Abstract
The 3’ untranslated region (3’UTR) of mRNA plays a key role in post-transcriptional regulation of gene expression. Most eukaryotic protein-coding genes express 3’UTR isoforms owing to alternative cleavage and polyadenylation (APA). The 3’UTR isoform expression profile of a cell changes in cell proliferation, differentiation, and stress conditions. Here we review the emerging theme of regulation of 3’UTR isoforms in cell metabolic reprogramming, focusing on cell growth and autophagy responses through the mTOR pathway. We discuss regulatory events that converge on the Cleavage Factor I complex, a master regulator of APA in 3’UTRs, and recent understandings of isoform-specific m6A modification and endomembrane association in determining differential metabolic fates of 3’UTR isoforms.
Introduction
Cleavage and polyadenylation (CPA) of precursor RNA is responsible for 3’ end maturation of almost all eukaryotic mRNAs 1,2. About 70-80% of protein-coding genes in the human genome harbor multiple CPA sites (also known as polyA sites or PASs), leading to alternative polyadenylation (APA) isoforms 3. Most APA sites are located in the last exon, resulting in expression of mRNA isoforms with different 3’UTR sizes. The 3’UTR isoform expression profile is highly variable across cell or tissue types 4-6. For example, brain and blood cells tend to express long and short 3’UTR isoforms, respectively 4. In addition, substantial, global 3’UTR size changes have been found in cell proliferation 7, cancer cell transformation 8, embryonic development 9, cell differentiation 10, and stress conditions 11,12. About 20% of human genes, typically those with large introns, also display intronic polyadenylation (or IPA) 3. IPA events are also dynamically controlled and often correlated with 3’UTR APA events 4-6, indicating common underlying regulatory mechanisms.
3’UTR APA isoforms are believed to have distinct mRNA metabolic fates, such as stability, translational efficiency, and subcellular localization, due to variable sequence and structure motifs embedded in different isoforms 13. For simplicity, the 3’UTR portion subject to alternative polyadenylation (APA) regulation is named alternative 3’UTR, or aUTR (Figure 1). Here we review recent advances in understanding mechanisms and consequences of 3’UTR isoform regulation in the context of cell metabolic reprogramming, including cell growth and autophagy. Readers are referred to other recent reviews for more comprehensive coverage of APA 14-16.
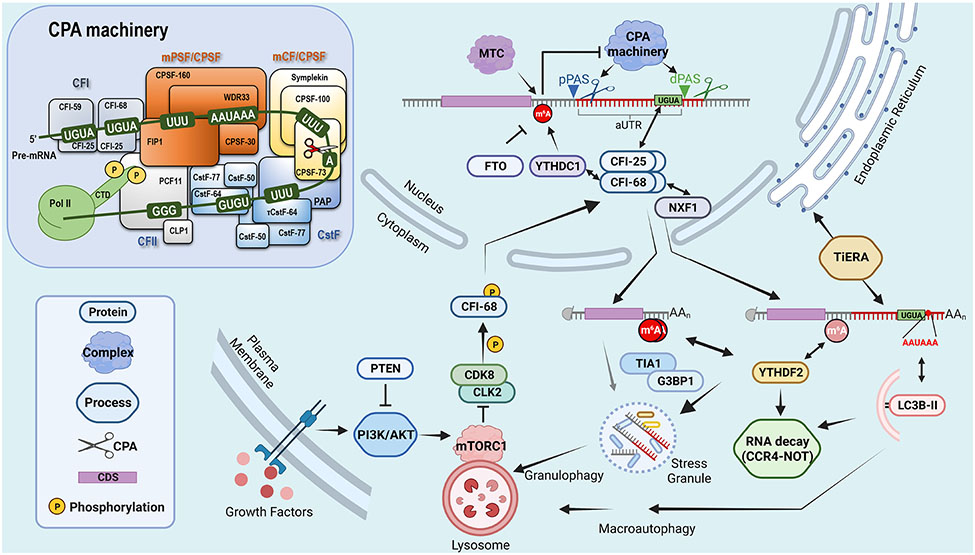
Figure 1.
Regulation of alternative polyadenylation (APA) site choice and differential mRNA metabolism between 3’UTR isoforms. A hypothetical gene with two 3’UTR APA sites are shown.
pPAS, proximal PAS; dPAS, distal PAS. MTC, m6A methyltransferase complex; LC3B-II, lipidated LC3B. CCR4-NOT carries out deadenylation of mRNA, often the first step for mRNA degradation. TiERA is translation-independent ER association. The core factors of CPA machinery and PAS motifs are also shown in the inset. Four subcomplexes in the machinery and two functional modules within CPSF are noted. CPSF-73 is the endonuclease responsible for precursor RNA cleavage, WDR33 and CPSF-30 collectively interact with the A[A/U]UAAA motif, and FIP1 binds to U-rich binding sequences. CstF exists as a dimer, each containing CstF-50, CstF-64/CstF-64τ and CstF-77. CstF-64 and CstF-64τ, two paralogs in the genome, have binding affinities to U-rich and GU-rich (GUGU or UGUG) motifs. CFI exists as a tetramer, comprising two molecules of CFI-25 together with two alternative larger subunits, While both CFI-59 and CFI-68 are RNA-binding proteins, CFI-25 has binding specificity to the UGUA motif. CFII includes CLP1 and PCF11, with the latter having binding affinity to G-rich RNA sequences 90.
