Abstract
Background:
CRISPR/Cas9 technology-mediated genome editing has significantly improved the targeted inactivation of genes in vitro and in vivo in many organisms. Neuropilins play crucial roles in zebrafish heart regeneration, heart failure in mice, and electrical remodeling after myocardial infarction in rats. But the cell-specific functions of nrp1 have not been described before. In this study, we have investigated the role of nrp1 isoforms, including nrp1a and nrp1b, in cardiomyocytes during cardiac injury and regeneration in adult zebrafish hearts.
Methods:
In this study, we have reported a novel CRISPR-based vector system for conditional tissue-specific gene ablation in zebrafish. Specifically, the cardiac-specific cardiac myosins light chain 2 (cmlc2) promoter drives Cas9 expression to silence the neuropilin-1(nrp1) gene in cardiomyocytes in a heat-shock inducible manner. This vector system establishes a unique tool to regulate the gene knockout in both the developmental and adult stages and hence, widens the possibility of loss-of-function studies in zebrafish at different stages of development and adulthood. Using this approach, we investigated the role of neuropilin isoforms nrp1a and nrp1b in response to cardiac injury and regeneration in adult zebrafish hearts.
Results:
We observed that both the isoforms (nrp1a and nrp1b) are upregulated after the cryoinjury. Interestingly, the nrp1b-knockout significantly delayed heart regeneration and impaired cardiac function in the adult zebrafish after cryoinjury, demonstrated by reduced heart rate, ejection fractions, and fractional shortening. In addition, we show that the knockdown of nrp1b but not nrp1a induces activation of the cardiac remodeling genes in response to cryoinjury.
Conclusions:
To our knowledge, this is the first study where we have reported a heat shock-mediated conditional knockdown of nrp1a and nrp1b isoforms using CRISPR/Cas9 technology in the cardiomyocyte in zebrafish, and furthermore have identified a crucial role for nrp1b isoform in zebrafish cardiac remodeling and eventually heart function in response to injury.
Keywords: CRISPR, Zebrafish, Cardiomyocytes, Regeneration, Neuropilin-1, Myocardial Infarction, Cryoinjury
Graphical Abstract
Introduction
CRISPR/Cas9 technology has become a powerful tool for targeted genome editing1; adapted from the bacterial immune mechanism2, it utilizes the advantage of the RNA-dependent detection of specific DNA sequences by Cas9 endonuclease. The designed guide RNA (gRNA) comprises a 3’ secondary structure that can interact with Cas9. It also contains a 20 bases long 5’ seed sequence that leads to targeting. The Cas9 screens the genome and cleaves it within sequences complementary to the seed sequence, which are immediately followed by the adjacent protospacer motif (PAM) NGG3. Previously, genes could efficiently be inactivated by transfection of gRNAs and Cas9 mRNA or DNA into bacteria, human, or mouse cells4–6.
Zebrafish (Danio rerio) has emerged as a popular model system for various biological studies, including cardiovascular diseases7–9. Specific characteristics such as transparent embryos, a short life cycle10, a highly conserved innate immune system with humans11, and ease of genetic manipulation12 make the zebrafish ideal for live imaging and dissecting new mechanisms regulating cardiovascular development. The application of the CRISPR/Cas9 tool in zebrafish is well documented. Previously, injection of gRNAs and Cas9 mRNA into one-cell stage embryos has been shown to produce indels at the target loci with comparatively high, although varying, rate of occurrence 13, 14. CRISPR-mediated mutations in zebrafish are heritable, allowing mutant strain’s speedy generation. Since gene deactivation by CRISPR/Cas9 tool is comprehensive and stable, this technology has provided an effective complementary method to morpholinos for loss-of-function studies in zebrafish, especially at subsequent stages of development. As the loss of some genes leads to embryonic lethality, there is a strong requirement to generate conditional, tissue-specific knockouts in the field. Previously, Ablain et al. reported a CRISPR-based vector system that enables stable, tissue-specific gene inactivation in vivo15. In this study, we report a novel CRISPR-based vector system that is modified from the previous vector described by Ablain et al. 15 for heat-inducible, cardiomyocyte-specific gene ablation in adult zebrafish. In this vector, the cardiac-specific, cardiac myosin light chain 2 (cmlc2) promoter drives the Cas9 expression to silence the neuropilin-1(nrp1) gene in cardiomyocytes in a heat-shock inducible manner. This vector system establishes a unique tool to knock out genes in both early and adult stages, expanding the likelihood of loss-of-function studies in zebrafish at different stages of development.
In contrast to mammals, adult zebrafish hearts can regenerate after injury. Neuropilins are co-receptors that have been reported previously to play crucial roles in zebrafish heart regeneration16, heart failure in mice17, and electrical remodeling after myocardial infarction in rats 18. But the cell-specific functions of nrp1 have not been described. In this study, we have investigated the role of nrp1 isoforms, including nrp1a and nrp1b, in cardiomyocytes during cardiac injury and regeneration in adult zebrafish hearts. Our results showed that both the isoforms (nrp1a and nrp1b) are upregulated in the adult zebrafish heart after the cryoinjury. Cardiomyocyte-specific nrp1b is required explicitly for cardiac repair in adult zebrafish.
In this study, we employed the novel, conditional CRISPR/Cas9 mediated tissue-specific gene inactivation system and showed that nrp1b isoform played a crucial role in adult zebrafish heart repair and function after injury. To our knowledge, this is the first study that described the heat shock inducible, conditional, and spatiotemporal knockout of tissue-specific genes in zebrafish. This tool can help study tissue-specific gene inactivation in a spatiotemporal manner.
Method
Data Availability Disclosure Statement
The authors declare that all supporting data and method descriptions are available within the article or from the corresponding authors upon reasonable request.
Zebrafish Maintenance
Wildtype zebrafish (Danio rerio) were maintained at the zebrafish core facility at Mayo Clinic, Jacksonville, FL. The fish room was held on light control of 14 h of light in a day at 29 °C. Adult zebrafish were kept in 2.75 L and 6 L tanks on a water recirculation rack system with a male-to-female ratio of 1:2. Adult zebrafish were fed a range of dry fish food. Adult male and female zebrafish were separated for one week before breeding. Embryos were harvested by breeding two females and one female. The animal experiments were performed as per the Mayo Clinic-approved IACUC protocol number A000018914–14.
Construction of a heat shock inducible tissue specific CRISPR vector for gene targeting in zebrafish
We derived the current vector from a previous study15. Briefly, we cloned the zebrafish Hsp70 promoter from the AB zebrafish wildtype strain cDNA followed by a NheI site restriction enzyme digestion and ligation into the pcmlc2:GFP destination vector 15, 19 using ClaI and KpnI enzymes. The gRNA scaffold used by the Joung lab 14 was modified to replace BsaI enzyme sites with BseRI sites, which were inserted at the start site of the U6 promoter as described earlier15. The 20-bp target sequence of interest was designed using the online tool CHOPCHOP (http://chopchop.cbu.uib.no/). The sgRNA was cloned into the gRNA scaffold. We combined this with the middle-entry vector containing the Cas9 (codon-optimized for zebrafish) developed by the Chen lab20. The Cas9 can be introduced under any promoter of interest into pcmlc2:GFP and U6:gRNA in a single cloning step by using multisite Gateway cloning technology21. The destination vector was gifted from Dr. Leonard I Zon (Addgene plasmid).
Heat shock treatment
Heat shock was performed by transferring the zebrafish from 28.5°C to 37°C embryo water and incubating at 37°C to induce the gRNA expression. Briefly, to induce the heat shock promoter expression, heat shock was induced by raising the water temperature from 28.5°C to 37°C over 15 min, then maintaining at 37°C for 30 min, and finally decreasing it to 28.5°C over the next 15 min in a programmable water bath as described22, 23.
mRNA and gRNA synthesis
Tol2 mRNA was produced by in vitro transcription from a pCS2 vector as described previously15,20 using mMESSAGE mMACHINE SP6 kit (Thermofisher Scientific). The gRNAs target sequence was identified by using the online tool CHOPCHOP 24, and was produced following established methods 14. Three target sequences were tested for each isoform. The Hsp70 and Cmlc2 promoters were amplified from the cDNA established from wild-type AB embryos using the following primers: Hsp70 forward: 5’-TCAGGGGTGTCGCTTGGTTATTTCC-3,’ and Hsp70 reverse; 5’-ATTTATACAATTTATGGTGCAATTG-3’, cmlc2 forward: 5’-GCTTAAATCAGTTGTGTTAAATAAGAGAC-3’, and cmlc2 reverse: 5’-GGTCACTGTCTTGCTTTGCTG-3’. The cDNA sequence was then cloned into a middle entry clone for Gateway and inserted into the pDest Tol2 pA2 vector between an SP6 promoter and a polyA sequence as described earlier 15.
Microinjections
20 pg of CRISPR DNA constructs and 20 pg of Tol2 mRNA were injected into one-cell stage wildtype zebrafish embryos (AB strain) by using the Harvard apparatus’s pico injector (PLI-100). After microinjection, embryos were raised in the embryo water at 28°C-29°C until adulthood.
Each component of the CRISPR construct was confirmed by sanger sequencing and restriction digestion before final assembly, as described in Ablain et al., 201515. The CRISPR editing was confirmed by Sanger sequencing and T7E1 mutagenesis assay. CRISPR/Cas9 mediated nrp1a and nrp1b ablation was confirmed by Western blotting.
