Abstract
Background:
Altered gut microbiota has been associated with cognitive dysfunction and Alzheimer's disease, but little is known among people living with HIV.
Objective:
To examine associations between gut microbiota and cognitive impairment among women with or without HIV.
Methods:
This is a cross-sectional study of 446 women (302 HIV+) who had completed a neuropsychological test battery and stool sample collected within 1 year. Gut microbiota composition was quantified using 16SV4 rRNA gene sequencing and microbial functional pathways were predicted using PICRUSt. Cognitive domains included attention, executive function, learning, memory, fluency, processing speed, and motor function. Cognitive impairment was defined as two or more domains with T scores<1 SD below mean. ANCOM-II was used to identify taxa and functional pathways associated with cognitive impairment, and the associations were further examined by multivariable logistic regression.
Results:
In overall sample, adjusting for multiple covariates including HIV status, we found that higher abundance of Methanobrevibacter, Odoribacter, Pyramidobacter, Eubacterium, Ruminococcus, and Gemmiger, and lower abundance of Veillonella were associated with cognitive impairment. The associations between these taxa and cognitive impairment were more profound in HIV+ women compared to HIV− women. Most associations with bacterial taxa were observed for learning and memory. We found accompanying microbial functional differences associated with cognitive impairment, including twelve enriched pathways and three depleted pathways.
Conclusion:
In women with or without HIV infection, this study identified multiple altered gut bacterial taxa and functional pathways associated with cognitive impairment, supporting the potential role of gut microbiota in cognitive dysfunction and Alzheimer's disease.
Keywords: HIV, Cognitive Impairment, Gut Microbiome, Alzheimer’s Disease, Human, Women
Introduction
With the availability of combination antiretroviral therapy (cART), the overall mortality of people living with Human Immunodeficiency Virus (HIV) has decreased greatly and more attention has shifted to aging related diseases among this population. When including asymptomatic disease, 31-47% of people living with HIV (PLWH) are estimated to have cognitive impairment. [1, 2] Among PLWH, the majority of individuals with cognitive impairment now demonstrates mild forms of disease. [1] A previous study in the Women’s Interagency Health Study (WIHS) found that HIV infection status was associated with significant cognitive deficits in processing speed, attention, verbal learning and delayed memory.[3] In addition, older PLWH are also at risk for Alzheimer's disease which might be due to compounding effects of HIV and aging.[4] The potential causes of cognitive impairment in PLWH have been suggested, including incomplete viral suppression in the central nervous system (CNS), neural injury due to viral proteins and inflammatory responses, neurotoxicity of ART, metabolic disorders and increased amyloid-β deposition in the brain. [5] However, the actual mechanism is still not well understood.
The gut-brain axis involves bi-directional communication between gut microbiota (GMB) and the CNS through neuronal, endocrine, and immune-mediated processes.[6-8] Studies have shown that GMB may play a part in the development of neurodegenerative diseases.[9, 10] It has been reported that, as compared with people with normal cognitive function, those with mild cognitive impairment and Alzheimer’s disease showed altered GMB profiles, including reduced Lachnospiraceae, Ruminococcaceae, Clostridiaceae, Mogibacteriaceae, Turicibacteaceae and Peptostreptococcaceae families, Lachnospira, Ruminiclostridium, Dialister, Clostridium and Bifidobacterium genera, and enriched Proteobacteria, Gammaproteobacteria, Enterobacteriaceae, Rikenellaceae, Alistipes, Prevotella, Odoribacter, and Barnesiella. [10-14] Microbial dysbiosis, such as increased abundance of pro-inflammatory bacteria and decreased abundance of anti-inflammatory bacteria, may influence the immune system and lead to local and systemic inflammation. [15] Translocation of bacteria and increased permeability of the gut epithelial barrier and blood-brain barrier result in neuroinflammatory response in the brain.[16] Additionally, microbial-produced bioactive metabolites, such as short-chain fatty acids (SCFA), serotonin, kynurenine and amyloids, play essential roles in neurotransmission and neuromodulation. [15, 16]
While prior evidence provides a basis for linking the gut-brain axis with cognitive disorders, the sample sizes of existing studies are relatively small (N<200) and most studies included participants in the hospital setting. Moreover, to the best of our knowledge, no study has investigated the relationship between GMB and cognitive impairment in the context of HIV infection, although PLWH are at risk for gut dysbiosis.[17] Thus, in this study, we examined associations between gut bacterial features (overall diversity, individual bacterial taxa and bacterial functional pathways) and cognitive impairment among 446 women with or without HIV from a community-based HIV cohort, the WIHS. Moreover, we hypothesized that some of the associations might be stronger in PLWH due to gut barrier dysfunction, which may enhance microbial translocation,[18] compared to those without HIV. Thus, we also explored potential effect modification by HIV serostatus on the association between GMB and cognitive impairment.
Materials and Methods
Study population
The WIHS is a prospective cohort study of women with and at risk for HIV since 1994, now part of the MACS WIHS Combined Cohort Study (MWCCS). [19-21] Participants were recruited from 10 cities in the United States were followed up every 6 months to collect biospecimens, medical history, medication use, health-related behaviors and anthropometry. To ensure comparability with HIV+ women, HIV− women who engaged in high risk behaviors for HIV were recruited.[19] Since 2009 to 2019, participants were administered a comprehensive neuropsychological testing every 2 years.[3] From 2016 to 2019, the Bronx, Brooklyn, and Chicago WIHS sites collected stool samples from participants.[22] Written informed consent was obtained from participants. The study was approved by institutional review boards at each site.
We included 466 WIHS participants who provided stool samples, which were obtained within about 1 year either before or after completion of a neuropsychological test battery. After excluding 11 samples with low sequencing depth (<2000 sequence reads per sample) and 9 samples with missing data >3 domains of a neuropsychological test, a total of 446 participants were in the study sample, 302 of whom were HIV+ women.
Cognitive function.
Participants completed a neuropsychological test battery assessing the following domains: learning (Hopkins Verbal Learning Test-Revised [HVLT-R]-total learning across trials 1 to 3); memory (HVLT-R-delay free recall); psychomotor speed (Symbol Digit Modalities Test, Stroop-Trial 2); attention/working (Letter Number Sequencing); motor function (Grooved Pegboard dominant and non-dominant hands); verbal fluency (letter and semantic); and executive function (Trail Making Test Part B and Stroop Test Interference Trial). [23] Timed outcomes were log transformed to normalize distributions and reverse scored, so higher equated to better performance. As previously described,[2, 3, 23] demographically-adjusted T scores were then derived for each cognitive domain using data from the HIV− women. Age, years of education, Wide Range Abilities Test (WRAT)-3 Reading Recognition subtest score, race/ethnicity, and number of prior neuropsychological test completions were included in the regression equations,[3] and domain-specific T-scores were then created. Most of the participants (94%) included in the study sample had scores available in all 7 domains. Impairment on each domain was defined as T-scores < 1 standard deviation (SD) below the mean of the HIV− women. [2, 23] If participants had two or more domains with impairment, they were considered to have global cognitive impairment. [2] The primary outcome of interest is global cognitive impairment. The secondary outcomes are impairment in each domain. In addition, we also created a global performance score by averaging domain-specific T-scores as a continuous global cognitive measure.
Stool sample collection and microbiome measurement.
