Abstract
Introduction:
The diagnosis of Alzheimer's disease (AD) requires the presence of amyloid and tau pathology, but it remains unclear how they affect the structural network in the pre-clinical stage. We aimed to assess differences in topological properties in cognitively normal (CN) individuals with varying levels of amyloid and tau pathology, as well as their association with AD pathology burden.
Methods:
A total of 68 CN individuals were included and stratified by normal/abnormal (−/+) amyloid (A) and tau (T) status based on positron emission tomography results, yielding three groups: A−T− (n = 19), A+T− (n = 28), and A+T+ (n = 21). Topological properties were measured from structural connectivity. Group differences and correlations with A and T were evaluated.
Results:
Compared with the A−T− group, the A+T+ group exhibited changes in the structural network topology. At the global level, higher assortativity was shown in the A+T+ group and was correlated with greater tau burden (r = 0.29, p = 0.02), while no difference in global efficiency was found across the three groups. At the local level, the A+T+ group showed disrupted topological properties in the left hippocampus compared with the A−T− group, characterized by lower local efficiency (p < 0.01) and a lower clustering coefficient (p = 0.014).
Conclusions:
The increased linkage in the higher level architecture of the white matter network reflected by assortativity may indicate increased brain resilience in the early pathological state. Our results encourage further investigation of the topological properties of the structural network in pre-clinical AD.
Impact statement
The present work explored the topological patterns of the brain structural network in subjects with pre-clinical Alzheimer's disease (AD) and their correlation with the two AD hallmarks, amyloid and tau. Results showed an increased linkage in the higher level architecture of the white matter network measured by assortativity, while global efficiency remained unchanged. The study provides a potential graph-based biomarker of brain connectivity for early identification of AD, and the results encourage further investigation of structural network properties.
Keywords: Alzheimer's disease, assortativity, diffusion tensor imaging, graph theory, structural connectivity, tractography
Introduction
Alzheimer's disease (AD) is characterized by two hallmarks, namely deposition of β-amyloid (Aβ) and accumulation of phosphorylated tau in neurofibrillary tangles. These features can appear years before the manifestation of clinical symptoms. Detecting AD-related pathological changes during the asymptomatic stage is crucial for understanding the disease mechanisms and progression, enabling potential interventions, and facilitating participant enrollment in clinical trials (Vogt et al., 2022).
At present, confirmation of Aβ and tau can only be achieved through postmortem examination (Scheltens et al., 2021). This has driven research efforts to identify in vivo imaging biomarkers that aid in AD diagnosis. In vivo detection of Aβ and tau is possible through positron emission tomography (PET) and cerebral spinal fluid analysis by lumbar puncture. Nonetheless, the widespread use of these techniques is hindered due to the limited availability of radioactive tracers, expensive cost, and inherent invasiveness. Structural magnetic resonance imaging (MRI) typically only reveals reduced hippocampal volume in the late stages of the disease, but this is not specific to AD since it can also be related to aging or other types of dementia (Pini et al., 2016).
Studies have demonstrated that a healthy brain network follows a highly efficient network topology with a combination of high segregation and integration (Bullmore and Bassett, 2011). This leads to a hypothesis of AD as a disconnection syndrome (Delbeuck et al., 2003), which suggests that alterations in communication between anatomically and functionally connected brain areas could lead to clinical symptoms in AD.
Graph theory provides a wide range of quantitative measures for characterization of the network architecture of the brain (Rubinov and Sporns, 2010), which can offer insights into the neurodegenerative process. Disruptions in structural network topology have been observed in AD (Lo et al., 2010), mild cognitive impairment (Jacquemont et al., 2017), subjective cognitive decline (Shu et al., 2018), pre-clinical AD (Pereira et al., 2018), and carriers of autosomal dominant AD mutations (Prescott et al., 2022). Additionally, these disruptions are associated with cognitive decline and AD pathology (King-Robson et al., 2021). These findings indicate that topological analysis could be a valuable tool for investigating changes in the structural network at an early stage.
The definition of AD has shifted from the clinical to biological construct in the research setting to reflect the underpinning neuropathology (Jack et al., 2018). However, with the updated research framework, the impact of Aβ and tau on the structural network topology in the pre-clinical stage is not yet fully understood. Thus, the present work aimed to study topological properties of the structural connectome in pre-clinical AD, as indexed by Aβ and tau PET imaging biomarkers.
We first investigated the differences in topological properties at both global and nodal levels in cognitively normal (CN) participants with varying levels of Aβ and tau burden. We then evaluated the relationship between network topological properties and the burden of Aβ and tau.
Materials and Methods
All data were acquired from the Alzheimer's Disease Neuroimaging Initiative 3 database (ADNI 3) (http://adni.loni.usc.edu/), which was launched in 2003 as a public–private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessments can be combined to assess the progression of mild cognitive impairment (MCI) and early AD.
Men and women aged 55–90 years across CN, mild cognitive impairment, and AD dementia groups were included in ADNI, where clinical diagnosis was made according to the criteria of the National Institute of Neurological and Communicative Disorders and Stroke–Alzheimer's Disease and Related Disorders Association (NINCDS-ADRAD) (McKhann et al., 2011). Due to the retrospective nature of the study, informed consent was waived.
