Abstract
Objective
We aimed to explore the prognostic value of Septin9 DNA methylation in breast cancer.
Methods
Breast cancer patients with and without recurrence or metastasis and matched non-breast cancer patients were screened retrospectively from 2014 to 2016. Bisulfite conversion and fluorescence quantitative methylation-specific polymerase chain reaction were used to detect the Septin9 methylation status and distribution levels in patient breast tissues.
Results
Septin9 DNA methylation was more frequent in breast cancer tissues than in non-breast cancer tissues, but was not significantly correlated with any relevant breast cancer patient clinicopathological characteristic. Septin9 methylation rates were higher in patients with recurrence or metastasis. Septin9 methylation, tumor size, lymph node status, and progesterone receptor (PR) expression could influence prognosis. Septin9 methylation was significantly associated with worse disease-free survival in breast cancer patients, with receiver operating characteristic curve analysis indicating that it had good prognostic ability, with an area under the curve (AUC) value of 0.719. The AUC values increased when Septin9 methylation was combined with tumor size, lymph node status, and PR to predict prognosis.
Conclusions
Septin9 DNA methylation was an independent predictors of breast cancer prognostic risk. This could possibly help improve comprehensive prognosis prediction methods when combined with other risk factors.
Keywords: Breast cancer, Septin9, DNA methylation, recurrence, metastasis, prognosis, disease-free survival
Introduction
Breast cancer is the most common malignancy in women and poses a serious health threat worldwide. The Global Burden of Disease study from 2019 estimates that there were 2 million breast cancer cases from 1990 to 2017, with an age-standardized incidence rate (ASIR) of 45.9 per 100,000 and age-standardized death rate (ASDR) of 14.1 per 100,000. 1 These rates both increased from the 2012 GLOBOCAN estimates (ASIR:43.1, ASDR:12.9). 2 Although the breast cancer screening processes and comprehensive treatment approaches are satisfactory, approximately one-third of patients still have poor outcomes. Tumor metastasis is the main cause of breast cancer related-death, but the core factors that promote the metastasis and growth of cancer cells from primary tumors to distant locations are still unclear.
Deaths can be prevented if breast cancer is detected early. Only a few proteins have been used to detect early-stage breast cancer, including carcinoma antigen 153 (CA153), carcinoembryonic antigen (CEA), and carcinoma antigen 125 (CA125). However, a lack of specificity and/or sensitivity has prevented these markers from having significant diagnostic value. Therefore, there is an urgent need to identify sensitive and specific molecular biomarkers for clinical practice. In the cancer research field, the influence of reversible epigenetic modifications in tumors has received increasing attention. DNA methylation is particularly associated with tumorigenesis. 3
Numerous studies have confirmed that the Septin9 gene is associated with various malignant tumors. In 2010, Septin9 was shown to be essential for pseudopod protrusion and tumor cell migration and invasion. 4 Septin9 can serve as an oncogene or tumor suppressor gene in a cancer type-dependent context. Recent studies5,6 have found that upregulated Septin9_i1 expression can accelerate breast cancer cell migration, while Septin9_i2 overexpression can inhibit their migration. However, expression of Septin9_i1 alone may not be sufficient for tumorigenesis. Thus far, Septin9_v2 promoter methylation has been identified. Septin9 DNA methylation has been used as a new tumor biomarker for colorectal cancer screening and diagnosis, which also plays an important role in prognostic recurrence monitoring.7,8 Additionally, both Matsui et al. 9 and Chen et al. 10 have suggested that Septin9_v2 expression in breast cancer is also regulated by promoter methylation. No relevant clinical research has examined if Septin9 DNA methylation can promote breast cancer invasion and metastasis. Therefore, to further clarify the role of Septin9 in breast cancer, we conducted retrospective screening and follow-up grouping to compare patients with or without recurrence or metastasis. We also aimed to explore the correlations between Septin9 DNA methylation and patient clinicopathological features.
Materials and methods
Study design
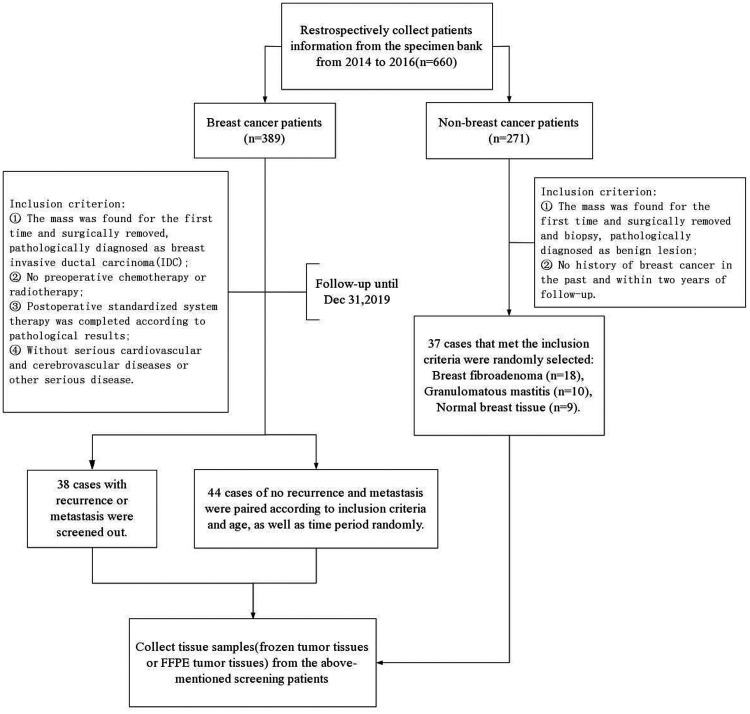
This study was a retrospective analysis of data from breast cancer patients who were treated in Guangdong Women and Children Hospital between 2014 and 2016. The reporting of this study conformed to STROBE guidelines. 11 The patients were selected based on their medical records, pathology reports, and follow-up results. The detailed screening flowchart is shown in Figure 1. The relevant clinical data of the patients are shown in Table 1. The corresponding tissue samples were analyzed for Septin9 DNA methylation levels. All patient details have been de-identified. The study was approved by the local ethics committee (reference number: 202001177) and all participants provided verbal informed consent before specimen collection according to institutional guidelines, which was documented in the specimen bank. Our institutional review board provided an exemption for this study because of its retrospective nature. The study was conducted in accordance with the Helsinki Declaration of 1975 as revised in 2013.
