Abstract
The complete Bacillus subtilis genome contains two genes with the potential to encode glutamate dehydrogenase (GlutDH) enzymes. Mutations in these genes were constructed and characterized. The rocG gene proved to encode a major GlutDH whose synthesis was induced in media containing arginine or ornithine or, to a lesser degree, proline and was repressed by glucose. A rocG null mutant was impaired in utilization of arginine, ornithine, and proline as nitrogen or carbon sources. The gudB gene was expressed under all growth conditions tested but codes for a GlutDH that seemed to be intrinsically inactive. Spontaneous mutations in gudB that removed a 9-bp direct repeat within the wild-type gudB sequence activated the GudB protein and allowed more-efficient utilization of amino acids of the glutamate family.
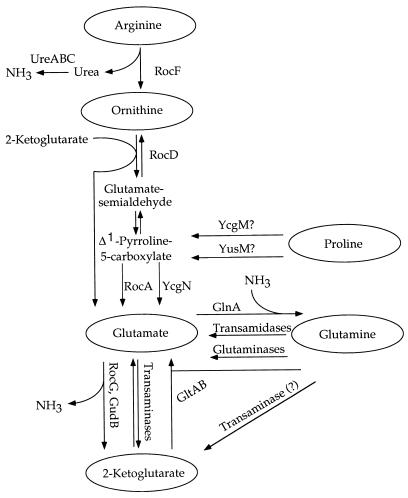
Interconversions of 2-ketoglutarate and glutamate are a major link between carbon and nitrogen metabolism in all living organisms. These reactions are catalyzed by several enzymes: glutamate synthase [2-ketoglutarate + glutamine + NAD(P)H → 2 × glutamate + NAD(P)], glutamate dehydrogenase (GlutDH) [2-ketoglutarate + NH3 + NAD(P)H ↔ glutamate + NAD(P)], and glutamate aminotransferases (transaminases) [2-ketoglutarate + amino acid ↔ glutamate + keto acid] (Fig. 1).
FIG. 1.
Pathways of utilization of amino acids of the glutamate family. Some enzymes are indicated by the names of their corresponding genes as follows UreABC, urease; RocF, arginase; RocD, ornithine aminotransferase; RocA and YcgN, Δ1-pyrroline-5-carboxylate dehydrogenases; YcgM and YusM, proline oxidases (proline dehydrogenases); RocG and GudB, glutamate dehydrogenases; GltAB, glutamate synthase; and GlnA, glutamine synthetase. Pathways of arginine and proline biosynthesis from glutamate are not shown.
Synthesis of Bacillus subtilis glutamate synthase, encoded by the gltAB operon, is positively regulated by GltC, a member of the LysR family of transcriptional regulators (5, 9), and negatively regulated by TnrA (4), a protein that also regulates other genes involved in nitrogen metabolism (47). Recently, we demonstrated that GltC activity is modulated by the activity of cellular GlutDH(s) and could be related to the rate of 2-ketoglutarate production (4). In order to understand GltC regulation, we sought to characterize mutations in genes coding for B. subtilis GlutDHs. The very existence of GlutDH activity in B. subtilis was a matter of some controversy (24, 27, 40), but the complete B. subtilis genome sequence (28) contains two genes highly similar to GlutDH-encoding genes of other organisms. The putative products of B. subtilis genes yweB (ipa-75d) (19), located at 331.0° on the chromosomal map (28) and here called rocG, and ypcA (41), located at 205.2° (28) and here called gudB, consist of 424 and 426 amino acids (aa), respectively, and are 74% identical to each other, 51 to 54% identical to GlutDHs from another gram-positive bacterium, Clostridium difficile (29), and an archaeon, Pyrococcus furiosus (14), and similar to many other hexameric GlutDHs from different organisms. The functions of the two B. subtilis genes were not known. In this work we analyzed involvement of these two genes in utilization of amino acids of the glutamate family. The rocG gene proved to encode the major catabolic GlutDH, while gudB seemed to encode an intrinsically inactive GlutDH. Spontaneous mutations in gudB generated active catabolic GlutDH.
MATERIALS AND METHODS
Bacterial strains and culture media.
Bacterial strains used in this study are listed in Table 1. B. subtilis strains were grown at 37°C in DS nutrient broth medium or in TSS minimal medium (16) with 0.5% glucose or 0.4% succinate as a carbon source and a 0.2% nitrogen source. The same media with the addition of agar were used for growth of bacteria on plates. L broth or L agar (31) was used for growth of Escherichia coli strains. The following antibiotics were used when appropriate: chloramphenicol (2.5 μg/ml), neomycin (2.5 to 5 μg/ml), phleomycin (0.25 μg/ml), tetracycline (15 μg/ml), or the combination of erythromycin (0.5 μg/ml) and lincomycin (12.5 μg/ml) for B. subtilis strains and ampicillin (50 to 100 μg/ml), kanamycin (25 μg/ml), or tetracycline (10 μg/ml) for E. coli strains.
TABLE 1.
Bacterial strains used in this study
| Organism and strain | Genotype | Source or reference |
|---|---|---|
| E. coli JM107 | endA1 gyrA96 thi hsdR17 (rk− mk+) supE44 relA1 λ− Δ(lac-proAB) e14−/F′ traD36 proAB lacIqlacZΔM15 | 48 |
| B. subtilis | ||
| SMY | Wild type | P. Schaeffer |
| BB1252 | rocA::pBB901 (neo) | SMY × pBB901 |
| BB1253 | ypdA::pBB902 (neo) | SMY × pBB902 |
| BB1267 | ΔrocG::ble | SMY × pBB918 |
| BB1268 | ΔgudB::tet | SMY × pBB920 |
| BB1271 | ΔrocG::ble ΔgudB::tet | BB1267 × DNA of BB1268 |
| BB1283 | ΔrocG::ble gudB1 | Spontaneous mutant of BB1267 |
| BB1284 | ΔrocG::ble gudB1 ypdA::pBB902 (neo) | BB1283 × pBB902 |
| BB1302 | gudB1 ypdA::pBB902 (neo) | SMY × DNA of BB1284 |
| BB1401 | ΔamyE::(gudB′-lacZerm) | SMY × pBB933 |
DNA manipulations and transformation.
