Abstract
Based upon our earlier studies (A. Tapias, A. R. Fernández de Henestrosa, and J. Barbé, J. Bacteriol. 179:1573–1579, 1997) we hypothesized that the regulatory sequence of the Rhizobium etli recA gene was TTGN11CAA. However, further detailed analysis of the R. etli recA operator described in the present work suggests that it may in fact be GAACN7GTAC. This new conclusion is based upon PCR mutagenesis analysis carried out in the R. etli recA operator, which indicates that the GAAC and GTAC submotifs found in the sequence GAACN7GTAC are required for the maximal stimulation of in vivo transcription and in vitro DNA-protein complex formation. This DNA-protein complex is also detected when the GAACN7GTAC wild-type sequence is modified to obtain GAACN7GAAC, GTACN7GTAC, or GAACN7GTTC. The wild-type promoters of the Rhizobium meliloti and Agrobacterium tumefaciens recA genes, which also contain the GAACN7GTAC sequence, compete with the R. etli recA promoter for the DNA-protein complex formation but not with mutant derivatives in any of these motifs, indicating that the R. etli, R. meliloti, and A. tumefaciens recA genes present the same regulatory sequence.
The recA gene has been isolated from a large number of bacteria (19). Its product is a multifunctional protein which is best characterized by its role in DNA recombination (12). RecA does, however, also play a central role in regulating the SOS response to DNA damage. Many proteins induced as part of this response help the cell survive, by either directly repairing damaged DNA or allowing the cell to tolerate DNA lesions until they can be repaired efficiently (29). LexA protein is the common transcriptional repressor of SOS-regulated genes, which include both recA and lexA themselves (6). Blockage to replication or damage to DNA generates an inducing signal which, with the help of Ssb protein, results in the activation of RecA coprotease functions (13, 14, 25). In its coprotease-proficient state, RecA mediates the efficient posttranslational cleavage and inactivation of the LexA repressor. Inducible DNA repair systems are apparently quite common and have been described in several bacteria (19).
The Escherichia coli LexA protein binds to a specific region situated upstream of the SOS genes. Alignment of various SOS-regulated genes generated the consensus sequence CTG(TA)5CAG, known as the E. coli SOS box (31). Similar E. coli-like SOS boxes have been identified in the promoter regions of many recA and lexA genes from gram-negative bacteria (7, 19). Likewise, and by comparing promoter regions of several din (damage inducible) genes, the GAACN7GTTC palindrome has been proposed as the Bacillus subtilis SOS box (3). Recent data indicate that, in fact, the B. subtilis SOS box is slightly larger and has a consensus sequence of CGAACRNRYGTTYC (33). Many gram-positive bacterial species, such as Mycobacterium tuberculosis, Mycobacterium leprae, Streptococcus pneumoniae, and Clostridium perfringens, among others, have a B. subtilis-like SOS box upstream of their recA gene (4, 9, 20, 21). B. subtilis and M. tuberculosis LexA-like proteins have been purified, and binding to their respective SOS boxes has been previously demonstrated (18, 20, 32). Other bacterial SOS boxes, such as that belonging to Rhizobium etli, remain to be elucidated.
We have previously reported that the R. etli recA gene is DNA damage inducible (5) and have demonstrated that the TTG and CAA motifs found in the TTGCGAGAGTGGAACAA (TTGN11CAA) sequence, upstream of the R. etli recA gene, are necessary for DNA damage-mediated induction (28). Interestingly, our data indicated that the TTG motif seemed to be less important than CAA. So, the effect of the substitution of the TTG motif, in both formation of a DNA-protein complex in vitro and recA DNA damage mediated in vivo, was weaker, which was a consequence of changing the CAA motif (28). By comparison, there was only a slight decrease in these two parameters when the TTG motif was changed. These findings therefore led us to systematically determine the role of each nucleotide base in the TTGN11CAA sequence, which appears to control R. etli recA gene expression.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The CNPAF512 wild-type R. etli strain employed in this study has been described previously (28). The wild-type strains of Rhizobium meliloti and Agrobacterium tumefaciens were 2021 and C58, respectively (28). All plasmid constructions and cloning experiments were performed in E. coli DH5α. R. etli, R. meliloti, and A. tumefaciens strains were grown at 30°C in PY, LB, and MG media (22), respectively, and E. coli cells were cultured at 37°C in LB (17). Antibiotics were added to the culture media at the appropriate concentration for each bacterial species (22).
General genetic techniques.
Plasmid DNA was transformed into competent E. coli cells as described by Silhavy et al. (27). Bacterial matings were performed with the E. coli S17 system as reported previously (22). Construction of recA-lacZ fusions and β-galactosidase assays were carried out as previously published (28).
DNA manipulations and sequencing.