Subjects
In this study, the selection criteria for the database were CN participants with availability of T1-weighted MRI, florbetapir (AV45) PET, flortaucipir (AV1451) PET, and diffusion tensor imaging at baseline. The diffusion imaging was further limited to acquisition using Siemens scanners with 48 diffusion directions to reduce the impact of scanner types on tractography and graph construction (Kurokawa et al., 2021). Demographic data such as age, sex, years of education, clinical dementia rating (CDR), mini-mental state examination (MMSE), and clinical diagnosis were recorded.
PET biomarkers and group classification
According to the National Institute on Aging and Alzheimer's Association 2018 Research Framework (Jack et al., 2018), the presence of Aβ determines an individual's placement on the Alzheimer's continuum. Specifically, the presence of Aβ without tau (A+T−) indicates Alzheimer's pathologic change, while the presence of both Aβ and tau (A+T+) is indicative of AD.
Following the recommendation, we categorized CN subjects into the following groups: (1) CN subjects with normal Aβ and tau (A−T−); (2) CN subjects with abnormal Aβ, but normal tau (A+T−); and (3) CN subjects with abnormal Aβ and tau (A+T+). The status of Aβ and tau was determined using florbetapir and flortaucipir PET values provided by the University of California, Berkeley. The florbetapir PET was used to assess global Aβ load.
The mean standardized uptake value ratio (SUVR) on florbetapir PET images was determined in anterior/posterior cingulate, lateral parietal, and lateral temporal regions using the whole cerebellum as the reference region. A cutoff value of 1.11 was applied to determine Aβ positivity (A+) (Landau et al., 2013). The flortaucipir PET SUVR values were used to quantify tau load.
A size-weighted mean flortaucipir SUVR was calculated in metatemporal regions, including the amygdala, entorhinal cortex, fusiform, and inferior temporal and middle temporal gyri, and normalized by inferior cerebellar gray matter. Tau positivity (T+) was thresholded at SUVR >1.23 (Jack et al., 2017).
MRI acquisition
All data were collected using a 3T Siemens scanner (Siemens Medical Solutions, Erlangen, Germany). Image protocols were standardized across ADNI study sites. Details of MRI parameters can be found at http://adni.loni.usc.edu/methods/documents/mri-protocols/. Three-dimensional, T1-weighted volumetric sequences were acquired with the following parameters: echo time = 2.98 ms, repetition time = 2300 ms, inversion time = 900 ms, flip angle = 10°, field of view = 208 × 240 × 256 mm, and acquired resolution = 1 × 1 × 1 mm.
Axial diffusion MRI data were acquired with 48 diffusion-encoding directions at b = 1000 s/mm2 and 6 nondiffusion-weighted b0 images. Other scanning parameters include resolution = 2 × 2 × 2 mm; time repetition = 7200–9600 ms; time echo = 56–82 ms; flip angle = 90°; and field of view = 232 × 232 × 160 mm.
Structural MRI preprocessing
T1-weighted images were corrected for head motion and intensity inhomogeneity, followed by nonbrain tissue removal using the pipeline implemented in FreeSurfer (version 7.1.1, http://surfer.nmr.mgh.harvard.edu/). The brain was parcellated into 68 cortical regions based on the Desikan–Killiany atlas (Desikan et al., 2006) as well as the following 7 subcortical regions in each hemisphere (Fischl et al., 2002): thalamus, caudate, putamen, pallidum, hippocampus, amygdala, and accumbens.
Considering that white matter hyperintensities (WMHs) shown on T2 FLAIR images are possible factors associated with vascular disease and may confound the fiber tracking algorithm (Reginold et al., 2018), we controlled for WMH volumes in the statistical analysis. WMH volumes were obtained from ADNI, which were estimated with a modified Bayesian probability structure through histogram fitting. Briefly, the likelihood estimates of each image were calculated through histogram segmentation and thresholding. Then, along with WMH priors (which were estimated from more than 700 individuals) and tissue constraints, the probabilities were thresholded at 3.5 standard deviations above the mean to create a binary WMH mask. Finally, the mask was back-transformed to native space for volume calculation. WMH volumes were available for all included subjects.
Diffusion MRI preprocessing
Diffusion-weighted imaging (DWI) volumes were processed using the FMRIB software library (FSL) toolbox and MRtrix3 (Tournier et al., 2019) to correct for noise, Gibbs ringing artifacts, head motion, and eddy current distortions. b vectors were rotated accordingly (Leemans and Jones, 2009). All nonbrain tissues were removed from the diffusion-weighted images using the FSL Brain Extraction Tool.
A field map was utilized to correct for echo planar imaging (EPI)-induced susceptibility artifacts, which usually caused geometric distortions at tissue–fluid interfaces. The field map was calculated from a double-echo gradient echo sequence by taking the difference between two phase images and dividing that by the echo time difference. Skull-stripped b0 images were registered to their respective T1 image based on white matter boundaries with the FSLs epi_reg tool to coregister DWI and T1 images. The resulting 3D transformation matrices and deformation fields were back-transformed and applied to the T1 image, so all images were coregistered in the diffusion space.
All processed images were visually inspected for quality control. Fifteen images were excluded from further analysis due to excessive distortion artifacts, and one image was excluded due to lack of a field map for EPI artifact correction.