The methods for plasmid isolation, agarose gel electrophoresis, use of restriction and DNA modification enzymes, DNA ligation, PCR, and electroporation of E. coli cells were as described by Sambrook et al. (34). B. subtilis chromosomal DNA was isolated by modification of a published procedure (16). Transformation of B. subtilis by chromosomal or plasmid DNA was performed by modification of the method of Anagnostopoulos and Spizizen (2).
Cloning of the rocG gene.
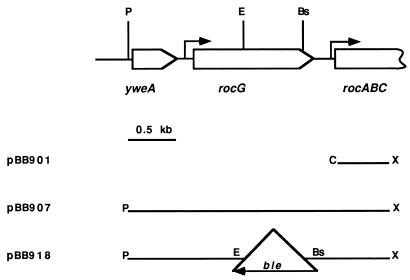
A PCR product corresponding to the 5′ end of rocA, the gene immediately downstream of rocG (Fig. 2), was synthesized by using rocA-specific oligonucleotides. The 0.52-kb ClaI-XhoI fragment derived from this product was cloned in pBB544, a derivative of pBluescript SK(−) (Stratagene, Inc.), containing a neomycin resistance marker (7). The resulting plasmid, pBB901, was integrated into the chromosome of B. subtilis SMY, creating strain BB1252. To clone DNA adjacent to the site of integration of pBB901, the chromosomal DNA of strain BB1252 was digested with PstI, self ligated, and introduced by electroporation into E. coli cells. The isolated plasmid, pBB907, had a 2.75-kb insert of chromosomal DNA carrying the entire coding regions of rocG and the upstream gene yweA.
FIG. 2.
Genetic map of the rocG region (19) and plasmids carrying different parts of this region. The restriction sites are abbreviated as follows: Bs, BstBI; C, ClaI; E, EcoRI; P, PstI; and X, XhoI. The ClaI and XhoI sites of the insert in pBB901 and derivative plasmids were constructed by PCR. All plasmids are derivatives of pBB544 (7). Transcription initiation sites for rocG (6) and rocABC (11) are shown by the arrows. The 1.4-kb ble cassette is not drawn to scale.
Cloning of the gudB gene.
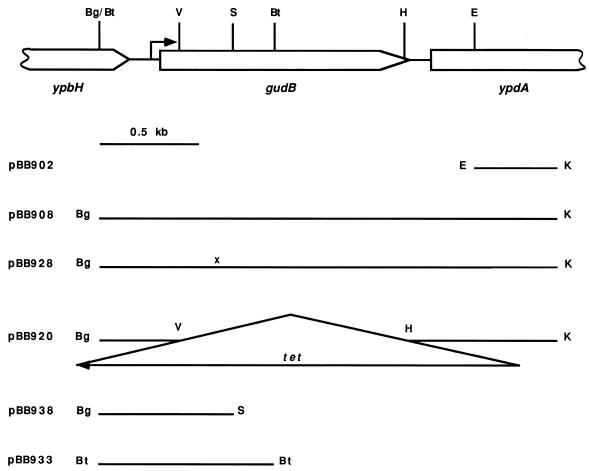
A PCR product corresponding to an internal part of the ypdA gene (Fig. 3) was synthesized by using ypdA-specific oligonucleotides. The 0.41-kb EcoRI-KpnI fragment derived from this product was cloned in pBB544 (7). The resulting plasmid, pBB902, was integrated into the chromosome of B. subtilis SMY. The chromosomal DNA of the resulting strain, BB1253, was digested with BamHI and BglII, self ligated, and introduced by electroporation into E. coli cells. The isolated plasmid, pBB908, had a 2.33-kb insert of chromosomal DNA carrying the entire coding region of gudB.
FIG. 3.
Genetic map of the gudB region (41) and plasmids carrying different parts of this region. The restriction sites are abbreviated as follows: Bg, BglII; Bt, BstYI; E, EcoRI; H, HindIII; K, KpnI; S, StyI; and V, EcoRV. Only relevant EcoRV, HindIII, and StyI sites are shown. The KpnI site of the insert in pBB902 and derivative plasmids was constructed by PCR. Plasmid pBB933 is a derivative of pJPM82 (7); other plasmids are derivatives of pBB544 (7). Construction of some plasmids is described in Materials and Methods; other plasmids were constructed by deleting or subcloning fragments of the gudB region. The location of the gudB transcription initiation site (see Results) is indicated by the arrow. The 1.9-kb tet cassette is not drawn to scale. x denotes the location of the gudB1 mutation within pBB928.
Construction of a rocG null mutant.
A deletion-insertion mutation within the rocG gene was created by replacing the 0.64-kb EcoRI-BstBI fragment of pBB907 (Fig. 2) with a 1.6-kb EcoRI-AccI ble cassette, excised from pJPM136 (25). (pJPM136 was constructed by cloning the 1.4-kb HincII-SspI fragment of pMK3 derivative [44rsqb;), carrying the ble gene of pUB110 [38] that determines resistance to phleomycin (Phlr), into the SmaI site of pJPM1 [33]. The orientation of the ble gene in this construction coincides with that of the lacZ gene.) The orientation of the ble gene in the resulting ΔrocG::ble plasmid, pBB918 (Fig. 2), is opposite that of the rocG gene. pBB918 was introduced into B. subtilis SMY, and Phlr Neos transformants, arising from double-crossover homologous recombination events, were selected. The replacement of the chromosomal rocG gene by the ΔrocG::ble allele in strain BB1267 was confirmed by comparing sizes of the PCR fragments from the wild-type and mutant rocG chromosomal loci.
Construction of a gudB null mutant.
A deletion-insertion mutation within the gudB gene was created by replacing the 1.16-kb EcoRV-HindIII fragment of pBB908 (Fig. 3) with a 1.9-kb BamHI-HindIII tet cassette, excised from pBEST307 (23). The orientation of the tet gene in the resulting ΔgudB::tet plasmid, pBB920 (Fig. 3), is opposite that of the gudB gene. pBB920 was introduced into B. subtilis SMY, and Tetr Neos transformants, arising from double-crossover homologous recombination events, were selected. The replacement of the chromosomal gudB gene by the ΔgudB::tet allele in strain BB1268 was confirmed by comparing sizes of the PCR fragments from the wild-type and mutant gudB chromosomal loci.