Figure 1.
Flowchart of the specific study population.
Table 1.
Correlations of Septin9 DNA methylation with clinicopathological features of breast cancer patients.
|
Septin9
|
||||
|---|---|---|---|---|
| Characteristics | Cases (%) | Cases with negative methylation | Cases with positive methylation (%) | P-value |
| All cases | 82 (100) | 39 | 43 (52.4) | – |
| Age, years | ||||
| <47 | 38 (46.3) | 20 | 18 (47.4) | 0.393 |
| ≥47 | 44 (53.7) | 19 | 25 (56.9) | |
| Menopausal status | ||||
| Premenopause | 51 (62.2) | 26 | 25 (49.0) | 0.426 |
| Postmenopause | 31 (37.8) | 13 | 18 (58.1) | |
| Tumor size (cm) | ||||
| ≤2 | 31 (37.8) | 16 | 15 (48.4) | 0.759 |
| >2 and ≤5 | 43 (52.4) | 20 | 23 (53.5) | |
| >5 | 9 (9.8) | 3 | 5 (55.6) | |
| Lymph node metastasis | ||||
| Negative | 36 (43.9) | 20 | 16 (44.4) | 0.437 |
| 1–3 Positive | 19 (23.2) | 8 | 11 (57.9) | |
| ≥4 Positive | 27 (32.9) | 11 | 16 (59.3) | |
| ER a | ||||
| Negative | 37 (45.1) | 20 | 17 (45.9) | 0.286 |
| Positive | 45 (54.9) | 19 | 26 (57.8) | |
| PR b | ||||
| Negative or low | 53 (64.6) | 27 | 26 (49.1) | 0.407 |
| High | 29 (35.4) | 12 | 17 (58.6) | |
| HER2 c | ||||
| Negative | 42 (51.2) | 22 | 20 (47.6) | 0.370 |
| Positive | 40 (48.8) | 17 | 23 (57.5) | |
| Ki-67 d | ||||
| Low | 16 (19.5) | 8 | 8 (50.0) | 0.828 |
| High | 66 (80.5) | 31 | 35 (53.0) | |
| Subtype (IHC) e | ||||
| Luminal A | 10 (12.2) | 5 | 5 (50.0) | 0.148 |
| Luminal B | 35 (42.7) | 14 | 21 (60.0) | |
| ERBB2+ | 22 (26.8) | 9 | 13 (59.1) | |
| Basal-like | 15 (18.3) | 11 | 4 (26.7) | |
| Histological grade | ||||
| I | 9 (11.0) | 3 | 6 (66.7) | 0.085 |
| II | 56 (68.3) | 24 | 32 (57.1) | |
| III | 17 (20.7) | 12 | 5 (29.4) | |
| Lympho-vascular invasion | ||||
| No | 61 (74.4) | 29 | 32 (52.5) | 0.995 |
| Yes | 21 (25.6) | 10 | 11 (52.4) | |
ER, estrogen receptor; PR, progesterone receptor; IHC, immunohistochemistry.
The positive threshold of the ER IHC test was set as ≥1%.
The positive threshold of the PR IHC test was set as ≥1%, with 20% as the threshold for high and low expression and the cut-off value of Luminal A and Luminal B.
A HER2 IHC score of 3+ was defined as positive.
The Ki-67 threshold was 30% for high and low expression.
Subtype (IHC) – from Guidelines and Norms for Diagnosis and Treatment of Breast Cancer by Chinese Anti-Cancer Association(CACA):
Luminal A: ER/PR-positive, PR-high, HER2-negative, Ki-67-low; Luminal B: b1, HER2-negative, ER/PR-positive, and PR-low or Ki-67-high; b2, HER2-positive, ER/PR-positive, any state of Ki-67; ERBB2+: ER and PR-negative and HER2-positive; Basal-like: ER, PR, and HER2-negative.
Tissue samples
Breast cancer and non-breast cancer tissue samples were obtained from the specimen bank and pathology department of Guangdong Women and Children Hospital. Frozen tumor tissues were stored at −80°C until use. Formalin-fixed paraffin-embedded (FFPE) tumor tissues were used in cases that lacked individual frozen tumor tissues. FFPE tumor tissues were sectioned by an experienced pathologist to ensure enough of the tumor component was included.
DNA extraction and bisulfite conversion
Genomic DNA was extracted from frozen tumor tissues using a centrifugal adsorption column and a unique buffer system in the TIANamp Genomic DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. In addition, 10 to 15 sections were cut from each FFPE tumor tissue. The TIANamp FFPE DNA Kit (Tiangen) was used to extract DNA from FFPE tumor tissues. Briefly, xylene was used to dewax and remove the paraffin, then special lysis conditions were applied to release DNA from the tissue sections. The DNA concentrations were measured using a Quawell Q5000 UV spectrophotometer (Quawell Technology, San Jose, CA, USA). Then, 500 ng of the extracted DNA was bisulfite-converted using a GS DNA Methylation Kit (GeneShine Biotechnology, Shanghai, China) according to the manufacturer’s recommendations. The bisulfite-modified DNA was stored at 2°C to 8°C for up to 24 hours or immediately used for qPCR analysis.