Stool samples were collected using a home-based self-collection kit containing RNAlater prepared in the laboratory of Dr. Robert D Burk at Albert Einstein College of Medicine. [22] In brief, stool sample was self-collected and placed in a supplied container including a stabilizer (RNAlater) and 0.5 mm diameter glass beads and instructed to shake the tube in order to mix the stool and the preservative which stabilizes DNA and RNA.[24] After collection, stool samples were stored at room temperature and mailed back to the lab through USPS. The lab froze the samples immediately at −80 °C upon receipt. As described previously, 16S rRNA V4 gene region amplification was performed on DNA extracted from stool samples using a bead-beating procedure by the MiSeq platform (Illumina, San Diego, CA) at Albert Einstein College of Medicine Sequencing Core.[25]
Bioinformatic analysis
Microbiome bioinformatics analyses were performed using the Quantitative Insights Into Microbial Ecology (QIIME2) software package (2019.10) with the Deblur pipeline.[26] The α-diversity indices (Shannon index and observed amplicon sequencing variant (ASV)) and β- diversity Jensen Shannon Divergence were calculated using QIIME2 and R phyloseq package after rarefication at 40 different sequencing depths (from 20 to 35,000 sequence reads per sample).[27] The functional potential of the GMB was imputed by PICRUSt (to calculate estimated relative abundances of KEGG ortholog groups).[28] We excluded samples with sequencing depths <2000 sequence reads per sample after the Deblur workflow.[25] After this exclusion, the lowest sequencing depth was 2507 in the study sample. Detailed quality control assessment was previously reported. [25]
Covariates
Data on age, race, education, annual income, health behaviors (recreational drug use and smoking status) and medication use, and blood samples were collected using standardized protocols at semiannual core study visits.[29] Recreational drugs include marijuana, crack, cocaine, heroin or injection drug use. Smoking status was assessed by current, ever or never smoking. ART include protease inhibitors, nucleoside and non-nucleoside reverse transcriptase inhibitors, and information on participant use of each type of drugs in the past six months was collected. HIV serostatus was ascertained using the enzyme linked immunosorbent assay method and confirmed by Western blot. Hepatitis C virus (HCV) infection was based on a serological test for antibodies or a nucleic acid test for viral RNA. Other HIV-related characteristics include cluster of differentiation 4+ (CD4+) cell count, HIV RNA and ART use. Undetectable HIV-1 viral load was defined as ⩽20 copies/mL.
Statistical analysis
Characteristics of HIV+ and HIV− women were compared using t-test for continuous variables and chi-squared test for categorical variables. Microbial α-diversity indices Shannon index and number of observed ASVs were compared by cognitive impairment using Wilcoxon rank-sum test. Permutational multivariate ANOVA (PERMANOVA) and principal-coordinate analysis (PCoA) were used to examine the differences in microbial β-diversity by cognitive impairment. For taxa and functional pathways analysis, we conducted the Analysis of Composition of Microbiomes (ANCOM-II), given its good control of false discovery rate (FDR), to identify candidates associated with global cognitive impairment.[30] We kept taxa that were present in at least 25% of samples with mean relative abundance > 0.01%. All models adjusted for age, race, education (below high school, high school and above high school), poverty (annual income ⩽$12000), recreational drug use, HIV status, HCV infection, site, antibiotic use, smoking (never, former and current), HIV viral load and ART use (only among HIV+ women). We included HIV viral load (detectable vs. undetectable) and ART use as categorical variables in which an additional level was created for HIV− women. These covariates were considered as potential confounders based on our previous analyses on GMB or cognitive function in the WIHS.[23, 31] We conducted ANCOM-II using raw count data and FDR threshold at 0.10, at multiple taxonomic levels including phylum (n=14), class (n=25), order (n=41), family (n=74), genus (n=168), and species (n=142), and for functional pathways (n=353). The analysis excluded unknown taxa at these taxonomic levels. An ANCOM-II detection level ≥ 0.60 indicates that the ratios of a taxon to at least 60% of other taxa were significantly different by cognitive impairment status, adjusting for multiple testing (FDR q<0.10). We examined multiple taxonomic levels to see whether the results were consistent within a taxonomic lineage, and we controlled for multiple testing by using FDR at each taxonomic level.
We used multivariable logistic regression to estimate the odds ratios (OR) of cognitive impairment by relative abundance of bacterial taxa or pathways identified in the ANCOM-II analysis of global cognitive impairment, adjusting for the same covariates as in the ANCOM-II models. Centered log-ratio (CLR) transformation in relative abundances of taxonomic units or functional pathways were used. A pseudocount of min (relative abundance)/2 was added to exact zero relative abundance before taking logs. As a secondary analysis, we also estimated the OR of impairment in each domain by relative abundance of the identified taxa or pathways associated with the primary outcome (i.e., overall cognitive impairment) in the ANCOM-II analysis. In addition to the primary analysis among all women, we also carried out the analysis stratified by HIV serostatus. To test potential effect modification by HIV serostatus, we included a product term of taxa and HIV serostatus in the regression models. For bacteria found to be associated with cognitive impairment, relative abundance between HIV+ and HIV− women was compared using Wilcoxon rank-sum test. We examined the associations of the identified taxa with global and domain-specific T scores as continuous cognitive outcomes, using linear regression. We also conducted sensitivity analysis among women who had stool sample collection and neuropsychological test at the same visit. We assessed correlations between identified taxa and MetaCyc pathways using CLR transformed relative abundance and spearman correlation coefficient. Analyses were performed using R 4.0.3. A two-sided P<0.05 was considered statistically significant in regression models.
Results
Table 1 shows characteristics of the 446 women (302 HIV +, mean age 53.1 years). Cognitive impairment was identified in 122 (27.4%) women. As compared with HIV− women, HIV+ women were slightly older, were more likely to be non-Hispanic white, current smoker and have education below high school, and were less likely to have recreational drug use and marijuana use. We observed a higher percent of global cognitive impairment and impairment in most domains except for memory among HIV+ women as compared with HIV− women. However, most of these differences were not statistically significant.
Table 1.