Mapping and cloning of the gudB1 mutation.
pBB902 (Fig. 3) was integrated into the chromosome of B. subtilis BB1283 (rocG gudB1), creating strain BB1284. The genetic linkage between the integrated plasmid and the gudB1 mutation was scored by the disappearance of the pale colony phenotype characteristic of rocG mutants (see Results) during subsequent transformation of BB1267 (rocG) cells to neomycin resistance by using BB1284 chromosomal DNA. The gudB1 allele of BB1284 was cloned as described above for the wild-type gudB allele, resulting in pBB928 (Fig. 3). The gudB mutations from other strains were cloned in a similar way.
Construction of a gudB-lacZ fusion.
The 0.89-kb BstYI fragment of pBB908 (Fig. 3) containing the 5′ part of gudB and 305 bp upstream of the gudB initiation codon was cloned at the BamHI site of pJPM82 (7). The resulting plasmid, pBB933 (Fig. 3), contained the gudB fragment fused to the promoterless E. coli lacZ gene in the proper orientation. Strain BB1401 (Table 1), carrying a gudB-lacZ transcriptional fusion integrated at the amyE locus, was isolated after transformation of strain SMY with pBB933, selecting for resistance to erythromycin and screening for loss of α-amylase production, which indicated a double-crossover homologous recombination event. The Amy phenotype was assayed with colonies grown overnight on tryptose blood agar base (Difco)–0.2% starch plates (16).
RNA isolation and primer extension.
Cells of B. subtilis were grown in TSS-glucose medium to late exponential phase. Pelleted cells from an 8-ml culture were resuspended in 0.1 ml of 20% sucrose–150 mM NaCl–1 mM EDTA and treated with lysozyme (0.4 mg/ml) for 20 min at 37°C. One milliliter of Trizol reagent (Gibco BRL, Life Technologies) was added, and RNA was extracted according to the manufacturer’s instructions. Primer extension experiments were performed by a modification of a previously described protocol (34) with the use of Superscript II reverse transcriptase (Gibco BRL, Life Technologies) and as primers, oligonucleotide oBB57 (5′-CTTATGTATTACGGTTTGGGTTG) or oBB60 (5′-CAATTCGTATACCTCTTCGGG), corresponding to positions +81 to +58 and +123 to +103, respectively, relative to the gudB initiation codon.
DNA sequencing.
Relevant parts of the gudB gene were sequenced by the dideoxy chain termination method of Sanger et al. (35) by using vector- or gudB-specific oligonucleotides as primers and a Sequenase reagent kit (Amersham Life Science) as recommended by the manufacturer. Plasmid double-stranded DNA to be sequenced was purified by a modification of an alkaline-lysis procedure (34) or by using a QIAprep Spin Miniprep Kit (Qiagen). DNA and protein sequences were analyzed by using DNA Strider and the BLAST program (1).
Enzyme assays.
β-Galactosidase activity was determined as described previously (8). The specific activity was expressed in Miller units (31).
To determine GlutDH activity, 30-ml cell cultures in the mid-exponential stage of growth were harvested, washed, and subjected to sonication in 3 ml of 50 mM Tris-Cl (pH 7.5)–20% glycerol–100 mM NaCl–1 mM EDTA–1 mM phenylmethylsulfonyl fluoride. Cell extracts were clarified by low-speed centrifugation, and 10- to 100-μl samples were assayed at room temperature in 1 ml of 55 mM Tris-Cl (pH 7.5)–2% glycerol–10 mM NaCl–100 mM NH4Cl–10 mM 2-ketoglutarate–0.2 mM NADH. Oxidation of NADH was monitored as the decrease in absorption at 340 nm. Nonspecific oxidation of NADH was determined from reactions lacking ammonia. One unit of GlutDH activity was defined as the amount needed to convert 1 nmol of NADH per mg of protein per min to NAD. The protein concentration was determined by the Bio-Rad Protein Assay with bovine serum albumin as a standard.
RESULTS
Characterization of a rocG mutant.
The yweB (ipa-75d) gene (19), which we had provisionally named gudA, was renamed rocG (6) because it belongs to the RocR regulon involved in utilization of arginine and ornithine (11, 17). rocG was cloned from the B. subtilis chromosome as a 2.8-kb PstI-XhoI fragment (Fig. 2), as described in Materials and Methods. A 0.6-kb internal fragment of rocG was replaced by the ble marker (Fig. 2) and the mutant ΔrocG::ble gene was substituted for the wild-type rocG allele in the chromosome (see Materials and Methods), resulting in strain BB1267 (Table 1).
BB1267 grew more slowly in nutrient broth medium (generation time of 52 min versus 36 min for the wild-type strain) and had a reduced cell yield (final optical density at 600 nm of 1.1 to 1.2 versus 2.3 to 2.5 for a wild-type strain). BB1267 lost the ability to utilize proline, ornithine, or arginine (Fig. 1) as sole carbon source in minimal medium and grew more slowly when proline or ornithine was utilized as sole nitrogen source (Table 2). BB1267 formed heat-resistant spores in nutrient broth medium at a normal frequency, although sporulating cells tended to form clumps and the course of sporulation and spore release was delayed (data not shown). Another insertion in the ipa-75d (rocG) gene was constructed by P. Glaser (18a) but had not been characterized. We used this mutation in our preliminary experiments and found that the mutant strain had the same phenotype as strain BB1267 (data not shown).
TABLE 2.
Growth of B. subtilis strains in TSS minimal medium
| Carbon source | Nitrogen source | Generation time of strain
|
||
|---|---|---|---|---|
| SMY (wild type) | BB1267 (ΔrocG::ble) | BB1284 (ΔrocG:: ble gudB1) | ||
| Glucose | Ammonia | 57 | 56 | 61 |
| Glutamine | 51 | NDa | ND | |
| Glutamate | 126 | 133 | 57 | |
| Proline | 80 | 110 | 60 | |
| Ornithine | 66 | 127 | 57 | |
| Arginine | 53 | 52 | 57 | |
| Proline | Proline | 92 | NGb | 90 |
| Ornithine | Ornithine | 82 | NG | 146 |
| Arginine | Arginine | 79 | NG | 138 |
ND, not done.
NG, no growth under these conditions.
Characterization of a ΔgudB::tet mutant.