Fluorescence quantitative methylation-specific polymerase chain reaction (qMS-PCR)
Quantitative PCR was performed for Septin9 methylation in the tissue samples on the ABI 7500 Real Time Fluorescence Quantitative PCR System (Thermo Fisher Scientific, Waltham, MA, USA). The reactions were 20 μL in volume and contained 11.5 μL PCR mix, 0.5 μL polymerase, and 8 μL bisulfite-converted template DNA. The methylation site investigated here maps upstream to 225 to 305 bp of exon 1 of the Septin9 transcript isoform v2 (GenBank accession No. NM_001113493.1). 12 The housekeeping gene β-actin (ACTB) was used as an internal reference gene. Septin9 DNA primer sequences: Forward: 5′-TTTAGTTAGCGCGTAGGGTTC-3′; Reverse: 5′-AACTAATAAACAACGAATCGCG-3′. ACTB primer sequences: Forward: 5′-ATAATAAAAAGGAGGTTGGAT-3′; Reverse; 5′-CTCCCRCAAAACAACCAC-3′. The probe sequences of Septin9 and ACTB were as follows: FAM-ACGCCCCCGACGAAACC-BHQ1 and VIC-CCACCTTACCCTAAACACTACAAC-BHQ1. The thermal cycling conditions were: 1 cycle at 94°C for 20 minutes, followed by 50 cycles of 62°C for 30 s, 55.5°C for 35 s, and 93°C for 30 s. The amplification results were determined by determining the cycle threshold (CT) value and ACTB and Septin9 amplification curves. DNA from breast cancer tissue was the positive control, DNA from non-breast cancer tissue was the negative control, and double distilled water (ddH2O) was the blank control. A schematic diagram of the CT values and amplification curve is shown in Figure 2. PCR amplification reactions were performed in triplicate for each sample and the 1/3 algorithm was used to assess the effectiveness. 13 Samples with the delta CT(Septin9−ACTB)<14 were judged as a positive result. Receiver operating characteristic (ROC) curve analysis was performed on the Septin9 methylation amplification results. Septin9 methylation scores were calculated using the following formula: 2[CT( ACTB )−CT( Septin 9 )] × 100. 14
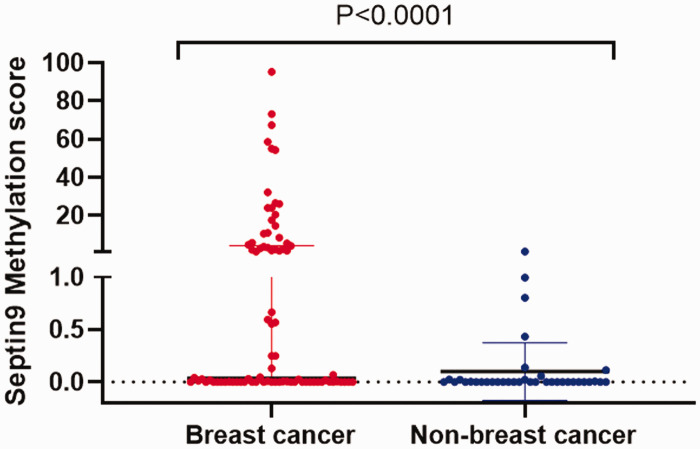
Figure 2.
Scatter plot distribution of Septin9 methylation scores in breast cancer and non-breast cancer tissues. P < 0.0001.
Follow-up and disease-free survival (DFS)
All patients abided by the follow-up principle from the Guidelines and Norms for Diagnosis and Treatment of Breast Cancer by Chinese Anti-Cancer Association (CACA). The cut-off date was 31 December 2019, ensuring that the follow-up time of each patient was ≥36 months. Breast ultrasound, mammography, and CT results were used to determine if local recurrence or metastasis occurred postoperatively. In this study, the patients with recurrence or metastasis included the following conditions: (1) simple recurrence (local recurrence or regional recurrence); (2) distant metastasis (single organ), mainly manifesting as liver, lung, bone, or brain metastasis; (3) distant metastasis (multiple organs), involving two or more organs simultaneously; and (4) both recurrence and metastasis together. DFS was defined as the time (months) from surgery to the appearance of recurrence or metastasis.
Statistical analysis
SPSS 16.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA) software were used to conduct statistical analyses and draw graphics. Non-normally distributed variables are represented as the median (interquartile range). The Mann–Whitney test was used as the non-parametric test. The chi-square test was used for statistical analysis of categorical variables. The Cox proportional hazards model was used to analyze the regression relationships between breast cancer prognosis and multiple risk factors, with the recurrence or metastasis status and DFS used as dependent variables. The statistically significant risk factors were screened out by the Forward LR method. The Kaplan–Meier (KM) method and log-rank tests were used to compare the DFS of breast cancer patients in different subgroups. ROC curve analysis was used and the area under the curve (AUC) value was calculated to evaluate the efficacy of different prognostic risk factors. P-values <0.05 were considered statistically significant.
Results
Septin9 DNA methylation comparison in breast cancer and non-breast cancer tissues
This study included 82 breast cancer patients, of which 38 patients had recurrence or metastasis and 44 patients did not have non-recurrence or metastasis. We also selected 37 patients with benign breast disease as a control group.
In the 37 non-breast cancer tissues, we found Septin9 methylation rates of 12.5% (2/16) in FFPE samples and 19.0% (4/21) in frozen samples. These were not statistically significantly different, suggesting that the FFPE tissue preparation did not affect methylation detection. Moreover, there was also no significant difference in Septin9 DNA methylation among breast fibroadenoma, granulomatous mastitis, and normal breast tissues (data not shown).
Of the 82 breast cancer tissues, 43 (52.4%) were methylated, while only 16.2% (6/37) of the non-breast cancer tissues were found to be methylated. Septin9 methylation was significantly more frequent in breast cancer tissues compared with non-breast cancer tissues (χ2 = 13.811, P = 0.000). Similarly, the median Septin9 methylation score of the 82 breast cancer samples was 0.03633 (0.0006225–4.202), which was significantly higher than 0.00023 (0.00010–0.02232) of the 37 non-breast cancer samples (Mann–Whitney test: P < 0.0001) (Figure 2).
Correlations between Septin9 DNA methylation in breast cancer tissues and patient clinicopathological features
The clinicopathological characteristics of the 82 breast cancer patients were collected and are summarized in Table 1. The mean age at diagnosis was 47.5 years (range, 21 to 70 years). However, the chi-square test showed that there was no significant correlation between Septin9 DNA methylation and any of the examined clinicopathological factors, including age, menopausal status, tumor size, lymph node metastasis, estrogen receptor (ER), progesterone receptor (PR), HER2, Ki-67, subtype, histological grade, or lympho-vascular invasion.
