Abstract
There has been increased interest in bacterial polyadenylation with the recent demonstration that 3′ poly(A) tails are involved in RNA degradation. Poly(A) polymerase I (PAP I) of Escherichia coli is a member of the nucleotidyltransferase (Ntr) family that includes the functionally related tRNA CCA-adding enzymes. Thirty members of the Ntr family were detected in a search of the current database of eubacterial genomic sequences. Gram-negative organisms from the β and γ subdivisions of the purple bacteria have two genes encoding putative Ntr proteins, and it was possible to predict their activities as either PAP or CCA adding by sequence comparisons with the E. coli homologues. Prediction of the functions of proteins encoded by the genes from more distantly related bacteria was not reliable. The Bacillus subtilis papS gene encodes a protein that was predicted to have PAP activity. We have overexpressed and characterized this protein, demonstrating that it is a tRNA nucleotidyltransferase. We suggest that the papS gene should be renamed cca, following the notation for its E. coli counterpart. The available evidence indicates that cca is the only gene encoding an Ntr protein, despite previous suggestions that B. subtilis has a PAP similar to E. coli PAP I. Thus, the activity involved in RNA 3′ polyadenylation in the gram-positive bacteria apparently resides in an enzyme distinct from its counterpart in gram-negative bacteria.
mRNA polyadenylation now appears to be a common property of all living cells, but until recently it had been extensively studied only in eukaryotes (reviewed in references 9, 41, and 49). In the eubacteria, polyadenylated mRNAs have been detected in both gram-positive and gram-negative organisms (reviewed in references 43 and 44). Although an Escherichia coli poly(A) polymerase (PAP) activity was first described over 30 years ago (4), a convincing demonstration that polyadenylation is a general feature of prokaryotic messages was difficult because the poly(A) tails are short, mRNA turnover is very rapid, and only a fraction of the mRNAs are polyadenylated. The recent purification of E. coli PAP I and the identification of its gene (7) has led to increased interest in bacterial polyadenylation. PAP I is a 50-kDa protein encoded by the pcnB gene, which has been implicated in the control of plasmid copy number (30, 31, 33). The polyadenylation of RNA I, a plasmid-encoded antisense RNA that regulates ColE1 plasmid replication, mediates its degradation via a 3′ exonucleolytic pathway (23, 50, 51). Poly(A)-mediated exonucleolytic degradation has also been described for the antisense RNAs that regulate R1 plasmid replication and partition (36, 45), and it is believed to be involved in the degradation of mRNA (8, 21, 22, 37).
A second putative PAP of 35 kDa (PAP II), encoded by the open reading frame f310, has been identified in E. coli (6, 27). Strains with a disruption of the gene encoding either PAP I or PAP II are viable, suggesting a possible functional redundancy. However, there is no obvious sequence homology between the two E. coli polymerases, and PAP II is not related to any other known protein sequence. We previously detected proteins related to PAP I in a variety of bacteria, including Bacillus subtilis, Desulfovibrio gigas, and Proteus mirabilis, using an antibody raised against the purified E. coli PAP I (39). PAP I is a member of the X polymerase family, which includes the eukaryotic PAPs as well as all of the known nucleotidyltransferases (34, 52). The PAP I and tRNA nucleotidyltransferase of E. coli have extensive homology in their N-terminal halves (35). Their similarity to the eukaryotic PAPs is limited to a small number of conserved residues that are critical for activity (34, 39a). The tRNA 3′ CCA-adding activity is specific to the tRNA nucleotidyltransferases (reviewed in reference 14). In eukaryotes, where the CCA is rarely encoded by the tRNA gene, the tRNA nucleotidyltransferase is essential (1). In bacteria, the 3′ CCA is generally encoded by the tRNA gene. Nevertheless, the E. coli CCA-adding enzyme has an important role in the repair of tRNA 3′ ends, and its inactivation significantly slows growth (11, 16, 53).
Although the E. coli tRNA nucleotidyltransferase can add CCA or repair ends (CA or A addition), we know of no evidence that it can add longer 3′-terminal extensions (Fig. 1A). In contrast, PAP I in vitro can add poly(A) tails several hundred nucleotides long to tRNA or other RNA substrates (Fig. 1B) (39). Although the principal role of PAP I is believed to be 3′ polyadenylation, PAP I and polynucleotide phosphorylase can repair partially damaged tRNA CCA ends in E. coli mutants that are deficient for CCA-adding activity (40). In this paper, we show that, like E. coli, other organisms from the β and γ subdivisions of the purple bacteria have two genes encoding proteins that can be classified as either a tRNA nucleotidyltransferase or PAP by protein sequence comparisons. However, prediction of the activity of Ntr proteins encoded by genes from more distantly related bacteria is not reliable. The gram-positive bacterium B. subtilis, whose genome has now been completely sequenced, contains a single gene encoding an Ntr protein that we show is a tRNA nucleotidyltransferase. The apparent lack of an Ntr protein with PAP activity is surprising because previous evidence suggested that B. subtilis has an activity similar to that of E. coli PAP I.