Restriction enzymes, T4 DNA ligase, T4 DNA polymerase, and a DIG DNA labelling and detection kit were purchased from Boehringer Mannheim. The conditions used for plasmid DNA extractions, restriction endonuclease digestion, agarose gel electrophoresis, and isolation and ligation of DNA fragments have been described elsewhere (23). The DNA sequence of the PCR-mutagenized fragments was determined by dideoxy sequencing (24) on an ALF Sequencer (Pharmacia Biotech). In all cases, the entire nucleotide sequence was determined on both DNA strands. The presence of the desired mutation in the various recA-lacZ fusions was determined by DNA sequencing of each fusion with a 5′-fluorescein-CGAACGGCCAGTGAATCCG-3′ primer, which extends from nucleotides +32 to +14 with respect to the translational starting point of the E. coli lacZ gene.
DNA probes.
The R. etli recA probes employed in retardation experiments were obtained by PCR mutagenesis using specific upper primers which contained the desired mutation (Fig. 1). All of these fragments were labelled with a 5′-end digoxigenin-labelled lower primer (Fig. 1). The introduction of the desired mutation was confirmed by DNA sequence analysis. The 5′-end digoxigenin-labelled primer used to obtain the upper fragment (see Fig. 8) was 5′-CCGCCTTCCGATGCCGAAGC-3′. The lower primers used were 5′-GCCCGTGTTCTATATTTG-3′ for the wild-type fragment and 5′-GCCCGTGTTCTATATTCGGATATCGA-3′ and 5′-GCCCGTCGGATATATTTG-3′ for the mutagenized derivatives.
FIG. 1.
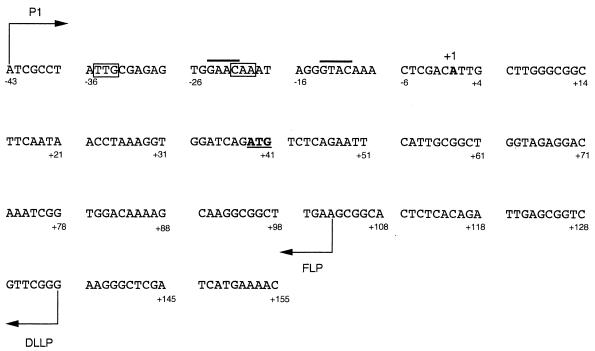
Nucleotide sequence of the 5′ end of the R. etli recA gene. P1 indicates the starting point of the upper primers used to obtain the recA fragments employed in this work. The transcriptional starting point (+1) is indicated in boldface type. The two halves of the TTGN11CAA palindrome which had been previously proposed as the control region of the R. etli recA gene are boxed. FLP and DLLP indicate the starting points of the lower primers used either to construct recA fusions or to obtain fragments employed in gel retardation experiments, respectively. The right and left submotifs of the GAACN7GTAC sequence proposed in this work as the regulatory element of the R. etli recA gene are overlined.
FIG. 8.
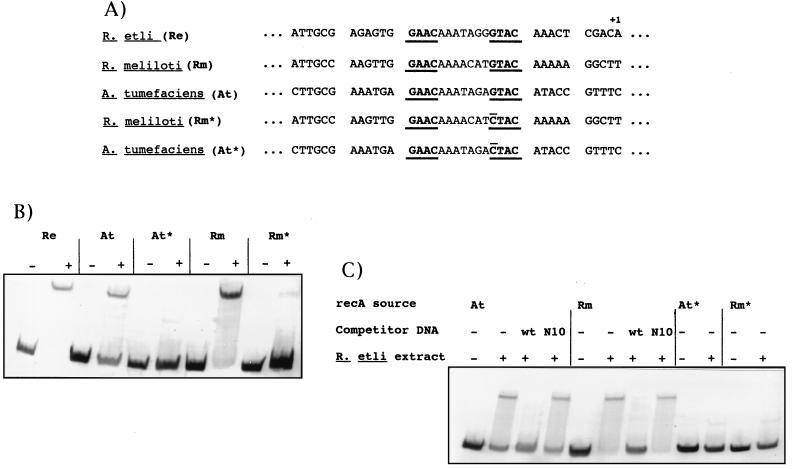
(A) Sequence of the upstream region of the R. etli, R. meliloti, and A. tumefaciens recA genes in which the GAACN7GTAC motif (which is underlined) is present. The sequences of mutant derivatives of both R. meliloti and A. tumefaciens recA genes obtained after PCR mutagenesis (labelled with an asterisk) are also shown. The mutational change introduced is overlined. As a landmark, the +1 position of the R. etli recA gene is indicated. (B) Gel mobility of both wild-type and mutant derivatives of the R. etli, R. meliloti, and A. tumefaciens recA promoters in the absence (−) or in the presence (+) of their respective crude cell extracts. (C) Electrophoretic mobility of both the wild-type and mutant derivative forms of R. meliloti and A. tumefaciens recA promoters in the presence of R. etli crude cell extract. The effect of the addition of either the wild type (wt) or a mutant form (N10) of the R. etli recA promoter on the gel mobility of the recA promoters of R. meliloti and A. tumefaciens is also shown. Strain abbreviations used in panels B and C are defined in panel A.