Comparison of Septin9 DNA methylation in breast cancer patients with or without recurrence/metastasis
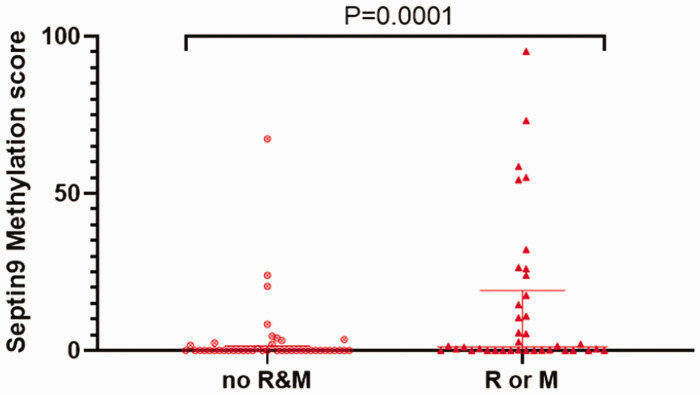
Among the 82 breast cancer cases, 38 patients (46.3%) had local recurrence or distant metastasis. Of these, 27 (71.1%) had Septin9 methylation. On the contrary, only 16 cases of 44 patients without recurrence and metastasis (36.4%) showed Septin9 methylation (χ2 = 9.838, P = 0.002). The mean DFS time of the 38 patients with recurrence or metastasis was 20.2 months, while the mean DFS time of the 44 non-recurrence/metastasis patients was 53.5 months. The patients with recurrence or metastasis had a significantly higher distribution of Septin9 methylation scores compared with the non-recurrence/metastasis group, with median values of 1.244 (0.008570–19.16) and 0.0055 (0.00000–1.431), respectively (Mann–Whitney test: P = 0.0001) (Figure 3). Moreover, among the 38 patients with recurrence or metastasis, there was no significant difference in Septin9 methylation by status (simple recurrence, distant metastasis, or recurrence combined with metastasis; data not shown).
Figure 3.
Scatter plot distribution of Septin9 methylation scores in breast cancer tissues between the patients without and with recurrence or metastasis. No R&M, non-recurrence and metastasis patients; R or M, patients with recurrence or metastasis. P = 0.0001.
Correlations between Septin9 DNA methylation in breast cancer tissues and patient prognosis
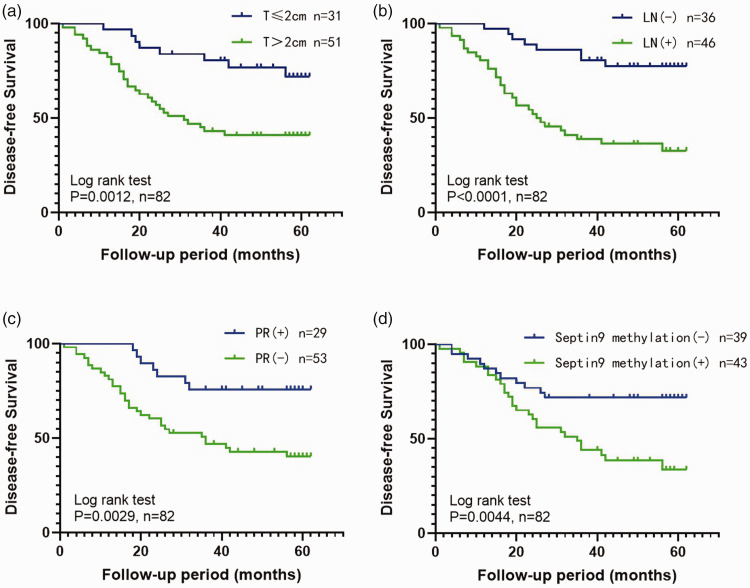
Univariate and multivariate Cox regression analysis of relevant clinicopathological features and Septin9 methylation status showed that Septin9 methylation, tumor size, lymph node status, and PR expression in breast cancer tissues could serve as independent prognostic predictors for breast cancer (P < 0.0001, Table 2). KM analysis also suggested that Septin9 DNA methylation was significantly associated with worse DFS in breast cancer patients (log-rank P = 0.0044. The 3-year cumulative survival probability of DFS in the Septin9 methylation-negative group was 71.8%, which was clearly higher than that of the Septin9 methylation-positive group by 48.8% (Figure 4).
Table 2.
Cox regression analysis of prognosis-related risk factors in 82 breast cancer patients.
| Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| Characteristics* | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
| Age, years (≥35 vs. <35) | 1.626 | 0.576–4.591 | 0.358 | |||
| Tumor size (≤2 cm vs. >2 cm) | 3.161 | 1.446–6.912 | 0.004 | 2.316 | 1.037–5.169 | 0.040 |
| Lymph node status (negative vs. positive) | 4.457 | 2.034–9.763 | 0.000 | 3.359 | 1.495–7.550 | 0.003 |
| ER a (negative vs. positive) | 1.974 | 1.040–3.746 | 0.037 | |||
| PR b (negative or low vs. high) | 3.232 | 1.422–7.348 | 0.005 | 3.464 | 1.515–7.922 | 0.003 |
| HER2 c (negative vs. positive) | 2.399 | 1.239–4.644 | 0.009 | |||
| Ki-67 d (low vs. high) | 3.707 | 1.139–12.063 | 0.030 | |||
| Histological grade (I vs. II–III) | 1.133 | 0.402–3.196 | 0.813 | |||
| Lympho-vascular invasion (no vs. yes) | 2.651 | 1.381–5.089 | 0.003 | |||
| Septin9 methylation (negative vs. positive) | 2.671 | 1.319–5.406 | 0.006 | 2.563 | 1.252–5.248 | 0.010 |
ER, estrogen receptor; PR, progesterone receptor; CI, confidence interval.