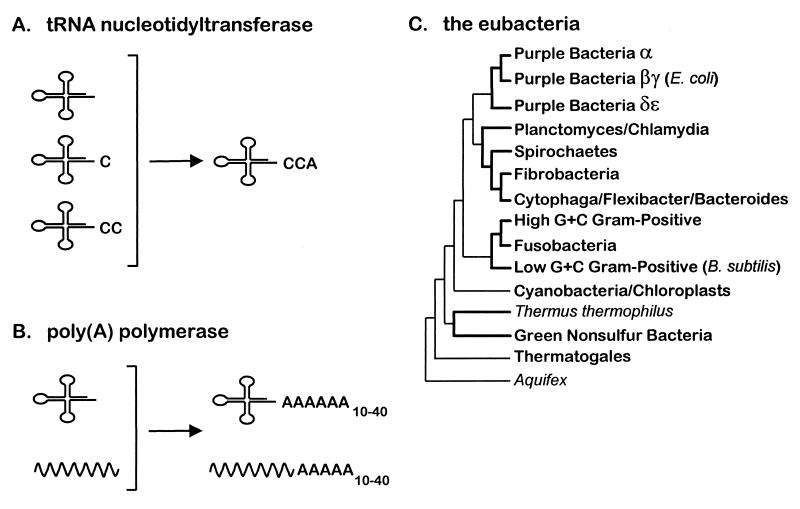
FIG. 1.
(A) Repair of tRNA CCA ends by nucleotidyltransferase; (B) 3′ poly(A) addition to tRNA or other RNA substrates by PAP; (C) phylogenetic tree showing the relationship of the major families of eubacteria (38). Thermus thermophilus and Aquifex are single organisms.
MATERIALS AND METHODS
The construction of pET11a-derived vectors for protein expression and the preparation of protein extracts was done as described previously (39). Briefly, the DNAs encoding the E. coli cca and B. subtilis papS open reading frames were PCR amplified with the oligonucleotide pairs 5′ATATGAAGATTTATCTGGTCGGTGGTGC3′ and 5′TCATTCAGGCTTTGGGCAAGCTTGTTCC3′ (E. coli) or 5′ATATGGAAAAAGTTTTTATCAAAGCACTTCC3′ and 5′TTAATGTTAGACCGCATGTCTTCAGCC3′ (B. subtilis) with genomic DNA from E. coli D10 (20) or B. subtilis QB936 (13). In the construction expressing the E. coli CCA-adding enzyme, we changed the GTG initiation codon to ATG. Protein expression was done in the BL21(DE3) strain with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) induction for 2 h. Cultures, concentrated fivefold, were suspended in a lysis buffer and sonicated. The extracts were treated with 10 μg of DNase I/ml and then adjusted to 0.8 M NH4Cl. The cell debris and the ribosomes were removed by centrifugation. The purities of the overexpressed proteins were estimated with a Coomassie blue-stained sodium dodecyl sulfate (SDS)-polyacrylamide gel as follows: E. coli PAP I, 10%; E. coli tRNA CCA-adding enzyme, 2%; and B. subtilis PapS, 30%. The low level of the CCA-adding enzyme was due to its inefficient expression. In the assays described below, 250 (PAP I), 1,250 (CCA-adding enzyme), or 80 ng (PapS) of total protein was used to give equivalent amounts of each overexpressed protein (25 ng). For ease of comparison, the values for normalized specific activities (see Table 2) are based on the estimated amounts of the overexpressed proteins.
TABLE 2.
Activity of the B. subtilis PapS protein
| Assay conditionsa | Substrates | Normalized specific activity (U/mg)b
|
||
|---|---|---|---|---|
| PAP I (E. coli) | CCA (E. coli) | PapS (B. subtilis) | ||
| tRNA, ATP | 1,510 | 910 | 880 | |
| CCA addition | tRNA, CTP | 0 | 1,530 | 1,490 |
| poly(A), ATP | 520 | 0 | 0 | |
| Polyadenylation | tRNA, ATP | 2,820 | 0 | 0 |
Incorporation of radioactive AMP or CMP was assayed by trichloroacetic acid precipitation under CCA addition or polyadenylation assay conditions (see Materials and Methods).
Values, based on the assay of protein extracts, have been normalized to the amount of overexpressed protein (see Materials and Methods). For each determination, a background, which ranged from 10- to 100-fold lower than the signal, was subtracted. The background reactions used an equivalent amount of total protein prepared from the BL21(DE3) strain with the pET11a expression vector. The units of activity are micromoles of AMP (or CMP) incorporated per hour in the CCA addition assay and nanomoles of AMP incorporated per 10-minute interval in the polyadenylation assay.