Characteristics of WIHS participants by HIV status
| All (N=446) | HIV+ (N=302) | HIV− (N=144) | P-value | |
|---|---|---|---|---|
| Age, years, mean (SD) | 53.1 (8.3) | 53.2 (7.8) | 52.8 (9.3) | 0.59 |
| Race/ethnicity, % | 0.30 | |||
| Non-Hispanic black | 323 (72.4) | 214 (70.9) | 109 (75.7) | |
| Hispanic | 92 (20.6) | 62 (20.5) | 30 (20.8) | |
| Non-Hispanic white | 19 (4.3) | 16 (5.3) | 3 (2.1) | |
| Other | 12 (2.7) | 10 (3.3) | 2 (1.4) | |
| Education attainment, % | 0.84 | |||
| Below high school | 180 (40.4) | 124 (41.1) | 56 (39.2) | |
| High school | 141 (31.7) | 93 (30.8) | 48 (33.6) | |
| Above high school | 124 (27.9) | 85 (28.1) | 39 (27.3) | |
| Years of education | 11.6 (3.0) | 11.7 (3.1) | 11.5 (3.0) | 0.56 |
| WRAT-3 reading subtest, mean (SD) | 87.8 (18.6) | 88.0 (19.1) | 87.5 (17.8) | 0.81 |
| Annual income ≤$12000, % | 235 (52.7) | 154 (51.0) | 81 (56.2) | 0.35 |
| Recreational drug use, % | 105 (23.5) | 66 (21.9) | 39 (27.1) | 0.27 |
| Marijuana use, % | 90 (20.2) | 56 (18.5) | 34 (23.6) | 0.26 |
| Cigarette use, % | 0.10 | |||
| Current smoker | 181 (40.6) | 74 (24.5) | 24 (16.7) | |
| Former smoker | 167 (37.4) | 114 (37.7) | 67 (46.5) | |
| Never smoker | 98 (22.0) | 114 (37.7) | 53 (36.8) | |
| Alcohol use>7 drinks/week, % | 30 (6.7) | 18 (6.0) | 12 (8.3) | 0.46 |
| Antibiotic use, % | 23 (5.2) | 15 (5.0) | 8 (5.6) | 0.97 |
| Hepatitis C virus antibody, % | 97 (21.8) | 70 (23.3) | 27 (18.8) | 0.34 |
| Among HIV seropositive | ||||
| CD4 count | 670 (492-923) | |||
| HIV-1 Viral load≤20 copies/ml, % | 224 (74.2) | |||
| ART use, % | 277 (91.7) | |||
| Site | 0.45 | |||
| Bronx | 185 (41.5) | 121 (40.1) | 64 (44.4) | |
| Brooklyn | 132 (29.6) | 95 (31.5) | 37 (25.7) | |
| Chicago | 129 (28.9) | 86 (28.5) | 43 (29.9) | |
| Cognitive impairment | 122 (27.4) | 89 (29.5) | 33 (22.9) | 0.18 |
| Impairment in domains | ||||
| Learning | 74 (16.6) | 55 (18.2) | 19 (13.2) | 0.23 |
| Memory | 73 (16.4) | 47 (15.6) | 26 (18.1) | 0.60 |
| Attention | 62 (14.6) | 45 (15.6) | 17 (12.5) | 0.48 |
| Executive function | 67 (15.1) | 54 (17.9) | 13 (9.2) | 0.02 |
| Motor function | 62 (14.1) | 44 (14.9) | 18 (12.5) | 0.60 |
| Speed | 62 (13.9) | 45 (14.9) | 17 (11.8) | 0.46 |
| Verbal function | 60 (13.5) | 46 (15.2) | 14 (9.7) | 0.15 |
Data are N (%) or mean (SD). SD: standard deviation. WRAT: Wide Range Abilities Test. HIV: human immunodeficiency virus. CD4: cluster of differentiation 4. ART: antiretroviral therapy. Recreational drug use included marijuana, crack, cocaine, heroin and injection drug use. P-values comparing characteristics by HIV status were obtained from t-test for continuous variables, Chi-squared test for categorical
Associations of gut microbiome alpha diversity and beta diversity with global cognitive impairment
Gut microbiome alpha diversity as measured by Shannon index and number of observed ASVs were higher among women with cognitive impairment as compared to those without (P≤0.001). The associations were consistent among HIV+ women. (Figure S1) No difference in alpha diversity by cognitive impairment status was found among HIV− women. (Figure S1) No difference in beta diversity as measured by Jensen-Shannon Divergence (JSD) was found in HIV+ or HIV− women using PERMANOVA and PCoA (R2=0.47%-0.59%, P>0.05, Figure S2).
Associations of microbial taxa with global cognitive impairment
A total of 7 genera and 5 species showed greater abundance while 1 genus showed lower abundance among women with cognitive impairment as compared to women without, using the ANCOM-II method adjusting for covariates (detection level ≥ 0.60, FDR q<0.10). The identified taxa included Methanobrevibacter, Pyramidobacter, Gemmiger, Ruminococcus, Eubacterium, Odoribacter, Veillonella, Pyramidobacter piscolens, Ruminococcus bromii, Ruminococcus cadillus, Gemmiger formicilis and Eubacterium biforme. (Figure 1) The relative abundances of Methanobrevibacter and Eubacterium biforme were lower while the relative abundance of Ruminococcus bromii was higher among HIV+ women as compared with HIV− women. (P<0.05, Figure S3)
Figure 1. Relative abundance of taxa identified using ANCOM2 by cognitive impairment.
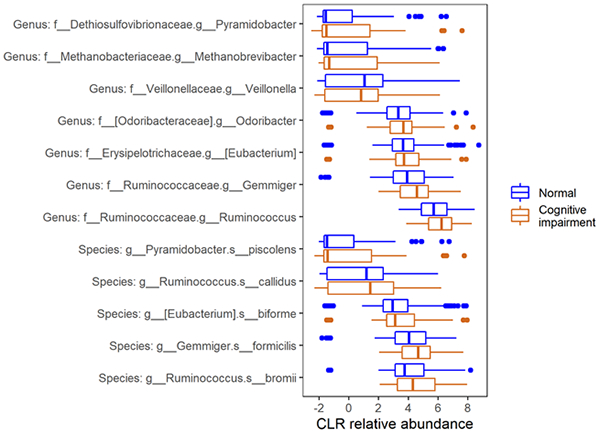
Taxa were identified using ANCOM-II adjusting for age, race, education, poverty, recreational drug use, HCV, site, antibiotics, smoking, viral load and ART use (among HIV+) as appropriate. Central-log-ratio-transformed relative abundance are shown among women with cognitive impairment and women with normal cognitive function. Taxa were ordered by relative abundance at genus and species level, respectively.
The ORs of cognitive impairment decreased with higher abundance of Veillonella while increased with higher abundance of other taxa. (Figure 2) The associations of most taxa, except for Methanobrevibacter and R. cadillus, with cognitive impairment, were more profound in HIV+ women, and there were significant interactions (P<0.03) for Eubacterium, Veillonella, E. biforme and R. bromii. (Figure 2) The results were generally consistent in analysis using global T score to define cognitive outcomes. (Figure S4) Sensitivity analysis among women with concurrent stool sample collection and cognitive assessment showed similar results. (Table S1)
Figure 2. Association between taxa abundance and cognitive impairment among all women and according to HIV serostatus.
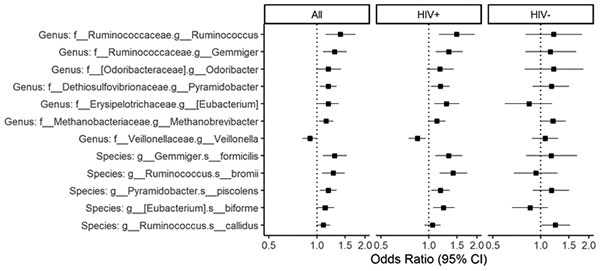
Taxa were identified using ANCOM-II adjusting for age, race, education, poverty, recreational drug use, HCV, site, antibiotics, smoking, viral load and ART use (among HIV+) as appropriate. Logistic regression was used to show OR of cognitive impairment by relative abundance of taxa, adjusting for above covariates used in ANCOM-II. Relative abundance were CLR-transformed. P for interaction<0.05 for Eubacterium, Veillonella, Eubacterium biforme and Ruminococcus bromii. Taxa were ordered by OR at genus and species level, respectively.