The ypcA gene (41) was renamed gudB because it encodes a second putative GlutDH of B. subtilis. gudB was cloned from the chromosome as a 2.3-kb BglII-KpnI fragment (Fig. 3) as described in Materials and Methods. A 1.2-kb internal fragment of gudB was replaced by the tet marker (Fig. 3) and the mutant ΔgudB::tet gene was substituted for the wild-type gudB allele in the chromosome (see Materials and Methods), resulting in strain BB1268 (Table 1).
No growth defects were detected for strain BB1268 in nutrient broth or in minimal medium (data not shown). The phenotype of a rocG gudB double null mutant was very similar to the phenotype of a rocG single mutant. We conclude that rocG encodes the major GlutDH of B. subtilis and that gudB contributes little to the total cellular GlutDH activity.
Isolation of gudB gain-of-function mutants.
Strain BB1267 (rocG) forms pale colonies on DS nutrient broth plates, probably due to lower growth yield in this medium. Wild-type-like papillae arising spontaneously at high frequency on the surface of these pale colonies were isolated. These pseudorevertants regained the ability to utilize proline, ornithine, or arginine as sole carbon source and also acquired the ability to utilize glutamate or glutamine as sole carbon source (a wild-type strain cannot grow with glutamate or glutamine as sole carbon source). Spontaneous mutants capable of utilizing glutamate or glutamine as sole carbon source were also directly isolated from a wild-type strain after prolonged incubation in liquid minimal medium containing one of these amino acids as sole carbon and nitrogen sources. The latter mutants formed wild-type-like colonies even when they carried deletions in rocG.
Four independent mutations (two from rocG pseudorevertants, one from a glutamate-utilizing strain, and one from a glutamine-utilizing strain) were mapped to the vicinity of the gudB locus (see Materials and Methods and data not shown). By complementation analysis using an integrative plasmid pBB938 (Fig. 3) and similar plasmids, the mutations were mapped to the 5′ part of the gudB gene, upstream of the StyI site (Fig. 3) (data not shown). In accordance with this result, no Gud+ pseudorevertants could be obtained from a ΔrocG ΔgudB double mutant. Since these results implied that the mutations were gain-of-function alleles of gudB, the chromosomal gudB regions of the four gudB mutants were cloned (see Materials and Methods) and sequenced. All mutations, called gudB1, turned out to be identical deletions of 9 bp in the 5′ coding part of the gudB gene. This region of the wild-type gudB gene (which was resequenced and found to be identical to the published sequence) contains a direct repeat of 9 nucleotides, GTGAAGGCG, corresponding to a direct repeat of 3 aa at positions 93 to 95 and 96 to 98 of the 426-aa GudB protein. The gudB1 deletion precisely eliminated one copy of the repeat in both the nucleotide and amino acid sequences (Fig. 4).
FIG. 4.
Partial alignment of the RocG, GudB, GudB1, and C. symbiosum GlutDH (45) sequences. Amino acids are numbered with respect to their positions in corresponding proteins. The amino acids comprising βc and α6 of C. symbiosum GlutDH (3) and the site of a deletion of 3 aa in GudB are underlined. Residues conserved in most GlutDHs (45) are in bold. C. symbiosum K89 and K113 are essential lysines involved in glutamate binding (42).
Characterization of gudB1 mutants.
Strain BB1284 containing the gudB1 and ΔrocG::ble mutations, in addition to its ability to utilize glutamate or glutamine as sole carbon source, also grew faster than a wild-type strain in glucose minimal medium with glutamate, proline, or ornithine as sole nitrogen source (Table 2). In contrast, this strain had a slight growth defect in glucose-ammonia minimal medium that was more pronounced on glucose-ammonia agar plates (Table 2 and data not shown). The growth phenotype of a gudB1 single mutant (strain BB1302) in glucose medium was very similar to the growth phenotype of strain BB1284 (data not shown); i.e., the activity of RocG did not affect the GudB1 phenotype.
GlutDH activities of wild-type and mutant strains.
GlutDH activity was determined in cell extracts in the presence of ammonia and 2-ketoglutarate as substrates (see Materials and Methods). The level of total GlutDH activity in cells grown in nutrient broth medium was low in early exponential phase but reached higher levels in the middle and late stages of exponential growth (Table 3 and data not shown). GlutDH activity was not detected in cells grown in nutrient broth medium in the presence of glucose (Table 3), in agreement with earlier results on total GlutDH activity (27). GlutDH activity was very similar in a wild-type strain and in a ΔgudB::tet mutant, but very little, if any, activity was detected in a ΔrocG::ble mutant or in a double rocG gudB null mutant (Table 3). Thus, RocG but not GudB contributed to GlutDH activity in wild-type cells grown in nutrient broth medium.
TABLE 3.
GlutDH activity of a wild-type strain and GlutDH mutants in nutrient broth mediuma
| Medium | GlutDH activity ofa:
|
||||
|---|---|---|---|---|---|
| SMY (wild type) | BB1268 (ΔgudB::tet) | BB1267 (ΔrocG:: ble) | BB1271 (ΔrocG::ble ΔgudB::tet) | BB1284 (ΔrocG::ble gudB1) | |
| DS (nutrient broth) | 346 | 440 | ≤45 | ≤45 | 447 |
| DS (nutrient broth) + 1% glucose | ≤45 | ≤45 | ≤45 | ≤45 | 250 |
Cells of the indicated strains were grown in DS medium to mid-exponential growth phase (optical density at 600 nm of 0.6 to 0.8). GlutDH activity was assayed and expressed in units as described in Materials and Methods. All numbers are averages of at least two experiments, and the mean errors did not exceed 30%.
In cells grown in minimal medium, RocG activity, tested in strain BB1268 (ΔgudB::tet), was very low whenever glucose was present, and detectable activity was seen only when proline, ornithine or arginine was added (Table 4). High RocG activity was observed in the presence of ornithine or arginine when succinate replaced glucose or when these amino acids served as sole carbon and nitrogen sources (Table 4). Intermediate activity was detected when proline served as sole carbon and nitrogen sources (Table 4). The mechanism of glucose repression of RocG and the SigL-dependent regulation of RocG by RocR will be detailed elsewhere (6). GlutDH activity in a wild-type strain was indistinguishable from activity in a ΔgudB::tet mutant (data not shown). No GlutDH activity was detected in rocG mutant strain BB1267 under any growth conditions (data not shown), reflecting very low activity or lack of activity of the wild-type GudB in minimal medium.