The following conditions were used to measure the specific activities (see Table 2). The tRNA nucleotidyltransferase assay was done as described previously (15). The protein extracts were diluted in 10 mM glycine (pH 9.4)–1 mg of Saccharomyces cerevisiae tRNA/ml. Reaction mixtures (200 μl) containing 50 mM glycine (pH 9.4), 5 mM MgCl2, 500 μM ATP, 10 μCi of [α-32P]ATP (or [α-32P]CTP), 150 μg of yeast tRNA or poly(A), and 25 ng of overexpressed protein were incubated for 10 min at 37°C. In the gel analysis of the tRNA addition products (see Fig. 4), the reaction mixtures were the same except that the volume was reduced to 20 μl. The PAP assay was done as described previously (39). The protein extracts were diluted in a solution containing 10 mM Tris-HCl (pH 7.5), 500 mM NaCl, 5% glycerol, 0.5% Triton X-100, 1 mM EDTA, and 1 mM dithiothreitol. Reaction mixtures (200 μl) containing 10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1% glycerol, 0.1% Triton X-100, 4 mM MgCl2, 200 μM ATP, 20 μCi of [α-32P]ATP, 4 μg of yeast tRNA, and 25 ng of the overexpressed protein were incubated for 15 min at 37°C.
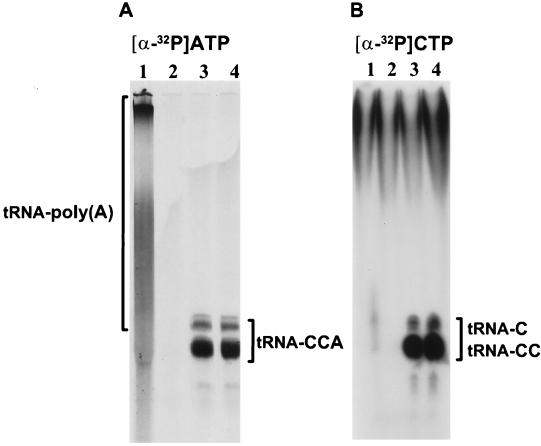
FIG. 4.
Polyacrylamide gel analysis of products under CCA assay conditions with yeast tRNA as the acceptor and [α-32P]ATP (A) or [α-32P]CTP (B) as the substrate. Lanes: 1, E. coli PAP I; 2, protein extract from BL21(DE3) with the pET11a expression vector; 3, E. coli tRNA nucleotidyltransferase; 4, B. subtilis PapS.
PCR amplifications of genomic DNA with inosine-containing oligonucleotides were done with 100 ng of B. subtilis DNA with the oligonucleotide pair 5′GTIGGIGGI(C/G)I(A/G)TI(C/A)GIGA(T/C)3′ and 5′(G/A)TTIAIIGTIA(A/G)ITCIC(G/T)IT3′. Control amplifications were done with the pET11a-pcnB plasmid and the same oligonucleotides. The reactions (50 μl) with Tub DNA polymerase (Amersham) were performed under the following conditions: 95°C, 30 s; 46°C, 3 min; 66°C, 40 s; 50 cycles.
RESULTS
Nucleotidyltransferase proteins in the eubacteria.
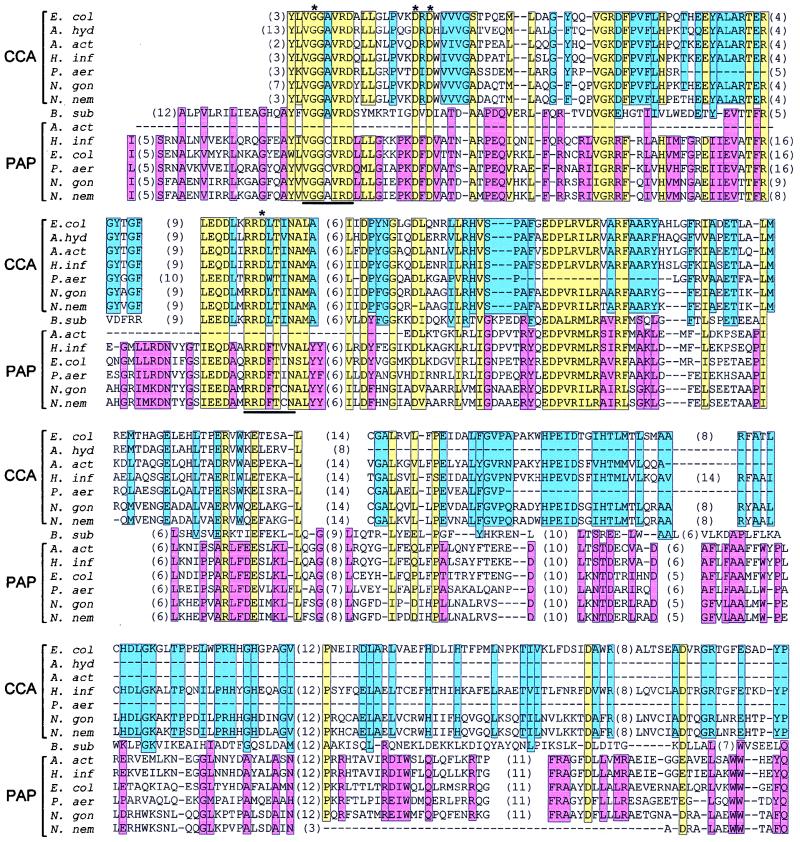
With many genomic sequencing projects finished or in progress, extensive comparisons among the eubacteria are now possible (Fig. 1C). Putative Ntr proteins have already been detected in bacteria such as Haemophilus influenzae, Acidaminococcus fermentans, Aeromonas hydrophyla, and B. subtilis (34, 52). We found 25 more proteins in a recent search (May 1998), and their number will surely increase as more genomes are completed. Table 1 summarizes the protein sequences detected in this search. All of these homologues share a conserved amino-terminal domain containing motifs that are characteristic of the Ntr protein family (34, 52). For the gram-negative organisms in the β and γ subdivisions of the purple bacteria, which include E. coli, two Ntr proteins were predicted. As indicated in Table 1, it was possible to classify the predicted proteins as either a PAP or tRNA nucleotidyltransferase (noted as CCA in Table 2) by comparison with the E. coli homologues. The alignment of these two groups of proteins is shown in Fig. 2. The remainder of the proteins identified in Table 1 cannot be reliably classified by sequence alignment. This is illustrated in Fig. 2, where the B. subtilis Ntr protein sequence is aligned with the tRNA nucleotidyltransferase and PAP homologues. The colored boxes indicate the conserved amino acids. There are 28 residues conserved between the B. subtilis homologue and the tRNA nucleotidyltransferases, 37 residues in common with the PAPs, and 55 residues in common with both groups of proteins. The somewhat-better alignment with the PAPs led to the prediction that this protein is a PAP, and its gene was named papS (29).