The clinicopathological parameters associated with prognosis are derived from Guidelines and Norms for Diagnosis and Treatment of Breast Cancer by Chinese Anti-Cancer Association (CACA).
The positive threshold of the ER IHC test was set as ≥1%.
The positive threshold of the PR IHC test was set as ≥1%, with 20% as the threshold for high and low expression.
A HER2 IHC score of 3+ was defined as positive.
The Ki-67 threshold was 30% for high and low expression.
Figure 4.
Kaplan–Meier survival curves (disease-free survival) were plotted according to the four groups of prognostic independent risk factors: (a) tumor size (T), (b) lymph node metastasis (LN), (c) progesterone receptor expression (PR), and (d) Septin9 methylation.
Prognostic predictive efficacy of the Septin9 methylation test and its combined risk factors
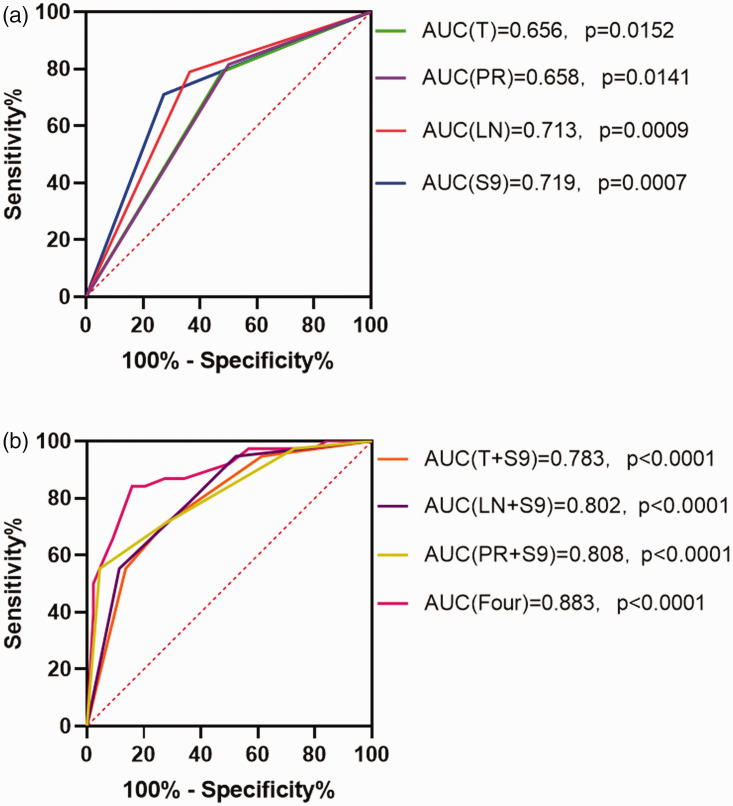
The sensitivity, specificity, positive predictive value, and negative predictive value of prognostic Septin9 DNA methylation detection in breast cancer tissues were 71.1%, 72.7%, 69.2%, and 74.4%, respectively. Its sensitivity value was lower than those of tumor size, lymph node status, and PR expression, which were 78.9%, 78.9%, and 81.6%, respectively. However, the sensitivity values when Septin9 DNA methylation was combined with tumor size, lymph node status, or PR expression were 94.7%, 94.7%, and 97.4%, respectively. Among the prognostic risk factors analyzed in the ROC curve comparison, Septin9 methylation in breast cancer tissues had an AUC value of 0.719 (95% confidence interval (CI) = 0.606–0.832, P = 0.0007). The AUC values of Septin9 methylation combined with tumor size, lymph node status, or PR expression for predicting prognosis were 0.783, 0.802, and 0.808, respectively (all P < 0.0001). Combining the four risk factors could bring the AUC value as high as 0.883 (95% CI = 0.848–0.958, P < 0.0001) (Figure 5).
Figure 5.
The receiver operating characteristic (ROC) curves plotted according to the four groups of prognostic independent risk factors. (a) The area under the ROC curve (AUC) values for tumor size (T), progesterone receptor expression (PR), lymph node metastasis (LN), and Septin9 methylation (S9) and (b) The AUC values for S9 with T, S9 with PR, S9 with LN, and all four together.
Discussion
Breast cancer is a serious health threat worldwide, with the main associated cause of death being tumor recurrence and metastasis. Therefore, identifying additional molecular biological factors that affect breast cancer recurrence and metastasis is of great significance. In recent years, Septin9 has been described as a potential biomarker for early screening, diagnosis, and prognosis of some malignant tumors, including breast cancer. 15 The Septin-9 protein belongs to a class of GTPases involved in many cellular processes. Human Septin9 is located on chromosome 17q25.3 and contains 17 exons. The gene encodes 18 unique transcripts through alternative splicing that code for 15 polypeptides, of which Septin9_v1, v2, v3, v4, v4*, and v5 are the most common. 16 Different tissues and organs can have varying expression patterns of these transcripts. Septin9_v2 promoter methylation was first shown in colorectal cancer and has been used as a clinical tumor biomarker. 17 Subsequently, two studies reported that Septin9_v2 expression was also epigenetically regulated in breast cancer by promoter methylation.9,10 However, these studies did not examine recurrence and metastasis of the disease and lacked further clinical investigation. Therefore, this was a key point of our research.
Matsui et al. 9 first reported Septin9_v2 promoter methylation in 53% (10/19) of the primary breast cancer tissues examined, but did not observe this in any normal breast tissue (0/19). Additionally, the results of Chen et al. 10 showed that 83.1% (49/59) of breast cancer tissues had Septin9 DNA methylation. In our study, Septin9 DNA methylation distribution was significantly different between the breast cancer and non-breast cancer groups. The overall positive rate of Septin9 DNA methylation in breast cancer tissues was 52.4% (43/82), which was similar to the findings of Matsui et al. Although its sensitivity value was not high, its specificity and positive predictive values were 83.8% and 87.8%, respectively. These results potentially reflect the effect of Septin9 DNA methylation on breast cancer progression. In addition, although Septin9 DNA methylation was observed in only 16.2% of non-breast cancer tissues and no significant difference was found between the non-breast cancer tissue sample types, its methylation did not exclude the long-term risk of breast cancer. Thus, Septin9 DNA methylation might have significance in early screening, which needs further exploration. Here, we also analyzed the relationships between Septin9 DNA methylation in breast cancer tissues and patient clinicopathological characteristics. However, Septin9 methylation had no significant correlation with any of the features examined.