TABLE 4.
GlutDH activity in minimal mediuma
| Carbon source | Nitrogen source | GlutDH activity of:
|
|
|---|---|---|---|
| RocG | GudB1 | ||
| Glucose | Ammonia | ≤45 | 159 |
| Glucose | Glutamate | ≤45 | 178 |
| Succinate | Glutamate | ≤45 | NDb |
| Glucose | Glutamine | ≤45 | ND |
| Succinate | Glutamine | ≤45 | ND |
| Glucose | Proline | 76 | 182 |
| Succinate | Proline | 67 | 391 |
| Proline | Proline | 246 | 465 |
| Glucose | Ornithine | 74 | 158 |
| Succinate | Ornithine | 792 | 430 |
| Ornithine | Ornithine | 769 | 633 |
| Glucose | Arginine | 97 | 106 |
| Succinate | Arginine | 998 | ND |
| Arginine | Arginine | 1,040 | 662 |
Cells were grown in TSS medium. GlutDH activity was assayed and expressed in units as described in Materials and Methods. Strains BB1268 (ΔgudB::tet) and BB1284 (ΔrocG::ble gudB1) were used for determination of RocG and GudB1 activities, respectively. All numbers are averages of at least two experiments, and the mean errors did not exceed 30%.
ND, not done.
Intermediate to high levels of GudB1-GlutDH activity could be measured in a ΔrocG::ble gudB1 strain, BB1284, both in nutrient broth and in minimal media (Tables 3 and 4); these activities were decreased two- to sixfold when glucose was present but varied little when cells were grown with different nitrogen sources.
In accordance with previous results on total GlutDH activity in B. subtilis cells (27), RocG and GudB1 were specific with respect to NADH and were not active with NADPH as substrate. With glutamate as substrate and NAD as cofactor, the catabolic activities of these enzymes were about 10-fold-less efficient than anabolic activities (data not shown). The inactivity of wild-type GudB, the specific requirements for RocG activation in minimal media, and the very low catabolic activity of RocG in vitro probably explain the failure to detect GlutDH activity in B. subtilis in several previous studies (reviewed in reference 27).
Expression of the gudB gene.
A gudB-lacZ transcriptional fusion containing the entire intergenic region between gudB and the preceding gene, including its putative transcription terminator (Fig. 3), was constructed (see Materials and Methods). This fusion was active in an otherwise wild-type strain, BB1401, in glucose-ammonia medium; expression of the fusion increased 1.7- to 3.7-fold when glutamate or related amino acids served as nitrogen source (Table 5). Changes in expression of the gudB-lacZ fusion under different growth conditions did not correlate perfectly with the activity of GudB1 measured under similar growth conditions (Table 4), but only relatively small variations in activity were seen in each case. Increasing the osmolarity of the medium by the addition of 0.5 M NaCl, which should lead to a highly elevated cellular proline pool (46), did not affect gudB expression (data not shown). The gudB-lacZ fusion was expressed in nutrient broth medium at a level similar to that in minimal medium and was not significantly affected by the presence of glucose (data not shown).
TABLE 5.
Expression of a gudB-lacZ fusion in minimal mediuma
| Carbon source | Nitrogen source | β-Galactosidase activity |
|---|---|---|
| Glucose | Ammonia | 50 |
| Glutamine | 102 | |
| Glutamate | 172 | |
| Proline | 183 | |
| Ornithine | 89 | |
| Arginine | 84 | |
| Proline | Proline | 181 |
| Ornithine | Ornithine | 162 |
| Arginine | Arginine | 144 |
Cells of strain BB1401 were grown in TSS medium. β-Galactosidase activity was assayed and expressed in Miller units as described in Materials and Methods. All numbers are averages of at least two experiments, and the mean errors did not exceed 20%.
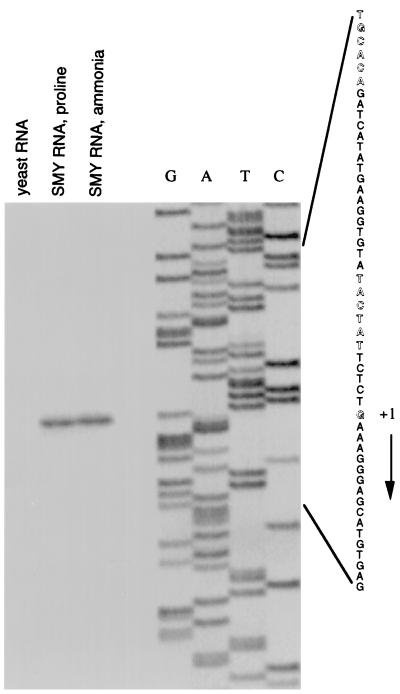
A primer extension experiment identified the putative transcription start point of gudB at position G (−46) with respect to the initiation codon (Fig. 5). Sequences similar to the −35 and −10 regions of a ςA-dependent promoter could be identified upstream of the gudB transcription start point (Fig. 5).
FIG. 5.
Primer extension analysis of gudB mRNA. Primer oBB60 was extended with reverse transcriptase by using total RNA from B. subtilis SMY grown in glucose minimal medium containing ammonia or proline as nitrogen source or from Saccharomyces cerevisiae (Sigma Chemical Company) as template. The sequence of the nontemplate strand of plasmid pBB917 deduced from sequencing reactions with oBB60 as primer is shown on the right. The apparent transcription start site of gudB and the −10 and the −35 regions of the likely gudB promoter are indicated by outlined letters. Primer oBB57 gave the same apparent gudB mRNA 5′ end (data not shown). The direction of transcription is shown by the arrow.
Attempts to identify the regulator of nitrogen source-dependent gudB expression have been unsuccessful so far. Expression of the gudB-lacZ fusion was affected less than twofold by mutations in genes known to be involved in nitrogen-source-dependent regulation, including sigL (13), tnrA (47), glnR (37), ahrC (30), gltC (9), or codY (39) (Table 6). In no case was nitrogen-source-dependent regulation eliminated. Mutations in ccpA (20), spo0A (21), sigB (32), or sigD (32) also had little or no effect on the regulation of gudB expression (Table 6). A mutation in abrB (43) reduced gudB-lacZ expression threefold in glucose-ammonia medium. gudB expression was not affected by ΔrocG::ble or ΔgudB::tet mutations; in a gudB1 strain, gudB-lacZ activity in glucose medium was reduced 2.4-fold (Table 6).