TABLE 1.
Putative Ntr proteins in the eubacteriaa
| Organism | Groupb | Genome sequence | Protein size (amino acids)c | Protein activityd | Reference |
|---|---|---|---|---|---|
| E. coli | PBγ | Complete | 473 | PAP | 5 |
| 412 | CCA | ||||
| H. influenzae | PBγ | Complete | 488 | PAP | 17 |
| 416 | CCA | ||||
| Pseudomonas aeruginosa | PBγ | Partial | 408* | PAP | University of Washington |
| 185* | CCA | ||||
| Actinobacillus actinomycetemcomitans | PBγ | Partial | 259* | PAP | University of Oklahoma |
| 207* | CCA | ||||
| Neisseria gonorrhoeae | PBβ | Partial | 470 | PAP | University of Oklahoma |
| 366* | CCA | ||||
| Neisseria meningitidis | PBβ | Partial | 375* | PAP | University of Oklahoma |
| 399 | CCA | ||||
| A. hydrophyla | GN | Partial | 203* | CCA | 26 |
| Francisella tularencis | GN | Partial | 84* | CCA | NCBIa |
| Chlamydia trachomatis | GN | Partial | 353* | ? | University of California |
| 290* | ? | ||||
| Helicobacter pylori | PBδɛ | Complete | 411 | ? | 48 |
| A. fermentans | GN | Partial | 343* | ? | 32 |
| Campylobacter jejuni | GN | Partial | 201* | ? | Sanger Center |
| Rickettsia prowazekii | GN | Partial | 149* | ? | University of Uppsala |
| Borrelia burgdorferi | SP | Complete | 410 | ? | 18 |
| B. subtilis | GP | Complete | 403 | ? | 29 |
| M. tuberculosis | GP | Complete | 482 | ? | Sanger Center |
| M. leprae | GP | Partial | 411 | ? | 19 |
| Streptococcus pyogenes | GP | Partial | 199* | ? | University of Oklahoma |
| Staphylococcus aureus | GP | Partial | 121* | ? | University of Oklahoma |
| Thermus thermophilus | Partial | 329* | ? | 3 | |
| Synechocystis | CB | Complete | 420 | ? | 28 |
| 949 | ? | ||||
| Aquifex | Complete | 512 | ? | 12 | |
| 845 | ? |
Putative Ntr proteins were identified by Blast (2) with the E. coli CCA-adding or PAP I protein sequence against databases described in published references or available at the following sites: University of Washington (www.genome.washington.edu), University of Oklahoma (www.genome.ou.edu), National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov), University of California (C. Fenner; ctgenome@socrates.berkeley.edu), Sanger Center (www.sanger.ac.uk), and University of Uppsala (www.unu.se).
PB, purple bacteria; GN, gram negative; SP, spirochetes; GP, gram positive; and CB, cyanobacteria.
Asterisks indicate that the protein was predicted from a partial coding sequence and the size is that of the incomplete protein.
For bacteria other than E. coli, PAP or tRNA nucleotidyltransferase (CCA) activity was predicted from the sequence. For bacteria that are only distantly related to E. coli, the activity could not be predicted (?).
FIG. 2.
Sequence alignment of the tRNA nucleotidyltransferase (CCA) and PAP I (PAP) homologues from members of the β and γ subdivisions of the purple bacteria (E. col, E. coli; A. hyd, A. hydrophila; A. act, Actinobacillus actinomycetemcomitans; H. inf, H. influenzae; P. aer, Pseudomonas aeruginosa; N. gon, Neisseria gonorrhoeae; N. nem, Neisseria meningitidis) with the related protein from B. subtilis (B. sub). The initial alignment was made with CLUSTAL (24, 25), and small realignments were made manually. The colored boxes indicate the positions of conserved amino acids of the CCA-adding family (blue), the PAP I family (pink), and both families (yellow). The asterisks show the conserved G-D-D-D residues that are the signature of the X polymerase family. The protein motifs used to design the degenerate inosine-containing oligonucleotides are underlined. The numbers in parentheses represent amino acids not shown in the alignment, and dashes represent gaps in the alignment.