Rmup (5′-GTTGGAACAAAACATGTAC-3′) and Atup (5′-ATGAGAACAAATAGAGTACATACC-3′) oligonucleotides were used as upper primers to obtain the R. meliloti and A. tumefaciens recA probes, respectively. The lower primers were the 5′-end digoxigenin-labelled oligonucleotides Rmlow (5′-CCGAACGAACGTTCGATCT-3′) and Atlow (5′-CCGAACGACCGTTCGATCT). Mutant R. meliloti and A. tumefaciens recA probes were obtained by using Rmup* (5′-GTTGGAACAAAACATCTACAAA-3′) and Atup* (5′-ATGAGAACAAATAGACTACATACC-3′) oligonucleotides, respectively. The positions of the mutant base in these oligonucleotides are underlined. In agreement with the R. meliloti and A. tumefaciens recA gene sequences (26, 29), the 5′ ends of the Rmup and Atup primers are 116 and 117 bases upstream of their respective translational starting point, and the 5′ ends of the Rmlow and Atlow primers are 56 bases downstream from this point.
When necessary, specific restriction sites were incorporated into the 5′ ends of the oligonucleotide primers to facilitate the subcloning of PCR fragments and the construction of lacZ fusions.
Electrophoretic mobility shift assays.
Crude cell extracts of members of the family Rhizobiaceae were obtained by sonication as previously reported (28). In all cases, the total protein concentration of these crude extracts was about 15 mg/ml. Binding mixtures were obtained by mixing the digoxigenin probe (10 ng) and crude cell extracts (180 μg of total protein) for 30 min at 30°C. Immediately thereafter, the samples were loaded on a native 5% polyacrylamide gel as previously described (28).
Samples were electrophoresed at 35 mA until the marker dyes had migrated approximately two-thirds of the total gel length (so as to avoid excessive protein-DNA dissociation). Complexes were transferred and detected as recommended by the supplier (Boehringer Mannheim).
Densitometrical analysis of the several retardation experiments was done by using the Molecular Analyst 1.5 (Bio-Rad) program. The reported binding activity represents the percentage of the total DNA applied to the gel that was bound. For every experiment, wild-type DNA was included in the set as an internal control. Under our assay conditions the binding activity of the wild-type probe was approximately 72% ± 7% (mean ± standard deviation) of total DNA. The wild-type binding activity was taken as 100%, and the mutant binding activity was expressed as a percentage of the wild-type activity. The presented data are the averages of three independent experiments.
RESULTS AND DISCUSSION
Effects of nucleotide substitution and addition in the TTGN11CAA sequence on electrophoretic mobility and inducibility of the R. etli recA promoter.
In order to further analyze the role of the TTG motif belonging to the TTGN11GAAC sequence cited above, several gel retardation experiments with R. etli recA promoter mutants containing single nucleotide changes in this TTG motif were performed. Figure 2 shows that none of these substitutions abolished the formation of the DNA-protein complex, although some changes in the TTG motif decreased its binding ability (Table 1).
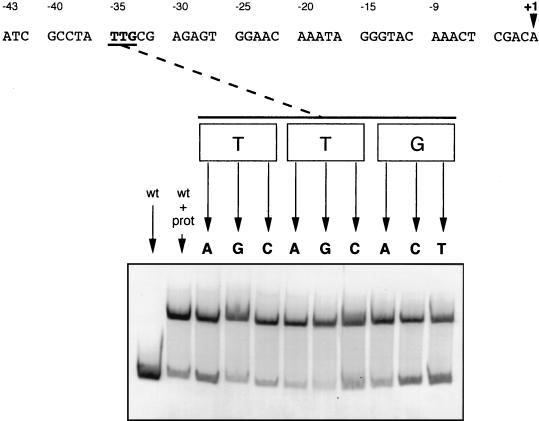
FIG. 2.
Effect of single nucleotide substitutions in the TTG motif of the TTGN11CAA palindrome upstream of the R. etli recA gene. The migration of the wild-type (wt) fragment either in the absence or in the presence of R. etli crude extract (prot) is shown as a control. The mobility of the mutant fragments is only shown in the presence of cellular extract.
TABLE 1.