TABLE 6.
Expression of a gudB-lacZ fusion in regulatory mutants in minimal mediuma
| Genotype | β-Galactosidase activity in the presence of indicated nitrogen source
|
|
|---|---|---|
| Ammonia | Proline | |
| Wild type | 50 | 183 |
| tnrA | 58 | 179 |
| glnR | 47 | 179 |
| gltC | NGb | 209 |
| codY | 39 | 124 |
| ahrC | 90 | 160 |
| abrB | 15 | 150 |
| ccpA | 55 | 119 |
| spo0A | 62 | 150 |
| sigL | 51 | 203 |
| sigB | 35 | 151 |
| sigD | 40 | 140 |
| ΔgudB::tet | 57 | 197 |
| ΔrocG::ble ΔgudB::tet | 57 | 221 |
| gudB1 | 21 | 75 |
Strain BB1401 (gudB′-lacZ) and its derivatives were grown in glucose minimal medium. β-Galactosidase activity was assayed and expressed in Miller units as described in Materials and Methods. All numbers are averages of at least two experiments, and the mean errors did not exceed 20%.
NG, no growth under these conditions.
DISCUSSION
GlutDH, a ubiquitous metabolic enzyme, catalyzes a reversible reaction that can carry out either the anabolic function of glutamate biosynthesis or the catabolic function of glutamate utilization. In some organisms, both functions are active under different physiological conditions (40). It has been known for many years, however, that B. subtilis has little or no anabolic GlutDH, since it possesses only one route of glutamate biosynthesis from ammonia, catalysis by glutamate synthase (36). Although in vitro the anabolic activities of RocG and GudB1 are much higher than their catabolic activities, under no conditions tested could either of these B. subtilis GlutDHs compensate in vivo for the defect in glutamate synthesis caused by a glutamate synthase mutation (4). Both RocG and GudB1 utilize NAD(H), rather than NADP(H), as is typical of catabolic enzymes (40). Thus, our data show that both rocG and gudB (or at least gudB1) code for catabolic GlutDHs, involved in utilization of glutamate and other amino acids of the glutamate family.
Our results explain the general inability of B. subtilis strains of 168 lineage, derived from strain SMY (10), to utilize glutamate or glutamine as sole carbon source (4, 22), despite the presence of two GlutDH genes. Production of one GlutDH, RocG, requires the presence of arginine, ornithine, or proline; this enzyme is not induced by glutamate or glutamine. The second GlutDH, GudB, while synthesized in the presence of glutamate or glutamine, has either undetectably low enzymatic activity or none at all. Only constitutive expression of RocG, for instance, in strains carrying a rocRc mutation (6, 18) or activation of GudB by an unusual gain-of-function mutation, as described herein, allows cells to utilize glutamate or glutamine as sole carbon source. It is likely that at least some reports of growth of B. subtilis strains with glutamate as sole carbon source were due to accumulation of gudB1-like mutations or mutations causing constitutive expression of rocG.
Cells that grow with glutamate, proline, ornithine, or arginine as sole nitrogen source need to produce ammonia in order to synthesize glutamine via the glutamine synthetase reaction (36). Catabolic GlutDHs could be responsible for ammonia production from all these amino acids (Fig. 1). Slow utilization of glutamate as sole nitrogen source (Table 2) again reflects the lack of induction of RocG by glutamate and the inactivity of wild-type GudB. Accordingly, the growth rate in glucose-glutamate medium is not affected by null mutations in rocG or gudB or both and is highly increased when GudB is made active by the gudB1 mutation. Aminotransferases, enzymes that transfer the amino group of glutamate to various keto acids, are likely responsible for the low rate of utilization of glutamate as sole nitrogen source in a wild-type strain. Under these conditions ammonia is probably produced by degradation of some of the amino acids generated by transamination.
The ability of proline or ornithine to provide intermediate growth rates as sole nitrogen source in glucose medium (Table 2) is probably due to induction of RocG. In accord with this conclusion, a rocG null mutant utilized proline or ornithine as sole nitrogen source at the same low rate as it utilized glutamate (Table 2). Less than full induction of RocG-GlutDH activity is sufficient for utilization of ornithine or proline as nitrogen source inasmuch as RocG is poorly expressed in glucose-ornithine or glucose-proline minimal medium (Table 4), but cells grow significantly faster with ornithine or proline than with glutamate as sole nitrogen source (Table 2). More-efficient utilization of ornithine and proline in the presence of glucose was provided by higher levels of activity of GudB1-GlutDH (Tables 2 and 4).
Arginine supports a high growth rate because, in addition to generating glutamate by the three-step roc pathway, it liberates urea, which can be degraded to ammonia by the action of urease (Fig. 1), even though the latter is not highly active under these conditions (12). A high growth rate with glutamine as sole nitrogen source (Table 2) is expected, since, in this case, the cells do not need to generate ammonia to synthesize glutamine and glutamate can be synthesized from glutamine by glutamate synthase, by transamidation reactions, or by two putative glutaminases, YbgJ and YlaM (28) (Fig. 1). Because the high growth rates conferred by glutamine or arginine as sole nitrogen source are mostly determined by GlutDH-independent pathways, it is not surprising that growth rates on these substrates were little affected by the lack of GlutDH in a rocG mutant (Table 2). In a double rocG ureC mutant, lacking urease activity, utilization of arginine as sole nitrogen source was severely impaired (data not shown).
The GlutDH activities of RocG and GudB1 correlated well with the ability of different strains to utilize amino acids of the glutamate family as sole nitrogen sources (Tables 2 and 4). For unknown reasons, little or no correlation was seen when these amino acids were utilized as sole carbon sources (Tables 2 and 4).