Associations of microbial taxa with impaired cognitive domains
For individual bacterial taxa, we found associations of most taxa with impairment in learning and memory, with directions consistent with those for global cognitive impairment. (Figure 3) There were also suggestive associations of taxa except for E.biforme and R.callidus with psychomotor speed and verbal function. (Figure 3) Most taxa were not associated with cognitive impairment in attention, executive function or motor function. (Figure 3) We also examined these associations in HIV+ and HIV− women separately, and most of these findings were predominantly observed in HIV+ women. (Table S2) When using domain T scores, the directions of the associations were largely consistent with the above findings in most domains except for executive function and motor function. (Figure S5) Higher abundance of Ruminococcus, R. bromii and Methanobrevibacter were associated with lower T scores for learning, memory, psychomotor speed and verbal function. In addition, higher abundance of Odoribacter, Gemmiger, G.formicillis, Pyramidobactor, P.piscolens were associated with lower T scores for memory. Unlike null findings with cognitive impairment in executive function and motor function, we found that most taxa were inversely associated with T scores for executive function and motor function. (Figure S5)
Figure 3. Association between taxa abundance and impairment in cognitive domains among all women.
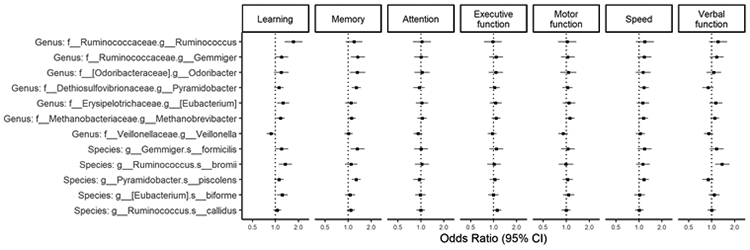
Taxa were identified using ANCOM-II adjusting for age, race, education, poverty, recreational drug use, HIV, HCV, site, antibiotics, smoking, viral load and art use (among HIV+) as appropriate. Logistic regression was used to show OR of cognitive impairment by relative abundance of taxa, adjusting for above covariates used in ANCOM-II. Relative abundance were CLR-transformed.
Associations of microbial metabolic pathways with global cognitive impairment
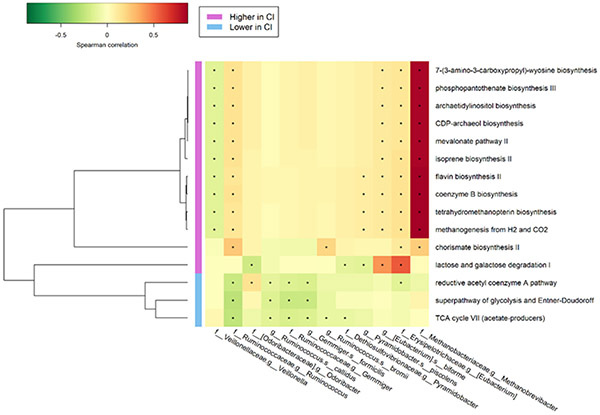
We identified 12 functional pathways that were enriched and 3 pathways that were depleted in women with cognitive impairment using ANCOM-II, adjusting for covariates. (Table S3) Ten of them, which were Archaea related pathways or methanogenesis pathways, were highly correlated with Methanobrevibacter (Spearman’s correlation coefficient>0.80). (Figure 4) Eubacterium had a strong correlation with lactose and galactose degradation I (r=0.56). (Figure 4) Veillonella was negatively correlated with most pathways, whereas reductive acetyl coenzyme A pathway, super pathway of glycolysis and Entner-Doudoroff, and tricarboxylic acid (TCA) cycle VII pathway were inversely correlated with most taxa. (Figure 4) The latter three pathways were mainly correlated with Ruminococcus and Gemmiger (r=−0.15 ~ −0.27). (Figure 4) In addition, they were inversely associated with cognitive impairment while other pathways were positively associated with cognitive impairment. (Table S3). The findings of super pathway of TCA cycle VII, and lactose and galactose degradation I pathways were only seen in HIV+ women while the findings of other pathways seemed to be present in both HIV+ and HIV− women. (Table S3)
Figure 4. Spearman’s correlation coefficients between taxa and pathways identified using ANCOM-II.
*: P<0.05 for spearman correlation coefficient.
Discussion
To the best of our knowledge, this is the first large observational study examining the associations between GMB and cognitive function in the context of HIV infection. In this cross-sectional study of middle-aged women with and without HIV who were comparable in demographic and socioeconomic status, we observed that higher microbial alpha diversity and abundances of 7 genera and 5 species were associated with cognitive impairment. Findings of increased risk for cognitive impairment with higher abundance of Pyramidobacter, Eubacterium, Ruminococcus, and Gemmiger, and lower abundance of Veillonella were mainly observed among HIV+ women, most of whom had undetectable viral load because of long-term ART use. Most of the identified taxa were related specifically to cognitive impairment in learning, memory and processing speed.
Previous studies have linked alterations in GMB with neurodegenerative disorders in animal models, and in humans through gut dysbiosis and inflammatory response. [10, 11, 13, 32-36] However, according to a recent review, findings were not consistent across published studies.[37] Although our findings suggest that GMB was altered among HIV+ women with cognitive impairment, especially in learning and memory domains, they are not fully consistent with published findings on correlates of cognitive impairment in the general population. We identified several genera that have been reported in literature in association with cognitive impairment, including Eubacterium, Odoribacter, Ruminococcus, and Veillonella. Eubacterium, Odoribacter and Ruminococcus produce SCFA through fermentation of carbohydrates while Veillonella utilizes lactate.[14, 38-41] The role of SCFA in inflammation has not been well elucidated, which may partially explain why there have been mixed findings in literature about the roles of these taxa in inflammation. SCFA are mainly believed to be anti-inflammatory, but may also exhibit multiple effects in leucocyte recruitment and chemokine production under different conditions and in different types of cells. SCFA may be pro-inflammatory when there is bacterial infection or damage of gut epithelium, or in microglial cells.[42] Furthermore, in contrast to the anti-inflammatory role of butyrate, acetate seems to be involved in cytokine production, which might be implicated in amyloid deposition in the brain relating to cognitive dysfunction and Alzheimer’s disease. [43]
Consistent with our findings, higher abundance of Odoribacter has been shown among patients with Alzheimer’s disease and mild cognitive impairment in comparison to people with normal cognitive function. [14, 44] Notably, its genes have been connected with Alzheimer’s disease pathway in Kyoto Encyclopedia of Genes and Genomes. [14, 45] In this study, Odoribacter was positively associated with impaired memory among HIV+ women, which has not been reported before, to the best of our knowledge. Evidence linking altered abundance of Eubacterium, Ruminococcus and Veillonella to cognitive impairment are mixed. Similar to what we found, some studies reported depleted Veillonella and enriched Eubacterium species, E. eligens, among patients with mild cognitive impairment,[13] while other studies reported opposite results for Veillonella with mild cognitive impairment, and E. eligens, E. hallii, and E. rectale among those with Alzheimer’s disease. [11, 14, 46] Interestingly, growth of E. rectale is stimulated by R. bromii, so it is not surprising to see both taxa showed positive association with cognitive impairment. [40, 47] Ruminococci are species with high abundance in the human intestines.[48] Some studies found lower abundance of Ruminococcus in patients with Alzheimer’s disease and mild cognitive impairment and a positive correlation of this genus with better naming function, and working memory. [11, 46] Yet, it has also been reported to be pro-inflammatory through its role in intestinal immune response.[49, 50] Nevertheless, findings from prior work and the current study support a link of GMB alteration (e.g., increased abundance of Odoribacter) with cognitive impairment and Alzheimer’s disease.