Effects of systematic single substitutions in the TTGN11CAA sequence on the formation of retarded complexes of the R. etli recA promotera
| Operator sequence | Nucleotide substitution | Binding activity (%) |
|---|---|---|
| Wild type | ||
| TTGCGAGAGTGGAACAA | 100 | |
| Mutated | ||
| TTGCGAGAGTGGAACAA | GTGCGAGAGTGGAACAA | 100 |
| ATGCGAGAGTGGAACAA | 88 | |
| CTGCGAGAGTGGAACAA | 100 | |
| TTGCGAGAGTGGAACAA | TGGCGAGAGTGGAACAA | 100 |
| TCGCGAGAGTGGAACAA | 96 | |
| TAGCGAGAGTGGAACAA | 100 | |
| TTGCGAGAGTGGAACAA | TTACGAGAGTGGAACAA | 100 |
| TTTCGAGAGTGGAACAA | 74 | |
| TTCCGAGAGTGGAACAA | 65 | |
| TTGCGAGAGTGGAACAA | TTGGGAGAGTGGAACAA | 94 |
| TTGAGAGAGTGGAACAA | 44 | |
| TTGTGAGAGTGGAACAA | 100 | |
| TTGCGAGAGTGGAACAA | TTGCGGGAGTGGAACAA | 52 |
| TTGCGTGAGTGGAACAA | 39 | |
| TTGCGCGAGTGGAACAA | 58 | |
| TTGCGAGAGTGGAACAA | TTGCGAGAGTAGAACAA | 86 |
| TTGCGAGAGTTGAACAA | 40 | |
| TTGCGAGAGTCGAACAA | 70 | |
| TTGCGAGAGTGGAACAA | TTGCGAGAGTGAAACAA | 70 |
| TTGCGAGAGTGTAACAA | —b | |
| TTGCGAGAGTGCAACAA | 10 | |
| TTGCGAGAGTGGAACAA | TTGCGAGAGTGGGACAA | 56 |
| TTGCGAGAGTGGTACAA | 34 | |
| TTGCGAGAGTGGCACAA | 17 | |
| TTGCGAGAGTGGAACAA | TTGCGAGAGTGGAGCAA | — |
| TTGCGAGAGTGGATCAA | 8 | |
| TTGCGAGAGTGGACCAA | 5 | |
| TTGCGAGAGTGGAACAA | TTGCGAGAGTGGAAGAA | 6 |
| TTGCGAGAGTGGAAAAA | — | |
| TTGCGAGAGTGGAATAA | — | |
| TTGCGAGAGTGGAACAA | TTGCGAGAGTGGAACGA | 100 |
| TTGCGAGAGTGGAACTA | 41 | |
| TTGCGAGAGTGGAACCA | 27 | |
| TTGCGAGAGTGGAACAA | TTGCGAGAGTGGAACAG | 82 |
| TTGCGAGAGTGGAACAT | 89 | |
| TTGCGAGAGTGGAACAC | 53 |
Positions affected by substitutions are underlined.
—, no band shift.
In contrast, a more-exhaustive mutational analysis of the TTGN11CAA sequence revealed that only nucleotide substitutions in positions −24, −23, −22, and −21 (corresponding to a GAAC motif) abolished or significantly decreased the recA promoter gel mobility (Table 1). Among these changes, those involving positions −22 and −21 appear to be the least permissive (Table 1). It is worth noting that the C of the GAAC tetranucleotide also belongs to the CAA motif of the TTGN11CAA sequence, which had been previously hypothesized as the binding site of the regulatory protein. These data convincingly show that the TTG motif is not important for binding.
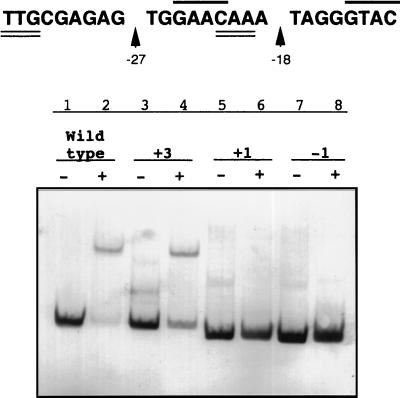
Moreover, results obtained strongly implicate the GAAC sequence. This inference is further supported by deletion and insertion analysis (Fig. 3). Thus, a three-base insertion at position −27 did not cause any loss in protein binding (Fig. 3). Nevertheless, either insertion or deletion of 1 nucleotide at position −18 (downstream from the GAAC submotif) is sufficient to suppress recA promoter retardation (Fig. 3). In addition, substitutions at position −24, −23, −22, or −21 also eliminate recA inducibility and produce a dramatic increase in the basal level of expression of the gene (Fig. 4).
FIG. 3.
Effect of changing the spacing between the palindromic submotifs found in TTGN11CAA (lanes 1 to 4) and the imperfect repeat present in GAACN7GTAC (lanes 5 to 8) upon the formation of retarded complexes of the R. etli recA promoter. Mutational analysis of the TTGN11CAA sequence was carried out by insertion of three bases (CCC) in its central region (position −27), whereas the GAACN7GTAC sequence was mutagenized by either the addition or deletion of one base in its central region (position −18). The mobility of the wild-type (lanes 1 and 2) and the mutagenized (lanes 3 to 8) fragments in the presence (lanes 2, 4, 6, and 8) or in the absence (lanes 1, 3, 5, and 7) of R. etli crude cell extract is shown. Fragments containing either the insertion or the deletion of 1 nucleotide (lanes 5 to 8) are slightly smaller than both the wild-type fragment (lanes 1 and 2) and the one carrying the insertion of 3 nucleotides (lanes 3 and 4) because of the presence of a SmaI restriction site in the 5′ end of the primers used to obtain these two fragments (see Materials and Methods).