Alternative splicing leads to numerous Septin9 mRNA transcripts that show varied expression patterns in different tissues. Septin9_v1 and Septin9_v2 are the most researched transcripts and closely related to breast cancer.18–20 Additionally, promoter methylation has only been validated for Septin9_v2 among the Septin9 family. Septin9 has been shown to be a tumor suppressor gene, with abnormal methylation patterns inhibiting its normal expression levels and causing it lose this function and ultimately promote cancer development. 21 Although not explored in our study, abnormal methylation of Septin9 DNA that can inhibit its normal expression pattern was verified by Matsui et al. 9 and Chen et al. 10 Matsui et al. also showed that there was a significant difference between the Septin9_v2 mRNA expression levels and methylation index. Septin9_v2 mRNA expression could be reactivated by demethylation treatment. 9 The results of the latter methylation study indicated Septin9 DNA methylation in 83.1% of the breast cancer tissues analyzed, with most of the corresponding low Septin9 expression observed in breast cancer tissues. 10
In clinical application, Septin9 DNA methylation detection could assist during colonoscopy-based diagnosis of colorectal cancer and thereby increase the positive diagnosis rate. Similarly, Lian et al.22–23 reported that a nomogram based on mammary ductoscopic indicators combined with breast duct lavage fluid RASSF1A methylation could improve the diagnostic sensitivity of nipple discharge, with a value of 88.5% (23/26). The significant difference of Septin9 DNA methylation levels in benign and malignant tumors in our study provided a theoretical basis for identifying new biomarkers. In future research, we will investigate if it can assist with breast ductoscopy and improve malignant breast tumor detection.
To explore if Septin9 DNA affects breast cancer patient prognosis, we examined the results of previous studies. Yeh et al. 24 demonstrated that inhibition of Septin9 expression in cancer cells can induce the epithelial-mesenchymal transition, which plays an important role in cancer metastasis. Conversely, another study 25 revealed that overexpression of Septin9 could lead to enhanced hypoxia inducible factor-1 (HIF-1) transcription, thereby increasing vascular endothelial growth factor (VEGF) expression levels and promoting angiogenesis. This can support tumor invasion and metastasis. Additionally, upregulation of Septin9_v1 could not only prevent the degradation of Jun N-terminal kinase (JNK) to inhibit cell apoptosis and promote cell proliferation, but could also promote the transformation of breast epithelial cells to breast stromal cells and thus support cancer development.26–27 Recently, extensive work by Verdier-Pinard et al. 6 indicated that upregulation of Septin9_i1 expression accelerated breast cancer cell migration rates, while overexpressing Septin9_i2 had the opposite effects. Increased cell migration can promote cancer invasion and metastasis. 28 These previous results suggest that both Septin9_v1 and Septin9_v2 are related to breast cancer recurrence and metastasis and are in a balance with each other. Methylation of the Septin9_v2 gene could break this balance and increase the metastatic potential of breast cancer.
According to the follow-up results, we divided 82 breast cancer patients into two groups: no R&M (non-recurrence and metastasis patients) and R or M (recurrence or metastasis patients). There were significant differences in Septin9 DNA methylation between the two groups. Intriguingly, Septin9 DNA methylation in breast cancer tissues was associated with recurrence or metastasis in patients according to survival analysis. The 3-year cumulative survival probability of DFS in the Septin9 DNA methylation-negative group was 71.8%, which was clearly higher than that of the Septin9 DNA methylation-positive group by 48.8%. This suggests that Septin9 DNA methylation-positive breast cancer was more aggressive. The hazard of recurrence or metastasis in Septin9 methylation-positive patients was 2.563 times that of Septin9 methylation-negative patients.
Historically, patient age, tumor size, lymph node status, hormone receptor status, HER2 status, Ki-67 expression status, histological grade, and lympho-vascular invasion have been used to assess breast cancer patient prognosis. 29 Most of these prognostic factors were statistically significant in our univariate Cox proportional hazard model. After considering Septin9 DNA methylation, both the univariate and multivariate Cox regression analyses suggested that it could be used as an independent prognostic indicator. Moreover, the sensitivity and specificity values of Septin9 methylation detection were 71.1% and 72.7%, respectively, providing the best overall performance for predicting prognosis. ROC curve analysis then indicated that the prediction accuracy of Septin9 methylation in breast cancer tissues was effective in the single index, which was improved by combining other prognostic risk factors. Collectively, our results suggest that the methylation of Septin9 DNA in breast cancer was significantly associated with postoperative disease recurrence or metastasis.
Overall, the biggest advantages of this study were the follow-up collection of prognostic information and clinical studies on the prognosis of breast cancer patients with long-term recurrence and metastasis. However, there were some limitations to our study. First, we only examined the indirect effect of Septin9 DNA methylation on breast cancer prognosis, while ignoring the direct effect of the Septin9_v1 and Septin9_v2 expression balance. Although it was clear that Septin9 methylation results in downregulated Septin9_v2 expression and loss of its anti-cancer effect, 30 it could not be excluded that single overexpression of Septin9_v1 also impacted prognosis.6,31 Future work should focus on the patients with recurrence or metastasis who did not have Septin9 DNA methylation to examine the different roles of Septin9 transcript variants. Second, this study lacks details regarding quality control, replicates, and the accuracy of the conducted analyses in the experimental methods for the methylation detection. Because there were very few studies on Septin9 methylation in breast cancer, we could only refer to other similar studies. Third, we did not provide experimental evidence for a relevant mechanism of action in this study. Lastly, although we used software to calculate an appropriate patient sample size, a larger sample size is still needed for validation and further research. The overall role of Septin9 DNA methylation in breast cancer patients with recurrence or metastasis still requires further exploration.