Characterization of the B. subtilis papS gene.
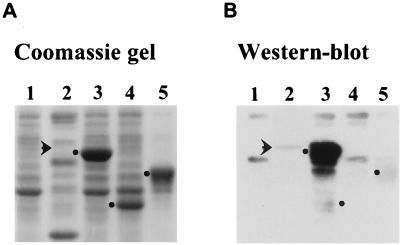
B. subtilis, a distant relative of E. coli, is a gram-positive organism that has been extensively studied. Because we were interested in characterizing PAP I-like enzymes, the papS gene was amplified by PCR and cloned, and the protein was expressed in E. coli. Using the genomic sequence, we chose the longest open reading frame, which begins with a methionine and encodes a protein of 403 amino acids. In the same manner, we cloned and expressed the E. coli CCA-adding enzyme, which, together with E. coli PAP I, served as a control for the following experiments. The overexpressed B. subtilis PapS protein migrated on SDS-polyacrylamide gels at the expected molecular mass of 45 kDa (Fig. 3A, lane 5). It was somewhat smaller than the E. coli PAP I, which migrates as a 50-kDa protein (Fig. 3A, lane 3). Figure 3B, a Western blot with an antibody against E. coli PAP I, shows the expected reaction with overexpressed PAP I (lane 3). In Fig. 3B, lanes 1 and 4, the protein that is somewhat smaller than the overexpressed PAP I is the endogenous E. coli PAP I that is processed by removing 17 amino acids from its amino terminus (7, 39). The faintly detected protein in Fig. 3B, lane 2, which is slightly larger than the overexpressed PAP I, is the B. subtilis protein that cross-reacts with the PAP I antibody (39). Note that PapS (Fig. 3B, lane 5), like the E. coli CCAase (Fig. 3B, lane 4), does not cross-react with the PAP I antibody, and it is smaller than the B. subtilis protein (Fig. 3B, lane 2). Thus, the papS gene product characterized here does not appear to be related to the B. subtilis protein that cross-reacts with the antibody against E. coli PAP I.
FIG. 3.
Expression and Western blot analysis of the B. subtilis PapS protein. Proteins were separated by SDS-polyacrylamide gel electrophoresis, and the gel was stained with Coomassie blue (A) or analyzed by Western blotting with the E. coli PAP I antibody (B). Lanes 1 and 2, E. coli and B. subtilis total proteins. Lanes 3 to 5, extracts of PAP I, E. coli tRNA nucleotidyltransferase, and B. subtilis PapS, respectively, were loaded to give similar amounts of each of the overexpressed proteins (see Materials and Methods). The dots in lanes 3 to 5 indicate the positions of the overexpressed proteins. The arrows in lanes 2 show the positions of a B. subtilis protein that cross-reacts with the PAP I antibody.
The activity of overexpressed PapS was measured under different assay conditions to discriminate between tRNA CCA addition and PAP activity. E. coli PAP I has detectable activity under both neutral and basic conditions (pHs 9 to 10), whereas the tRNA CCA-adding enzyme is only active under basic conditions (our data and reference 15). In these assays, either crude yeast tRNA, which contains molecules missing part or all of the CCA terminus, or poly(A) was used as the acceptor. The results shown in Table 2 indicate that the B. subtilis papS gene product has the same properties as the E. coli tRNA nucleotidyltransferase, with a comparable specific activity (published values for the E. coli enzyme range from 1,000 to 8,000 U/mg, depending on the preparation [14]). Like the E. coli CCA-adding enzyme, PapS adds AMP or CMP specifically to the tRNA acceptor, and it has no detectable activity under the polyadenylation assay conditions. As expected, PAP I, which is active under both assay conditions, only adds AMP and can use tRNA or poly(A) as an acceptor.
In Fig. 4, the products of the reaction under the CCA assay conditions with yeast tRNA and either [α-32P]ATP (Fig. 4A) or [α-32P]CTP (Fig. 4B) were characterized by electrophoresis on a denaturing polyacrylamide gel. E. coli PAP I (Fig. 4, lanes 1) elongated the tRNA, adding at least 400 AMP residues (Fig. 4A), but it is unable to incorporate CMP (Fig. 4B). Figure 4, lanes 2, contains a control mock preparation from cells without protein overexpression. Figure 4, lanes 3 and 4, shows the CCA-adding activity of the E. coli and B. subtilis enzymes, respectively. The molecules in the yeast tRNA population are extended to CCA (Fig. 4A) or to C and CC (Fig. 4B). The background in the high-molecular-weight regions of all the lanes in Fig. 4B is specific to the [α-32P]CTP label. The radiolabeled tRNA population in Fig. 4, lanes 3 and 4, is heterogeneous due to the size distribution of the yeast tRNAs (72 to 95 nucleotides [46]). Comparable profiles were observed when crude wheat germ tRNA was employed to characterize the CCA-adding enzyme from Sulfolobus shibatae (52). In all the data shown in Table 2 and Fig. 4, the activity of the B. subtilis PapS protein is indistinguishable from that of the E. coli tRNA nucleotidyltransferase. Thus, we conclude that PapS is a tRNA CCA-adding enzyme.