Core CPA factors mediate APA regulation
The PAS is defined by surrounding motifs 17 (Figure 1). In mammals, upstream PAS motifs include the A[A/U]UAAA hexamer or their close variants, U-rich sequences, and UGUA sequences; downstream PAS motifs include U-rich, GU-rich, and G-rich sequences 17 . The cleavage reaction typically takes place after an adenosine 3,18. The strength of a PAS is determined by all these motifs in a combinatory fashion 19,20. In general, due to motif composition differences, distal PASs, especially those at the 3’-most position of a gene, are stronger than proximal PASs 19. For example, the canonical motifs AAUAAA and UGUA in particular are conspicuously enriched at the 3’-most PASs 3.
The mammalian CPA machinery is composed of over 20 core proteins 1,21,22(Figure 1), most of which are engaged in forming four sub-complexes, including the Cleavage and Polyadenylation Specificity Factor (CPSF), the Cleavage stimulation Factor CstF, the Cleavage Factor I (CFI, also known as CFIm) and the Cleavage Factor II (CFII, also known as CFIIm). CPSF comprises two functional modules 21, namely, the mammalian polyadenylation specificity factor (mPSF), composed of CPSF-160, CPSF-30, WDR33, and FIP1, and the mammalian cleavage factor (mCF), composed of CPSF-100, CPSF-73, and Symplekin. CPSF proteins have binding affinities to the core PAS motifs, while CstF, CFI and CFII factors have affinities to auxiliary motifs for CPA (illustrated in the CPA machinery inset of Figure 1). In addition, RBBP6, Poly(A) polymerase (PAP), Poly(A) binding protein PAPN1, and RNA polymerase II (Pol II) are also key components of the CPA machinery.
Dysregulation of several CPA factors have been implicated in human developmental diseases and cancers 23-26. One contributing reason to disease etiology is that the expression level of several core CPA factors has a substantially impact on APA site selection 27-31. For example, decreased expression levels of FIP1, CstF-64/CstF-64τ or PCF11 all lead to a global shift to distal APA site usage, resulting in 3’UTR lengthening for most genes 27,28. Conversely, overexpression (OE) of PCF11 results in an opposite trend 32,33. The underlying reason for these global APA site shifts is the fact that proximal APA sites are typically weak, making their usage highly responsive to the CPA activity. On the other hand, these findings highlight the importance of individual CPA factors in determining the overall CPA activity.
By contrast, KD of CFI-25 or CFI-68 leads to drastic 3’UTR shortening for nearly every gene that has APA sites in the 3’UTR 27,29,34,35. Since the UGUA motif, which CFI binds, is enriched at distal PASs, CFI is believed to have an enhancing role biased to distal PAS usage. Interestingly, OE of CFI-25 or CFI-68 has only a limited effect on 3’UTR size globally 34, indicating that these proteins are likely to be in sufficient amounts under normal conditions. The mechanism by which CFI enhances CPA was revealed by Zhu et al., who showed that the arginine-serine repeat (RS)-like domain of CFI-59/68 directly interacts with the arginine-aspartate/glutamate (RE/D) domain of FIP1 36. Therefore, CFI could help recruit FIP1-contianing CPSF for CPA 35. Notably, despite protein sequence similarities between CFI-59 and CFI-68, the former has a much weaker function in enhancing CPA than the latter 37, which explains why CFI-59 KD does not lead to widespread 3’UTR shortening as does KD of CFI-25 or CFI-68 27,34,35.
In addition to its direct function in CPA, CFI-68 has been implicated in mRNA nuclear export. The protein has the property of shuttling between nucleus and cytoplasm 38 and physically interacts with the nuclear export factor NXF1 38,39. Notably, NXF1 KD also leads to global 3’UTR shortening as well as cumulation of Pol II around the 3’ end of genes, indicating that nuclear export is tightly connected with CPA, plausibly through CFI-68 39. Related to this, a recent study by Tang et al. showed that long 3’UTR isoforms can be further processed into short 3’UTR isoforms in the nuclear matrix 40, opening the possibility that some of the 3’UTR shortening effect of CFI-25 or -68 KD might be due to decreased nuclear export of long 3’UTR isoforms, which allows more time for CPA at proximal PASs. All in all, it is now well established that CFI-25/68 are master regulators of the 3’UTR size of mRNAs.