We also identified three genera that have not been previously related to neurocognitive disorders in prior literature, to the best of our knowledge, namely, Gemmiger, Methanobrevibacter and Pyramidobacter. G. formicilis has been observed to be lower in abundance among men with HIV who progressed to acquired immunodeficiency syndrome (AIDS) comparing to those who stayed AIDS-free for 10 years without using antiretroviral therapy.[51] It has also been associated with longer survival and developing colitis when using ipilimumab to treat patients with metastatic melanoma.[52] P.piscolens has been enriched in patients with chronic periodontitis, ischemic stroke, and low-set rectal cancer patients after FOLFOX treatment.[53-55] Pyramidobacter belongs to phylum Synergistes, which may play a pathogenic role in infections.[56, 57] Methanobrevibacter is a dominant archaea commonly found in healthy people, with M.smithii being the major species.[58, 59] M.smithii facilitates production of acetate, butyrate and ATP by removing dihydrogen from host gut environment.[58] It has been implicated in obesity, severe acute malnutrition in children, colorectal cancer, anorexia, inflammatory bowel disease, irritable bowel disease, diverticulosis, constipation and periodontitis.[58, 60] These three genera appear to share the common link to disease pathology involving immune reactions and inflammatory response. We speculate that they may play a role in the development of cognitive disorders in PLWH, who may be subject to chronic immune activation.[61] Future research are needed to understand the underlying mechanisms.
Among PLWH, HIV virus causes gut mucosa damage and translocation of proinflammatory microbial products such as lipopolysaccharide, which induces the release of cytokines in CNS, resulting in neuroinflammation.[61, 62] In line with this, we found that the associations between gut microbial features and cognitive impairment were more profound among HIV+ women. However, it should be noted that the non-significant results in HIV− women might be due to relatively smaller sample size in this group. In addition, our prior work has found enriched Ruminococcus genus in HIV+ women compared to HIV− women.[31] In the current study, we observed enriched abundance of R. bromii associated with both HIV infection and cognitive impairment. This supports a hypothesis that HIV infection may lead to altered GMB profile, which subsequently contributes to the development of cognitive impairment. Furthermore, PLWH are more prone to have systemic inflammation and are at higher risk of aging related disease such as cardiovascular disease, and neurological disorders compared to those without HIV.[61-64] Of note, among the genera we found, Eubacterium, Methanobrevibacter, and Pyramidobacter, have also been associated with aging.[65] Aside from a distinct study population of PLWH, another explanation for inconsistent findings between this study and other existing studies could be the different measures of cognitive functions and endpoints used across studies. Some studies used Alzheimer’s disease and dementia as endpoints, which are more severe forms of cognitive impairment. Even among studies investigating mild cognitive impairment, most studies did not assess cognitive function using seven domains. A majority of women in the present study only demonstrated mild cognitive impairment. Such conflicting results by disease severity has been seen in Bacteroides and E. eligens, which were found to be depleted in patients with Alzheimer’s disease but increased among people with mild cognitive impairment. [13, 14, 35, 36]
Several papers examined potential mechanisms through analysis of microbial gene pathways. They have identified increased glycan biosynthesis and metabolism, transport and catabolism, and vitamin B metabolism, and depleted butyrate biosynthesis pathways, transcription and membrane transport.[11, 14, 46] In our analysis, most pathways related to cognitive impairment were connected to Methanobrevibacter. For example, Coenzyme B and tetrahydromethanopterin are coenzymes in methanogensis while wyosine, CDP-archaeol and archaetidylinositol biosynthesis pathways are related to archaea, which are methanogens.[66] We found pathways depleted in cognitive impairment related to generation of cell energy and precursors of metabolites, including TCA cycle VII (acetate producers) and superpathway of glycosis and Entner-Doudoroff, and pathways enriched in cognitive impairment related to carbohydrate degradation pathway (lactose and galactose degradation I). In addition, we found that pathways related to biosynthesis, including the mevalonate pathway, an amino acid related pathway (chorismate biosynthesis II) and a vitamin related pathway (flavin biosynthesis II), were enriched among women with cognitive impairment. These pathways play fundamental roles in biosynthesis of isoprenoids (mevalonate pathway), aromatic amino acids such as phenylalanine, tryptophan and tyrosine, indole, vitamin K, and folate (chorismate pathway).[66] Riboflavin is an essential nutrient that mammals cannot synthesize, and has been linked with potential antioxidant and neuroprotective effects.[67, 68] Similar to our findings, a recent study also reported worse functioning in memory domains in association with increased bacterial vitamin metabolism related pathways including riboflavin, vitamin B6, folic acid, vitamin B1, and vitamin B12.[46] The authors hypothesized that as a result of bacteria competing for vitamins, host uptake of vitamins were limited. [46] In contrast, it is possible that bacterial vitamin pathways are enriched in cognitive impairment because the host may be vitamin deficient, resulting in cognitive impairment and necessitating bacterial synthesis of vitamins. More studies are needed to replicate our findings and explore these hypotheses.
Strengths and Limitations
To the best of our knowledge, this is the first study with a considerable sample size to examine GMB alterations in association with cognitive impairment among women living with HIV, with a comparison group of uninfected women. We conducted comprehensive assessment of cognitive functions of seven domains and explored microbiome composition change in relation to these functions in each domain. We included a comparison group of women at risk of HIV, who were comparable in demographics, socio-economic status and health-related behaviors as women with HIV. Such a design allowed us to evaluate and interpret the findings more meaningfully. We examined both microbiota compositional changes and metagenomic prediction of functional changes, which may shed light on the roles of GMB in cognitive disorders among PLWH.
Limitations included a cross-sectional study design, and thus we were unable to assess temporal relationships and establish causation between GMB and cognitive impairment. Future studies are needed to examine GMB in association with changes in cognitive outcomes. Second, stool sample collection was not always concurrent with cognitive function measurement. However, most (80%) of the stool sample collections were completed within 6 months of cognitive data collection, which is nondifferential by HIV status. Our sensitivity analysis has also shown consistent findings among those with GMB data collected at the same visit as cognitive assessment. Third, microbial functional pathways examined in our analysis were inferred based on 16S rRNA gene taxonomic data, and thus require confirmation in the future using shotgun metagenomic sequencing. Fourth, the study may lack power to test effect modification by HIV status due to a relatively smaller sample size of the HIV− group. Cognitive impairment was also defined based on data from this small group of HIV− women in the current analysis. Moreover, we did not collect diet and lifestyle data and cannot account for influences of these factors on GMB and cognitive function. Finally, generalization of the results from this study may be restricted to women living with HIV in the urban areas of northeast US, who were engaged in HIV research and under long-term HIV care.