Is there another gene encoding an Ntr protein in B. subtilis?
The B. subtilis genome apparently contains only one gene encoding an Ntr protein, which we have now identified as a CCA-adding enzyme. However, a second gene may have been missed because of an error in the genomic sequence. About 4,200 genes have been identified in B. subtilis, and it has been estimated that another 100 to 200 genes will be identified as the genomic sequence is corrected (29). We tried two approaches to identify a second gene. First, we screened a λGT11 expression library, derived from B. subtilis (47), using our antibody against E. coli PAP I. Over 80,000 plaques were examined, but we failed to detect a protein clearly related to E. coli PAP I. In a second approach, we used a pair of inosine-containing oligonucleotides designed to hybridize to the regions encoding two highly conserved protein motifs in the eubacterial Ntr proteins (Fig. 2). Pilot experiments demonstrated that these primers could amplify DNA fragments of the correct size with templates containing the cloned E. coli pcnB or cca genes. In Fig. 5, a 260-bp DNA fragment amplified from the B. subtilis genomic DNA (lane 1), which was slightly smaller than the 290-bp DNA fragment from the cloned E. coli pcnB gene (lane 2), was detected. The genomic PCR product had the size predicted for the known B. subtilis papS gene, and restriction digestion confirmed this identification (data not shown). Under the PCR conditions employed here, several products of 450 bp or larger were detected due to the hybridization of the degenerate oligonucleotides to nonspecific sites. These products are too large to correspond to an Ntr coding sequence. Thus, we could not detect another gene by this method.
FIG. 5.
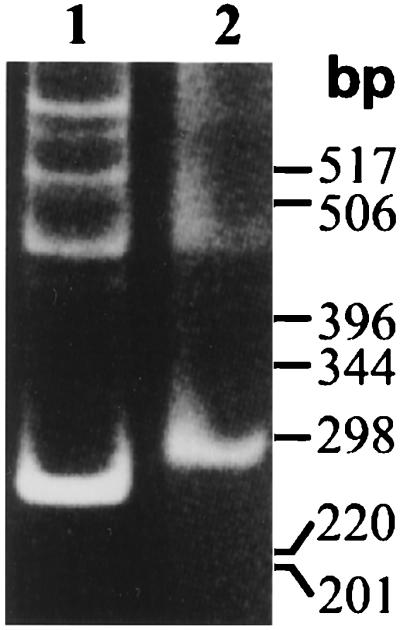
PCR amplification of the papS gene from B. subtilis genomic DNA with degenerate inosine-containing oligonucleotides designed to bind to DNA encoding two of the most highly conserved motifs in the eubacterial Ntr protein family (Fig. 2). Lane 1, B. subtilis genomic DNA; lane 2, cloned E. coli pcnB gene (in pET11a).
DISCUSSION
Our antiserum against E. coli PAP I was previously shown to detect proteins in several other gram-negative bacteria: Yersinia pseudotuberculosis, Erwinia carotovora, P. mirabilis, and D. gigas (39). We also detected a related protein in B. subtilis, a member of the low-G+C gram-positive family of bacteria (Fig. 1C). By Western blotting, the B. subtilis protein, with a mass of 55 kDa, reacted weakly with the antibody against E. coli PAP I, requiring a 10-fold-longer exposure for detection by chemiluminescence (39). The 45-kDa protein characterized here, which is the B. subtilis tRNA nucleotidyltransferase, does not react with the antibody against E. coli PAP I. The apparent lack of a PAP I homologue in B. subtilis suggests that the weak signal detected with the antibody against E. coli PAP I is due to a cross-reaction with an unrelated protein.
The genome of Mycobacterium tuberculosis, another gram-positive bacterium, has recently been completely sequenced, and like that of B. subtilis, it contains a single gene encoding a putative Ntr protein (Table 1). We have expressed and characterized the closely related protein from Mycobacterium leprae and found that it is also a tRNA nucleotidyltransferase (unpublished results). The finding that B. subtilis and M. tuberculosis each contain a single gene encoding a CCA-adding enzyme suggests that PAP I is either an ancient enzyme which has been lost in certain bacteria or that it a new enzyme in the purple bacteria arising from a recent duplication of the tRNA nucleotidyltransferase gene. It should be feasible to distinguish between these two possibilities when there is more information about the distribution of the CCA-adding and PAP I-like enzymes among the eubacteria.
Most of our understanding of RNA processing and degradation in the eubacteria comes from the study of E. coli. Of the proteins predicted by genomic sequencing, B. subtilis lacks RNase E, an endonuclease known to be important in E. coli rRNA processing and mRNA decay. Nevertheless, an RNase E-like activity has been suggested based on the study of the endonucleolytic processing of a tRNA synthetase message (10). When the tRNA synthetase gene from B. subtilis was transferred and expressed in E. coli, the message was processed at the same sites as in B. subtilis in an RNase E-dependent reaction. It was also correctly processed in vitro with purified RNase E. A related observation is that two PAP activities, suggested to be similar to E. coli PAP I and PAP II, have been described in B. subtilis (42). Thus, the failure to detect clearly discernible RNase E and PAP homologues in B. subtilis is unexpected, raising the possibility that these activities reside in proteins distinct from their counterparts in gram-negative bacteria.