White matter tractography
We performed probabilistic tractography with MRtrix3. The signal expected for a single-fiber white matter population (so-called response functions) was calculated for constrained spherical deconvolution to estimate fiber orientation distributions (FODs). The FOD maps were passed to the iFOD2 algorithm for fiber tracking. Seeding points were determined dynamically using the spherical-deconvolution informed filtering of the tractogram model (Smith et al., 2015). Anatomical constraints were applied to ensure minimal contamination from spurious streamline trajectories through gray matter (Smith et al., 2012). A total of 1 million streamlines were generated for better reproducibility (Roine et al., 2019). Other default tracking parameters included a step size of 1 mm, minimum fiber length of 4 mm, and FOD cutoff of 0.1. Finally, the SIFT2 algorithm was applied on all tractograms to filter implausible streamlines by assigning weights to each streamline. This step can improve the biological plausibility of the final connectome reconstruction (Smith et al., 2015).
Network construction and graph theoretical analysis
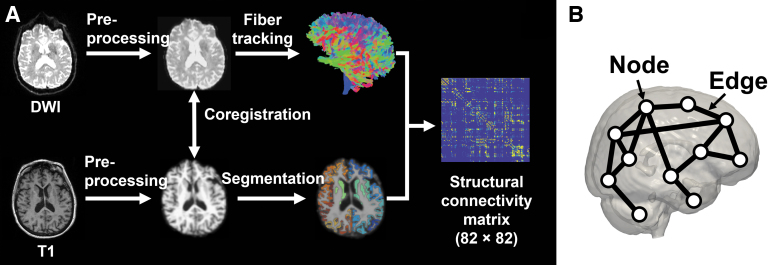
Graph theory was used to analyze topological changes in the brain network. In such analysis, the brain network is represented by a graph with nodes and edges. Nodes refer to brain regions, while edges refer to the connection between them. In this study, we defined nodes as the 82 regions segmented by FreeSurfer (68 cortical +14 subcortical regions) and edges as white matter links between these regions. Therefore, an 82 × 82 connectivity matrix was built for each subject. An overview of the methodology for building structural networks is shown in Figure 1.
FIG. 1.
(A) Flowchart of the brain network construction from DWI. (B) Illustration of the structural brain network. The nodes in the network are represented by circles, which correspond to anatomical regions. The edges, depicted as connecting lines between the circles, represent the connections between these regions. DWI, diffusion-weighted imaging.
To compare the network topology across different subjects and minimize the number of spurious connections in each network, we thresholded and binarized the connectivity matrices to ensure the same number of edges in all subjects. Fixed-density thresholding can yield a more reproducible and consistent network architecture while maintaining sensitivity to disease effects (De Brito Robalo et al., 2022). The network density is defined by the fraction of the number of edges over the maximal possible number of edges (i.e., 82 × 81/2 = 3321 possible edges in our case). All self-connections were excluded.
Considering the lack of a definitive method to select a single threshold, we chose to binarize the graph over a range of density threshold levels (5–9%, in the step of 0.2%) and estimated the global network properties at each threshold value. For global network characteristics, we analyzed global efficiency and assortativity. Global efficiency is a measure of network integration and it is calculated by the average inverse of the shortest path in the network. Assortativity is a measure of brain resilience and refers to the tendency of nodes to link with other similar nodes (Franciotti et al., 2019; Miraglia et al., 2022). A positive value indicates that nodes tend to link to other nodes with the same or similar degree.
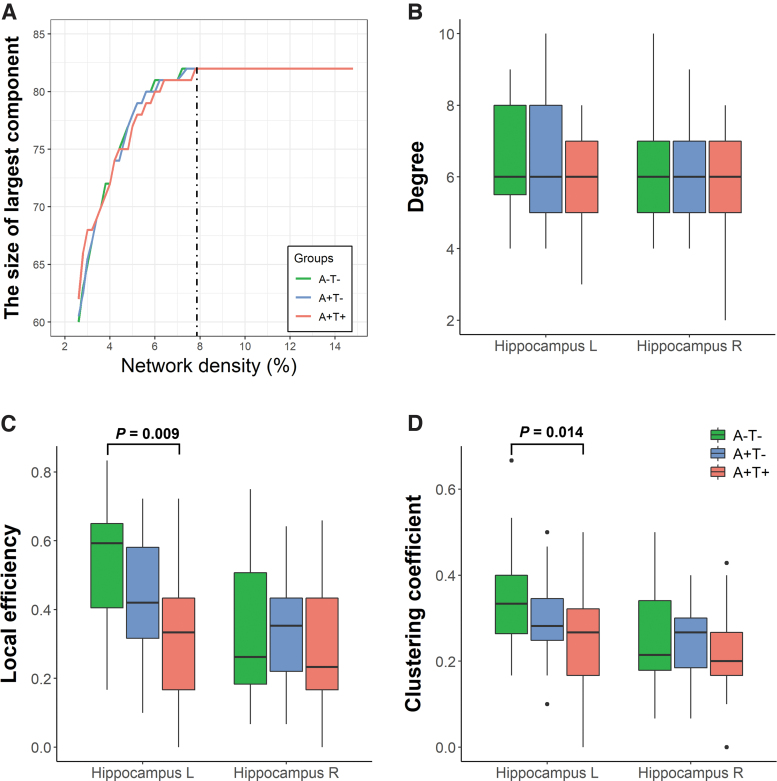
For the local network analysis, we constructed structural networks at a specific threshold of 7.8% (Fig. 3A). This threshold ensures that all regions are included in the network while minimizing the number of false-positive connections. For nodal characteristics, we focused on the hippocampus and assessed various nodal topological metrics such as degree, local efficiency, and clustering coefficient.