3’UTR isoform regulation by the mTOR pathway
The mammalian target of rapamycin (mTOR) pathway is a key player in cell proliferation and growth 41. In keeping with the notion that 3’UTR isoform expression is coupled with cell proliferation and differentiation 7,42, activation of mTOR was found to cause 3’UTR shortening in multiple human and mouse cells 43. Based on selective KD of Raptor or Rictor, components of the two distinct mTOR complexes in the cell, the same study found that mTORC1, not mTORC2, is the one responsible for 3’UTR isoform regulation. Interestingly, genes with certain functions, such as protein processing in endoplasmic reticulum and ubiquitin-mediated proteolysis, appeared to be particularly affected by mTORC1-mediated 3’UTR shortening 43,44. Notably, similar Gene Ontology terms, such as protein transport and cellular response to stress, were among the top biological processes associated with CFI-25/68-regulated APA events in HEK293 cells 34, indicating a potential role of CFI in mTORC1-mediated APA. In the same vein, a recent phosphoproteomics study using HEK 293 cells with OE or KD of CFI-25 or CFI-68 highlighted the potential roles of CFI-25/68 in regulation of several kinases involved in cell metabolism 34, such as ULK1 and ERK1/2 45.
The PI3K/AKT signaling pathway plays a major role in mTORC1 regulation 46. PTEN, a tumor suppressor that is frequently mutated in cancers 46,47, is a key regulator of PI3K/AKT signaling. The PTEN gene produces multiple 3’UTR isoforms with variable contributions to the overall protein expression 48. CFI-68 and CFI-59 were recently found to have opposing roles in PTEN regulation 47, with the former promoting the expression of longer 3’UTR isoforms and the latter functioning in the opposite direction. The authors further indicated that many factors in the PI3K/AKT signaling pathway are subject to similar 3’UTR isoform regulations by CFI-68 and CFI-59, raising the possibility of a concerted regulatory scheme. One of the reasons that 3’UTR isoform regulation is critical for PTEN expression is because of the multiple miRNA target sites embedded in its 3’UTRs 49. Interestingly, the general 3’UTR shortening in cancer cells, which could remove miRNA target sites from 3’UTRs of mRNAs in a global fashion, was found to enhance the functions of the miRNAs that target PTEN mRNAs, a phenomenon also known as competing endogenous RNA or ceRNA 50. Another facet of 3’UTR APA regulation of PTEN comes from the Mayr group, who found that deletion of an enhancer region at the PTEN promoter alters the relative expression levels of 3’UTR isoforms in breast cancer cell line MCF7 51. Whether CFI factors are involved in this enhancer-mediated 3’UTR APA scheme of PTEN would be interesting to explore in the future.
mTORC1 is inhibited in cells under nutrient starvation, eliciting catabolic processes such as autophagy 52. Using an autophagy-induced Drosophila model, involving overexpression of the key autophagy gene Atg1 (homolog of ULK1/2 in mammals) in the eye, Tang et al. identified multiple CPA factors as autophagy regulators, including CFI-68, CstF-64, WDR33, CPSF-160 and PCF11 53. The authors delineated a mTORC1-CDK8/CLK2 (DOA in Drosophila)-CFI-68 axis in autophagy regulation, in which inhibition of mTORC1 leads to phosphorylation of the RS domain CFI-68 by CDK8/CLK2, resulting in nuclear localization of CFI-68 and enhanced distal APA site selection. Importantly, this mechanism regulates both Atg1 and another autophagy gene Atg8a (homolog of LC3 genes in mammals) by switching short 3’UTR isoform expression to long 3’UTR isoform expression in starvation. Because mRNA stability of these long 3’UTR isoforms is greater than that of short 3’UTR isoforms, CFI-68-mediated lengthening of 3’UTR in starvation increases their protein expression and thus enhances their functions in autophagy response 53. Interestingly, in the Tang et al. study, CPSF and CstF KDs showed similar phenotypes to CFI-68 KD, while PCF11 KD showed an opposite trend. It remains to be addressed whether the distinct function of PCF11 in the autophagy context is due to different directions of APA events for the same genes 27 or different sets of genes are regulated by PCF11 32,33. It is also worth noting that hyperphosphorylation of the RS domain of CFI-68 was found to inhibit its interaction with FIP1 in human cells 37. Therefore, further studies are needed to reconcile two seemingly opposing consequences of RS domain phosphorylation of CFI-68.
3’UTR regulation has also been studied for other genes involved in autophagy. Using a murine pro-B cell line Ba/F3 that encodes U2AF35S34F, a mutant, oncogenic form of the core splicing factor U2AF35, Park et al. showed that CFI-59 and CFI-68 interact differently with U2AF35S34F, leading to a switch to distal 3’UTR PAS usage in the autophagy gene ATG7 54. Because its long 3’UTR isoform is less efficiently translated than the short 3’UTR isoform, the APA regulation results in downregulation of ATG7 protein and hence autophagy defects, which predispose cells to additional mutations that cause cell transformation.