T7E1 Mutagenesis Assay
T7E1 assay was performed as reported15. Briefly, genomic DNA was extracted from 2-day-old embryos using the HotSHOT method. A fragment of approximately 400 bp was amplified from genomic DNA using the following primers. For nrp1a, forward: 5’-TGTTTGTTTTTGGTTTGTCAGC-3’, and reverse: 5’-CTTGTGTCGTACTCTATGCAACG-3’. For nrp1b: forward: 5’-CAGCACACAAGGGTTACCATAA-3’, and reverse: 5’-GTCAATGCATCACTATGGCAGTAACCC-3’.
Whole-mount in situ hybridization and antibody labeling
We cloned approximately 460 bp-long fragments from the 3’ end of the nrp1a and nrp1b gene and cloned them into the PCR-TOPO 4 vector (Invitrogen) using primers (nrp1a forward: 5’-GCGCTCTTTCGGTTTCCTTC-3’, nrp1a reverse: 5’-TGAAGAGCTGGTTTCCCGAC-3’,nrp1b forward: 5’- ATCCCAACAACCTGGAGTGC-3’ and nrp1b reverse: 5’- GGCGTCCAGCCATTTTCAAG-3’) and synthesized both the sense and anti-sense RNA probes by in vitro transcription. In situ, hybridization was performed following established protocols25. The cryosection was stained with the cardiomyocyte-specific myosin heavy chain (MYH6) antibody labeled with Alexa-488 using the standard protocol.
For the FISH experiment, the nrp1a and nrp1b probes were labeled with Cy3 using the HyperScribe™ T7 High Yield Cy3 RNA Labeling Kit (APExBio, MA) as outlined in the manufacturer’s protocol. The in situ hybridization procedure was carried out using the Cy3 labeled probes as described previously25, followed by immunofluorescence labeling with CM-specific MYH6 antibody or F-actin-specific Phalloidin-Alexa 488 (Thermofisher) following the published protocol26.
Reagents and Antibodies
The anti-NRP1, anti-VE-Cadherin, and anti-BrdU antibodies were purchased from Cell Signaling (Danvers, MA). BrdU (Bu20a) Mouse mAb #5292, Neuropilin-1 (D62C6) Rabbit mAb #3725, VE-Cadherin (D87F2) XP® Rabbit mAb #2500. Anti-MYH6 antibody was purchased from Abcam (Anti-MYH6 antibody [3–48 G5C7] (ab207926). Alexa Fluor™ 488 Phalloidin (A12379) was purchased from Thermofisher Scientific. Anti-beta-actin- antibody (MABT825) was purchased from Millipore Sigma.
Imaging
Embryos were mounted in 0.8% low melting point agarose containing tricaine (160 mg/L) for imaging. The images were acquired using an Olympus dissecting microscope, Zeiss Axio vision microscope, Raman microscope, and LSM 880 Confocal microscope.The in-situ images in Fig S4 were captured by EVOS M5000 microscope at 63x magnification. The images in the figures were processed by Adobe Photoshop to adjust the contrast.
Cryoinjury
Zebrafish aged 10–12 months were anesthetized in 0.1% tricaine (Sigma Aldrich) and placed in a damp sponge on the ventral side. As described previously, a small incision was made through the chest with iridectomy scissors to access the heart27. The ventricular wall was directly frozen by a stainless steel cryoprobe precooled in liquid nitrogen and applied for 23–25 seconds. The tip of the cryoprobe was approximately 6 mm long with a diameter of 0.8 mm. In order to avoid the complete freezing of the heart by the probe, fish water at room temperature was added to the head of the cryoprobe. Post-cryo-injury, the zebrafish were transferred to the normal fish water to recover and were then allowed to regenerate for various times at water temperatures 28°C-29°C.
ECG recordings and acquisition
At least 3 ECGs were recorded from each zebrafish at every time point. To reduce the biological variations, the same animals were used for recordings before the injury and at the following time points after surgery. The zebrafish were anesthetized by immersion in 0.015% tricaine solution for 60 seconds. Anesthetized zebrafish were placed ventral side up in a sponge. Each ECG was recorded for 60–120 seconds using iWORX ECG instrument (iWorx.Inc) by following the previous method 28, and then the zebrafish recovered in water free of tricaine.
Echocardiogram
Echocardiography was performed on conscious zebrafish using a Vevo 2100 and 3100 high-resolution imaging system with an MS-700S transducer (VisualSonics). Zebrafish were sedated in 0.015% tricaine and immobilized on a sponge immersed in fish water. Imaging was performed on the short axis (perpendicular to the fish) and the long axis (parallel to the fish) using B-mode using the Vevo 2100 and 3100 Cardiac Package, taking the average measurement of three consecutive contractions as described earlier29.
RNA extraction
Zebrafish heart ventricles were dissected and quickly frozen in liquid nitrogen. This was followed by thorough homogenization in 700 ml QIAzol reagent (Invitrogen, Carlsbad, USA) using a motor cordless homogenizer (Kimble, 749540–0000, USA) at maximum speed. Chloroform (250 μl) was added to the homogenized embryo/tissue, followed by shaking vigorously for 15 s and incubating at room temperature for 5 min. The samples were then centrifuged at 13,000 rpm for 10 min at 4°C. The upper aqueous phase containing RNA was carefully transferred to a new tube without disturbing the interface. RNA was precipitated by adding an equal volume of 70% ethanol and loaded onto a spin column from an RNeasy mini kit (Qiagen, Valencia, USA) according to the manufacturer’s instructions.
First-strand cDNA synthesis and RT PCR
Total RNA (1 μg) was reverse transcribed to produce cDNA using a superscript cDNA Synthesis kit (Biorad) as described in the manual. In all cases, the qRT-PCRs were performed using SYBR green PCR master mix. Standard reactions (10 μl) were assembled as follows: 5 μl of SYBR green qPCR supermix-with Rox (Invitrogen), 1 μl of forward primer (5 μM), 1 μl of reverse primer (5 μM), 1 μl of the template and 20 μl nuclease-free water. Templates were 1:10 diluted cDNA samples, and in the case of negative controls, cDNAs were replaced by nuclease-free water. All real-time assays were carried out in triplicate using an Applied Biosystems QuantStudio real-time PCR platform using the primers listed in Table 1. Forty amplification cycles were performed, with each cycle consisting of 94 °C for 15 s followed by 60 °C for 1 min. The primers used are listed in Supplemental Table 1.
Histology
For Hematoxylin and Eosin staining, the heart tissue was fixed in 4% PFA overnight at 4°C and submitted to the core facility at Mayo Clinic, Jacksonville. Briefly, the tissues were dehydrated and embedded in paraffin blocks. Sections were cut at the thickness of 5 μm, rehydrated, and stained with Mayer’s Hematoxylin for 12 minutes. The nuclear staining was differentiated for 5 seconds in 0.37% HCl prepared in 70% ethanol, and the slides were washed in running tap water for 10 minutes. The protein staining was obtained by incubating for 10 minutes in 0.1% Eosin Y solution in water with a drop of acidic acid, followed by a rapid wash in water. The tissue sections were dehydrated in a water/ethanol series, cleared in xylol, and mounted in Entellan medium (Merck). All consecutive sections of 5 hearts per group were photographed for morphometric analysis using an Aperio scanner (Leica). The infarct region and the intact myocardium were demarcated, and the sizes were measured using ImageJ software. The percentage of the infarct size relative to the entire ventricle was calculated.
Statistics
All analyses were performed using GraphPad Prism 9.0 (GraphPad Software Inc., La Jolla, CA, USA). Experiments were routinely repeated at least three times. All values are expressed as the mean ± SEM. Normality and the logarithmic test were performed using GraphPad software to confirm the normal distribution of the data. The homogeneity of variance was analyzed using Levene’s test. Statistical differences were determined to be significant at p < 0.05. Statistical significance was evaluated with Prism 9.0 software by using a t-test or One-way/two-way ANOVA (multiple comparisons).
Results:
nrp1a and nrp1b are expressed in the heart of zebrafish embryos and adult zebrafish.
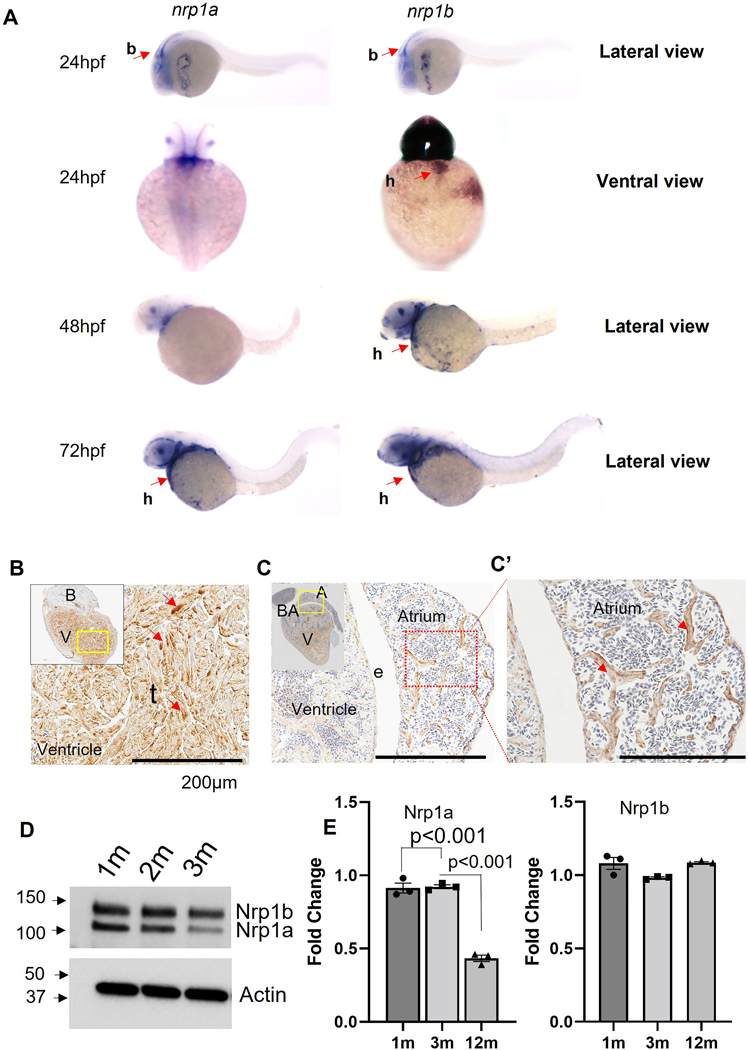
We first checked nrp1a and nrp1b mRNA levels in the developing zebrafish hearts using in situ hybridization. Both nrp1a and nrp1b were observed to be expressed in the brain of zebrafish embryos at 24h and 48h post fertilization (Fig 1A). The nrp1b isoform was expressed in the heart of 48 hpf and 72 hpf embryos (Fig 1A). The nrp1a isoform also showed expression in the hearts at 72 hpf (Fig 1A). As shown in Fig 1B, the immunostaining shows that nrp1 isoforms, including nrp1a and nrp1b, were ubiquitously expressed in the ventricles and the trabecular muscles in the cardiac atrium and ventricle of 12-month-old zebrafish.