FIG. 3.
(A) The graph shows the largest connected component size of the network in A−T− (green line), A+T− (blue line), and A+T+ (red line) groups as a function of network density. The largest component size tends to increase with higher density. The dashed line indicates the lowest density threshold (7.8%) in which networks of the 3 groups included all nodes (i.e., 82 regions). Local topological properties of the hippocampus such as degree (B), local efficiency (C), and clustering coefficient (D) were calculated under the density threshold of 7.8%. Significant differences in local efficiency and clustering coefficient in the left hippocampus were found between the A+T+ and A−T− groups. The box plots show the median and interquartile range. A−/A+, negative/positive amyloid-PET; L, left; PET, positron emission tomography; R, right; T−/T+, negative/positive tau-PET.
Degree is calculated as the number of links that are connected to a particular node. Local efficiency reflects the extent of integration between immediate neighbors of a given node (Fornito et al., 2016) and it is calculated as the average shortest path length within the node's neighborhood. The clustering coefficient is calculated as the fraction of a node's neighbors that are neighbors with each other. All network topological metrics were calculated by the Brain Connectivity Toolbox in MATLAB (Rubinov and Sporns, 2010).
Statistical analyses
The normality of continuous variables was first examined using the Shapiro–Wilk test, in which the assumption of normality was violated. WMH was log-transformed for subsequent analysis due to its skewness. For continuous variables with normal distribution, an analysis of variance, followed by Scheffe's tests, was conducted for group comparison. For continuous variables with skewed distribution, the Kruskal–Wallis test along with the Mann–Whitney U test for post hoc comparisons was performed. The chi-square test was employed to analyze categorical variables.
An analysis of covariance was performed with control of age, sex, years of education, and WMH volume to evaluate group differences in network metrics. A false discovery rate was used to adjust false-positive results in multiple comparisons. If the results were significant, then post hoc comparisons were performed with the Bonferroni method.
Pearson's partial correlation coefficient was utilized by controlling for age, sex, years of education, and WMH volume to assess the associations of network topological metrics with global Aβ load and tau burden in the temporal metaregion of interest (ROI). Network metrics were calculated under the density threshold of 7.8% to reduce the number of comparisons. Correlation was examined in the whole sample as well as within each group.
All statistical analyses were performed using SPSS 26.0 (Armonk, NY). Statistical tests were two-sided, with the significance level set to p < 0.05.
Results
Demographics
After selection and image quality control, a total of 68 CN participants were included and classified as A−T− in 19 cases, A+T− in 28 cases, and A+T+ in 21 cases according to amyloid and tau PET imaging. The demographic details are presented in Table 1. The groups were comparable in terms of sex (p = 0.13), age (p = 0.35), years of education (p = 0.42), CDR (p = 0.35), MMSE (p = 0.34), WMH volumes (p = 0.65), and hippocampal volume (p = 0.88). However, they differed significantly in terms of global amyloid-PET SUVR and tau-PET SUVR (all with p < 0.001).
Table 1.
Demographics
| A−T− (n = 19) | A+T− (n = 28) | A+T+ (n = 21) | p | |
|---|---|---|---|---|
| Sex: F/M | 14/5 | 13/15 | 14/7 | 0.13 |
| Age, years | 72.7 (7.5) | 74.3 (8.5) | 76.2 (6.0) | 0.35 |
| Education, years | 15.9 (2.2) | 16.6 (2.6) | 16.9 (2.4) | 0.42 |
| CDR | 0 (0, 0) | 0 (0, 0) | 0 (0, 0) | 0.35 |
| MMSE | 29 (28, 30) | 29 (29, 30) | 29 (27, 30) | 0.34 |
| WMH volumes, cm3 | 1.7 (0.5, 4.1) | 2.0 (0.7, 9.6) | 1.8 (0.6, 10.3) | 0.65 |
| Global amyloid-PET SUVR | 1.02 (0.99, 1.04) | 1.17 (1.14, 1.30) | 1.35 (1.19, 1.47) | <0.001a,b |
| Tau-PET SUVR | 1.16 (1.11, 1.19) | 1.16 (1.12, 1.20) | 1.27 (1.25, 1.34) | <0.001b,c |
| Hippocampal volume, cm3 | 8.0 (0.8) | 8.0 (0.9) | 7.9 (0.8) | 0.88 |
Values are expressed as mean with standard deviation for normally distributed data, otherwise expressed as median with interquartile range.
Statistically significant difference between A−T− and A+T− (p < 0.001).
Statistically significant difference between A−T− and A+T+ (p < 0.001).
Statistically significant difference between A+T− and A+T+ (p < 0.001).
A, amyloid; CDR, clinical dementia rating; MMSE, mini-mental state examination; PET, positron emission tomography; SUVR, standardized uptake value ratio; T, tau; WMH, white matter hyperintensity.