ACKNOWLEDGMENTS
This research was supported by the Centre National de la Recherche Scientifique (CNRS), with additional funding from the Association pour la Recherche sur le Cancer (ARC). L.C.R. is a predoctoral fellow of the ARC.
REFERENCES
- 1.Aebi M, Kirchner G, Chen J-Y, Vijayraghavan U, Jacobson A, Martin N C, Abelson J. Isolation of a temperature-sensitive mutant with altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem. 1990;265:16216–16220. [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ashby M K, Bergquist P L. Cloning and sequence of IS1000 from Thermus thermophilus HB8. Plasmid. 1990;24:1–11. doi: 10.1016/0147-619x(90)90020-d. [DOI] [PubMed] [Google Scholar]
- 4.August J T, Ortiz J, Hurwitz J. Ribonucleic acid-dependent ribonucleotide incorporation. J Biol Chem. 1962;237:3786–3793. [PubMed] [Google Scholar]
- 5.Blattner F R, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Cao G-J, Pogliano J, Sarkar N. Identification of the coding region for a second poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:11580–11585. doi: 10.1073/pnas.93.21.11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao G-J, Sarkar N. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci USA. 1992;89:10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coburn G A, Mackie G A. Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3′-exonucleases dependent on oligoadenylation and RNA secondary structure. J Biol Chem. 1996;271:15776–15781. doi: 10.1074/jbc.271.26.15776. [DOI] [PubMed] [Google Scholar]
- 9.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 10.Condon C, Putzer H, Luo D, Grunberg-Manago M. Processing of the Bacillus subtilis thrS leader mRNA is RNase E-dependent in Escherichia coli. J Mol Biol. 1997;268:235–242. doi: 10.1006/jmbi.1997.0971. [DOI] [PubMed] [Google Scholar]
- 11.Cudny H, Lupski J R, Godson G N, Deutscher M P. Cloning, sequencing, and species relatedness of the Escherichia coli cca gene encoding the enzyme tRNA nucleotidyltransferase. J Biol Chem. 1986;261:6444–6449. [PubMed] [Google Scholar]
- 12.Deckert G, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 13.Dedonder R A, Lepesant J, Lepesant-Kejzlarova J, Billault A, Steinmetz M, Kunst F. Construction of a kit of reference strains for rapid genetic mapping in Bacillus subtilis. Appl Env Microbiol. 1977;33:989–993. doi: 10.1128/aem.33.4.989-993.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deutscher M P. tRNA nucleotidyltransferase. In: Boyer P D, editor. The enzymes. 3rd ed. Vol. 15. New York, N.Y: Academic Press; 1982. pp. 183–215. [Google Scholar]
- 15.Deutscher M P. Transfer RNA nucleotidyltransferase. Methods Enzymol. 1990;181:434–439. doi: 10.1016/0076-6879(90)81141-g. [DOI] [PubMed] [Google Scholar]
- 16.Deutscher M P, Foulds J, McClain W H. Transfer ribonucleic acid nucleotidyl-transferase plays an essential role in the normal growth of Escherichia coli and in the biosynthesis of some bacteriophage T4 transfer ribonucleic acids. J Biol Chem. 1974;249:6696–6699. [PubMed] [Google Scholar]
- 17.Fleischmann R D, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 18.Fraser C M, et al. Genomic sequence of a lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:539–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 19.Fsihi H, De Rossi E, Salazar L, Cantoni R, Labo M, Riccardi G, Akiff H E, Eiglmeier K, Bergh S, Cole S T. Gene arrangement and organization in an approximately 76 kb fragment encompassing the oriC region of the chromosome of Mycobacterium leprae. Microbiology. 1996;142:3147–3161. doi: 10.1099/13500872-142-11-3147. [DOI] [PubMed] [Google Scholar]
- 20.Gesteland R F. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966;16:67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- 21.Hajnsdorf E, Braun F, Haugel-Nielsen J, Regnier P. Polyadenylation destabilizes the rpsO mRNA of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:3973–3977. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haugel-Nielsen J, Hajnsdorf E, Regnier P. The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J. 1996;15:3144–3152. [PMC free article] [PubMed] [Google Scholar]
- 23.He L, Söderbom F, Gerhart E, Wagner H, Binnie U, Binns N, Masters M. PcnB is required for the rapid degradation of RNAI, the antisense RNA that controls the copy number of ColE1 related plasmids. Mol Microbiol. 1993;9:1131–1142. doi: 10.1111/j.1365-2958.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- 24.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 25.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignment on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 26.Jahagirdar R, Howard S P. Isolation and characterization of a second exe operon required for extracellular protein secretion in Aeromonas hydrophila. J Bacteriol. 1994;176:6819–6826. doi: 10.1128/jb.176.22.6819-6826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalapos M P, Cao G-J, Kushner S R, Sarkar N. Identification of a second poly(A) polymerase in Escherichia coli. Biochem Biophys Res Commun. 1994;198:459–465. doi: 10.1006/bbrc.1994.1067. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko, T., et al. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3(Suppl.):109–136. [DOI] [PubMed]