The metabolic fate of 3’UTR isoforms through LC3B and membrane association
Because the 3’UTR is a hotbed for RNA motifs involved in post-transcriptional regulation, 3’UTR size changes could substantially alter mRNA metabolism. While most studies have shown that short 3’UTR isoforms tend to be more stable 3,55 and have higher translational efficiencies 56 than long 3’UTRs, there are many exceptions 57. This is due likely to the fact that 3’UTRs can harbor both stabilizing and destabilizing motifs 14-16,58. In addition, while long 3’UTR size could have higher affinities to UPF1, a key player of nonsense-mediated decay (NMD) 59,60, a recent study using long-read sequencing of NMD substrates does not support the view that long 3’UTR size per se activates NMD 61. A recent study by Hwang et al., however, indicates that long 3’UTRs may be indeed generally unstable in autophagy conditions 62. Lipidation of the autophagy regulator LC3B through conjugation with phosphatidylethanolamine is a commitment step in the canonical macroautophagy pathway 63,64. LC3B-II, the lapidated form of the protein, plays a key role in formation of phagophore, leading to autophagosome maturation 65-67. Hwang et al. found that LC3B is a bona fide RNA-binding protein with specificity to AAUAAA 62, revealing a new mRNA degradation pathway under autophagy conditions. This mechanism, termed LC3B-mediated mRNA decay (LMD), involves interactions of LC3B with mRNAs and subsequent mRNA deadenylation through the CCR4-NOT complex. LMD takes place before lysosome fusion and is thus distinct from lysosome-mediated RNA degradation within the lysosome 68. Importantly, LC3B lipidation enhances LMD, raising the possibility that membrane-associated mRNAs may be more prone to this degradation pathway. In addition, because the number of AAUAAA generally correlates with 3’UTR size and is more enriched at distal PASs used by long 3’UTR isoforms (our unpublished data), the LMD pathway might differentiate 3’UTR isoforms in mRNA stability control during autophagy (Figure 1).
In addition to stability control, 3’UTRs are critical for mRNA localization, especially in polarized cells, such as neurons 69. However, it is increasingly clear that 3’UTRs could alter mRNA distribution even in cells not highly polarized. By comparing 3’UTR isoforms in membrane and cytosol fractions of mouse C2C12 myoblast cells and differentiated myotubes, Cheng et al. found that long 3’UTRs promote endoplasmic reticulum (ER) association in a translation-independent manner 70, a mechanism dubbed translation independent ER association (TiERA). 3’UTR length, GC content and structural properties play deterministic roles in TiERA potentials. It is noteworthy that the membrane fraction they isolated contained other organelles as well, leaving open the question as to whether 3’UTRs could also help recruit mRNAs to other organelles. On this note, association of mRNAs with endosomes was found to help localize translating mRNAs in neuronal cells 71. On the other hand, membrane-associated mRNAs may subject them to a different set of regulatory mechanisms than those in the cytosol. For example, membrane-associated mRNAs may be more prone to LMD or lysosome-mediated mRNA degradation. Related to this, stress granules (SGs), membrane-less protein-RNA aggregates through liquid-liquid phase separation under stress conditions 72, could be formed in both cytosol and ER. Removal of SGs through lysosomes, also known as granulophagy, has implications in many human diseases, including cancer and neuronal degeneration 73. Importantly, 3’UTR size is an important feature for mRNA recruitment into SGs 74 and long 3’UTRs isoforms were found to be preferentially degraded after SG-inducing arsenic stress in mouse C2C12 myoblast cells and NIH3T3 fibroblast cells 11. Therefore, it is possible that, due to the differential association with organelles and membrane-less granules, 3’UTR isoforms have distinct metabolic fates during and after stress.
The connection between m6A and 3’UTR isoform expression
Methylation of the N6 position of adenosine (m6A) is a widespread modification of mRNA carried out by the m6A methyltransferase complex (MTC) 75. Recent studies have shown that m6A levels globally increase upon mTORC1 activation, leading to heightened m6A-mediated mRNA metabolism 76 77, promoting cell proliferation and growth as well as suppressing autophagy 78. m6A levels are typically enriched near the stop codon in the last exon 79,80 and are associated with increased mRNA degradation 81 and higher translational efficiency 82, functions that are mediated by m6A cytoplasmic readers, such as YTHDF proteins 83.
Using anti-m6A RNA immunoprecipitation (RIP) and human H1 embryonic stem cells, Molinie et al. found that short 3’UTR isoforms are more likely to have higher levels of m6A than long 3’UTR isoforms from the same gene 84. In addition, Yue et al. found that the m6A motif GGACU was enriched, albeit modestly, around the proximal PAS site in genes with multiple APA sites in the last exon 85. These findings support the notion that 3’UTR isoforms could have different m6A contents, leading to distinct metabolic fates.