Global network properties
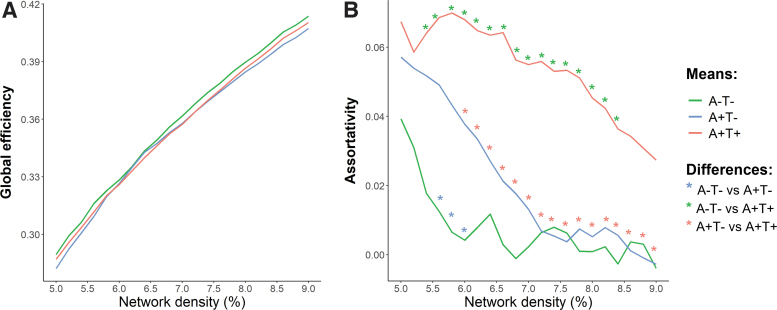
Results from the comparison of global network metrics among groups are presented in Figure 2 and Supplementary Table S1. Overall, no significant differences were observed in global efficiency across groups at different densities, while after controlling for age, sex, years of education, and WMH volume, a significant difference in assortativity was found.
FIG. 2.
Global topological properties. Mean values of global efficiency (A) and assortativity (B) are shown for A−T− (green), A+T− (blue), and A+T+ (red) groups at different network densities. *Indicates statistical differences between A−T− and A+T− (blue) groups, between A−T− and A+T+ (green) groups, and between A+T− and A+T+ (red) groups. Further details on the statistical differences are shown in Supplementary Table S1. A, amyloid; T, tau.
Specifically, the A+T+ group exhibited increased assortativity compared with the A−T− group and A+T− group at the density range of 5.4–8.4% and 6.0–9.0%, respectively. Additionally, the A+T− group showed higher assortativity than the A−T− group at the density range of 5.6–6.0%.
Local network properties in the hippocampus
In Figure 3, comparisons among different groups of topological properties in the hippocampus are presented, such as degree, local efficiency, and clustering coefficient calculated under the density threshold of 7.8%, in which all 82 nodes were included in the largest component of the 3 groups.
In the left hippocampus, significant differences in network properties were observed across groups for local efficiency [F (2, 61) = 4.845, p = 0.011] and clustering coefficient [F (2, 61) = 4.513, p = 0.015], but results were comparable in terms of degree [F (2, 61) = 1.815, p = 0.171]. The A+T+ group showed lower local efficiency (p = 0.009) and a lower clustering coefficient (p = 0.014) than the A−T− group. Conversely, no significant differences were found across groups in the right hippocampus (all p values >0.05).
Association of network measures with Aβ and tau burden
As significant differences were observed in the assortativity, local efficiency, and clustering coefficient of the left hippocampus across the three groups, we conducted further analysis to explore their relationship with global Aβ load and tau burden in the temporal meta-ROI under the density threshold of 7.8%.
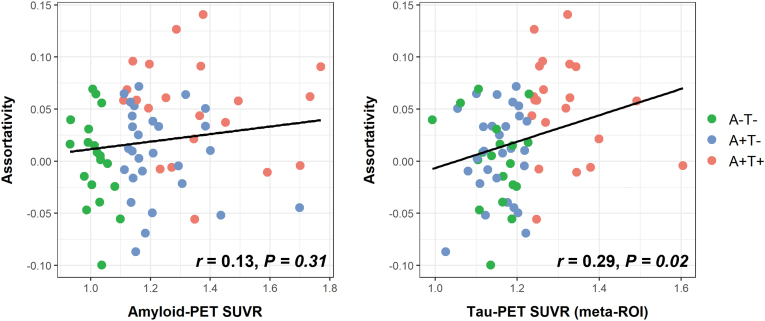
The results showed that in the entire sample, an association was observed between assortativity and tau load (Fig. 4), with higher assortativity correlated with a greater tau-PET SUVR (r = 0.29, p = 0.02). However, this correlation was not observed in the subgroups (Table 2). No correlation was found between assortativity and amyloid load (r = 0.13, p = 0.31). Nodal metrics, including local efficiency and clustering coefficient, did not show any association with either amyloid or tau load (Supplementary Table S2).
FIG. 4.
Scatter plot showing the correlation between assortativity and global amyloid-PET SUVR (left) and between assortativity and tau-PET SUVR in the temporal meta-ROI (right). Temporal meta-ROI includes amygdala, entorhinal, fusiform, and inferior temporal and middle temporal gyri. ROI, region of interest; SUVR, standardized uptake value ratio.
Table 2.
Associations Between Assortativity and Amyloid and Tau
| Cohorts | Amyloid-PET |
Tau-PET |
||
|---|---|---|---|---|
| r | p | r | p | |
| Whole sample | 0.13 | 0.31 | 0.29 | 0.02 |
| A−T− | −0.38 | 0.16 | −0.17 | 0.53 |
| A+T− | −0.25 | 0.15 | 0.14 | 0.52 |
| A+T+ | −0.12 | 0.66 | −0.01 | 0.97 |
Discussion
This study aims to describe the structural network changes in the pre-clinical stage of Alzheimer's continuum and their association with amyloid and tau load. The study showed no significant difference in terms of global efficiency across the groups, but higher assortativity was observed in the A+T+ group when compared with the A+T− and A−T− groups, indicating a more robust network organization and higher brain resilience.
