 1993;90:2266–2270. doi: 10.1073/pnas.90.6.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liyanage H, Palmer D A, Ullrich M, Bender C L. Characterization and transcriptional analysis of the gene cluster for coronafacic acid, the polyketide component of the phytotoxin coronatine. Appl Environ Microbiol. 1995;61:3843–3848. doi: 10.1128/aem.61.11.3843-3848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liyanage H, Penfold C, Turner J, Bender C L. Sequence, expression and transcriptional analysis of the coronafacate ligase-encoding gene required for coronatine biosynthesis by Pseudomonas syringae. Gene. 1995;153:17–23. doi: 10.1016/0378-1119(94)00661-b. [DOI] [PubMed] [Google Scholar]
- 25.Lynch A S, Lin E C C. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol. 1996;178:6238–6249. doi: 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell R E. Coronatine biosynthesis: incorporation of l-[U-14C]isoleucine and l-[U-14C]threonine into the 1-amido-1-carboxy-2-ethylcyclopropyl moiety. Phytochemistry. 1985;24:247–249. [Google Scholar]
- 27.Mitchell R E, Young S A, Bender C L. Coronamic acid, an intermediate in coronatine biosynthesis by Pseudomonas syringae. Phytochemistry. 1994;35:343–348. [Google Scholar]
- 28.Mohr C D, Martin D W, Konyescni W M, Govan J R W, Lory S, Deretic V. Role of the far-upstream sites of the algD promoter and the algR and rpoN genes in environmental modulation of mucoidy in Pseudomonas aeruginosa. J Bacteriol. 1990;172:6576–6580. doi: 10.1128/jb.172.11.6576-6580.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pabo C O, Sauer T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 30.Palmer D A, Bender C L. Effects of environmental and nutritional factors on the production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl Environ Microbiol. 1993;59:1619–1626. doi: 10.1128/aem.59.5.1619-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer D A, Bender C L, Sharma S. Use of Tn5-gusA5 to investigate environmental and nutritional effects on gene expression in the coronatine biosynthetic gene cluster of Pseudomonas syringae pv. glycinea. Can J Microbiol. 1997;43:517–525. doi: 10.1139/m97-074. [DOI] [PubMed] [Google Scholar]
- 32.Pao G M, Tam R, Lipschitz L S, Saier M H. Response regulators: structure, function, and evolution. Res Microbiol. 1994;145:356–362. doi: 10.1016/0923-2508(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 33.Parkinson J S, Kofoid E C. Communication modules in bacterial signalling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 34.Parry R J, Mhaskar S V, Lin M-T, Walker A E, Mafoti R. Investigations of the biosynthesis of the phytotoxin coronatine. Can J Chem. 1994;72:86–99. [Google Scholar]
- 35.Peñaloza-Vázquez A, Rangaswamy V, Ullrich M, Bailey A M, Bender C L. Use of translational fusions to the maltose binding protein to produce and purify proteins in Pseudomonas syringae and assess their activity in vivo. Mol Plant-Microbe Interact. 1996;9:637–641. doi: 10.1094/mpmi-9-0637. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Martín J, Rojo F, DeLorenzo V. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol Rev. 1994;58:268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reitzer L J, Movsas B, Magasanik B. Activation of glnA transcription by nitrogen regulator I (NRI)-phosphate in Escherichia coli: evidence for a long-range physical interaction between NRI-phosphate and RNA polymerase. J Bacteriol. 1989;171:5512–5522. doi: 10.1128/jb.171.10.5512-5522.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riggs P. Expression and purification of maltose-binding protein fusions. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1994. pp. 16.6.1–16.6.13. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Steffen P, Goyard S, Ullmann A. Phosphorylated BvgA is sufficient for transcriptional activation of virulence-regulated genes in Bordetella pertussis. EMBO J. 1996;14:633–741. [PMC free article] [PubMed] [Google Scholar]
- 41.Strauss J K, Maher L J., III DNA bending by asymmetric phosphate neutralization. Science. 1994;266:1829–1834. doi: 10.1126/science.7997878. [DOI] [PubMed] [Google Scholar]
- 42.Tang L, Grimm A, Zhang Y-X, Hutchinson C R. Purification and characterization of the DNA-binding protein Dnr1, a transcriptional factor of daunorubicin biosynthesis in Streptomyces peucetius. Mol Microbiol. 1996;22:801–813. doi: 10.1046/j.1365-2958.1996.01528.x. [DOI] [PubMed] [Google Scholar]
- 43.Ullrich M, Bender C L. The biosynthetic gene cluster for coronamic acid, an ethylcyclopropyl amino acid, contains genes homologous to amino acid activating enzymes and thioesterases. J Bacteriol. 1994;176:7574–7586. doi: 10.1128/jb.176.24.7574-7586.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ullrich M, Guenzi A C, Mitchell R E, Bender C L. Cloning and expression of genes required for coronamic acid (2-ethyl-1-aminocyclopropane-1-carboxylic acid), an intermediate in the biosynthesis of the phytotoxin coronatine. Appl Environ Microbiol. 1994;60:2890–2897. doi: 10.1128/aem.60.8.2890-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ullrich M, Peñaloza-Vázquez A, Bailey A M, Bender C L. A modified two-component regulatory system is involved in temperature-dependent biosynthesis of the Pseudomonas syringae phytotoxin coronatine. J Bacteriol. 1995;177:6160–6169. doi: 10.1128/jb.177.21.6160-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van den Eede G, Deblaere R, Goethals K, Montagu M V, Holsters M. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol Plant-Microbe Interact. 1992;5:228–234. doi: 10.1094/mpmi-5-228. [DOI] [PubMed] [Google Scholar]
- 47.Walker M S, DeMoss J A. NarL-phosphate must bind to multiple upstream sites to activate transcription from the narG promoter of Escherichia coli. Mol Microbiol. 1994;14:633–641. doi: 10.1111/j.1365-2958.1994.tb01302.x. [DOI] [PubMed] [Google Scholar]
- 48.Young S A, Park S K, Rodgers C, Mitchell R E, Bender C L. Physical and functional characterization of the gene cluster encoding the polyketide phytotoxin coronatine in Pseudomonas syringae pv. glycinea. J Bacteriol. 1992;174:1837–1843. doi: 10.1128/jb.174.6.1837-1843.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]