FIG. 4.
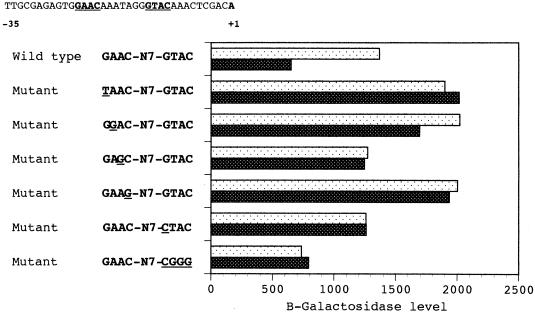
Effects of mutations in the GAAC and GTAC submotifs of the GAACN7GTAC sequence upon the transcription of the R. etli recA gene, as measured by β-galactosidase activity in recA-lacZ fusions. β-Galactosidase activities of all fusions were measured in the absence of mitomycin C (open bars with black dots) or 180 min after its addition at 1.6 μg/ml (solid bars with white dots). The activities (in units) are the mean data from three independent assays. Values were reproducible to within an error of ±10%.
Identification of R. etli recA regulatory sequence.
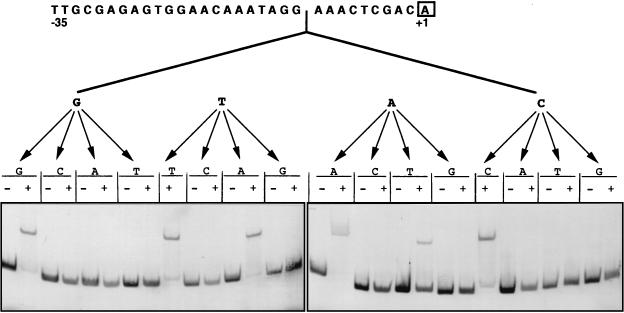
The data presented above raised the possibility that at least one regulatory element of the recA gene is located downstream from the GAAC motif. Inspection of the surrounding region revealed the presence of a GTAC sequence (positions −13 to −10 in Fig. 1) seven bases downstream from the GAAC tetranucleotide described above (Table 1 and Fig. 4). These two sequences (GAAC and GTAC) are potentially variants of a symmetrical GAACN7GTTC palindrome. To test this hypothesis, the role of the GTAC submotif was further characterized (Fig. 5). Our findings indicate that replacing G and C with any base eliminates recA promoter mobility. Moreover, promoter mobility is lost whether T and A are replaced by either G or C (Fig. 5). The retardation band is also detected when the T and A of this GATC motif are changed to A and T, respectively (Fig. 5). These results suggest that the recA promoter can also be shifted when the second submotif is either GAAC or GTTC, although GTTC appeared to bind less well.
FIG. 5.
Effects of systematic variation on the submotif GTAC found in the GAACN7GTAC sequence upon the electrophoretic mobility of the R. etli recA promoter. The symbols + and − refer to the presence or absence of R. etli crude protein extract, respectively.
Substitution within the GTAC submotif resulted in a loss of recA promoter inducibility in vivo, although the basal level in this case was practically the same as that exhibited by the wild type (Fig. 4). This reduction in derepressed level may be attributed to the fact that the GTAC tetranucleotide overlaps the predicted −10 region of the recA promoter (28). Hence, an increase in expression due to the absence of negative regulation would be neutralized by a decrease in the efficiency of transcription. Similar behavior has been described in other inducible genes (2). However, the inducibility of the recA gene is lost and the basal level is increased to values obtained in DNA damage-induced wild-type cells only when the G-to-C change is introduced into the GTAC motif (Fig. 4). This fact clearly demonstrates that the inducibility of the R. etli recA gene is dependent upon the GTAC motif.