The association between assortativity and tau load, but not amyloid load, suggests that the brain may possess resilience to tau pathology. Additionally, the left hippocampus of A+T+ individuals exhibited lower local efficiency and a lower clustering coefficient. Our results encourage further investigation of structural network properties in pre-clinical AD.
Global network properties
At the global scale, efficiency did not differ significantly across groups of CN individuals with varying levels of AD pathology, while assortativity was higher in the A+T+ group than in the A−T− and A+T− groups. Assortativity describes how nodes in a complex network tend to link with other similar nodes. A positive assortativity coefficient implies that nodes in a network are more likely to connect to nodes with similar degrees, indicating enhanced information processing efficiency and greater resistance to random attacks or deliberate removal of central nodes (Sun et al., 2020). Therefore, higher assortativity in the A+T+ group suggests that their brain networks are more robust and resilient.
Brain resilience refers to an individual's ability to cope with pathology while remaining CN (Arenaza-Urquijo and Vemuri, 2018). Our findings support this notion by showing increased assortativity in groups with greater pathology, while maintaining comparable global efficiency. Thus, the network measure could complement identification of pre-clinical AD.
Assortativity measured from structural connectivity was rarely reported. In a single study by Coninck et al. (2020), increased assortativity was found in subjects with AD. Our results further demonstrate that the increased assortativity can be detected at the pre-clinical stage. In other AD studies, assortativity has usually been measured from functional connectivity using functional MRI (Bahrami and Hossein-Zadeh, 2015; Luo et al., 2021) and electroencephalography (Fathian et al., 2022; Franciotti et al., 2019; Kim et al., 2022). However, comparisons with these studies are not valid because structural connectivity tends to form an assortative network, while functional connectivity tends to form a disassortative network (Lim et al., 2019).
In the present study, there was no significant difference in global efficiency between CN participants with different levels of amyloid and tau pathology. However, previous studies have reported lower global efficiency in individuals with mild cognitive impairment (Bergamino et al., 2022; Jacquemont et al., 2017), subjective cognitive decline (Shu et al., 2018), and pre-clinical AD (Fischer et al., 2015; Pereira et al., 2018) and CN individuals with an autosomal dominant AD mutation (Prescott et al., 2022).
Some studies suggested that amyloid alone may not be sufficient to disrupt the structural network topology during the pre-clinical stage (Pereira et al., 2018). The lack of decreased global efficiency in the A+T+ group in this study suggests that deposition of both amyloid and tau may not be sufficient to alter global efficiency.
Local network properties in the hippocampus
At the nodal scale, the A+T+ group showed a lower clustering coefficient and local efficiency than the A−T− group. The clustering coefficient measures the proportion of neighbors of a node that are connected to each other, and a decrease in this coefficient could indicate reduced connectivity within the local neighborhood or increased connectivity between the node and remote nodes (Jacquemont et al., 2017). In the left hippocampus, the degree, or the number of connected links, remained the same, so the lower clustering coefficient suggests reduced connectivity within the neighborhood.
The decrease resulted specifically for the left hippocampus, which is more susceptible to AD pathogenesis (Shi et al., 2009). The reduction in local efficiency indicates a loss of local integration around the left hippocampus, and this has also been observed in the amnestic MCI converter (Sun et al., 2019). Therefore, changes in the topology of the hippocampus could potentially serve as biomarkers for monitoring the early accumulation of AD pathology substrates.
Limitation
This study has several limitations. First, the size of the cohort is relatively small, leading to less statistical power to distinguish between groups. While multicenter diffusion images were available in ADNI, the reliability of the graph theory metrics could be reduced by including different scanners (Kurokawa et al., 2021). We limited the data to Siemens scanners with 48 diffusion directions to minimize such impact, resulting in the small sample size.
Second, we did not further classify subjects based on the neurodegeneration profile due to the small sample size. Neurodegeneration factors such as decrease of fluorodeoxyglucose and temporal atrophy may correlate with the disrupted structural network more than amyloid in pre-clinical AD (Pereira et al., 2018) and MCI (Jacquemont et al., 2017). We controlled the neurodegeneration effect in the complementary analysis and arrived at the same conclusion. Further studies may explore the complete spectrum of pre-clinical AD, assessing additional neurodegeneration factors.
Third, probabilistic tractography was utilized to construct connectomes, and this method suffers from severe false-positive connections (Sarwar et al., 2019). We applied the stringent thresholding method to maximize the likelihood of identifying true white matter connections (Buchanan et al., 2020).
Conclusions
This study investigated the topological properties of the brain's structural network in the pre-clinical stage of Alzheimer's continuum and their correlation with amyloid and tau burden. We found preserved global efficiency across groups, but increased assortativity correlating with the tau burden. Such increased linkage in higher level architecture of the white matter network supports the notion of increased brain resilience in the early disease state and may be a response to tau pathology.
Our study provides a potential graph-based biomarker of brain connectivity for early identification of pre-clinical AD and encourages further investigation of changes in the topological properties of the structural brain network in the early stage of AD.
Supplementary Material
Acknowledgments
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data, but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Data collection and sharing for this project were funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012).
ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; Euroimmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer, Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics.
The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory of NeuroImaging at the University of Southern California.