A key question concerning m6A deposition is whether APA site selection process could influence the level of m6A in 3’UTR. The VIRMA component of MTC was found to physically interact with the CFI-25/68 complex 85. Interestingly, while VIRMA KD globally suppresses m6A enrichment in the 3’UTR and around the stop codon, CFI-25 KD leads to expression of short 3’UTR isoforms with increased m6A in these regions 85. This finding indicates that CFI controls both the size and m6A content of 3’UTR. The connection between m6A and 3’UTR isoform expression is further supported by the observations that long 3’UTR isoforms are relatively more abundant after ablations of m6A eraser FTO 86 or nuclear m6A reader YTHDC1 87. Notably, YTHDC1, which is required for embryo viability and germline development in mice 87, was found to physically interact with CFI-68, SRSF3, and SRSF7 in HEK293T cells 87. The authors indicated that these interactions support a role of m6A in regulation of APA and splicing. Another study by Chen et al., however, showed that YTHDC1 could also interact with FIP1, thereby hindering its interaction with CPSF-30 88 and hence causing PAS usage suppression. Therefore, while it is indisputable that m6A and 3’UTR isoform expression are well connected, the detailed mechanism(s) for the interplay between m6A deposition and APA site selection, especially concerning the role of YTHDC1, still require further experimentation.
Conclusions and future perspectives
3’UTR isoform regulation is increasingly appreciated as a widespread mechanism that modulates gene expression. A growing number of physiological and pathological conditions have been found to impact 3’UTR isoform expression in the cell, such as cell growth, differentiation, and stress. The mTORC1-CFI axis is emerging as a key signaling pathway leading to 3’UTR size control. The consequences of 3’UTR size changes could be multifold. Endomembrane association, AAUAAA-mediated LMD, and 3’UTR m6A content are recently uncovered mechanisms that can differentiate the metabolic fates for 3’UTR isoforms. These studies raise many interesting questions that require further investigation. Due to space limitation, we list three of them here:
The mTORC1-CFI axis needs to be further examined in different metabolic and stress conditions. Because CFI factors have a global effect on almost every gene containing 3’UTR APA sites, how specific genes are targeted for 3’UTR size regulation needs to be addressed. In addition, why CFI-59 and CFI-68, two CFI proteins with similar sequences, appear to have substantial differences APA regulation and how these two proteins have differential impacts on cell signaling require further investigation.
Long 3’UTR isoforms tend to be enriched on ER and perhaps other organelles as well. The underlying mechanism(s) are still elusive. Whether some RBPs can be lipidated, similar to LC3B, and serve as receptors for 3’UTR sequences or structures need to be explored. In addition, the difference in mRNA metabolism for cytosolic vs. membrane-bound mRNAs is an interesting subject to study, especially in cells with markedly different 3’UTR isoform expression patterns, such as blood cells and neurons.
The m6A content has a clear role in mRNA metabolism. The cause and consequence of m6A content difference between 3’UTR isoforms need more rigorous examination. The role of YTHDC1 in 3’UTR isoform regulation could be further analyzed by comparing nascent pre-mRNAs and newly made mature RNAs. To what extent YTHDF proteins, which can promote stress granule formation 89, contribute to differential recruitment of 3’UTR isoforms with different m6A contents to stress granules needs to be firmly established.
ACKNOWLEDGMENTS
We thank Chengyu Liang, and members of BT lab for helpful discussions. This work was funded by NIH grants GM084089 and GM129069 to BT.