Conclusions
This study provides evidence for the associations of the GMB with cognitive impairment among women with HIV. More studies are needed to validate these findings, understand the underlying mechanisms, and assess the potential of the GMB to become biomarkers and/or therapeutic targets in cognitive diseases including Alzheimer’s disease among PLWH. Further examinations of metabolites such as SCFA, and enzymes in bacterial functional pathways may deepen the understanding of the complicated biological mechanisms linking the GMB with cognitive function.
Supplementary Material
Acknowledgements
Data in this manuscript were collected by the Women’s Interagency HIV Study (WIHS), now the MACS/WIHS Combined Cohort Study (MWCCS).
The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH). MWCCS (Principal Investigators): Atlanta CRS (Ighovwerha Ofotokun, Anandi Sheth, and Gina Wingood), U01-HL146241; Baltimore CRS (Todd Brown and Joseph Margolick), U01-HL146201; Bronx CRS (Kathryn Anastos, David Hanna, and Anjali Sharma), U01-HL146204; Brooklyn CRS (Deborah Gustafson and Tracey Wilson), U01-HL146202; Data Analysis and Coordination Center (Gypsyamber D’Souza, Stephen Gange and Elizabeth Topper), U01-HL146193; Chicago-Cook County CRS (Mardge Cohen and Audrey French), U01-HL146245; Chicago-Northwestern CRS (Steven Wolinsky), U01-HL146240; Northern California CRS (Bradley Aouizerat, Jennifer Price, and Phyllis Tien), U01-HL146242; Los Angeles CRS (Roger Detels and Matthew Mimiaga), U01-HL146333; Metropolitan Washington CRS (Seble Kassaye and Daniel Merenstein), U01-HL146205; Miami CRS (Maria Alcaide, Margaret Fischl, and Deborah Jones), U01-HL146203; Pittsburgh CRS (Jeremy Martinson and Charles Rinaldo), U01-HL146208; UAB-MS CRS (Mirjam-Colette Kempf, Jodie Dionne-Odom, and Deborah Konkle-Parker), U01-HL146192; UNC CRS (Adaora Adimora and Michelle Floris-Moore), U01-HL146194. The MWCCS is funded primarily by the National Heart, Lung, and Blood Institute (NHLBI), with additional co-funding from the Eunice Kennedy Shriver National Institute Of Child Health & Human Development (NICHD), National Institute On Aging (NIA), National Institute Of Dental & Craniofacial Research (NIDCR), National Institute Of Allergy And Infectious Diseases (NIAID), National Institute Of Neurological Disorders And Stroke (NINDS), National Institute Of Mental Health (NIMH), National Institute On Drug Abuse (NIDA), National Institute Of Nursing Research (NINR), National Cancer Institute (NCI), National Institute on Alcohol Abuse and Alcoholism (NIAAA), National Institute on Deafness and Other Communication Disorders (NIDCD), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute on Minority Health and Health Disparities (NIMHD), and in coordination and alignment with the research priorities of the National Institutes of Health, Office of AIDS Research (OAR). MWCCS data collection is also supported by UL1-TR000004 (UCSF CTSA), UL1-TR003098 (JHU ICTR), UL1-TR001881 (UCLA CTSI), P30-AI-050409 (Atlanta CFAR), P30-AI-073961 (Miami CFAR), P30-AI-050410 (UNC CFAR), P30-AI-027767 (UAB CFAR), and P30-MH-116867 (Miami CHARM).
The authors gratefully acknowledge the contributions of the study participants and dedication of the staff at the MWCCS sites.
Funding
This study was supported by the National Heart, Lung, and Blood Institute (NHLBI) R01HL140976. Other related funding sources included K01HL129892, K01HL137557, K01HL160146, R01HL126543, R01 HL132794, R01HL083760 and R01HL095140 from the NHLBI, R01MD011389 from the National Institute on Minority Health and Health Disparities, U01 AI035004 and the Einstein-Rockefeller-CUNY Center for AIDS Research (P30AI124414) funded by the National Institute of Allergy and Infectious Diseases , Einstein Cancer Research Center (P30CA013330) funded by National Cancer Institute, and the Feldstein Medical Foundation Research Grant to Q.Q..
Footnotes
Conflict of Interest/Disclosure Statement
The authors have no conflict of interest to report.
Data Availability
The data supporting the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
- [1].Heaton RK, Clifford DB, Franklin DR Jr., Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I, Group C (2010) HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 75, 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sacktor N, Skolasky RL, Seaberg E, Munro C, Becker JT, Martin E, Ragin A, Levine A, Miller E (2016) Prevalence of HIV-associated neurocognitive disorders in the Multicenter AIDS Cohort Study. Neurology 86, 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Maki PM, Rubin LH, Valcour V, Martin E, Crystal H, Young M, Weber KM, Manly J, Richardson J, Alden C, Anastos K (2015) Cognitive function in women with HIV: findings from the Women's Interagency HIV Study. Neurology 84, 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rubin LH, Sundermann EE, Moore DJ (2019) The current understanding of overlap between characteristics of HIV-associated neurocognitive disorders and Alzheimer's disease. J Neurovirol 25, 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Heaton RK, Franklin DR, Ellis RJ, McCutchan JA, Letendre SL, Leblanc S, Corkran SH, Duarte NA, Clifford DB, Woods SP, Collier AC, Marra CM, Morgello S, Mindt MR, Taylor MJ, Marcotte TD, Atkinson JH, Wolfson T, Gelman BB, McArthur JC, Simpson DM, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I, Group C, Group H (2011) HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. J Neurovirol 17, 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cryan JF, Dinan TG (2012) Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 13, 701–712. [DOI] [PubMed] [Google Scholar]
- [7].Mayer EA, Knight R, Mazmanian SK, Cryan JF, Tillisch K (2014) Gut microbes and the brain: paradigm shift in neuroscience. J Neurosci 34, 15490–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Westfall S, Lomis N, Kahouli I, Dia SY, Singh SP, Prakash S (2017) Microbiome, probiotics and neurodegenerative diseases: deciphering the gut brain axis. Cell Mol Life Sci 74, 3769–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen Y, Fang L, Chen S, Zhou H, Fan Y, Lin L, Li J, Xu J, Chen Y, Ma Y, Chen Y (2020) Gut Microbiome Alterations Precede Cerebral Amyloidosis and Microglial Pathology in a Mouse Model of Alzheimer's Disease. Biomed Res Int 2020, 8456596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Vogt NM, Kerby RL, Dill-McFarland KA, Harding SJ, Merluzzi AP, Johnson SC, Carlsson CM, Asthana S, Zetterberg H, Blennow K, Bendlin BB, Rey FE (2017) Gut microbiome alterations in Alzheimer's disease. Sci Rep 7, 13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu P, Wu L, Peng G, Han Y, Tang R, Ge J, Zhang L, Jia L, Yue S, Zhou K, Li L, Luo B, Wang B (2019) Altered microbiomes distinguish Alzheimer's disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain Behav Immun 80, 633–643. [DOI] [PubMed] [Google Scholar]