Figure 1. Nrp1 expression in the zebrafish embryo and the adult heart.
(A). Representative In-situ hybridization image showing nrp1a and nrp1b expression in 1 dpf, 36 hpf, 2 dpf, and 3 dpf zebrafish embryo (Red arrowheads indicate the expression in the heart. (B, C, and C’) Immunohistochemistry image showing Nrp1 expression in adult zebrafish heart (Yellow and the red dotted box indicates the area under focus in the enlarged image in B, C, and C’, and the red arrowhead indicates the NRP1 expression as indicated by the staining), (D) Western blot showing the NRP1a and NRP1b expression in adult heart at 1, 3 and 12 months (m). (E) Quantification of D. Error bars represents the mean ± SEM from three experiments. Scale bar = 200μm. b, brain; h, heart; e, endothelium; t, trabecula. Statistical significance was evaluated with Prism 9.0 software using one-way ANOVA with Sidak’s multiple comparison test.
Next, Western blot analysis of the protein lysates prepared from 1-, 3-, and 12-months old zebrafish heart ventricles confirms that both the nrp1 isoforms are expressed in adult hearts. The Western blot showed two bands at ∼130-kDa and 150-kDa, corresponding to Nrp1a [916 amino acids (aa)] and Nrp1b (959 aa), respectively (Fig 1D). As shown in Fig 1E, the quantification of the Western bands showed a differential protein expression level of the Nrp1a isoforms at the three stages of development (1 mo, 3 mo, and 12 mo). The nrp1b isoform showed consistent expression levels in all three stages analyzed. To test the specificity of the NRP1 antibody used in our study, we have also performed Western blotting to check the specific NRP1 knockdown in mouse CM treated with NRP1 shRNA. We tested two shRNAs to target mouse ventricle cardiomyocytes (HL-1). As shown in Figure S1A, immunofluorescence staining confirmed the NRP1 knockdown in the cardiomyocytes after shRNA treatment. Western blot analysis also confirms the NRP1 downregulation after shRNA treatment (Fig S1 B–C). We also confirmed the expression of NRP1 in cardiomyocytes using both adult zebrafish heart ventricle specimens and cultured mouse ventricle cardiomyocytes (HL-1) (Fig S2 and S3), which were co-stained with cardiomyocyte-specific marker myosin heavy chain 6 (MYH6). Furthermore, in situ, hybridization using isoform-specific probes and the immunofluorescent staining with CM-specific MYH6 antibody was performed to examine the CM-specific expression of these two isoforms. The nrp1a and nrp1b probes (purple-blue) displayed overlapping expression with the CM marker MYH6 (green) in the zebrafish heart section (Fig S4A and B). To further confirm the CM-specific expression of these isoforms, fluorescence in situ hybridization (FISH) was performed by using Cy3 labeled nrp1a and nrp1b probes (sense and anti-sense) followed by immunofluorescent staining with F-actin labeling phalloidin and MYH6 specific antibody. Our results confirmed the expression of these isoforms in the cardiomyocytes, as shown by the co-staining of cardiomyocyte-MYH6 in green and nrp1 isoforms probe in red (Fig S5A and B). Bright red punctates in sections of both isoforms were noticed (Fig S5A and B). Detailed analysis under higher magnification (100x) suggests these red punctuates are outside the nucleus (Fig S6 A). We also noticed that this red color punctuates are less evident in the nrp1b probe stained sections. We will investigate this sub-cellular localization of nrp1 isoforms in detail in our future studies. Using the sense probe, we did not observe any specific fluorescent signals (Figure S6B). These results collectively support the expression of both nrp1a and nrp1b in CMs. However, a detailed analysis will be required to confirm and verify the difference in the protein level at different stages of development, such as the early developmental patterns of these genes. It will be interesting to analyze the protein and mRNA expression levels of the isoforms at various developmental stages and in adulthood, which will help explain the roles of the individual isoforms at the corresponding stages.
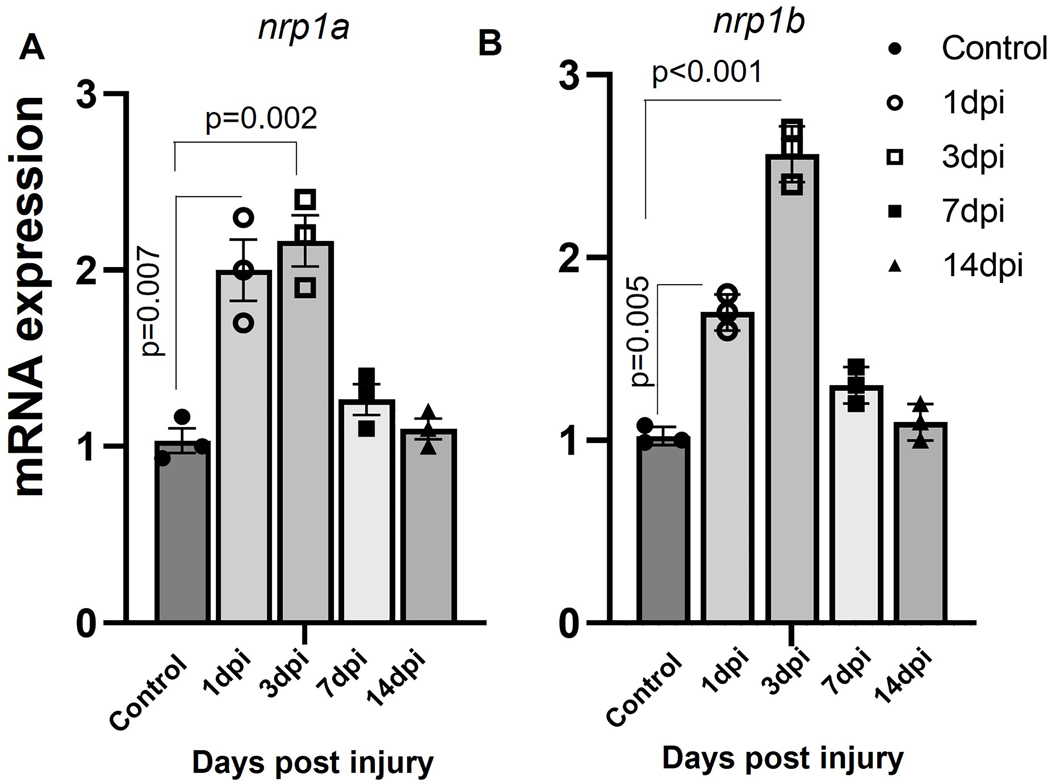
nrp1a and nrp1b are upregulated after cryoinjury in zebrafish adult hearts.
Next, we quantified how cryoinjury regulated the nrp1a and nrp1b mRNA levels in ventricles using qPCR. The nrp1a and nrp1b mRNA levels were upregulated on day 1, continued to increase 3–5 folds on day 3, and restored to normal levels at day 14 post cryoinjury (Fig. 2A and B). In situ hybridization analysis shows cardiomyocyte-specific expression of both nrp1a and nrp1b (Fig S4 B), confirming an enriched expression of cardiomyocyte-specific nrp1a and nrp1b levels after the injury.
Figure 2. Differential expression of nrp1a and nrp1b in the injured heart.
(A). RT PCR showing the upregulation of nrp1a mRNA expression after injury in zebrafish adult heart. (B) qRT PCR showing the upregulation of nrp1b mRNA expression in adult zebrafish hearts after cryoinjury. Briefly, the RNA was isolated from both the control and injured heart after 1d, 3d, 7d, and 14-day after cryoinjury, and the relative mRNA expression was analyzed. The error bars represent the mean ± SEM. The experiments were repeated three times. Statistical significance was evaluated with Prism 9.0 software using one-way ANOVA with Sidak’s multiple comparisons tests.