- 29.Kunst F, et al. The complete genome sequence of the Gram-positive bacterium B. subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 30.Liu J D, Parkinson J S. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J Bacteriol. 1989;171:1254–1261. doi: 10.1128/jb.171.3.1254-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 32.Mack M, Bendrat K, Zelder O, Eckel E, Linder D, Buckel W. Location of the two genes encoding glutaconate coenzyme A-transferase at the beginning of the hydroxyglutarate operon in Acidaminococcus fermentans. Eur J Biochem. 1994;226:41–51. doi: 10.1111/j.1432-1033.1994.tb20024.x. [DOI] [PubMed] [Google Scholar]
- 33.March J B, Collom M D, Hart-Davis D, Oliver I R, Master M. Cloning and characterization of an Escherichia coli gene, pcnB, affecting plasmid copy number. Mol Microbiol. 1989;3:903–910. doi: 10.1111/j.1365-2958.1989.tb00239.x. [DOI] [PubMed] [Google Scholar]
- 34.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and a catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 35.Masters M, March J B, Oliver I R, Collins J F. A possible role for the pcnB gene product of Escherichia coli in modulating RNA:RNA interactions. Mol Gen Genet. 1990;220:341–344. doi: 10.1007/BF00260507. [DOI] [PubMed] [Google Scholar]
- 36.Mikkelsen N D, Gerdes K. Sok antisense RNA from plasmid R1 is functionally inactivated by RNase E and polyadenylated by poly(A) polymerase I. Mol Microbiol. 1997;26:311–320. doi: 10.1046/j.1365-2958.1997.5751936.x. [DOI] [PubMed] [Google Scholar]
- 37.O’Hara E B, Chekanova J A, Ingle C A, Kushner Z R, Peters E, Kushner S R. Polyadenylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen G J, Woese C R, Overbeek R. The wind of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–3. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raynal L C, Krisch H M, Carpousis A J. Bacterial poly(A) polymerase: an enzyme that modulates RNA stability. Biochimie. 1996;78:390–398. doi: 10.1016/0300-9084(96)84745-6. [DOI] [PubMed] [Google Scholar]
- 39a.Raynal, L. C., and A. J. Carpousis. Unpublished data.
- 40.Reuven N B, Zhou Z, Deutscher M P. Functional overlap of tRNA nucleotidyltransferase, poly(A) polymerase I, and polynucleotide phosphorylase. J Biol Chem. 1997;272:33255–33259. doi: 10.1074/jbc.272.52.33255. [DOI] [PubMed] [Google Scholar]
- 41.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar B, Cao G J, Sarkar N. Identification of two poly(A) polymerases in B. subtilis. Biochem Mol Biol Int. 1997;41:1045–1050. doi: 10.1080/15216549700202111. [DOI] [PubMed] [Google Scholar]
- 43.Sarkar N. Polyadenylation of mRNA in bacteria. Microbiology. 1996;12:3125–3133. doi: 10.1099/13500872-142-11-3125. [DOI] [PubMed] [Google Scholar]
- 44.Sarkar N. Polyadenylation of mRNA in prokaryotes. Annu Rev Biochem. 1997;66:173–197. doi: 10.1146/annurev.biochem.66.1.173. [DOI] [PubMed] [Google Scholar]
- 45.Söderbom F, Binnie U, Masters M, Wagner E G H. Regulation of plasmid R1 replication: PcnB and RNase E expedite the decay of the antisense RNA, copA. Mol Microbiol. 1997;26:493–504. doi: 10.1046/j.1365-2958.1997.5871953.x. [DOI] [PubMed] [Google Scholar]
- 46.Söll D. Transfer RNA: an RNA for all seasons. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1993. pp. 157–184. [Google Scholar]
- 47.Suh J-W, Boylan S A, Price C W. Gene for the alpha subunit of B. subtilis RNA polymerase maps in the ribosomal protein gene cluster. J Bacteriol. 1986;168:65–71. doi: 10.1128/jb.168.1.65-71.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomb J F, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 49.Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 50.Xu F, Cohen S N. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature. 1995;374:180–183. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]
- 51.Xu F, Lin-Chao S, Cohen S N. The Escherichia coli pcnB gene promotes adenylation of antisense RNA I of ColE1-type plasmids in vivo and degradation of RNA I decay intermediates. Proc Natl Acad Sci USA. 1993;90:6756–6760. doi: 10.1073/pnas.90.14.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yue D, Maizels N, Weiner A M. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: characterisation of the hyperthermophile Sulfolobus shibatae. RNA. 1996;2:895–908. [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu L, Deutscher P M. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J. 1987;6:2473–2477. doi: 10.1002/j.1460-2075.1987.tb02528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]