Conclusions
Our study provides new evidence that Septin9 DNA methylation is involved in breast cancer development and is associated with disease recurrence or metastasis. We found this methylation to be an independent predictive indicators for assessing prognostic risk, which can be combined with other prognostic risk factors to improve the comprehensive prognostic ability. This is therefore a potential new auxiliary predictor of poor prognosis in breast cancer patients. However, further large-scale studies and longer follow-up periods are still needed to verify the clinical significance of Septin9 methylation.
Supplemental Material
Supplemental material, sj-pdf-1-imr-10.1177_03000605231220827 for Septin9 DNA methylation is associated with breast cancer recurrence or metastasis by Shao-Ling Zhang, Hai-Jing Yu, Zhen-Qiang Lian, Jian Wan, Si-Mei Xie, Wen Lei, Qiu-Ping Chen, Liang Zhang and Qi Wang in Journal of International Medical Research
Acknowledgements
We thank Hong-Yi Gao and Hui-Bin Li from the pathology department of Guangdong Women and Children Hospital for providing the FFPE tumor tissues examined in this study.
Author contributions: SLZ and HJY acquired the specimens, performed the experiments, reviewed the literature, collected relevant clinical data, performed the statistical analyses, interpreted the results, and drafted the manuscript. QW, LZ, ZQL, and JW helped design the study, interpret the results, and revise the manuscript. SMX assisted with specimen acquisition and clinical data collection. WL and QPC helped with performing the experiments and interpreting the results. All authors read and approved the final manuscript.
The authors declare that there is no conflict of interest.
Funding: This research was supported by national public welfare project of revision of breast hyperplasia regulations of China (No. 201502027) and the project of preliminary discussion on the subject of DNA methylation molecular markers related to the recurrence and metastasis of triple-negative breast cancer (No. A2021063). These funding parties had no influence on the study design, data collection, analysis, or interpretation.
ORCID iDs: Shao-Ling Zhang https://orcid.org/0000-0003-0801-0783
Hai-Jing Yu https://orcid.org/0000-0001-7557-826X
Data availability statement
All data supporting this study will be available from the corresponding author upon reasonable request.
References
- 1.Global Burden of Disease Cancer Collaboration; Fitzmaurice C, Abate D, Abbasi N, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. Jama Oncol 2019; 5: 1749–1768. DOI: 10.1001/jamaoncol.2019.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghoncheh M, Pournamdar Z, Salehiniya H. Incidence and Mortality and Epidemiology of Breast Cancer in the World. Asian Pac J Cancer Prev 2016; 17: 43–46. Journal Article. DOI: 10.7314/apjcp.2016.17.s3.43. [DOI] [PubMed] [Google Scholar]
- 3.Kresovich JK, Gann PH, Erdal S, et al. Candidate gene DNA methylation associations with breast cancer characteristics and tumor progression. Epigenomics-Uk 2018; 10: 367–378. Journal Article; Research Support, N.I.H., Extramural. DOI: 10.2217/epi-2017-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shankar J, Messenberg A, Chan J, et al. Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res 2010; 70: 3780–3790. Journal Article; Research Support, Non-U.S. Gov't. DOI: 10.1158/0008-5472.CAN-09-4439. [DOI] [PubMed] [Google Scholar]
- 5.Verdier-Pinard P, Salaun D, Bouguenina H, et al. Septin 9_i2 is downregulated in tumors, impairs cancer cell migration and alters subnuclear actin filaments. Sci Rep-Uk 2017; 7: 44976. Journal Article; Research Support, Non-U.S. Gov't. DOI: 10.1038/srep44976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng Y, Cao Y, Liu L, et al. SEPT9_i1 regulates human breast cancer cell motility through cytoskeletal and RhoA/FAK signaling pathway regulation. Cell Death Dis 2019; 10: 720. Journal Article. DOI: 10.1038/s41419-019-1947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Chen PM, Liu RB. Advance in plasma SEPT9 gene methylation assay for colorectal cancer early detection. World J. Gastro Oncol 2018; 10: 15–22. Journal Article; Review. DOI: 10.4251/wjgo.v10.i1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J, Fei F, Zhang M, et al. The role of (m)SEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. Bmc Cancer 2019; 19: 450. Journal Article. DOI: 10.1186/s12885-019-5663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsui S, Kagara N, Mishima C, et al. Methylation of the SEPT9_v2 promoter as a novel marker for the detection of circulating tumor DNA in breast cancer patients. Oncol. Rep 2016; 36: 2225–2235. Journal Article. DOI: 10.3892/or.2016.5004. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Zhou C, Liu W, et al. Methylated septin 9 gene for noninvasive diagnosis and therapy monitoring of breast cancer(Article). Transl Cancer Res 2018; 7: 587–599. [Google Scholar]
- 11.Von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med 2007; 147: 573–577. Guideline; Journal Article; Research Support, Non-U.S. Gov't. DOI: 10.7326/0003-4819-147-8-200710160-00010. [DOI] [PubMed] [Google Scholar]
- 12.Lyu J, Chen J, Zhang X, et al. Septin 9 Methylation in Nasopharyngeal Swabs: A Potential Minimally Invasive Biomarker for the Early Detection of Nasopharyngeal Carcinoma. Dis Markers 2020; 2020: 7253531. DOI: 10.1155/2020/7253531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song L, Yu H, Jia J, et al. A systematic review of the performance of the SEPT9 gene methylation assay in colorectal cancer screening, monitoring, diagnosis and prognosis. Cancer Biomark 2017; 18: 425–432. Journal Article; Review; Systematic Review. DOI: 10.3233/CBM-160321. [DOI] [PubMed] [Google Scholar]