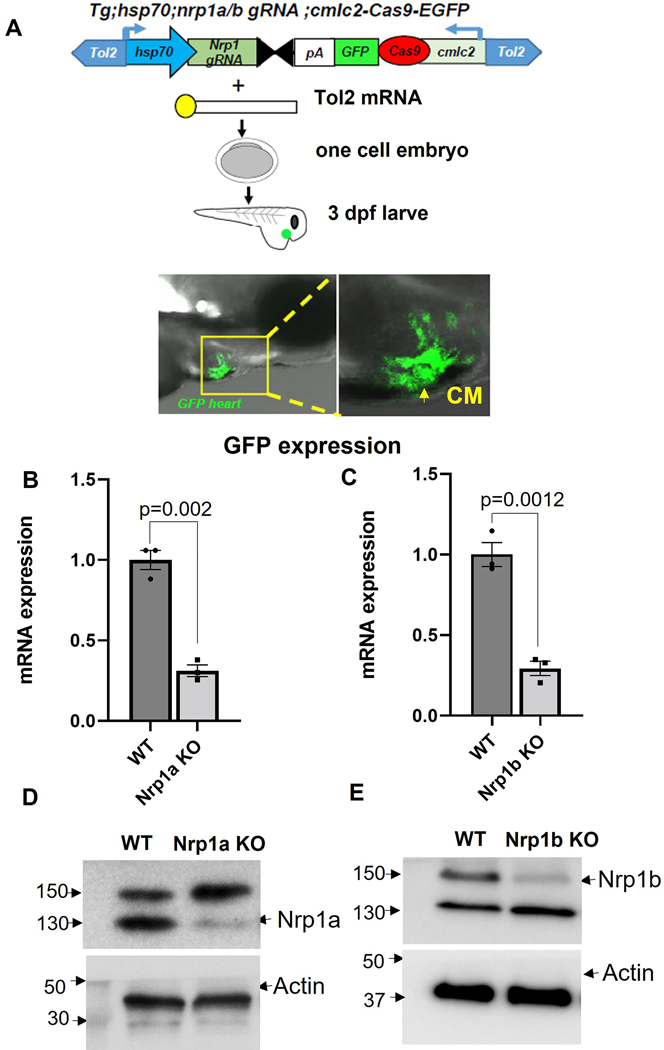
Construction of an integrative CRISPR vector for gene targeting in zebrafish heart
To study the spatiotemporal role of Nrp1 isoforms in the zebrafish heart, we developed a conditional tissue-specific CRISPR-Cas9 system. We developed a Tol2 integrative vector by modifying a previous vector developed by Ablain et al., 2015. As shown in Figure 3A, a zebrafish heat shock promoter (Hsp70) drives the expression of a nrp1a or nrp1b gRNA scaffold, into which zebrafish codon-optimized Cas9 flanked by two nuclear localization signals are cloned 20. The sgRNA is placed under the control of the Hsp70 promoter, which was cloned by using the Gateway cloning technology21, and green fluorescence protein (GFP) was identified downstream of the Cas9, which serves as a transgenesis marker19. Here cardiac myosin light chain (cmlc2) promoter drives the expression of the Cas9 flanked by GFP to induce the cardiomyocyte-specific expression of Cas9 mRNA (Fig 3A).
Figure 3. Outline of the CRISPR-Cas9 mediated nrp1a and nrp1b knockout.
(A). Schematic showing nrp1a and nrp1b targeted Crispr-Cas9 construct, and tol2 mRNA mediated integration into the 1–2 cell zebrafish embryo confirmed by the green fluorescence protein expression in the heart. (B and C) Relative mRNA expression of nrp1a and nrp1b showing nrp1a and nrp1b knockdown after heat shock induction in adult zebrafish heart. (D and E) Western blotting showed nrp1a and nrp1b protein expression knockdown in the adult zebrafish heart at 3 months (3m). Hsp70; heat shock protein promoter, which drives nrp1 gRNA expression, and cmlc2; Cardiomyocyte specific, cardiac myosin light chain 2 promoters which drive the Cas9 expression in the heart. The green fluorescence protein expression in the heart confirms cas9 expression. Tol2; Tol2 transposase, SV40 polyA sequence (pA). The qPCR and the western experiments were repeated at least three times (biological replicates), and the error bars indicate the mean ± SEM. Statistical significance was evaluated with Prism 9.0 software by using a student’s t-test.
AB zebrafish strain embryos were injected with vectors, along with Tol2 mRNA, at 1 cell stage. The fluorescent signal was observed specifically in the heart as early as 24 hpf, and it becomes more prominent at 3 dpf, as shown in Fig 3A. We did not observe any toxicity upon expression of our vectors in embryos apart from that usually associated with the microinjection of DNA plasmids at the one-cell stage.
The CRISPR vector allows tissue-specific gene disruption in zebrafish.
To determine the efficiency of gene targeting, we first performed the T7E1 mutagenesis assay30. In whole embryos, significant mutation rates at the nrp1a and nrp1b target locus were only detected with the vector containing a gRNA against nrp1a or b and the Hsp70 promoter (Fig S7A–B). Injection of vectors expressing Cas9 under the control of the cardiomyocyte-specific cmlc2 promoters and with the Hsp70 promoter driving the expression of a gRNA against nrp1a and nrp1b disrupts the nrp1 gene and protein expression after 72 hours post heat shock induction as depicted by the mRNA expression (Fig 3 B and C), and protein level (Fig 3 D and E). Sanger sequencing confirmed the genome editing (Fig S7B). Briefly, we applied heat shock to zebrafish by placing them in 37°C water bath for 30 minutes as described in the method, followed by tissue collection 72 hours later. As shown in Fig S5A, nrp1a knockout zebrafish have diminished nrp1a expression in the CMs. Similarly, nrp1b knockout zebrafish have reduced nrp1b levels in the CMs (Fig S5B). These data indicate heat shock successfully induced ablation of the nrp1a and nrp1b genes in the adult zebrafish heart.
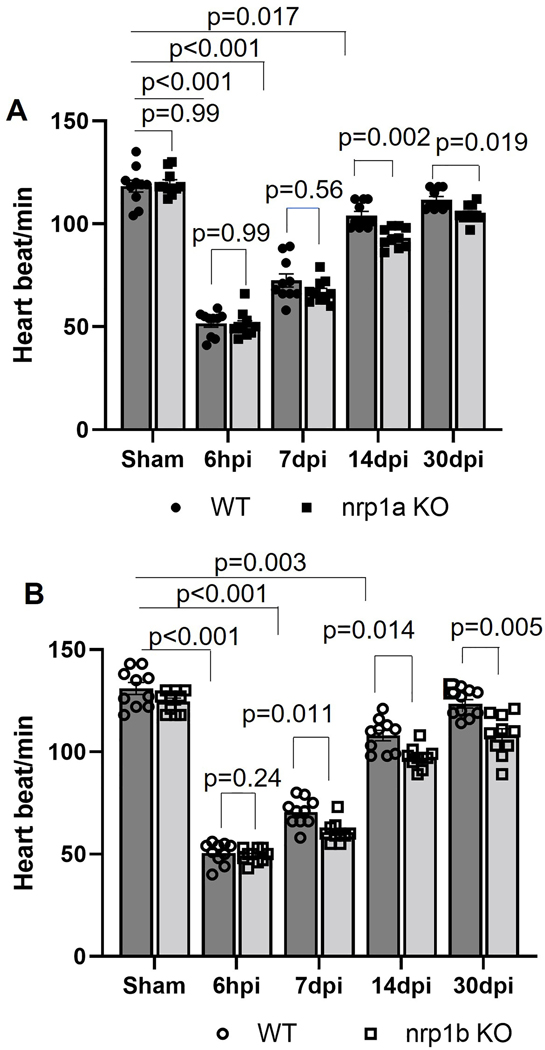
nrp1a and nrp1b abrogation have different effects on zebrafish cardiac function after cryoinjury in the adult heart.
To determine whether nrp1a and nrp1b abrogation alter heart function, we next utilized a noninvasive electrocardiogram (ECG) to analyze the heart rate in the 10 −12-month-old zebrafish before and after cryoinjury. The mean R-R intervals were used to evaluate the heart rate variation. In the WT zebrafish, decreased heartbeats were observed at 6 hpi and slowly recovered to normal levels at 14 dpi (Fig 4 A–B). Deletion of nrp1a reduced the heart rate at 7 dpi and 14 dpi but did not significantly affect the heartbeats at 30 days post cryoinjury as compared to the wildtype (Fig. 4A). Although nrp1b deficient zebrafish showed a similar level of heartbeats at baseline and at 6 hpi, their heart rates were significantly reduced at 7 dpi, 14 dpi, and 30 dpi, compared to WT zebrafish (Fig. 4B). The heat shock is known to affect the heart rate31. However, in the current study, we applied heat shock for 30 min to induce the NRP1 gRNA expression, and the functional analysis was carried out at least 3 days later when zebrafish are likely to recover from heat shock.
Figure 4. Heart rate analysis in 6-month-old WT and NRP1 KO zebrafish.
(A). Graph showing the heart rate recorded from N=10, WT, and N=10, nrp1a KO fish. (B). Graph showing the heart rate recorded from N=10, WT, and N=10 nrp1b KO zebrafish embryo. The bars represent the average values, and the error bars represent the mean ± SEM. Statistical significance was evaluated with Prism 9.0 software by using repeated measures of two-way ANOVA with Sidak’s tests (multiple comparisons).