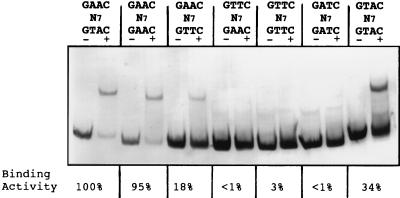
All together, these findings prompted us to create all the possible variants of this sequence in which the two internal bases were A or T and the combination had either a direct or inverted repeat of the two half sites. The recA promoter fragment containing either GTTCN7GTTC, GTTCN7GAAC or GATCN7GATC did not bind any protein (see Fig. 7). In contrast, however, converting the sequence to GAACN7GTTC and GTACN7GTAC reduced the extent of gel retardation (Fig. 6). It is worth noting that the GAACN7GAAC sequence bound nearly as well as the wild type. The reason why band retardation is found with the GAACN7GAAC sequence but not with the GTTCN7GTTC sequence, despite the fact that the former would have GTTCN7GTTC on the bottom strand, is so far unknown. However, and such as happens in the B. subtilis SOS box (33), some internal bases of the direct repeat are perhaps involved in DNA-protein binding. Interestingly, the GAACN7GTTC sequence presents the same two palindromic submotifs as the B. subtilis SOS box (3), although in the latter case there are only 4 nucleotides between these two submotifs. Our results indicate, nevertheless, that addition or deletion of just 1 nucleotide between the two submotifs abolishes the DNA gel mobility shift (Fig. 3). To our knowledge, the B. subtilis SOS box has been mutagenized in vitro at several internal and surrounding positions. Some of these changes eliminated LexA binding, whereas others reduced binding (32, 33). Whether the spacing between the two half sites can be altered is, however, still unknown.
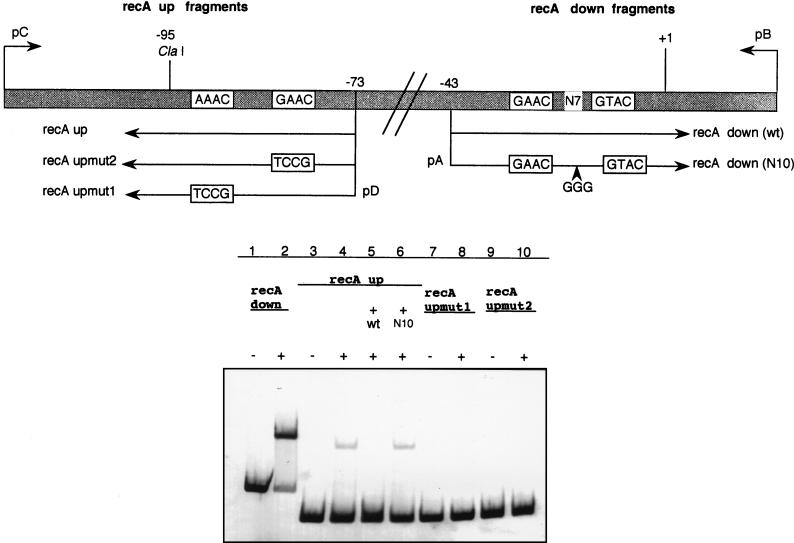
FIG. 7.
(A) Diagram showing the upstream region of the GAACN7GTAC sequence present in the R. etli recA promoter. The starting points of the upper (pC and pD) and lower (pA and pB) primers used to obtain the recA up and recA down fragments are indicated. Note that this figure is not drawn to scale. wt, wild type. (B) Electrophoretic mobilities of the recA down, recA up, recA upmut1, and recA upmut2 fragments in the absence (lanes 1, 3, 7, and 9) or in the presence (lanes 2, 4, 8, and 10) of R. etli crude extract. The effect of the addition of either a 10-fold excess of the unlabelled wild-type recA down fragment (lane 5) or 100-fold excess of the unlabelled recA down (N10) mutant fragment (lane 6) on the migration of the wild-type recA up fragment is also shown.
FIG. 6.
Mobility shift of the R. etli recA promoter carrying direct or inverted repeat variants of the paired half sites of the GAACN7GTAC motif. The resultant combinations of 5′ and 3′ submotifs obtained after PCR mutagenesis are indicated above each lane. The symbols + and − refer to the presence or absence of R. etli crude protein extract, respectively. The mean binding activity of each fragment in three independent experiments is also shown.
The R. etli recA operator contains two potential repressor binding sites.
The mutational experiments described above demonstrate that several derivatives of the GAACN7GTAC sequence are specifically recognized by the protein which regulates the R. etli recA gene. One of these derivatives coincides with the AAACN7GAAC sequence, which is centered at −85 bp in the R. etli recA promoter. Therefore, the protein-binding ability surrounding this upstream region was analyzed. Figure 7 indicates that a DNA-protein complex, presenting a binding activity of 8%, is indeed formed when this fragment is used. This complex is specific and apparently results from the binding of the same protein which recognizes the GAACN7GTAC sequence found in the downstream sequence, as binding disappears when a DNA fragment containing this sequence is used as a competitor, but not when GAACN10GTAC is used (Fig. 7). Furthermore, replacement of either the AAAC or GAAC submotif of the AAACN7GAAC motif with a TCCG tetranucleotide eliminates DNA-protein complex formation (Fig. 7). The in vivo functional significance of this second binding site is unknown. Although binding occurs at this sequence, DNA damage-mediated induction still occurs from recA promoter fragments lacking the AAACN7GAAC sequence (Fig. 4). Our data suggests that the protein controlling recA gene expression (which in all likelihood is a homolog of the LexA-like proteins) is able to bind to GAACN7GTAC and some variants found in the R. etli chromosome.