Alignment of RocG and GudB sequences showed a single significant gap that coincided with the tandem repeat of 3 aa which is deleted in the gudB1 mutant (Fig. 4). The same 3-aa gap is seen in pairwise alignments of the wild-type GudB sequence with sequences of other GlutDHs in GenBank (Fig. 4 and data not shown), indicating that the wild-type gudB sequence contains an insertion of 3 aa (9 bp) with respect to the common ancestral GlutDH sequence. The mechanism by which this 3-aa (9-bp) insertion occurred is unclear. Elimination of this insertion in the spontaneous gudB1 mutants could arise by strand slippage mispairing during DNA replication (15). A similar 9-bp deletion within a stretch of DNA containing a near perfect repeat of 9 bp in the torS gene of Escherichia coli has been described (26). Interestingly, the latter deletion was also a gain-of-function mutation and led to formation of a constitutively active protein (26). Maintenance of the 9-bp insertion in the wild-type gudB sequence could be explained by the slight growth defect of gudB1 mutants in glucose-ammonia medium, presumably because the glutamate-synthesizing activity of glutamate synthase is overwhelmed by the glutamate-degrading activity of GudB1.
The GudB and GudB1 sequences were modelled against the available three-dimensional structures of GlutDH from Clostridium symbiosum (3, 42) and P. furiosus (49). The 3-aa insertion within GudB was localized to helix α6 of GlutDH (Fig. 4), and it likely affects both the amino acid composition of this helix and its relative orientation with respect to the upstream strand βC. Both βC and α6 form part of the substrate binding and catalytic pocket of GlutDH and could be involved in intersubunit interactions (3, 42).
In summary, wild-type B. subtilis has two genes for GlutDH, one of which (rocG) encodes an enzyme that is induced by arginine, ornithine, or proline and contributes to use of these compounds as carbon or nitrogen sources. The second GlutDH gene (gudB) encodes a protein with very low or no enzymatic activity. A frequently occurring spontaneous mutation renders this enzyme active, suggesting that natural or laboratory conditions in which B. subtilis strains of Marburg/168 lineage have been grown select for emergence and maintenance of the inactive form of GudB.
ACKNOWLEDGMENTS
We are grateful to K. Matsuno, M. Débarbouillé, S. Fisher, R. Gardan, C. Kumamoto, G. Rapoport, C. Alén, C. Jourlin, N. Mani, and M. Ratnayake-Lecamwasam for helpful discussions and for reading the manuscript; to S. Fisher, P. Glaser, and L. Wray for gifts of strains; and to M. Berne, Tufts University Protein and Nucleic Acid Analysis Facility, for synthesis of oligonucleotides.
This work was supported by U.S. Public Health Service grant GM36718.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnastopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker P J, Britton K L, Engel P C, Farrants G W, Lilley K S, Rice D W, Stillman T J. Subunit assembly and active site location in the structure of glutamate dehydrogenase. Proteins Struct Funct Genet. 1992;12:75–86. doi: 10.1002/prot.340120109. [DOI] [PubMed] [Google Scholar]
- 4.Belitsky, B. R. Unpublished data.
- 5.Belitsky B R, Sonenshein A L. Mutations in GltC that increase Bacillus subtilis gltA expression. J Bacteriol. 1995;177:5696–5700. doi: 10.1128/jb.177.19.5696-5700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belitsky, B. R., and A. L. Sonenshein. Unpublished data.
- 7.Belitsky B R, Gustafsson M C U, Sonenshein A L, von Wachenfeldt C. An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J Bacteriol. 1997;179:5448–5457. doi: 10.1128/jb.179.17.5448-5457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belitsky B R, Janssen P J, Sonenshein A L. Sites required for regulation of Bacillus subtilis glutamate synthase expression. J Bacteriol. 1995;177:5686–5695. doi: 10.1128/jb.177.19.5686-5695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohannon D E, Sonenshein A L. Positive regulation of glutamate biosynthesis in Bacillus subtilis. J Bacteriol. 1989;171:4718–4727. doi: 10.1128/jb.171.9.4718-4727.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burkholder P R, Giles N H. Induced biochemical mutations in Bacillus subtilis. Am J Bot. 1947;33:345–348. [PubMed] [Google Scholar]
- 11.Calogero S, Gardan R, Glaser P, Schweizer J, Rapoport G, Débarbouillé M. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J Bacteriol. 1994;176:1234–1241. doi: 10.1128/jb.176.5.1234-1241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz-Ramos H, Glaser P, Wray L V, Jr, Fisher S H. The Bacillus subtilis ureABC operon. J Bacteriol. 1997;179:3371–3373. doi: 10.1128/jb.179.10.3371-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Débarbouillé M, Martin-Verstraete I, Kunst F, Rapoport G. The Bacillus subtilis sigL gene encodes an equivalent of ς54 from Gram-negative bacteria. Proc Natl Acad Sci USA. 1991;88:9092–9096. doi: 10.1073/pnas.88.20.9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggen R I L, Geerling A C M, Waldkötter K, Antranikian G, de Vos W M. The glutamate dehydrogenase-encoding gene of the hyperthermophilic archaeon Pyrococcus furiosus: sequence, transcription and analysis of the deduced amino acid sequence. Gene. 1993;132:143–148. doi: 10.1016/0378-1119(93)90527-a. [DOI] [PubMed] [Google Scholar]
- 15.Farabaugh P J, Schmeissner U, Hofer M, Miller J H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978;126:847–863. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- 16.Fouet A, Sonenshein A L. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J Bacteriol. 1990;172:835–844. doi: 10.1128/jb.172.2.835-844.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardan R, Rapoport G, Débarbouillé M. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J Mol Biol. 1995;249:843–856. doi: 10.1006/jmbi.1995.0342. [DOI] [PubMed] [Google Scholar]
- 18.Gardan R, Rapoport G, Débarbouillé M. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol Microbiol. 1997;24:825–837. doi: 10.1046/j.1365-2958.1997.3881754.x. [DOI] [PubMed] [Google Scholar]
- 18a.Glaser, P. Unpublished data.