- 14.Bu Q, Wang S, Ma J, et al. The clinical significance of FAM19A4 methylation in high-risk HPV-positive cervical samples for the detection of cervical (pre)cancer in Chinese women. Bmc Cancer 2018; 18: 1182. DOI: 10.1186/s12885-018-4877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J, Zheng MY, Li YW, et al. Structure and function of Septin 9 and its role in human malignant tumors. World J Gastro Oncol 2020; 12: 619–631. Journal Article; Review. DOI: 10.4251/wjgo.v12.i6.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Connolly D, Abdesselam I, Verdier-Pinard P, et al. Septin roles in tumorigenesis. Biol Chem 2011; 392: 725–738. Journal Article; Research Support, Non-U.S. Gov't; Review. DOI: 10.1515/BC.2011.073. [DOI] [PubMed] [Google Scholar]
- 17.Yang X, Xu ZJ, Chen X, et al. Clinical value of preoperative methylated septin 9 in Chinese colorectal cancer patients. World J Gastroentero 2019; 25: 2099–2109. Journal Article. DOI: 10.3748/wjg.v25.i17.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connolly D, Yang Z, Castaldi M, et al. Septin 9 isoform expression, localization and epigenetic changes during human and mouse breast cancer progression. Breast Cancer Res 2011; 13: R76. Journal Article; Research Support, Non-U.S. Gov't. DOI: 10.1186/bcr2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connolly D, Hoang HG, Adler E, et al. Septin 9 amplification and isoform-specific expression in peritumoral and tumor breast tissue. Biol Chem 2014; 395: 157–167. Journal Article; Research Support, N.I.H., Extramural; Research Support, Non-U.S. Gov't. DOI: 10.1515/hsz-2013-0247. [DOI] [PubMed] [Google Scholar]
- 20.Diesenberg K, Beerbaum M, Fink U, et al. SEPT9 negatively regulates ubiquitin-dependent downregulation of EGFR. J Cell Sci 2015; 128: 397–407. Journal Article; Research Support, Non-U.S. Gov't. DOI: 10.1242/jcs.162206. [DOI] [PubMed] [Google Scholar]
- 21.Toth K, Galamb O, Spisak S, et al. The influence of methylated septin 9 gene on RNA and protein level in colorectal cancer. Pathol Oncol Res 2011; 17: 503–509. Journal Article. DOI: 10.1007/s12253-010-9338-7. [DOI] [PubMed] [Google Scholar]
- 22.Lian ZQ, Wang Q, Zhang AQ, et al. A nomogram based on mammary ductoscopic indicators for evaluating the risk of breast cancer in intraductal neoplasms with nipple discharge. Breast Cancer Res Tr 2015; 150: 373–380. Journal Article; Research Support, Non-U.S. Gov't. DOI: 10.1007/s10549-015-3320-8. [DOI] [PubMed] [Google Scholar]
- 23.Lian ZQ, Wang Q, Li WP, et al. Screening of significantly hypermethylated genes in breast cancer using microarray-based methylated-CpG island recovery assay and identification of their expression levels. Int J Oncol 2012; 41: 629–638. Journal Article; Research Support, Non-U.S. Gov't. DOI: 10.3892/ijo.2012.1464. [DOI] [PubMed] [Google Scholar]
- 24.Yeh YT, Hur SS, Chang J, et al. Matrix stiffness regulates endothelial cell proliferation through septin 9. Plos One 2012; 7: E46889. Journal Article; Research Support, N.I.H., Extramural; Research Support, Non-U.S. Gov't. DOI: 10.1371/journal.pone.0046889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amir S, Wang R, Matzkin H, et al. MSF-A interacts with hypoxia-inducible factor-1alpha and augments hypoxia-inducible factor transcriptional activation to affect tumorigenicity and angiogenesis. Cancer Res 2006; 66: 856–866. Journal Article; Research Support, Non-U.S. Gov't; Research Support, U.S. Gov't, Non-P.H.S. DOI: 10.1158/0008-5472.CAN-05-2738. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez ME, Peterson EA, Privette LM, et al. High SEPT9_v1 expression in human breast cancer cells is associated with oncogenic phenotypes. Cancer Res 2007; 67: 8554–8564. Journal Article; Research Support, N.I.H., Extramural; Research Support, U.S. Gov't, Non-P.H.S. DOI: 10.1158/0008-5472.CAN-07-1474. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez ME, Makarova O, Peterson EA, et al. Up-regulation of SEPT9_v1 stabilizes c-Jun-N-terminal kinase and contributes to its pro-proliferative activity in mammary epithelial cells. Cell Signal 2009; 21: 477–487. Journal Article; Research Support, N.I.H., Extramural; Research Support, U.S. Gov't, Non-P.H.S. DOI: 10.1016/j.cellsig.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawson CD, Ridley AJ. Rho GTPase signaling complexes in cell migration and invasion. J Cell Biol 2018; 217: 447–457. Journal Article; Research Support, Non-U.S. Gov't; Review. DOI: 10.1083/jcb.201612069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breast Cancer Committee of Chinese Anti-Cancer Association. Guidelines and Norms for the Diagnosis and Treatment of Breast Cancer of the Chinese Anti-Cancer Association (2019 Edition). China Oncology 2019; 8: 609–680. DOI: 10.19401/j.cnki.1007-3639.2019.08.009. [Google Scholar]
- 30.Thomas ML, Marcato P. Epigenetic Modifications as Biomarkers of Tumor Development, Therapy Response, and Recurrence across the Cancer Care Continuum. Cancers (Basel) 2018; 10: 101. Journal Article; Review. DOI: 10.3390/cancers10040101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson EA, Stanbery L, Li C, et al. SEPT9_i1 and genomic instability: mechanistic insights and relevance to tumorigenesis. Gene Chromosome Canc 2011; 50: 940–949. Journal Article; Research Support, N.I.H., Extramural; Research Support, Non-U.S. Gov't; Research Support, U.S. Gov't, Non-P.H.S. DOI: 10.1002/gcc.20916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-imr-10.1177_03000605231220827 for Septin9 DNA methylation is associated with breast cancer recurrence or metastasis by Shao-Ling Zhang, Hai-Jing Yu, Zhen-Qiang Lian, Jian Wan, Si-Mei Xie, Wen Lei, Qiu-Ping Chen, Liang Zhang and Qi Wang in Journal of International Medical Research
Data Availability Statement
All data supporting this study will be available from the corresponding author upon reasonable request.