REFERENCE
- 1.Shi Y & Manley JL The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes & development 29, 889–897 (2015). 10.1101/gad.261974.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proudfoot NJ Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science (New York, N.Y.) 352, aad9926 (2016). 10.1126/science.aad9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang R, Zheng D, Yehia G & Tian B A compendium of conserved cleavage and polyadenylation events in mammalian genes. Genome research 28, 1427–1441 (2018). 10.1101/gr.237826.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang R & Tian B APAlyzer: a bioinformatics package for analysis of alternative polyadenylation isoforms. Bioinformatics (Oxford, England) 36, 3907–3909 (2020). 10.1093/bioinformatics/btaa266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Lee JY & Tian B Biased alternative polyadenylation in human tissues. Genome biology 6, R100 (2005). 10.1186/gb-2005-6-12-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smibert P et al. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell reports 1, 277–289 (2012). 10.1016/j.celrep.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandberg R, Neilson JR, Sarma A, Sharp PA & Burge CB Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science (New York, N.Y.) 320, 1643–1647 (2008). 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayr C & Bartel DP Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 (2009). 10.1016/j.cell.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji Z, Lee JY, Pan Z, Jiang B & Tian B Progressive lengthening of 3' untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proceedings of the National Academy of Sciences of the United States of America 106, 7028–7033 (2009). 10.1073/pnas.0900028106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng LC et al. Widespread transcript shortening through alternative polyadenylation in secretory cell differentiation. Nature communications 11, 3182 (2020). 10.1038/s41467-020-16959-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng D et al. Cellular stress alters 3'UTR landscape through alternative polyadenylation and isoform-specific degradation. Nature communications 9, 2268 (2018). 10.1038/s41467-018-04730-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollerer I et al. The differential expression of alternatively polyadenylated transcripts is a common stress-induced response mechanism that modulates mammalian mRNA expression in a quantitative and qualitative fashion. RNA (New York, N.Y.) 22, 1441–1453 (2016). 10.1261/rna.055657.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayr C Evolution and biological roles of alternative 3'UTRs. Trends in cell biology 26, 227–237 (2016). 10.1016/j.tcb.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitschka S & Mayr C Context-specific regulation and function of mRNA alternative polyadenylation. Nat Rev Mol Cell Biol (2022). 10.1038/s41580-022-00507-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber AJ & Zavolan M Alternative cleavage and polyadenylation in health and disease. Nature reviews. Genetics 20, 599–614 (2019). 10.1038/s41576-019-0145-z [DOI] [PubMed] [Google Scholar]
- 16.Tian B & Manley JL Alternative polyadenylation of mRNA precursors. Nature reviews. Molecular cell biology 18, 18–30 (2017). 10.1038/nrm.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian B & Graber JH Signals for pre-mRNA cleavage and polyadenylation. Wiley interdisciplinary reviews. RNA 3, 385–396 (2012). 10.1002/wrna.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen F, MacDonald CC & Wilusz J Cleavage site determinants in the mammalian polyadenylation signal. Nucleic acids research 23, 2614–2620 (1995). 10.1093/nar/23.14.2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Y, Miura RM & Tian B Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics (Oxford, England) 22, 2320–2325 (2006). 10.1093/bioinformatics/btl394 [DOI] [PubMed] [Google Scholar]
- 20.Bogard N, Linder J, Rosenberg AB & Seelig G A Deep Neural Network for Predicting and Engineering Alternative Polyadenylation. Cell 178, 91–106.e123 (2019). 10.1016/j.cell.2019.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y, Hamilton K & Tong L Recent molecular insights into canonical pre-mRNA 3'-end processing. Transcription 11, 83–96 (2020). 10.1080/21541264.2020.1777047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A, Clerici M, Muckenfuss LM, Passmore LA & Jinek M Mechanistic insights into mRNA 3'-end processing. Curr Opin Struct Biol 59, 143–150 (2019). 10.1016/j.sbi.2019.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis AG et al. Alternative polyadenylation dysregulation contributes to the differentiation block of acute myeloid leukemia. Blood 139, 424–438 (2022). 10.1182/blood.2020005693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gennarino VA et al. NUDT21-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation. eLife 4, e10782 (2015). 10.7554/eLife.10782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Prisco N et al. Alternative polyadenylation alters protein dosage by switching between intronic and 3'UTR sites. Science advances 9, eade4814 (2023). 10.1126/sciadv.ade4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnadottir GA et al. Population-level deficit of homozygosity unveils CPSF3 as an intellectual disability syndrome gene. Nature communications 13, 705 (2022). 10.1038/s41467-022-28330-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W et al. Systematic profiling of poly(A)+ transcripts modulated by core 3' end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS genetics 11, e1005166 (2015). 10.1371/journal.pgen.1005166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lackford B et al. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J 33, 878–889 (2014). 10.1002/embj.201386537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin G, Gruber AR, Keller W & Zavolan M Genome-wide Analysis of Pre-mRNA 3' End Processing Reveals a Decisive Role of Human Cleavage Factor I in the Regulation of 3' UTR Length. Cell reports 1, 753–763 (2012). 10.1016/j.celrep.2012.05.003 [DOI] [PubMed] [Google Scholar]