- [12].Ling Y, Gong T, Zhang J, Gu Q, Gao X, Weng X, Liu J, Sun J (2020) Gut Microbiome Signatures Are Biomarkers for Cognitive Impairment in Patients With Ischemic Stroke. Front Aging Neurosci 12, 511562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Guo M, Peng J, Huang X, Xiao L, Huang F, Zuo Z (2021) Gut Microbiome Features of Chinese Patients Newly Diagnosed with Alzheimer's Disease or Mild Cognitive Impairment. J Alzheimers Dis 80, 299–310. [DOI] [PubMed] [Google Scholar]
- [14].Haran JP, Bhattarai SK, Foley SE, Dutta P, Ward DV, Bucci V, McCormick BA (2019) Alzheimer's Disease Microbiome Is Associated with Dysregulation of the Anti-Inflammatory P-Glycoprotein Pathway. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shabbir U, Arshad MS, Sameen A, Oh DH (2021) Crosstalk between Gut and Brain in Alzheimer's Disease: The Role of Gut Microbiota Modulation Strategies. Nutrients 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sochocka M, Donskow-Lysoniewska K, Diniz BS, Kurpas D, Brzozowska E, Leszek J (2019) The Gut Microbiome Alterations and Inflammation-Driven Pathogenesis of Alzheimer's Disease-a Critical Review. Mol Neurobiol 56, 1841–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bandera A, De Benedetto I, Bozzi G, Gori A (2018) Altered gut microbiome composition in HIV infection: causes, effects and potential intervention. Curr Opin HIV AIDS 13, 73–80. [DOI] [PubMed] [Google Scholar]
- [18].Zevin AS, McKinnon L, Burgener A, Klatt NR (2016) Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS 11, 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bacon MC, von Wyl V, Alden C, Sharp G, Robison E, Hessol N, Gange S, Barranday Y, Holman S, Weber K, Young MA (2005) The Women's Interagency HIV Study: an observational cohort brings clinical sciences to the bench. Clin Diagn Lab Immunol 12, 1013–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Adimora AA, Ramirez C, Benning L, Greenblatt RM, Kempf MC, Tien PC, Kassaye SG, Anastos K, Cohen M, Minkoff H, Wingood G, Ofotokun I, Fischl MA, Gange S (2018) Cohort Profile: The Women's Interagency HIV Study (WIHS). Int J Epidemiol 47, 393–394i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].MACS/WIHS Combined Cohort Study, https://statepi.jhsph.edu/mwccs/,
- [22].Wang Z, Peters BA, Usyk M, Xing J, Hanna DB, Wang T, Post WS, Landay AL, Hodis HN, Weber K, French A, Golub ET, Lazar J, Gustafson D, Kassaye S, Aouizerat B, Haberlen S, Malvestutto C, Budoff M, Wolinsky SM, Sharma A, Anastos K, Clish CB, Kaplan RC, Burk RD, Qi Q (2022) Gut Microbiota, Plasma Metabolomic Profiles, and Carotid Artery Atherosclerosis in HIV Infection. Arterioscler Thromb Vasc Biol 42, 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rubin LH, Maki PM, Springer G, Benning L, Anastos K, Gustafson D, Villacres MC, Jiang X, Adimora AA, Waldrop-Valverde D, Vance DE, Bolivar H, Alden C, Martin EM, Valcour VG, Women's Interagency HIVS (2017) Cognitive trajectories over 4 years among HIV-infected women with optimal viral suppression. Neurology 89, 1594–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72, 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Peters BA, Xue X, Wang Z, Usyk M, Santoro N, Sharma A, Anastos K, Tien PC, Golub ET, Weber KM, Gustafson D, Kaplan RC, Burk R, Qi Q (2021) Menopausal status and observed differences in the gut microbiome in women with and without HIV infection. Menopause 28, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hanna DB, Post WS, Deal JA, Hodis HN, Jacobson LP, Mack WJ, Anastos K, Gange SJ, Landay AL, Lazar JM, Palella FJ, Tien PC, Witt MD, Xue X, Young MA, Kaplan RC, Kingsley LA (2015) HIV Infection Is Associated With Progression of Subclinical Carotid Atherosclerosis. Clin Infect Dis 61, 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kaul A, Mandal S, Davidov O, Peddada SD (2017) Analysis of Microbiome Data in the Presence of Excess Zeros. Front Microbiol 8, 2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang Z, Usyk M, Sollecito CC, Qiu Y, Williams-Nguyen J, Hua S, Gradissimo A, Wang T, Xue X, Kurland IJ, Ley K, Landay AL, Anastos K, Knight R, Kaplan RC, Burk RD, Qi Q (2020) Altered Gut Microbiota and Host Metabolite Profiles in Women With Human Immunodeficiency Virus. Clin Infect Dis 71, 2345–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gareau MG (2014) Microbiota-gut-brain axis and cognitive function. Adv Exp Med Biol 817, 357–371. [DOI] [PubMed] [Google Scholar]
- [33].Frohlich EE, Farzi A, Mayerhofer R, Reichmann F, Jacan A, Wagner B, Zinser E, Bordag N, Magnes C, Frohlich E, Kashofer K, Gorkiewicz G, Holzer P (2016) Cognitive impairment by antibiotic-induced gut dysbiosis: Analysis of gut microbiota-brain communication. Brain Behav Immun 56, 140–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kohler CA, Maes M, Slyepchenko A, Berk M, Solmi M, Lanctot KL, Carvalho AF (2016) The Gut-Brain Axis, Including the Microbiome, Leaky Gut and Bacterial Translocation: Mechanisms and Pathophysiological Role in Alzheimer's Disease. Curr Pharm Des 22, 6152–6166. [DOI] [PubMed] [Google Scholar]
- [35].Saji N, Murotani K, Hisada T, Tsuduki T, Sugimoto T, Kimura A, Niida S, Toba K, Sakurai T (2019) The relationship between the gut microbiome and mild cognitive impairment in patients without dementia: a cross-sectional study conducted in Japan. Sci Rep 9, 19227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Saji N, Niida S, Murotani K, Hisada T, Tsuduki T, Sugimoto T, Kimura A, Toba K, Sakurai T (2019) Analysis of the relationship between the gut microbiome and dementia: a cross-sectional study conducted in Japan. Sci Rep 9, 1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Spichak S, Bastiaanssen TFS, Berding K, Vlckova K, Clarke G, Dinan TG, Cryan JF (2021) Mining microbes for mental health: Determining the role of microbial metabolic pathways in human brain health and disease. Neurosci Biobehav Rev 125, 698–761. [DOI] [PubMed] [Google Scholar]
- [38].Hiippala K, Barreto G, Burrello C, Diaz-Basabe A, Suutarinen M, Kainulainen V, Bowers JR, Lemmer D, Engelthaler DM, Eklund KK, Facciotti F, Satokari R (2020) Novel Odoribacter splanchnicus Strain and Its Outer Membrane Vesicles Exert Immunoregulatory Effects in vitro. Front Microbiol 11, 575455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cattaneo A, Cattane N, Galluzzi S, Provasi S, Lopizzo N, Festari C, Ferrari C, Guerra UP, Paghera B, Muscio C, Bianchetti A, Volta GD, Turla M, Cotelli MS, Gennuso M, Prelle A, Zanetti O, Lussignoli G, Mirabile D, Bellandi D, Gentile S, Belotti G, Villani D, Harach T, Bolmont T, Padovani A, Boccardi M, Frisoni GB, Group I-F (2017) Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging 49, 60–68. [DOI] [PubMed] [Google Scholar]