The GAACN7GTAC motif is present and functional in R. meliloti and A. tumefaciens recA genes.
The results obtained in our previous studies suggest that R. etli, R. meliloti, and A. tumefaciens possess a common LexA-like repressor, since their recA genes are DNA damage inducible when introduced into any of these three bacterial species (22). As a consequence, we have hypothesized that the promoter of the recA gene of these microorganisms should contain the same regulatory sequence. Indeed, Fig. 8A reveals that the GAACN7GTAC motif is found upstream of the R. meliloti and A. tumefaciens recA genes. To test for the presence of specific binding to these sites, each recA gene fragment was incubated with a crude cell lysate made from the same strain. A stable DNA-protein complex was observed with fragments containing either the R. meliloti or A. tumefaciens recA genes when they were incubated with their respective crude cell lysates (Fig. 8B). In contrast, mutagenesis of these GAACN7GTAC motifs abolished the DNA-protein complex formation in both recA promoters (Fig. 8B). We were interested in determining whether these complexes were due to the binding of a protein homologous to that which gave rise to the band retardation in the R. etli extract and have analyzed the effect of the crude cell extracts on the mobility of R. meliloti and A. tumefaciens recA promoters. Figure 8C indicates that the mobility of the wild-type R. meliloti and A. tumefaciens recA promoters is shifted in the presence of the R. etli extract. Moreover, the retarded bands disappear when a wild-type R. etli recA promoter is used as a competitor but not when an unlabelled R. etli recA promoter carrying an insertion of three bases between the GAAC and GTAC motifs is used (Fig. 8C). Furthermore, mutant forms of the R. meliloti and A. tumefaciens GAACN7GTAC sequence did not show any change in electrophoretic mobility in the presence of the R. etli extract (Fig. 8C). Together, these data indicate that the recA genes of R. etli, R. meliloti, and A. tumefaciens possess the same damage-inducible regulatory sequence. Isolation and characterization of other DNA damage-inducible genes of these organisms is, however, required to determine whether the GAACN7GTAC sequence is the SOS box of the Rhizobiaceae family or whether it is only involved in the regulation of their recA genes.
ACKNOWLEDGMENTS
This work was funded by grants PB94-0687 and PB97-0194 of the Dirección General de Investigación Científica y Técnica of Spain (DGICYT) and partially supported by the Comissionat per Universitats i Recerca de la Generalitat de Catalunya. Angels Tapias was a recipient of a predoctoral fellowship from the Ministerio de Educación y Ciencia. We gratefully acknowledge the help of the Direcció General d’Universitats de la Generalitat de Catalunya for different grants for the purchase of equipment.
We are deeply indebted to Roger Woodgate for his comments on the manuscript and general helpful suggestions. We are grateful to Maria Aguilar for editorial assistance.
REFERENCES
- 1.Carra J H, Schleif R F. Variation of half-site organization and DNA looping by AraC protein. EMBO J. 1993;12:35–44. doi: 10.1002/j.1460-2075.1993.tb05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang C H, Winans S C. Resection and mutagenesis of the Agrobacterium tumefaciens virG gene. J Bacteriol. 1996;178:4717–4720. doi: 10.1128/jb.178.15.4717-4720.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheo D L, Bayles K W, Yasbin R E. Cloning and characterization of DNA damage-inducible promoter regions from Bacillus subtilis. J Bacteriol. 1991;173:1696–1703. doi: 10.1128/jb.173.5.1696-1703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis E O, Sedgwick S G, Colston M J. Novel structure of the recA locus of Mycobacterium tuberculosis implies processing of the gene product. J Bacteriol. 1991;173:5653–5662. doi: 10.1128/jb.173.18.5653-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández de Henestrosa A R, Barbé J. Autoregulation and kinetics of induction of the Rhizobium phaseoli recA gene. Mutat Res. 1994;308:99–107. doi: 10.1016/0027-5107(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 6.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: American Society for Microbiology Press; 1995. [Google Scholar]
- 7.Garriga X, Calero S, Barbé J. Nucleotide sequence analysis and comparison of the lexA genes from Salmonella typhimurium, Erwinia carotovora, Pseudomonas aeruginosa and Pseudomonas putida. Mol Gen Genet. 1992;236:125–134. doi: 10.1007/BF00279651. [DOI] [PubMed] [Google Scholar]
- 8.Ho Y S, Rosenberg M. Characterization of a third, cII-dependent, coordinately activated promoter on phage λ involved in lysogenic development. J Biol Chem. 1985;260:11838–11844. [PubMed] [Google Scholar]