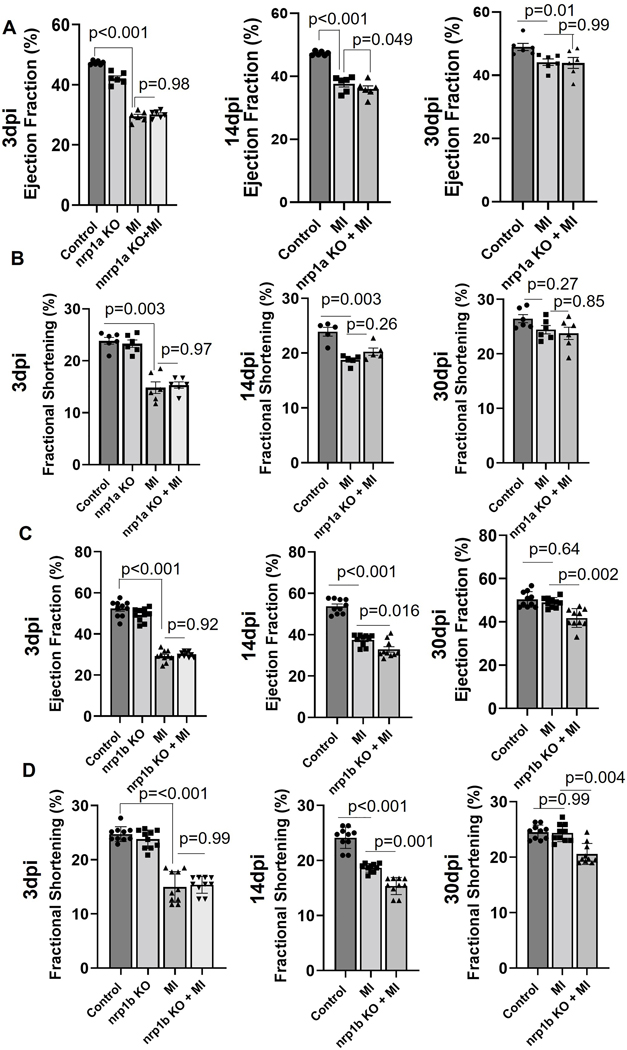
Next, we analyzed the cardiac function by echocardiography. As shown in Fig 5 (A–B), we observed that in the WT zebrafish, both ejection fraction (EF) and fractional shortening (FS) were reduced at 3 dpi and then recovered to normal levels at 30 dpi, indicating successful cardiac regeneration post-injury. Abrogation of the nrp1a isoform in the zebrafish heart does not affect the cardiac function 30 day post-injury, as shown by the similar level of EF and FS, compared with WT zebrafish. However, nrp1b deficient zebrafish showed delayed recovery of cardiac function and significantly reduced EF and FS at 30 dpi, compared to the WT group (Fig 5 C–D). Our results suggested that cardiomyocyte specific-nrp1b is required to fully recover after zebrafish cryoinjury.
Figure 5. Heart function analysis in adult WT and nrp1a and nrp1b KO zebrafish.
(A). Graph showing the ejection fraction (EF), recorded from N=6, WT, and nrp1a KO fish. and (B). Graph showing the fraction shortening (FS) recorded from N=6, WT, and nrp1b KO zebrafish embryo. CRISPR-Cas9 mediated nrp1b knockout delayed the cardiac function recovery after injury in adult zebrafish heart. (C). Graph showing the EF, recorded from N=10, WT, and nrp1b KO fish. (D). Graph showing the FS recorded from N=10, WT, and nrp1b KO zebrafish embryo. The bars represent the average value, and the error bars indicate the mean ± SEM. Statistical significance was evaluated with Prism 9.0 software by using one-way ANOVA with Sidak’s multiple comparisons tests.
nrp1b ablation results in increased expression of cardiac remodeling genes in adult zebrafish hearts after cryoinjury.
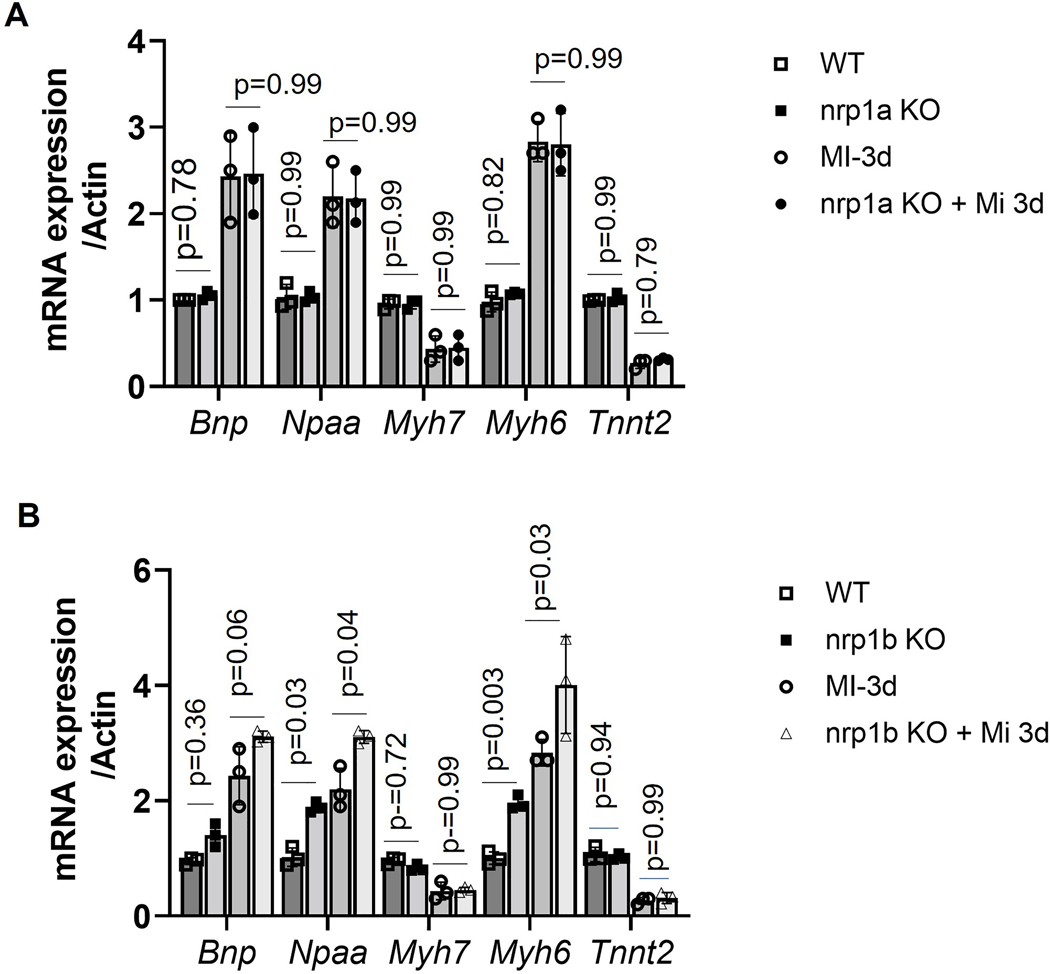
Previously, we have shown that Nrp1 ablation in mouse cardiomyocytes upregulates fetal gene expression32. To investigate the effects of nrp1a and nrp1b expression on fetal gene expression in 8 – 9 month-old zebrafish heart post cryoinjury, we measured the cardiac hypertrophy and stress markers, including the brain natriuretic peptide (Bnp), atrial natriuretic factor (Anf/Nppa), Myh6, and the Troponin gene (Tnnt2). As shown in Fig 6 A, the expression of Bnp, Nppa, and Myh6 were not changed in the nrp1a knockout group as compared to the WT, at both baselines or after cryoinjury. On the other hand, we observed that in the nrp1b knockout zebrafish, the expression of Bnp, Myh6, and Nppa was upregulated at both baselines and after cryoinjury compared to the WT zebrafish (Fig 6 B). Knockout of nrp1a or nrp1b did not alter the expression level of tnnt2 (Fig. 6B). These results suggest that deletion of nrp1b but not nrp1a aggravated the expression of cardiac remodeling genes after cryoinjury.
Figure 6. nrp1a and nrp1b abrogation results in differential cardiac remodeling gene expression in adult zebrafish hearts.
nrp1a and nrp1b have different regulatory effects on cardiac remodeling in adult zebrafish hearts. (A) mRNA expression of cardiac remodeling genes in heart isolated from the adult 8–9-month-old wildtype (WT) and nrp1a KO zebrafish after 3 days post cryoinjury (3dpi). (B). mRNA expression of cardiac remodeling genes in heart isolated from the adult 8–9-month-old wt and nrp1b KO zebrafish heart after 3dpi. The qRT PCR experiments were repeated three times (N=3 hearts were combined to isolate the RNA in each group), and the error bars indicate the mean ± SEM. Statistical significance was evaluated with Prism 9.0 software by using two-way ANOVA with Sidak’s multiple comparisons tests.
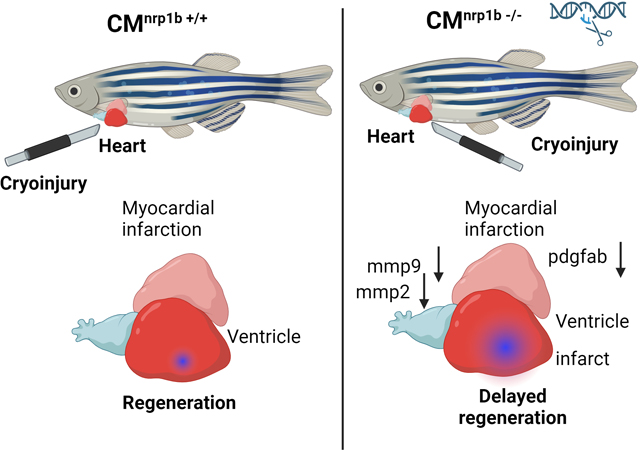
nrp1b ablation delays cardiac regeneration in zebrafish.