Contributor Information
Collaborators: the Alzheimer's Disease Neuroimaging Initiative
Authors' Contributions
Q.C. was involved in conceptualization, data curation, methodology, software, investigation, formal analysis, and writing—original draft. J.A. was involved in conceptualization, investigation, supervision, and writing—review and editing. M.D. was involved in writing—review and editing. L.S. and Y.-X.W. were involved in conceptualization and writing—review and editing. W.C. was involved in conceptualization, supervision, project administration, and writing—review and editing.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported by a direct grant from the Chinese University of Hong Kong [Project Code: 2020.016(4054595)].
Supplementary Material
References
- Arenaza-Urquijo EM, Vemuri P. Resistance vs resilience to Alzheimer disease. Neurology 2018;90(15):695–703; doi: 10.1212/WNL.0000000000005303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami M, Hossein-Zadeh G-A. Assortativity Changes in Alzheimer's Diesease: A Resting-State FMRI Study. In: 2015 23rd Iranian Conference on Electrical Engineering. IEEE: New York, NY; 2015; pp. 141–144. [Google Scholar]
- Bergamino M, Schiavi S, Daducci A, et al. Analysis of brain structural connectivity networks and white matter integrity in patients with mild cognitive impairment. Front Aging Neurosci 2022;14:793991; doi: 10.3389/fnagi.2022.793991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan CR, Bastin ME, Ritchie SJ, et al. The effect of network thresholding and weighting on structural brain networks in the UK Biobank. Neuroimage 2020;211:116443; doi: 10.1016/j.neuroimage.2019.116443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore ET, Bassett DS. Brain graphs: Graphical models of the human brain connectome. Annu Rev Clin Psychol 2011;7(1):113–140; doi: 10.1146/annurev-clinpsy-040510-143934 [DOI] [PubMed] [Google Scholar]
- Coninck JCP, Ferrari FAS, Reis AS, et al. Network properties of healthy and Alzheimer brains. Phys A Stat Mech Appl 2020;547:124475; doi: 10.1016/j.physa.2020.124475 [DOI] [Google Scholar]
- De Brito Robalo BM, Vlegels N, Leemans A, et al. Impact of thresholding on the consistency and sensitivity of diffusion MRI-based brain networks in patients with cerebral small vessel disease. Brain Behav 2022;12(5):e2523; doi: 10.1002/brb3.2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbeuck X, Van der Linden M, Collette F. Alzheimer's disease as a disconnection syndrome? Neuropsychol Rev 2003;13(2):79–92; doi: 10.1023/a:1023832305702 [DOI] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006;31(3):968–980; doi: 10.1016/j.neuroimage.2006.01.021 [DOI] [PubMed] [Google Scholar]
- Fathian A, Jamali Y, Raoufy MR, et al. The trend of disruption in the functional brain network topology of Alzheimer's disease. Sci Rep 2022;12(1):14998; doi: 10.1038/s41598-022-18987-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer FU, Wolf D, Scheurich A, et al. Altered whole-brain white matter networks in preclinical Alzheimer's disease. Neuroimage Clin 2015;8:660–666; doi: 10.1016/j.nicl.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 2002;33(3):341–355; doi: 10.1016/S0896-6273(02)00569-X [DOI] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Bullmore E.. Fundamentals of Brain Network Analysis. Academic Press: Cambridge, MA; 2016. [Google Scholar]
- Franciotti R, Falasca NW, Arnaldi D, et al. Cortical network topology in prodromal and mild dementia due to Alzheimer's disease: Graph theory applied to resting state EEG. Brain Topogr 2019;32(1):127–141; doi: 10.1007/s10548-018-0674-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Bennett DA, Blennow K, et al. NIA-AA research framework: Toward a biological definition of Alzheimer's disease. Alzheimers Dement 2018;14(4):535–562; doi: 10.1016/j.jalz.2018.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Wiste HJ, Weigand SD, et al. Defining imaging biomarker cut points for brain aging and Alzheimer's disease. Alzheimers Dement 2017;13(3):205–216; doi: 10.1016/j.jalz.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont T, De Vico Fallani F, Bertrand A, et al. Amyloidosis and neurodegeneration result in distinct structural connectivity patterns in mild cognitive impairment. Neurobiol Aging 2017;55:177–189; doi: 10.1016/j.neurobiolaging.2017.03.023 [DOI] [PubMed] [Google Scholar]
- Kim J-G, Kim H, Hwang J, et al. Differentiating amnestic from non-amnestic mild cognitive impairment subtypes using graph theoretical measures of electroencephalography. Sci Rep 2022;12(1):6219; doi: 10.1038/s41598-022-10322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Robson J, Wilson H, Politis M, et al. Associations between amyloid and tau pathology, and connectome alterations, in Alzheimer's disease and mild cognitive impairment. J Alzheimers Dis 2021;82(2):541–560; doi: 10.3233/JAD-201457 [DOI] [PubMed] [Google Scholar]
- Kurokawa R, Kamiya K, Koike S, et al. Cross-scanner reproducibility and harmonization of a diffusion MRI structural brain network: A traveling subject study of multi-B acquisition. Neuroimage 2021;245:118675; doi: 10.1016/j.neuroimage.2021.118675 [DOI] [PubMed] [Google Scholar]