- 9.Johnston J L, Sloan J, Fyfe J A M, Davies J K, Rood J I. The recA gene from Clostridium perfringens is induced by methyl methanosulphonate and contains an upstream Cheo box. Microbiology. 1997;143:885–890. doi: 10.1099/00221287-143-3-885. [DOI] [PubMed] [Google Scholar]
- 10.Karlin S, Weinstock G M, Bendel V. Bacterial classifications derived from RecA protein sequence comparisons. J Bacteriol. 1995;177:6881–6893. doi: 10.1128/jb.177.23.6881-6893.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kessler B, de Lorenzo V, Timmis K N. Identification of a cis-acting sequence within the Pm promoter of the TOL plasmid which confers XylS-mediated responsiveness to substituted benzoates. J Mol Biol. 1993;230:699–703. doi: 10.1006/jmbi.1993.1189. [DOI] [PubMed] [Google Scholar]
- 12.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieberman H B, Witkin E M. DNA degradation, UV sensitivity and SOS-binding protein: effects of mutations and treatments that alter levels of exonuclease V or recA protein. Mol Gen Genet. 1983;190:92–100. doi: 10.1007/BF00330329. [DOI] [PubMed] [Google Scholar]
- 14.Little J W. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–422. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- 15.Makino K, Amemura M, Kawamoto T, Kimura S, Shinagawa H, Nakata A, Suzuki M. DNA binding of PhoB and its interaction with RNA polymerase. J Mol Biol. 1996;259:15–26. doi: 10.1006/jmbi.1996.0298. [DOI] [PubMed] [Google Scholar]
- 16.Michiels J, Broek A V, Vanderleyden J. Molecular cloning and nucleotide sequence of the Rhizobium phaseoli recA gene. Mol Gen Genet. 1991;228:486–490. doi: 10.1007/BF00260644. [DOI] [PubMed] [Google Scholar]
- 17.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 18.Miller M C, Resnick J B, Smith B T, Lovett C M. The Bacillus subtilis dinR gene codes for the analogue of Escherichia coli LexA. J Biol Chem. 1996;271:33502–33508. [PubMed] [Google Scholar]
- 19.Miller R V, Kokjohn T A. General microbiology of recA. Environmental and evolutionary significance. Annu Rev Microbiol. 1990;44:365–394. doi: 10.1146/annurev.mi.44.100190.002053. [DOI] [PubMed] [Google Scholar]
- 20.Movahedzadeh F, Colston M J, Davis E O. Determination of DNA sequences required for regulated Mycobacterium tuberculosis RecA expression in response to DNA-damaging agents suggests that two modes of regulation exist. J Bacteriol. 1997;179:3509–3518. doi: 10.1128/jb.179.11.3509-3518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearce B J, Naughton A M, Campbell E A, Masure H R. The rec locus, a competence-induced operon in Streptococcus pneumoniae. J Bacteriol. 1995;117:86–93. doi: 10.1128/jb.177.1.86-93.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riera J, Fernández de Henestrosa A, Garriga X, Tapias A, Barbé J. Interspecies regulation of the recA gene of gram-negative bacteria lacking an E. coli-like SOS operator. Mol Gen Genet. 1994;245:523–527. doi: 10.1007/BF00302266. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sassanfar M, Roberts J W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 26.Selbitschka W, Arnold W, Priefer U B, Rottschäter T, Schmidt M, Simon R, Pühler A. Characterization of recA genes and recA mutants of Rhizobium meliloti and Rhizobium leguminosarum biovar viciae. Mol Gen Genet. 1991;229:86–95. doi: 10.1007/BF00264217. [DOI] [PubMed] [Google Scholar]
- 27.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusion. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 28.Tapias A, Fernández de Henestrosa A R, Barbé J. Characterization of the promoter of the Rhizobium etli recA gene. J Bacteriol. 1997;179:1573–1579. doi: 10.1128/jb.179.5.1573-1579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker G C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wardham H, McPherson M J, Harris C A, Sharma E, Sastry G R K. Molecular analysis of the recA gene of Agrobacterium tumefaciens C58. Gene. 1992;121:133–136. doi: 10.1016/0378-1119(92)90171-k. [DOI] [PubMed] [Google Scholar]
- 31.Wertman K F, Mount D. Nucleotide sequence binding specificity of the LexA repressor of Escherichia coli K-12. J Bacteriol. 1985;163:376–384. doi: 10.1128/jb.163.1.376-384.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winterling K W, Levine A S, Yasbin R E, Woodgate R. Characterization of DinR, the Bacillus subtilis SOS repressor. J Bacteriol. 1997;179:1698–1703. doi: 10.1128/jb.179.5.1698-1703.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winterling K W, Chafin D, Hayes J J, Sun J, Levine A S, Yasbin R E, Woodgate R. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J Bacteriol. 1998;180:2201–2211. doi: 10.1128/jb.180.8.2201-2211.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]