To further confirm the effect of cardiomyocyte-specific nrp1a and nrp1b on cardiac regeneration, 6-month-old WT, nrp1a, and nrp1b knockout zebrafish were subjected to cryoinjury, and heart ventricles were analyzed for scar area as shown in the trichome staining at 3 dpi, 14dpi, and 30 dpi, which was expressed as relative fold of the area of the ventricle (Fig. 7). At 3 dpi and 14 dpi, deletion of nrp1a or nrp1b did not significantly affect the scar sizes. However, at 30 dpi, nrp1a knockout zebrafish showed similar scar areas with control zebrafish (p=0.038, Fig. 7A and B), both of which have nearly resolved scars, whereas nrp1b deficient zebrafish exhibited significantly increased scar areas compared with WT group ( 5.2% ± 1.48% vs. 3.7% ± 0.98%). These results indicate that cardiomyocyte-specific nrp1b is required to resolve the scar and cardiac regeneration.
Figure 7. Analysis of the infarct size in nrp1a and nrp1b KO zebrafish after cryoinjury.
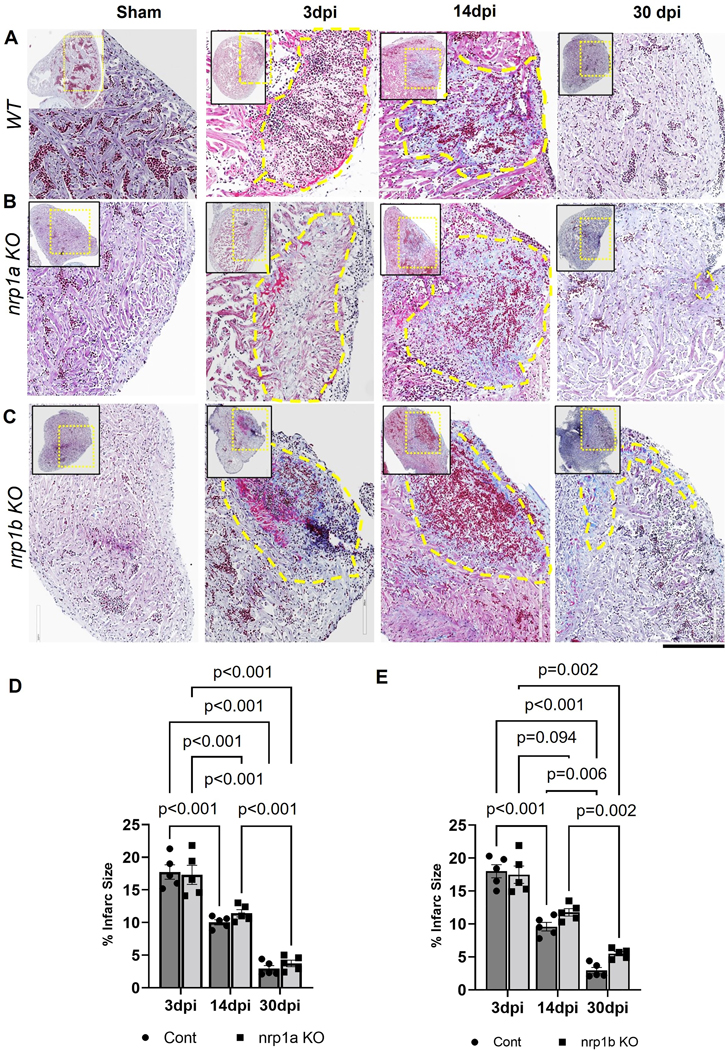
(A-C) Representative heart cross-section of Sham, 3 dpi, 14 dpi, and 30 dpi heart from the top of the ventricle (left upper corner) to the ventricular apex (right bottom corner) labeled to trichome staining to visualize the healthy myocardium in purple, fibrin in red and collagen in blue in WT, nrp1a KO, and nrp1b KO zebrafish. (D) Quantification of the infarct area in WT vs. nrp1a KO zebrafish. (E) Quantification of the infarct area in WT vs nrp1b KO zebrafish. The yellow dotted box indicates the region of the heart analyzed in the enlarged image, and the yellow dashed regions indicate the infarct area. v, ventricle; ba, bulbus arteriosus; a, atrium. The bars indicate the average values, and the error bars indicate the mean ± SEM. The quantifications were performed by measuring the infarct area from five samples at each time point. Statistical significance was evaluated with Prism 9.0 software by two-way ANOVA with Sidak’s multiple comparisons test.
nrp1b abrogation inhibits the proliferation of cardiomyocytes.
BrdU proliferation assay and PCNA staining in the nrp1a and nrp1b knockout fish after cryoinjury were performed to investigate further the role of nrp1b depletion in the proliferation. Our result confirmed that nrp1b abrogation significantly reduced the number of proliferating cells as compared to the nrp1a and wildtype fish heart, whereas nrp1a and wildtype zebrafish heart show no significant difference in proliferation (Figure S8). We also check the effect of nrp1a and nrp1b ablation on neovascularization. As shown in Fig S9 A and B, nrp1a and nrp1b ablations reduced the number of new vessels formed in the injury site.
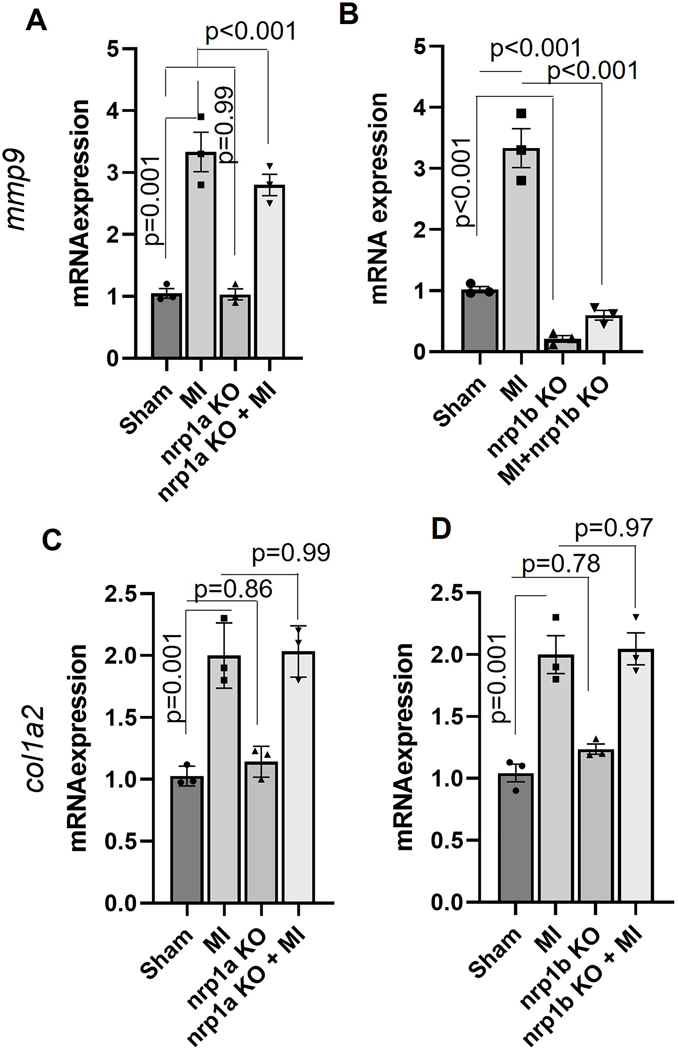
nrp1b ablation abrogates the MMP9 gene expression in zebrafish hearts.
Previously, it has been shown that matrix metalloproteinases (MMPs) activity is critical during the inflammatory phase of heart regeneration33. To see whether nrp1 ablation alters the expression of MMPs, we measured the mRNA expression of mmp2, mmp9 and mmp13 in the RNA samples isolated from the nrp1a and nrp1b knockout zebrafish hearts after three day post cryoinjury. While we did not observe significant changes in the expression of mmp9 when nrp1a was downregulated (Fig 9A), there was a significantly reduced mmp9 mRNA level in nrp1b isoform knockout zebrafish after cryoinjury, compared to the control cryoinjury group (Fig 9B). We observed that mmp2 but not mmp13 expression was reduced in the nrp1b knockout zebrafish heart (Fig S10 A). Additionally, we have tested the mRNA expression of various markers, including vegfa, vegfr2, vegfr1, vegfc, pdgfab, tgfb1, and vimentin at early timepoint (1dpi). We observed increased expression of vegfr2, vegfa, vegfc, pdgfab and tgfb1 at 1dpi in the wild type. We also show upregulation of vimentin expression at 1dpi in the wild-type zebrafish. We evaluate the expression of these genes to study the role of angiogenesis and the expression of genes associated with epithelial-to-mesenchymal transition, which are implicated in the regeneration process16. Nrp1b depletion was found to inhibit the upregulation of pdgfab, vimentin, and tgfb1 after injury at 1dpi (Fig S10 B–E).
To confirm the direct regulation of MMP9 by NRP1, we have transfected mouse ventricle cardiomyocyte (HL-1) cells with NRP1 shRNA and analyzed the mRNA expression of MMP9. We did observe reduced RNA expression of MMP9 in the NRP1 shRNA transfected cells (Fig S11A). Previous studies have suggested that regeneration-associated cardiac fibrosis influences the heart regeneration process. In this study, we measured the expression of the other ECM gene Col1a2 (collagen synthesis gene), a well-known fibrosis marker. We observed that nrp1a or nrp1b knockout did not significantly affect the expression of Col1a2 mRNA. These results indicate that upregulation of mmp9 and reduced expression of vimentin, vegfr2, pdgfab, and tgfb1 are involved in the delayed cardiac regeneration of nrp1b deficient zebrafish.
nrp1a and nrp1b isoforms do not affect the expression of each other.