- 30.Jenal M et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149, 538–553 (2012). 10.1016/j.cell.2012.03.022 [DOI] [PubMed] [Google Scholar]
- 31.Di Giammartino DC et al. RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3' UTRs. Genes & development 28, 2248–2260 (2014). 10.1101/gad.245787.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamieniarz-Gdula K et al. Selective Roles of Vertebrate PCF11 in Premature and Full-Length Transcript Termination. Molecular cell 74, 158–172.e159 (2019). 10.1016/j.molcel.2019.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R, Zheng D, Wei L, Ding Q & Tian B Regulation of Intronic Polyadenylation by PCF11 Impacts mRNA Expression of Long Genes. Cell reports 26, 2766–2778 e2766 (2019). 10.1016/j.celrep.2019.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh S et al. CFIm-mediated alternative polyadenylation remodels cellular signaling and miRNA biogenesis. Nucleic acids research 50, 3096–3114 (2022). 10.1093/nar/gkac114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y et al. Molecular Mechanisms for CFIm-Mediated Regulation of mRNA Alternative Polyadenylation. Mol Cell 69, 62–74 e64 (2018). 10.1016/j.molcel.2017.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukherjee S, Graber JH & Moore CL Macrophage differentiation is marked by increased abundance of the mRNA 3' end processing machinery, altered poly(A) site usage, and sensitivity to the level of CstF64. Frontiers in immunology 14, 1091403 (2023). 10.3389/fimmu.2023.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y et al. Molecular Mechanisms for CFIm-Mediated Regulation of mRNA Alternative Polyadenylation. Molecular cell 69, 62–74.e64 (2018). 10.1016/j.molcel.2017.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruepp MD et al. Mammalian pre-mRNA 3' end processing factor CF I m 68 functions in mRNA export. Molecular biology of the cell 20, 5211–5223 (2009). 10.1091/mbc.E09-05-0389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S et al. The mRNA Export Receptor NXF1 Coordinates Transcriptional Dynamics, Alternative Polyadenylation, and mRNA Export. Molecular cell 74, 118–131 e117 (2019). 10.1016/j.molcel.2019.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang P et al. Alternative polyadenylation by sequential activation of distal and proximal PolyA sites. Nature structural & molecular biology 29, 21–31 (2022). 10.1038/s41594-021-00709-z [DOI] [PubMed] [Google Scholar]
- 41.Laplante M & Sabatini DM mTOR signaling in growth control and disease. Cell 149, 274–293 (2012). 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji Z & Tian B Reprogramming of 3' untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PloS one 4, e8419 (2009). 10.1371/journal.pone.0008419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang JW et al. mRNA 3'-UTR shortening is a molecular signature of mTORC1 activation. Nature communications 6, 7218 (2015). 10.1038/ncomms8218 [DOI] [PubMed] [Google Scholar]
- 44.Chang JW et al. An integrative model for alternative polyadenylation, IntMAP, delineates mTOR-modulated endoplasmic reticulum stress response. Nucleic acids research 46, 5996–6008 (2018). 10.1093/nar/gky340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez-Lopez N, Athonvarangkul D, Mishall P, Sahu S & Singh R Autophagy proteins regulate ERK phosphorylation. Nat Commun 4, 2799 (2013). 10.1038/ncomms3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manning BD & Toker A AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405 (2017). 10.1016/j.cell.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tseng HW et al. Distinct, opposing functions for CFIm59 and CFIm68 in mRNA alternative polyadenylation of Pten and in the PI3K/Akt signalling cascade. Nucleic acids research 50, 9397–9412 (2022). 10.1093/nar/gkac704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W et al. Distinct regulation of alternative polyadenylation and gene expression by nuclear poly(A) polymerases. Nucleic acids research 45, 8930–8942 (2017). 10.1093/nar/gkx560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thivierge C et al. Alternative polyadenylation confers Pten mRNAs stability and resistance to microRNAs. Nucleic Acids Res 46, 10340–10352 (2018). 10.1093/nar/gky666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park HJ et al. 3' UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nature genetics 50, 783–789 (2018). 10.1038/s41588-018-0118-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwon B et al. Enhancers regulate 3' end processing activity to control expression of alternative 3'UTR isoforms. Nature communications 13, 2709 (2022). 10.1038/s41467-022-30525-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao J, Zhai B, Gygi SP & Goldberg AL mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci U S A 112, 15790–15797 (2015). 10.1073/pnas.1521919112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang HW et al. The TORC1-Regulated CPA Complex Rewires an RNA Processing Network to Drive Autophagy and Metabolic Reprogramming. Cell metabolism 27, 1040–1054 e1048 (2018). 10.1016/j.cmet.2018.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park SM et al. U2AF35(S34F) Promotes Transformation by Directing Aberrant ATG7 Pre-mRNA 3' End Formation. Molecular cell 62, 479–490 (2016). 10.1016/j.molcel.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guvenek A et al. Neuronal Cells Display Distinct Stability Controls of Alternative Polyadenylation mRNA Isoforms, Long Non-Coding RNAs, and Mitochondrial RNAs. Frontiers in genetics 13, 840369 (2022). 10.3389/fgene.2022.840369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Floor SN & Doudna JA Tunable protein synthesis by transcript isoforms in human cells. eLife 5, e10921 (2016). 10.7554/eLife.10921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spies N, Burge CB & Bartel DP 3' UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome research 23, 2078–2090 (2013). 10.1101/gr.156919.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garneau NL, Wilusz J & Wilusz CJ The highways and byways of mRNA decay. Nature reviews. Molecular cell biology 8, 113–126 (2007). 10.1038/nrm2104 [DOI] [PubMed] [Google Scholar]