- 19.Glaser P, Kunst F, Arnaud M, Coudart M-P, Gonzales W, Hullo M-F, Ionescu M, Lubochinsky B, Marcelino L, Moszer I, Presecan E, Santana M, Schneider E, Schwezer J, Vertes A, Rapoport G, Danchin A. Bacillus subtilis genome project: cloning and sequencing of the 97 kb region from 325° to 333°. Mol Microbiol. 1993;10:371–384. [PubMed] [Google Scholar]
- 20.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of α-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 21.Hoch J A. spo0 genes, the phosphorelay, and the initiation of sporulation. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 747–755. [Google Scholar]
- 22.Iijima T, Diesterhaft M D, Freese E. Sodium effect of growth on aspartate and genetic analysis of a Bacillus subtilis mutant with high aspartase activity. J Bacteriol. 1977;129:1440–1447. doi: 10.1128/jb.129.3.1440-1447.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itaya M. Construction of a novel tetracycline resistance gene cassette useful as a marker on the Bacillus subtilis chromosome. Biosci Biotech Biochem. 1992;56:685–686. doi: 10.1271/bbb.56.685. [DOI] [PubMed] [Google Scholar]
- 24.Jahns T. Occurrence of cold-labile NAD-specific glutamate dehydrogenase in Bacillus species. FEMS Microbiol Lett. 1992;96:187–192. doi: 10.1016/0378-1097(92)90401-9. [DOI] [PubMed] [Google Scholar]
- 25.Janssen, P. J., and J. P. Mueller. Unpublished data.
- 26.Jourlin C, Bengrine A, Chippaux M, Méjean V. An unorthodox sensor protein (TorS) mediates the induction of the tor structural genes in response to trimethylamine N-oxide in Escherichia coli. Mol Microbiol. 1996;20:1297–1306. doi: 10.1111/j.1365-2958.1996.tb02648.x. [DOI] [PubMed] [Google Scholar]
- 27.Kane J F, Wakim J, Fischer R S. Regulation of glutamate dehydrogenase from Bacillus subtilis. J Bacteriol. 1981;148:1002–1005. doi: 10.1128/jb.148.3.1002-1005.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine K M, Dusterhoft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Henaut A, Hilbert H, Holsappel S, Hosono S, Hullo M F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee S M, Levine A, Liu H, Masuda S, Mauel C, Medigue C, Medina N, Mellado R P, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O’Reilly M, Ogawa K, Ogiwara A, Oudega B, Park S H, Parro V, Pohl T M, Portetelle D, Porwollik S, Prescott A M, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror S J, Serror P, Shin B S, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Tognoni A, Tosato V, Uchiyama S, Vandenbol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler E, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa H F, Zumstein E, Yoshikawa H, Danchin A. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 29.Lyerly D M, Barroso L A, Wilkins T D. Identification of the latex test-reactive protein of Clostridium difficile as glutamate dehydrogenase. J Clin Microbiol. 1991;29:2639–2642. doi: 10.1128/jcm.29.11.2639-2642.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller C M, Baumberg S, Stockley P G. Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol Microbiol. 1997;26:37–48. doi: 10.1046/j.1365-2958.1997.5441907.x. [DOI] [PubMed] [Google Scholar]
- 31.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 32.Moran C P., Jr . RNA polymerase and transcription factors. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 653–667. [Google Scholar]
- 33.Mueller J P, Bukusoglu G, Sonenshein A L. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J Bacteriol. 1992;174:4361–4373. doi: 10.1128/jb.174.13.4361-4373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schreier H J. Biosynthesis of glutamine and glutamate and assimilation of ammonia. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 281–298. [Google Scholar]
- 37.Schreier H J, Brown S W, Hirschi K D, Nomellini J F, Sonenshein A L. Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J Mol Biol. 1989;210:51–63. doi: 10.1016/0022-2836(89)90290-8. [DOI] [PubMed] [Google Scholar]
- 38.Semon D, Movva N R, Smith T F, El Alama N, Davies J. Plasmid-determined bleomycin resistance in Staphylococcus aureus. Plasmid. 1987;17:45–63. doi: 10.1016/0147-619x(87)90007-2. [DOI] [PubMed] [Google Scholar]
- 39.Slack F J, Serror P, Joyce E, Sonenshein A L. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol Microbiol. 1995;15:689–702. doi: 10.1111/j.1365-2958.1995.tb02378.x. [DOI] [PubMed] [Google Scholar]
- 40.Smith E L, Austen B M, Blumenthal K M, Nyc J F. Glutamate dehydrogenases. In: Boyer P D, editor. The enzymes. 3rd ed. XI. New York, N.Y: Academic Press; 1975. pp. 293–367. [Google Scholar]
- 41.Sorokin A, Azevedo V, Zumstein E, Galleron N, Ehrlich S D, Serror P. Sequence analysis of the Bacillus subtilis chromosome region between the serA and kdg loci cloned in a yeast artificial chromosome. Microbiology. 1996;142:2005–2016. doi: 10.1099/13500872-142-8-2005. [DOI] [PubMed] [Google Scholar]
- 42.Stillman T J, Baker P J, Britton K L, Rice D W. Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis. J Mol Biol. 1993;234:1131–1139. doi: 10.1006/jmbi.1993.1665. [DOI] [PubMed] [Google Scholar]
- 43.Strauch M A. AbrB, a transition state regulator. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 757–764. [Google Scholar]
- 44.Sullivan M A, Yasbin R E, Young F E. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- 45.Teller J K, Smith R J, McPherson M J, Engel P C, Guest J R. The glutamate dehydrogenase gene of Clostridium symbiosum. Cloning by polymerase chain reaction, sequence analysis and overexpression in Escherichia coli. Eur J Biochem. 1992;206:151–159. doi: 10.1111/j.1432-1033.1992.tb16912.x. [DOI] [PubMed] [Google Scholar]
- 46.Whatmore A M, Chudek J A, Reed R H. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J Gen Microbiol. 1990;136:2527–2535. doi: 10.1099/00221287-136-12-2527. [DOI] [PubMed] [Google Scholar]
- 47.Wray L V, Jr, Ferson A E, Rohrer K, Fisher S H. TnrA, a transcriptional factor required for global nitrogen regulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:8841–8845. doi: 10.1073/pnas.93.17.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 49.Yip K S P, Stillman T J, Britton K L, Artymiuk P J, Baker P J, Sedelnikova S E, Engel P C, Pasquo A, Chiaraluce R, Consalvi V, Scandurra R, Rice D W. The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures. Structure. 1995;3:1147–1158. doi: 10.1016/s0969-2126(01)00251-9. [DOI] [PubMed] [Google Scholar]