Analysis of the mRNA and protein level of each isoform when the other is abrogated does not display significant changes in the expression of one isoform upon the removal of another isoform (Figure S11B and C). However, the current results do not exclude the possibility that cellular localization may change.
Discussion
To increase the potential applications of the effective CRISPR-Cas technology, we have developed a targeting vector that allows tissue-specific inactivation of a spatiotemporal gene by modifying a previously developed vector15. We show that the CRISPR-Cas9-mediated knockout technology can be spatially controlled in zebrafish, and our vector system allows for the generation of stable zebrafish lines with heart-specific gene knockout. We have exploited this technology to overcome the challenges in the embryonic lethality associated with the global knockout of essential genes, which confounds the analysis of their functions in animal models. Moreover, since the approach is Tol2 transposon technology-based, our approach may apply to model organisms other than zebrafish.
Both the nrp1a and nrp1b mRNAs were expressed in the zebrafish heart with different levels across the development, and both of the isoforms were strongly upregulated in the zebrafish heart 1–3-days after cryoinjury. These expressions were shown to coincide with the epicardial activation time, which occurs very early following cardiac injury16. It was observed that the upregulation of nrp1 isoforms was maintained for 3–7-days after the cryoinjury, further supporting the notion that nrp1 is involved in the early phase of regenerating heart post cryoinjury. Lowe et al. have suggested that these isoforms play an essential role in the epicardial activation phase, which occurs during the first 3–7-days of regeneration 16. Our finding did observe an important role of nrp1a in the early time point (3–14 dpi) of heart regeneration, as reported by Lowe et al. 2019. But we show the role of cardiac-specific nrp1b isoform, which has not been examined before in the process of heart regeneration in zebrafish hearts. We noticed that the cardiac-nrp1b isoform showed a more detrimental effect on regeneration than the nrp1a isoform.
One major difference is that Lowe et al. used the global nrp1a deficient zebrafish model, whereas we used the heat inducible-cardiomyocyte-specific nrp1a knockout zebrafish, suggesting that the role of nrp1a in cardiac regeneration is spatiotemporal dependent. Given that impaired cardiac regeneration is only observed in the global but not cardiomyocyte-specific nrp1a knockout zebrafish model, it is likely that the non-cell autonomous role of nrp1a is required for cardiac regeneration. On the other hand, we have utilized a cardiomyocyte-specific nrp1a and nrp1b knockout system, which restrict the nrp1 deletion in the cardiomyocyte in an inducible manner. Moreover, we did observe similar inhibitory effects of both nrp1a, and nrp1b ablation in the cryoinjury-induced neovascularization, which was also reported by Lowe et al.16. Our results suggest nrp1b/MMP mediated regulation in the injured heart. Our study advocates that tissue-specific ablation of nrp1a and nrp1b isoforms will help to understand their cell-specific function in cardiac regeneration after cryoinjury.
Analysis of CRISPR-Cas9-mediated nrp1a and nrp1b knockout zebrafish provided direct evidence that nrp1b is required for zebrafish heart regeneration. As previously described 16, the nrp1a knockout fish did not display any morphological defect or pathological phenotype. Similarly, we did not observe any lethal phenotype in either nrp1a or nrp1b adult knockout fish. It was previously reported that ablation of nrp1 using morpholino oligomers produces a lethal phenotype in zebrafish embryos34. Lowe et al. and we have shown that the spatiotemporal knockdown of nrp1 in adult zebrafish didn’t cause lethality, allowing us to explore the functions of these genes in adult cardiac repair after injury. Following cardiac injury, the CRISPR-Cas9-mediated, cardiomyocyte-specific nrp1a and nrp1b knockout fish demonstrated a different effect on heart regeneration. The nrp1b knockout significantly lowered regenerative function compared to WT controls and the nrp1a knockout group. The delayed and incomplete removal of fibrin deposits essential for the scar resolution process in the knockout zebrafish demonstrated the importance of nrp1b for heart regeneration. The delayed wound closure in nrp1b fish probably indicates a failure of the myocardium to migrate efficiently towards the subepicardial layer and to prevent fibrosis after cryoinjury, as suggested by Lowe et al., 201516. These findings support that nrp1b is required for zebrafish heart regeneration following cryoinjury. We have examined the mRNA expression of both isoforms in the nrp1a and nrp1b knockout zebrafish hearts. We did not observe the expression level change of one isoform when the other was deleted. This suggests no compensatory role of one isoform on the other. It is possible that cardiomyocyte-specific double deletion of nrp1a and nrp1b would have an even more harmful role in the heart regeneration and function of zebrafish. Furthermore, our data show that nrp1b regulates the mmp9 expression and thus indicating a possible role of mmp9 in fibrosis during the repair process in heart regeneration. Cardiomyocyte-specific NRP1 depletion in mice is accompanied by the upregulation of cardiac remodeling genes, including ANF, BNP βMHC17. Positive regulation of MMP9 by NRP1 in trophoblast cell proliferation and migration is known35. NRP1 ablation results in decreased protein and mRNA levels of MMP935.
Moreover, the role of the myofibroblasts is essential in post-MI healing. MMP9 promotes cardiac fibroblast migration, increased collagen production, increases angiogenic factors, and stimulates the transition of cardiac fibroblasts to myofibroblasts36. ECM produced by myofibroblasts contributes to tissue replacement and scar formation after the MI. Importantly, MMP9 has been associated with cardiac remodeling, and targeted deletion of mmp9 in mice leads LV remodeling37–39. Mechanistically, mmp9 inhibition resulted in reduced enzymatic activity, which is responsible for ECM breakdown. In zebrafish, mmp9 and mmp13 are shown to play key roles in the inflammatory phase of heart regeneration following cryoinjury33. A recent study from Xu et al. showed an increase in MMP enzymatic activity and elevated expression of MMP9 and MMP13 in the injured area of hearts from as early as 1 day post-cryoinjury (1 dpi)33. Inhibition of MMP9 resulted in impaired heart regeneration, as indicated by the larger scar and reduced numbers of proliferating cardiomyocytes33. Together these results and our data suggested nrp1b regulation of MMP9 in the cardiac remodeling in regeneration post cryoinjury. These findings warrant further investigation into the nrp1/MMP axis in regulating cardiac regeneration post-injury. One limitation of this study is that our current staining of nrp1a and nrp1b isoforms did not provide conclusive evidence to establish a different expression pattern of these isoforms. It is also possible that the different location of NRP1 isoforms in distinct cardiomyocyte populations may be involved in the different phenotypic outcome upon knockdown of nrp1a or nrp1b, which is out of the scope of our study”. Investigating the molecular inside into the role of these isoforms in the phenotypic outcome may offer novel perspectives for future studies in the field.
Supplementary Material
Figure 8. nrp1b abrogation inhibits mmp9 expression in adult zebrafish hearts.
(A-B) Graph showing the relative mRNA expression of mmp9 in (A) WT and nrp1a KO. (B) WT and nrp1b KO zebrafish heart after cryoinjury. (C-D). Graph showing the relative mRNA expression of col1a2 in (C) WT and nrp1a KO (D) WT and nrp1b KO zebrafish heart after cryoinjury. Error bars represent the mean ± SEM, and the qRT PCR experiments were repeated three times. Statistical significance was evaluated with Prism 9.0 software by using one-way ANOVA.
Highlights:
Our studies reported a novel heat shock inducible-CRISPR-based vector system for conditional, tissue-specific gene ablation in zebrafish.
This vector system establishes a unique tool for tissue-specific and spatiotemporal regulation of genes in both the developmental and adult stages.
We observed that both the neuropilin-1 isoforms (nrp1a and nrp1b) are upregulated after the cryoinjury in the heart. However, the cardiac-specific nrp1b-knockout significantly altered heart regeneration and reduced nature.
To our knowledge, this is the first study where we have reported a heat shock-mediated conditional knockdown of nrp1a and nrp1b isoforms using CRISPR/Cas9 technology in the cardiomyocyte in zebrafish and identified a crucial role for nrp1b isoform in zebrafish heart function in response to injury.
Acknowledgment
The authors thank Dr. Leonard I. Zon’s laboratory, Boston Children’s hospital for providing the CRISPR vector clones. We thank Dr. Pritam Das for reviewing the manuscript and providing his helpful suggestions. We thank Dr. Lewis-Tuffin Laura, Cellular Imaging and Flow Cytometry Specialist, Mayo Clinic Florida Cytometry and Cell Imaging Laboratory for her help in Confocal imaging. We thank Brandy Edenfield, Mayo Clinic Florida, cancer Biology Histology Core for her support in immunostaining.
Funding
This work was supported by the National Institutes of Health [HL140411 and CA78383-20 to D. Mukhopadhyay, HL148339 to Y. Wang], Florida Department of Health Cancer Research Chair’s Fund Florida [grant number#3J-02 to D. Mukhopadhyay]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Nonstandard Abbreviations and Acronyms
- CRISPR/Cas9
Clustered regularly interspaced short palindromic repeats/clustered regularly interspaced short-palindromic repeat-associated 9
- PAM
protospacer adjacent motif
- ECG
electrocardiogram
- PFA
paraformaldehyde
- ShRNA
short hairpin RNA
- CM
cardiomyocyte
- FISH
Fluorescence in situ hybridization
- EF
ejection fraction
- FS
fractional shortening
- ECM
extracellular matrix
- RT-PCR
Real-time Polymerase chain reaction
- KO
knockout
- NRP1
neuropilin −1
- MI
myocardial infarction
- BrdU
Bromodeoxyuridine (5-bromo-2’-deoxyuridine
- EC
endothelial cell
Footnotes
Disclosures: None.