- Landau SM, Lu M, Joshi AD, et al. Comparing positron emission tomography imaging and cerebrospinal fluid measurements of β-amyloid. Ann Neurol 2013;74(6):826–836; doi: 10.1002/ana.23908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans A, Jones DK. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn Reson Med 2009;61(6):1336–1349; doi: 10.1002/mrm.21890 [DOI] [PubMed] [Google Scholar]
- Lim S, Radicchi F, van den Heuvel MP, et al. Discordant attributes of structural and functional brain connectivity in a two-layer multiplex network. Sci Rep 2019;9(1):2885; doi: 10.1038/s41598-019-39243-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C-Y, Wang P-N, Chou K-H, et al. Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer's disease. J Neurosci 2010;30(50):16876–16885; doi: 10.1523/jneurosci.4136-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Sun T, Ma C, et al. Alterations of brain networks in Alzheimer's disease and mild cognitive impairment: A resting state FMRI study based on a population-specific brain template. Neuroscience 2021;452:192–207; doi: 10.1016/j.neuroscience.2020.10.023 [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association Workgroups on Diagnostic Guidelines for Alzheimer's Disease. Alzheimers Dement 2011;7(3):263–269; doi: 10.1016/j.jalz.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miraglia F, Vecchio F, Pappalettera C, et al. Brain connectivity and graph theory analysis in Alzheimer's and Parkinson's disease: The contribution of electrophysiological techniques. Brain Sci 2022;12(3):402; doi: 10.3390/brainsci12030402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JB, van Westen D, Stomrud E, et al. Abnormal structural brain connectome in individuals with preclinical Alzheimer's disease. Cereb Cortex 2018;28(10):3638–3649; doi: 10.1093/cercor/bhx236 [DOI] [PubMed] [Google Scholar]
- Pini L, Pievani M, Bocchetta M, et al. Brain atrophy in Alzheimer's disease and aging. Ageing Res Rev 2016;30:25–48; doi: 10.1016/j.arr.2016.01.002 [DOI] [PubMed] [Google Scholar]
- Prescott JW, Doraiswamy PM, Gamberger D, et al. Diffusion tensor MRI structural connectivity and PET amyloid burden in preclinical autosomal dominant Alzheimer disease: The DIAN cohort. Radiology 2022;302(1):143–150; doi: 10.1148/radiol.2021210383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginold W, Sam K, Poublanc J, et al. Impact of white matter hyperintensities on surrounding white matter tracts. Neuroradiology 2018;60(9):933–944; doi: 10.1007/s00234-018-2053-x [DOI] [PubMed] [Google Scholar]
- Roine T, Jeurissen B, Perrone D, et al. Reproducibility and intercorrelation of graph theoretical measures in structural brain connectivity networks. Med Image Anal 2019;52:56–67; doi: 10.1016/j.media.2018.10.009 [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 2010;52(3):1059–1069; doi: 10.1016/j.neuroimage.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Sarwar T, Ramamohanarao K, Zalesky A. Mapping connectomes with diffusion MRI: Deterministic or probabilistic tractography? Magn Reson Med 2019;81(2):1368–1384; doi: 10.1002/mrm.27471 [DOI] [PubMed] [Google Scholar]
- Scheltens P, De Strooper B, Kivipelto M, et al. Alzheimer's disease. Lancet 2021;397(10284):1577–1590; doi: 10.1016/S0140-6736(20)32205-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Liu B, Zhou Y, et al. Hippocampal volume and asymmetry in mild cognitive impairment and Alzheimer's disease: Meta-analyses of MRI studies. Hippocampus 2009;19(11):1055–1064; doi: 10.1002/hipo.20573 [DOI] [PubMed] [Google Scholar]
- Shu N, Wang X, Bi Q, et al. Disrupted topologic efficiency of white matter structural connectome in individuals with subjective cognitive decline. Radiology 2018;286(1):229–238; doi: 10.1148/radiol.2017162696 [DOI] [PubMed] [Google Scholar]
- Smith RE, Tournier J-D, Calamante F, et al. Anatomically-constrained tractography: improved diffusion MRI streamlines tractography through effective use of anatomical information. Neuroimage 2012;62(3):1924–1938; doi: 10.1016/j.neuroimage.2012.06.005 [DOI] [PubMed] [Google Scholar]
- Smith RE, Tournier J-D, Calamante F, et al. SIFT2: Enabling dense quantitative assessment of brain white matter connectivity using streamlines tractography. Neuroimage 2015;119:338–351; doi: 10.1016/j.neuroimage.2015.06.092 [DOI] [PubMed] [Google Scholar]
- Sun M, Xie H, Tang Y. Directed network defects in Alzheimer's disease using Granger causality and graph theory. Curr Alzheimer Res 2020;17(10):939–947; doi: 10.2174/1567205017666201215140625 [DOI] [PubMed] [Google Scholar]
- Sun Y, Bi Q, Wang X, et al. Prediction of conversion from amnestic mild cognitive impairment to Alzheimer's disease based on the brain structural connectome. Front Neurol 2019;9; doi: 10.3389/fneur.2018.01178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier JD, Smith R, Raffelt D, et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage 2019;202:116137; doi: 10.1016/j.neuroimage.2019.116137 [DOI] [PubMed] [Google Scholar]
- Vogt NM, Hunt JFV, Adluru N, et al. Interaction of amyloid and tau on cortical microstructure in cognitively unimpaired adults. Alzheimers Dement 2022;18(1):65–76; doi: 10.1002/alz.12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.