- [40].Ze X, Duncan SH, Louis P, Flint HJ (2012) Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6, 1535–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Loomba R, Ling L, Dinh DM, DePaoli AM, Lieu HD, Harrison SA, Sanyal AJ (2021) The Commensal Microbe Veillonella as a Marker for Response to an FGF19 Analog in NASH. Hepatology 73, 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Vinolo MA, Rodrigues HG, Nachbar RT, Curi R (2011) Regulation of inflammation by short chain fatty acids. Nutrients 3, 858–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Marizzoni M, Cattaneo A, Mirabelli P, Festari C, Lopizzo N, Nicolosi V, Mombelli E, Mazzelli M, Luongo D, Naviglio D, Coppola L, Salvatore M, Frisoni GB (2020) Short-Chain Fatty Acids and Lipopolysaccharide as Mediators Between Gut Dysbiosis and Amyloid Pathology in Alzheimer's Disease. J Alzheimers Dis 78, 683–697. [DOI] [PubMed] [Google Scholar]
- [44].Ren T, Gao Y, Qiu Y, Jiang S, Zhang Q, Zhang J, Wang L, Zhang Y, Wang L, Nie K (2020) Gut Microbiota Altered in Mild Cognitive Impairment Compared With Normal Cognition in Sporadic Parkinson's Disease. Front Neurol 11, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kanehisa Laboratories. 2018. Alzheimer disease - reference pathway + Odoribacter splanchnicus. Date Accessed: 1/16/2022. URL: https://www.kegg.jp/kegg-bin/showpathway?category=Odoribacter%20splanchnicus&categorytype=species&mapno=05010. Database Provider: Kyoto Encyclopedia of Genes and Genomes [Google Scholar]
- [46].Arnoriaga-Rodriguez M, Mayneris-Perxachs J, Burokas A, Contreras-Rodriguez O, Blasco G, Coll C, Biarnes C, Miranda-Olivos R, Latorre J, Moreno-Navarrete JM, Castells-Nobau A, Sabater M, Palomo-Buitrago ME, Puig J, Pedraza S, Gich J, Perez-Brocal V, Ricart W, Moya A, Fernandez-Real X, Ramio-Torrenta L, Pamplona R, Sol J, Jove M, Portero-Otin M, Maldonado R, Fernandez-Real JM (2020) Obesity Impairs Short-Term and Working Memory through Gut Microbial Metabolism of Aromatic Amino Acids. Cell Metab 32, 548–560 e547. [DOI] [PubMed] [Google Scholar]
- [47].Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM (2019) Dynamics of Human Gut Microbiota and Short-Chain Fatty Acids in Response to Dietary Interventions with Three Fermentable Fibers. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].La Reau AJ, Meier-Kolthoff JP, Suen G (2016) Sequence-based analysis of the genus Ruminococcus resolves its phylogeny and reveals strong host association. Microb Genom 2, e000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Henke MT, Kenny DJ, Cassilly CD, Vlamakis H, Xavier RJ, Clardy J (2019) Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn's disease, produces an inflammatory polysaccharide. Proc Natl Acad Sci U S A 116, 12672–12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chen SJ, Chen CC, Liao HY, Lin YT, Wu YW, Liou JM, Wu MS, Kuo CH, Lin CH (2022) Association of Fecal and Plasma Levels of Short-Chain Fatty Acids With Gut Microbiota and Clinical Severity in Parkinson Disease Patients. Neurology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chen Y, Lin H, Cole M, Morris A, Martinson J, McKay H, Mimiaga M, Margolick J, Fitch A, Methe B, Srinivas VR, Peddada S, Rinaldo CR (2021) Signature changes in gut microbiome are associated with increased susceptibility to HIV-1 infection in MSM. Microbiome 9, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, Boselli L, Routier E, Cassard L, Collins M, Vaysse T, Marthey L, Eggermont A, Asvatourian V, Lanoy E, Mateus C, Robert C, Carbonnel F (2017) Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol 28, 1368–1379. [DOI] [PubMed] [Google Scholar]
- [53].Deng ZL, Szafranski SP, Jarek M, Bhuju S, Wagner-Dobler I (2017) Dysbiosis in chronic periodontitis: Key microbial players and interactions with the human host. Sci Rep 7, 3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Li N, Wang X, Sun C, Wu X, Lu M, Si Y, Ye X, Wang T, Yu X, Zhao X, Wei N, Wang X (2019) Change of intestinal microbiota in cerebral ischemic stroke patients. BMC Microbiol 19, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Li J, Li J, Lyu N, Ma Y, Liu F, Feng Y, Yao L, Hou Z, Song X, Zhao H, Li X, Wang Y, Xiao C, Zhu B (2020) Composition of fecal microbiota in low-set rectal cancer patients treated with FOLFOX. Ther Adv Chronic Dis 11, 2040622320904293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Vartoukian SR, Palmer RM, Wade WG (2007) The division "Synergistes". Anaerobe 13, 99–106. [DOI] [PubMed] [Google Scholar]
- [57].Horz HP, Citron DM, Warren YA, Goldstein EJ, Conrads G (2006) Synergistes group organisms of human origin. J Clin Microbiol 44, 2914–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gaci N, Borrel G, Tottey W, O'Toole PW, Brugere JF (2014) Archaea and the human gut: new beginning of an old story. World J Gastroenterol 20, 16062–16078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dridi B, Henry M, El Khechine A, Raoult D, Drancourt M (2009) High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS One 4, e7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Camara A, Konate S, Tidjani Alou M, Kodio A, Togo AH, Cortaredona S, Henrissat B, Thera MA, Doumbo OK, Raoult D, Million M (2021) Clinical evidence of the role of Methanobrevibacter smithii in severe acute malnutrition. Sci Rep 11, 5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hong S, Banks WA (2015) Role of the immune system in HIV-associated neuroinflammation and neurocognitive implications. Brain Behav Immun 45, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Deeks SG, Tracy R, Douek DC (2013) Systemic effects of inflammation on health during chronic HIV infection. Immunity 39, 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Feinstein MJ, Hsue PY, Benjamin LA, Bloomfield GS, Currier JS, Freiberg MS, Grinspoon SK, Levin J, Longenecker CT, Post WS (2019) Characteristics, Prevention, and Management of Cardiovascular Disease in People Living With HIV: A Scientific Statement From the American Heart Association. Circulation 140, e98–e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Canizares S, Cherner M, Ellis RJ (2014) HIV and aging: effects on the central nervous system. Semin Neurol 34, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wu L, Zeng T, Zinellu A, Rubino S, Kelvin DJ, Carru C (2019) A Cross-Sectional Study of Compositional and Functional Profiles of Gut Microbiota in Sardinian Centenarians. mSystems 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Caspi R, Billington R, Keseler IM, Kothari A, Krummenacker M, Midford PE, Ong WK, Paley S, Subhraveti P, Karp PD (2020) The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res 48, D445–D453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Saedisomeolia A, Ashoori M (2018) Riboflavin in Human Health: A Review of Current Evidences. Adv Food Nutr Res 83, 57–81. [DOI] [PubMed] [Google Scholar]
- [68].McNeill G, Jia X, Whalley LJ, Fox HC, Corley J, Gow AJ, Brett CE, Starr JM, Deary IJ (2011) Antioxidant and B vitamin intake in relation to cognitive function in later life in the Lothian Birth Cohort 1936. Eur J Clin Nutr 65, 619–626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.