- 59.Hogg JR & Goff SP Upf1 senses 3'UTR length to potentiate mRNA decay. Cell 143, 379–389 (2010). 10.1016/j.cell.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fischer JW, Busa VF, Shao Y & Leung AKL Structure-Mediated RNA Decay by UPF1 and G3BP1. Molecular cell 78, 70–84 e76 (2020). 10.1016/j.molcel.2020.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karousis ED, Gypas F, Zavolan M & Mühlemann O Nanopore sequencing reveals endogenous NMD-targeted isoforms in human cells. Genome biology 22, 223 (2021). 10.1186/s13059-021-02439-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hwang HJ et al. LC3B is an RNA-binding protein to trigger rapid mRNA degradation during autophagy. Nature communications 13, 1436 (2022). 10.1038/s41467-022-29139-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sou YS, Tanida I, Komatsu M, Ueno T & Kominami E Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J Biol Chem 281, 3017–3024 (2006). 10.1074/jbc.M505888200 [DOI] [PubMed] [Google Scholar]
- 64.Abildgaard MH, Brynjolfsdottir SH & Frankel LB The Autophagy-RNA Interplay: Degradation and Beyond. Trends Biochem Sci 45, 845–857 (2020). 10.1016/j.tibs.2020.07.007 [DOI] [PubMed] [Google Scholar]
- 65.Hansen M, Rubinsztein DC & Walker DW Autophagy as a promoter of longevity: insights from model organisms. Nat Rev Mol Cell Biol 19, 579–593 (2018). 10.1038/s41580-018-0033-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li X, He S & Ma B Autophagy and autophagy-related proteins in cancer. Mol Cancer 19, 12 (2020). 10.1186/s12943-020-1138-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johansen T & Lamark T Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J Mol Biol 432, 80–103 (2020). 10.1016/j.jmb.2019.07.016 [DOI] [PubMed] [Google Scholar]
- 68.Frankel LB, Lubas M & Lund AH Emerging connections between RNA and autophagy. Autophagy 13, 3–23 (2017). 10.1080/15548627.2016.1222992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tushev G et al. Alternative 3' UTRs Modify the Localization, Regulatory Potential, Stability, and Plasticity of mRNAs in Neuronal Compartments. Neuron 98, 495–511.e496 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Cheng LC et al. Alternative 3' UTRs play a widespread role in translation-independent mRNA association with the endoplasmic reticulum. Cell reports 36, 109407 (2021). 10.1016/j.celrep.2021.109407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cioni JM et al. Late Endosomes Act as mRNA Translation Platforms and Sustain Mitochondria in Axons. Cell 176, 56–72 e15 (2019). 10.1016/j.cell.2018.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nicchitta CV An emerging role for the endoplasmic reticulum in stress granule biogenesis. Semin Cell Dev Biol (2022). 10.1016/j.semcdb.2022.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang B et al. ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol Cell 74, 742–757 e748 (2019). 10.1016/j.molcel.2019.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khong A et al. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Molecular cell 68, 808–820.e805 (2017). 10.1016/j.molcel.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang H et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567, 414–419 (2019). 10.1038/s41586-019-1016-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cho S et al. mTORC1 promotes cell growth via m(6)A-dependent mRNA degradation. Molecular cell 81, 2064–2075 e2068 (2021). 10.1016/j.molcel.2021.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Villa E et al. mTORC1 stimulates cell growth through SAM synthesis and m(6)A mRNA-dependent control of protein synthesis. Molecular cell 81, 2076–2093 e2079 (2021). 10.1016/j.molcel.2021.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tang HW et al. mTORC1-chaperonin CCT signaling regulates m(6)A RNA methylation to suppress autophagy. Proceedings of the National Academy of Sciences of the United States of America 118 (2021). 10.1073/pnas.2021945118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meyer KD et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149, 1635–1646 (2012). 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ke S et al. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes & development 29, 2037–2053 (2015). 10.1101/gad.269415.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399 (2015). 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi H et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res 27, 315–328 (2017). 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Molinie B et al. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nature methods 13, 692–698 (2016). 10.1038/nmeth.3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yue Y et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov 4, 10 (2018). 10.1093/nar/gky438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bartosovic M et al. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic acids research 45, 11356–11370 (2017). 10.1093/nar/gkx778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kasowitz SD et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS genetics 14, e1007412 (2018). 10.1371/journal.pgen.1007412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen L et al. Nuclear m(6) A reader YTHDC1 suppresses proximal alternative polyadenylation sites by interfering with the 3' processing machinery. EMBO reports 23, e54686 (2022). 10.15252/embr.202254686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fu Y & Zhuang X m(6)A-binding YTHDF proteins promote stress granule formation. Nat Chem Biol 16, 955–963 (2020). 10.1038/s41589-020-0524-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schafer P et al. Reconstitution of mammalian cleavage factor II involved in 3' processing of mRNA precursors. RNA (New York, N.Y.) 24, 1721–1737 (2018). 10.1261/rna.068056.118 [DOI] [PMC free article] [PubMed] [Google Scholar]