Bibliography
- 1.Sander JD; Joung JK, CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014, 32 (4), 347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M; Chylinski K; Fonfara I; Hauer M; Doudna JA; Charpentier E, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.) 2012, 337 (6096), 816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sternberg SH; Redding S; Jinek M; Greene EC; Doudna JA, DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507 (7490), 62–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho SW; Kim S; Kim JM; Kim JS, Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013, 31 (3), 230–2. [DOI] [PubMed] [Google Scholar]
- 5.Mali P; Esvelt KM; Church GM, Cas9 as a versatile tool for engineering biology. Nature Methods 2013, 10 (10), 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L; Ran FA; Cox D; Lin S; Barretto R; Habib N; Hsu PD; Wu X; Jiang W; Marraffini LA; Zhang F, Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.) 2013, 339 (6121), 819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen CT; Lu Q; Wang Y; Chen JN, Zebrafish as a model for cardiovascular development and disease. Drug Discov Today Dis Models 2008, 5 (3), 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González-Rosa JM, Zebrafish Models of Cardiac Disease: From Fortuitous Mutants to Precision Medicine. Circ Res 2022, 130 (12), 1803–1826. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson RN; van Eeden FJ, The zebrafish as a model of vascular development and disease. Prog Mol Biol Transl Sci 2014, 124, 93–122. [DOI] [PubMed] [Google Scholar]
- 10.Driever W; Stemple DL; Schier AF; Solnica-Krezel L, Zebrafish: genetic tools for studying vertebrate development. Trends in genetics : TIG 1994, 10 5, 152–9. [DOI] [PubMed] [Google Scholar]
- 11.Lieschke GJ; Trede NS, Fish immunology. Current Biology 2009, 19 (16), R678–R682. [DOI] [PubMed] [Google Scholar]
- 12.Lawson, Nathan D; Wolfe, Scot A, Forward and Reverse Genetic Approaches for the Analysis of Vertebrate Development in the Zebrafish. Developmental Cell 2011, 21 (1), 48–64. [DOI] [PubMed] [Google Scholar]
- 13.Gagnon JA; Valen E; Thyme SB; Huang P; Ahkmetova L; Pauli A; Montague TG; Zimmerman S; Richter C; Schier AF, Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. PloS one 2014, 9 (5), e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang WY; Fu Y; Reyon D; Maeder ML; Tsai SQ; Sander JD; Peterson RT; Yeh JR; Joung JK, Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 2013, 31 (3), 227–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ablain J; Durand EM; Yang S; Zhou Y; Zon LI, A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell 2015, 32 (6), 756–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe V; Wisniewski L; Sayers J; Evans I; Frankel P; Mercader-Huber N; Zachary IC; Pellet-Many C, Neuropilin 1 mediates epicardial activation and revascularization in the regenerating zebrafish heart. Development (Cambridge, England) 2019, 146 (13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y; Cao Y; Yamada S; Thirunavukkarasu M; Nin V; Joshi M; Rishi MT; Bhattacharya S; Camacho-Pereira J; Sharma AK; Shameer K; Kocher J-PA; Sanchez JA; Wang E; Hoeppner LH; Dutta SK; Leof EB; Shah V; Claffey KP; Chini EN; Simons M; Terzic A; Maulik N; Mukhopadhyay D, Cardiomyopathy and Worsened Ischemic Heart Failure in SM22-α Cre-Mediated Neuropilin-1 Null Mice. Arteriosclerosis, thrombosis, and vascular biology 2015, 35 (6), 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen HZ; Xie P; Zhang F; Ma Y; Li YL; Xu SK, Neuropilin 1 ameliorates electrical remodeling at infarct border zones in rats after myocardial infarction. Auton Neurosci 2018, 214, 19–23. [DOI] [PubMed] [Google Scholar]
- 19.Kwan KM; Fujimoto E; Grabher C; Mangum BD; Hardy ME; Campbell DS; Parant JM; Yost HJ; Kanki JP; Chien CB, The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 2007, 236 (11), 3088–99. [DOI] [PubMed] [Google Scholar]
- 20.Jao LE; Wente SR; Chen W, Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proceedings of the National Academy of Sciences of the United States of America 2013, 110 (34), 13904–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartley JL; Temple GF; Brasch MA, DNA cloning using in vitro site-specific recombination. Genome Res 2000, 10 (11), 1788–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoji W; Sato-Maeda M, Application of heat shock promoter in transgenic zebrafish. Development, Growth & Differentiation 2008, 50 (6), 401–406. [DOI] [PubMed] [Google Scholar]
- 23.Hoeppner LH; Phoenix KN; Clark KJ; Bhattacharya R; Gong X; Sciuto TE; Vohra P; Suresh S; Bhattacharya S; Dvorak AM; Ekker SC; Dvorak HF; Claffey KP; Mukhopadhyay D, Revealing the role of phospholipase Cβ3 in the regulation of VEGF-induced vascular permeability. Blood 2012, 120 (11), 2167–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montague TG; Cruz JM; Gagnon JA; Church GM; Valen E, CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 2014, 42 (Web Server issue), W401–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thisse C; Thisse B, High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 2008, 3 (1), 59–69. [DOI] [PubMed] [Google Scholar]
- 26.He J; Mo D; Chen J; Luo L, Combined whole-mount fluorescence in situ hybridization and antibody staining in zebrafish embryos and larvae. Nature Protocols 2020, 15 (10), 3361–3379. [DOI] [PubMed] [Google Scholar]
- 27.Chablais F; Veit J; Rainer G; Jaźwińska A, The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC developmental biology 2011, 11, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan J; Li H; Bu H; Jiao K; Zhang AX; Le T; Cao H; Li Y; Ding Y; Xu X, Aging-associated sinus arrest and sick sinus syndrome in adult zebrafish. PloS one 2020, 15 (5), e0232457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang Y; Sun Y; Luo C; Gu J; Shi Z; Lu G; Silvestre J-S; Chen Z, Evaluation of cardiac dysfunction in adult zebrafish using high frequency echocardiography. Life Sciences 2020, 253, 117732. [DOI] [PubMed] [Google Scholar]
- 30.Bai H; Liu L; An K; Lu X; Harrison M; Zhao Y; Yan R; Lu Z; Li S; Lin S; Liang F; Qin W, CRISPR/Cas9-mediated precise genome modification by a long ssDNA template in zebrafish. BMC Genomics 2020, 21 (1), 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Icoglu Aksakal F; Ciltas A, The impact of ultraviolet B (UV-B) radiation in combination with different temperatures in the early life stage of zebrafish (Danio rerio). Photochem Photobiol Sci 2018, 17 (1), 35–41. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y; Cao Y; Yamada S; Thirunavukkarasu M; Nin V; Joshi M; Rishi MT; Bhattacharya S; Camacho-Pereira J; Sharma AK; Shameer K; Kocher JP; Sanchez JA; Wang E; Hoeppner LH; Dutta SK; Leof EB; Shah V; Claffey KP; Chini EN; Simons M; Terzic A; Maulik N; Mukhopadhyay D, Cardiomyopathy and Worsened Ischemic Heart Failure in SM22-α Cre-Mediated Neuropilin-1 Null Mice: Dysregulation of PGC1α and Mitochondrial Homeostasis. Arteriosclerosis, thrombosis, and vascular biology 2015, 35 (6), 1401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu S; Webb SE; Lau TCK; Cheng SH, Matrix metalloproteinases (MMPs) mediate leukocyte recruitment during the inflammatory phase of zebrafish heart regeneration. Scientific Reports 2018, 8 (1), 7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martyn U; Schulte-Merker S, Zebrafish neuropilins are differentially expressed and interact with vascular endothelial growth factor during embryonic vascular development. Developmental Dynamics 2004, 231 (1), 33–42. [DOI] [PubMed] [Google Scholar]
- 35.Yang X; Chen D; He B; Cheng W, NRP1 and MMP9 are dual targets of RNA-binding protein QKI5 to alter VEGF-R/ NRP1 signalling in trophoblasts in preeclampsia. J Cell Mol Med 2021, 25 (12), 5655–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y; Xu F; Chen J; Shen X; Deng Y; Xu L; Yin J; Chen H; Teng F; Liu X; Wu W; Jiang B; Guo DA, Matrix metalloproteinase-9 induces cardiac fibroblast migration, collagen and cytokine secretion: inhibition by salvianolic acid B from Salvia miltiorrhiza. Phytomedicine 2011, 19 (1), 13–9. [DOI] [PubMed] [Google Scholar]
- 37.Bradham WS; Moe G; Wendt KA; Scott AA; Konig A; Romanova M; Naik G; Spinale FG, TNF-alpha and myocardial matrix metalloproteinases in heart failure: relationship to LV remodeling. Am J Physiol Heart Circ Physiol 2002, 282 (4), H1288–95. [DOI] [PubMed] [Google Scholar]
- 38.Ducharme A; Frantz S; Aikawa M; Rabkin E; Lindsey M; Rohde LE; Schoen FJ; Kelly RA; Werb Z; Libby P; Lee RT, Targeted deletion of matrix metalloproteinase-9 attenuates left ventricular enlargement and collagen accumulation after experimental myocardial infarction. J Clin Invest 2000, 106 (1), 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindsey ML; Escobar GP; Dobrucki LW; Goshorn DK; Bouges S; Mingoia JT; McClister DM Jr.; Su H; Gannon J; MacGillivray C; Lee RT; Sinusas AJ; Spinale FG, Matrix metalloproteinase-9 gene deletion facilitates angiogenesis after myocardial infarction. Am J Physiol Heart Circ Physiol 2006, 290 (1), H232–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all supporting data and method descriptions are available within the article or from the corresponding authors upon reasonable request.